Abstract
Recently there have been explosive discoveries of new long noncoding RNAs (lncRNAs) obtained by progress in the technology of second-generation sequencing. Genome scale analysis of transcriptome, in conjunction with studies on chromatin modifications at the epigenetic level, identified lncRNAs as a novel type of non-coding transcripts whose length is longer than 200 nucleotides. These transcripts are later found as major participants in various physiological processes and diseases, especially in human cancers. LncRNAs have been found to function as novel types of oncogenes and tumor suppressors during cancer progression through various mechanisms, which endow them with the potential of serving as reliable biomarkers and novel therapeutic targets for cancers.
Keywords: Long noncoding RNA, cancers, biomarkers, therapeutic targets
Introduction
In recent years, there has been an explosive growth in the identification of long noncoding RNAs (lncRNAs). Identification of new lncRNAs has been facilitated by progress in the technology of second-generation sequencing. This has allowed genome-wide analyses of the transcriptome and chromatin modifications at the epigenetic level. Previous research has shown the majority of the human genome produces a great number of transcripts without protein coding potential [1, 2]. LncRNAs are one such class of non-coding transcripts. They are operationally defined as RNA molecules longer than 200 nucleotides that do not appear to have canonical protein-coding potential, i.e. they contain no classic open reading frame (ORF) [3–8]. Considering that nearly 70% of the human genome is transcribed to RNA products, among which only a minority encodes proteins [2], the number of lncRNA genes appears very large. Following the initial cloning of lncRNAs such as XIST [9] and H19 [10, 11] from cDNA libraries, two independent groups reported that the number of lncRNA genes is no less than that of protein-coding genes by using the tiling array technology [12, 13]. Progress in tiling arrays [12–15], analysis of chromatin epigenetic modification [16], computational analysis of cDNA libraries [17, 18], and RNA sequencing [1, 19–21], have demonstrated that thousands of lncRNAs are widely expressed in human with noticeable specificity in tissue distribution. Recently, the GENCODE consortium (version 27) has reported 27,908 manually annotated and evidence-supported human lncRNAs originating from 15,778 gene loci [1, 2].
Many studies have revealed universal features of lncRNAs to help better understand this new class of transcripts. First, lncRNAs are independent transcriptional units. Many of the first identified lncRNAs were located in genomic regions that did not contain any previously known genes, for example, lncRNAs HOTAIR and H19 [14, 22]. Now it is widely recognized that lncRNAs are unlikely to exist as “transcriptional noise” or “gene trash” as perceived before. Second, lncRNA transcripts contain fewer exons than mRNAs and utilize canonical splicing sites. Third, lncRNA transcripts are under weaker selection pressures than protein-coding ones during evolution, and many are primate-specific. It is reasonable to conclude that lncRNAs bear more possibility in nucleic acid variations during evolution because they have no need to preserve sequences critical for coding conserved amino acids of functional protein domains. Under such weaker selection constraints, primates might have flexibly evolved a set of specific lncRNAs to help accommodate unique physiological functions of the species. Fourth, lncRNA transcription is under the control of epigenetic modifications of chromatin similar to that for protein-coding mRNAs. Many lncRNAs were initially identified by large-scale sequencing of cDNA libraries, and subsequently characterized by transcriptional signatures from RNA pol II binding and epigenetic modifications of chromatin [3, 14]. The transcription of lncRNAs is regulated by DNA modifications and histone codes similar to that for protein-coding mRNAs. Fifth, lncRNAs and protein-coding mRNAs also utilize the same transcriptional machinery. Transcription of the majority of lncRNAs is RNA pol II-dependent. LncRNA transcripts usually have a 5’ terminal methylguanosine cap and are polyadenylated. There is also evidence showing certain lncRNAs deficient in 3’ poly(A) tails, which are probably produced from RNA pol III promoters [23, 24]. Additionally, the splicing of nucleolar RNAs generates a subset of lncRNAs lacking in 5’ caps and 3’ poly(A) tails [25]. This suggests that lncRNAs share the same molecular machinery for biogenesis with protein-coding mRNAs. Crosstalk and reciprocal regulation in the transcription of lncRNAs and protein-coding mRNAs may exist. Last, lncRNA expression is comparatively low and highly tissue-specific. Comprehensive analysis of human lncRNAs revealed that lncRNAs are generally expressed at lower levels than protein-coding genes, and they manifest more tissue-specific expression patterns. For example, a large number of lncRNAs are expressed specifically in the brain [1].
Molecular mechanisms of lncRNAs
Despite the numerous functions lncRNAs possess, they utilize some common working mechanisms. These common mechanisms involve control of epigenetic modifications of chromatin, gene transcription, and mRNA stability, as well as protein translation [16, 19, 26–30]. First, lncRNAs usually regulate gene expression by modulating epigenetic modifications of chromatin. LncRNA transcripts can form scaffold structures and mediate the interaction between chromatin and epigenetic modification factors. For example, lncRNA HOTAIR recruits the silencing complexes PRC2 and LSD1/CoREST/REST to targeted DNA regions for H3K27 methylation and H3K4 demethylation [27], and consequently represses the expression of a large amount of HOTAIR target genes, such as HOXD cluster genes.
Second, lncRNAs promote or repress gene expression at the transcriptional level through various mechanisms. A specific group of lncRNAs termed enhancer RNAs (eRNAs) have been suggested to upregulate gene expression by acting as transcriptional enhancers [19, 28]. On the other hand, lncRNAs can effect on transcription factors to control gene expression. The lncRNA NRON (non-coding repressor of NFAT) suppresses nucleocytoplasmic shuttling of the transcription factor NFAT, leading to repression on downstream gene transcription [30]. In some cases, lncRNAs function as decoys for transcription factors or compete for DNA binding sites with transcription factors. For example, the lncRNA PANDA can sequester NF-YA protein away from its target genes involved in apoptosis [31]. Interestingly, lncRNAs also regulate gene expression by directly affecting the organization of the nuclear compartment. Examples are MALAT1 and TUG1, who reside in distinct nuclear structures of interchromatin granules and Polycomb bodies respectively, can cooperatively control relocation of growth-control genes between the two nuclear compartments, and mediate the assembly of coactivators or corepressors to regulate gene expression in response to growth signals [32].
Third, in addition to the functions in epigenetic and transcriptional control on gene expression, lncRNAs also have proven activities in post-transcriptional regulation, affecting the processing, stability and translational efficiency of mRNAs. MALAT1 was reported to modulate the nuclear localization and levels of phosphorylated Ser/Arg splicing factors, which directly affects the alternative splicing pattern of target gene pre-mRNAs [33]. Like protein-coding genes, some lncRNAs and transcripts from pseudogenes act as miRNA sponges to sequester miRNAs away from target transcripts and promote the stabilization of mRNA molecules [29, 34, 35]. LncRNAs also exert their functions in translational control. The association of lincRNA-p21 and the general translation repressor Rck was shown to repress the translational efficiency of β-Catenin and JUNB mRNAs [26]. Similarly, a translational regulatory lncRNA (treRNA) was also reported to inhibit the translation of E-Cadherin mRNA [36].
Meanwhile, in addition to the non-coding functions supported by a vast amount of evidence as mentioned, recent studies implied that lncRNAs may also perform their functions by translation into proteins. Using ribosome profiling or ribosome footprinting methods, these studies showed that lncRNAs are physiologically associated with ribosomes, and the pattern of ribosome protection suggested the possibility of lncRNA transcripts in translating into short peptides [37–40]. Furthermore, Schier et al. identified a 58-amino acid peptide encoded by a previously annotated non-coding RNA Toddler in zebrafish, and established its essential role in embryonic signaling [41]. Nevertheless, whether lncRNAs play their biological roles through encoding short peptides still remains a controversial question [42]. It will be an attractive field to help fully elucidate the working mechanism of lncRNAs.
LncRNAs in human cancers
Although the biological functions of lncRNAs have not been fully explored, more and more studies indicate that a most prominent function of lncRNAs is their regulatory roles in the development of human cancers. LncRNAs are active participants in tumor development by widely modulating cancer cell proliferation, apoptosis, migration, metabolism, and differentiation [6–8, 15, 31, 43–45]. The lncRNA HOTAIR promotes metastasis of cancers through PRC2-mediated epigenetic modification on target genes [15]. Correspondingly, increased expression of HOTAIR correlates with poor prognosis of many types of cancers [46, 47]. ANRIL represents another typical example of oncogenic lncRNA. ANRIL expression is upregulated in prostate cancer. Its association with both CBX7/PRC1 [48] and SUZ12/PRC2 [49] leads to repression of the tumor suppressor INK4A/INK4B. The lncRNA FAL1 with amplification in gene copy number showed increased expression levels in several types of cancers. FAL1 enhances the growth rate of cancer cells by interacting with BMI1/PRC1 to suppress p21 expression [50]. Similarly, the lncRNA DANCR, which is widely overexpressed in many types of human cancers, was found to promote cancer progression partly by modulating p21 expression [51]. A TP53-regulated lncRNA, LINP1, enhances double-strand DNA break repair through the non-homologous end joining pathway, and desensitizes triple-negative breast cancer cells to radiotherapy [52]. The lncRNA ceruloplasmin (NRCP) modulates cancer metabolism and promotes cancer growth by increasing the expression of genes related to glycolysis [45]. In addition, a large number of lncRNAs were also found to be associated with tumorigenesis, such as PCAT-1 [20], PANDA [31], MALAT-1 [53], lincRNA-p21 [44], and LINC00673 [54], etc.
It is worth noticing that genomic alterations and transcriptional deregulation of lncRNA genes are widely present in cancer cells. The somatic copy number alterations (SCNAs) of lncRNA genes across 5860 tumor samples from 13 tumor types from the Cancer Genome Atlas project were extensively analyzed. It was found that a high frequency of copy number gain and loss existed in an average of 13.16% and 13.53% of lncRNA genes, respectively [55]. In line with the genome-wide landscape of SCNAs, individual lncRNAs with copy number variation were identified. For example, the oncogenic lncRNA FAL1 with amplification in gene copy number shows increased expression levels in many types of cancers [50]. Focal amplifications containing both the lncRNA SAMMSON and the melanoma-specific oncogene MITF were located to chromosome 3p13 and 3p14. SAMMSON was characterized as a critical player in promoting melanoma cell growth and survival [56]. In addition to SCNAs, single nucleotide polymorphism (SNP) is another common genomic alteration in lncRNA genes associated with cancer risk and clinical outcomes. Genome-wide studies have indicated frequent overlap between lncRNAs and cancer-related SNPs [55, 57]. The tumor suppressor lncRNA LINC00673 antagonizes pancreatic cancer cell proliferation by promoting degradation of the oncogene PTPN11. A germline variant of LINC00673 has impaired activity and sensitizes cells to malignant transformation [54]. NBAT-1 is an lncRNA that controls neuroblastoma cell proliferation and invasion. Interestingly, an SNP in the intron of NBAT-1 indicates high risk and poor clinical outcome of neuroblastoma [58]. Besides genomic alterations, transcriptional deregulation of lncRNA genes is another mechanism responsible for their differential expression patterns in cancers. A study showed that DNA methylation patterns in the promoter regions of lncRNA genes are intrinsically distinct in cancerous and normal tissues [55]. Furthermore, lncRNA transcription is under control of important oncogenes or tumor suppressors. A subset of lncRNA genes is either suppressed (such as CONCR and LINP1) or promoted (such as lincRNA-p21 and DINO) by TP53 [44, 52, 59, 60]. The oncogenes RAS and MYC are also involved in transcriptional regulation of lncRNAs such as Orilnc1 and DANCR [51, 61].
LncRNAs may serve as biomarkers and therapeutic targets for human cancers
The explosion in lncRNA research has evoked great enthusiasm for the possibility of lncRNAs serving as diagnostic markers and therapeutic targets for human cancer. Hopefully, the unique features of lncRNAs, i.e. their differential expression patterns in cancer, temporal and tissue specificity, and diverse biological functions, will be advantages taken in clinical applications. The first inspiring case of lncRNA as a diagnostic marker is the Food and Drug Administration-approved testing of PCA3 for detection of prostate cancer [62]. Together with the traditional serum prostate specific antigen (PSA) testing, PCA3 testing has made diagnosis of this disease more accurate and specific. Potential lncRNA diagnostic markers for other types of cancers have been reported, including hepatocellular carcinoma and gastric cancer [63, 64]. In addition, lncRNA have been widely investigated as prognostic markers for cancer patients. Previous studies have revealed a correlation between lncRNA HOTAIR expression and colorectal cancer recurrence, lymph node metastasis, and poor prognosis [46, 65]. On the other hand, lncRNAs are considered ideal biomarkers due to their presence in body fluids, such as in the form of exosome-containing RNA [66–68], which enables noninvasive diagnosis of cancers. Although numerous studies have shown differential expression of lncRNAs in tumors compared to normal tissues, extensive research and clinical data are needed to confirm the consistency of lncRNA expression in tumor tissue and body fluids for diagnostic accuracy and specificity. Despite limited progress in the development of lncRNA biomarkers so far, the diversity in biological functions of lncRNAs endows them with great potential in serving as therapeutic targets for human cancers. A supportive case is lncRNA MALAT1, an oncogene first discovered in metastatic lung adenocarcinoma patients [53] and later found highly expressed in lung, breast and prostate cancers [69]. It plays critical roles in promoting proliferation and metastasis of cancer cells. Animal studies indicated that silencing MALAT1 expression by antisense oligonucleotide (ASO) efficiently impaired the tumor growth and metastatic ability of breast and lung cancer cells, respectively [70, 71], implying a potential therapy for cancers by manipulating oncogenic lncRNA expression.
Strategies for developing targeted therapy for cancers with lncRNAs
As critical players in tumor development, lncRNAs represent promising therapeutic targets for treating cancers. However, targeting lncRNA molecules is different from targeting protein-coding transcripts (Figure 1). First, the lack of protein products encoded by lncRNAs means that targeting methods are restricted to RNA molecules only. However, methods targeting RNA are currently limited. Second, unlike proteins with conserved domains and specific conformations, which serve as good targeting sites by small-molecule drugs, the three-dimensional structures of lncRNAs have been poorly explored. Furthermore, it has been speculated that lncRNA functions are independent on conserved three-dimensional structures, as they show relatively low evolutionary conservation of sequence compared to protein-coding genes. This makes structure-based drug design and screening difficult for lncRNA targets. Third, the working mechanism and regulation network of most lncRNAs have not been fully understood. This increases difficulties in developing specific targeting strategies for lncRNAs.
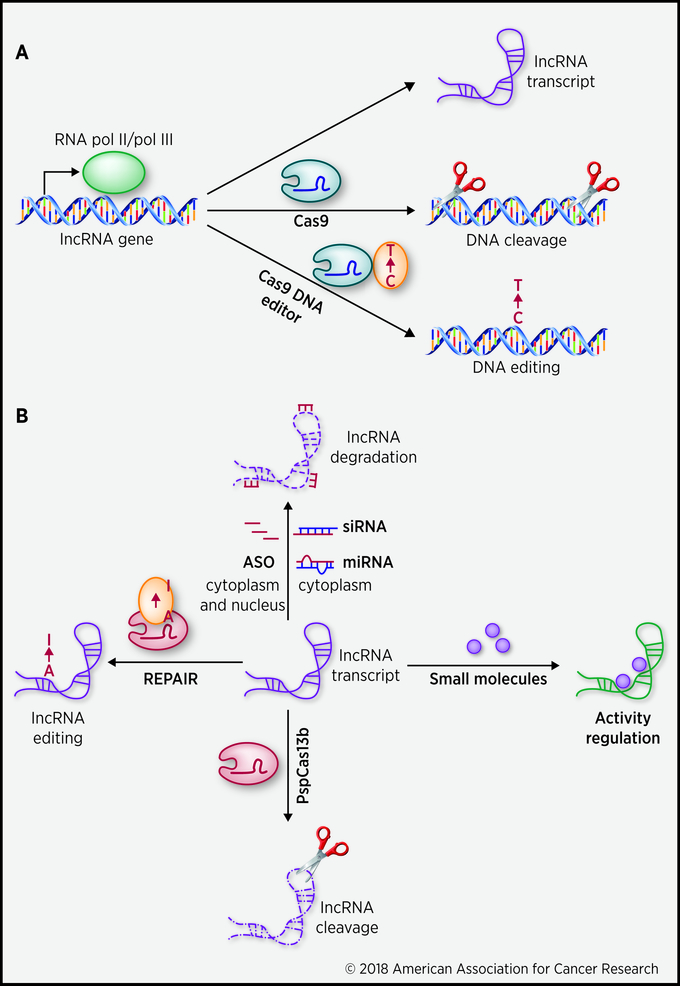
Figure 1. Strategies for development of lncRNA-targeted therapy for cancer treatment.
A. LncRNA genes are transcribed by RNA pol II/pol III to give rise to lncRNA transcripts (top). The Cas9 system can be applied to target and cleave DNA fragments from lncRNA genes (middle). And the Cas9 DNA editor system is capable of editing specific DNA bases (e.g. C to T) of lncRNA genes to correct tumorigenic SNP (bottom). B. lncRNA transcripts can be targeted by multiple ways. Antisense oligonucleotides (ASO), small interference RNA (siRNA), and microRNA (miRNA) represent the most popular methods to knockdown the expression level of lncRNA (top). The mechanism of ASO, siRNA and miRNA involves base pairing with lncRNA which leads to the degradation of lncRNA. Alternatively, lncRNA expression level can also be modulated by the PspCas13b system by its ability in RNA-guided recognition of RNA targets and its RNase activity (bottom). A modified Cas13 system with nucleotide editing activity (e.g. A to I), named REPAIR, can be further used to edit specific nucleotides in lncRNA, hence change the biological functions of lncRNA rather than cleave lncRNA molecules (left). Small molecules that can potentially bind to lncRNA transcripts may also be a prospective way to regulate the structural conformation and biological activities of lncRNAs (right).
Despite the mentioned problems, diverse targeting methods for lncRNAs are under investigation (Table 1). Using small interfering RNA (siRNA) and antisense oligonucleotide (ASO) to induce loss-of-function effect on lncRNAs represents the most common strategy. Similar strategies for targeting protein-coding genes have proven successful, and the systemic delivery methods have been widely explored [72–74]. It has been well proved that both siRNA and ASO function by base pairing with mRNA to form double-stranded RNAs or RNA-DNA hybrids, resulting in degradation of target mRNA by the RNA interference (RNAi) mechanism or by RNase H activity. This method has been adapted for knocking down lncRNAs with as potent effect as for mRNA silencing. For example, MALAT1 expression was suppressed by ASO to impair the tumor growth and metastatic ability of breast and lung cancer cells [70, 71]. Furthermore, siRNA and ASO are usually subjected to certain modifications in order to enhance their stability, meanwhile retaining targeting specificity and alleviating the interferon induction effect. Such modifications include adding 2-nt 3’overhangs to siRNA, 2′-O-methyl to siRNA and ASO, and locked nucleic acid (LNA) to ASO. To overcome poor stability and increase intracellular uptake of siRNA and ASO, delivery methods such as lipid-based carriers, polymersomes, and biocompatible nanoparticles can be employed. Last, the subcellular localization of lncRNAs should be considered when adapting the siRNA/ASO strategy. For lncRNAs located in the cytoplasm, both siRNA and ASO have satisfactory silencing effects. But for those located in the nucleus, ASO would be a better choice than siRNA due to the lack of RNAi machinery in the nuclear compartment. Meanwhile, considering that some lncRNAs function as sponges for microRNAs (miRNAs) to alleviate the suppressive effects of miRNAs on endogenous target mRNAs [34, 54], targeting lncRNAs by miRNAs would also be a promising strategy. The prominent advantages of using miRNAs include their compatibility with the endogenous regulatory machinery for lncRNAs, and the well-established delivery methods [74, 75].
Table 1. Strategies for targeting lncRNA in cancer therapy.
| Strategies | Effects | Advantage | Limitation |
|---|---|---|---|
|
siRNA, ASO, miRNA |
RNA degradation |
Specific, potent effect |
Stability, delivery |
| Cas9 | Gene knockout, DNA editing, gene mutation, etc. |
Versatile and long-term effect |
Genomic context dependent, PAM requirement, targeting efficiency, in vivo delivery |
|
| |||
| PspCas13b | RNA cleavage |
Programmable targeting, less sequence constraint |
Targeting efficiency, in vivo delivery |
|
| |||
|
REPAIR (dCas13-ADAR) |
RNA editing | Programmable editing |
Targeting efficiency, in vivo delivery |
| Small molecules | RNA binding | Convenient in vivo delivery |
Non-specificity, non-programmable targeting |
siRNA: small interfering RNA; ASO: antisense oligonucleotide; miRNA: microRNA; Cas9: CRISPR-associated 9; PspCas13b: CRISPR-associated 13 from Prevotella sp. P5-125; REPAIR: RNA Editing for Programmable A to I Replacement; dCas13: catalytically inactive PspCas13b; ADAR: adenosine deaminase acting on RNA
Besides traditional methods using siRNA, ASO and miRNA, the recently characterized CRISPR (clustered regularly interspaced short palindromic repeats)-associated deoxyribonuclease Cas9 gene editing system may be employed to achieve loss-of-function effects on lncRNAs. In this system, the Cas9 endonuclease is targeted to specific DNA sequences with a protospacer adjacent motif (PAM) by single-guide RNAs (sgRNAs) that are artificially designed [76–79]. This system has been widely applied for targeted DNA cleavage. For oncogenic lncRNA genes with well-characterized genomic localization, Cas9 can be directed by a pair of specifically designed sgRNAs targeting the 5’ and 3’ ends of the genes to generate knockout loci. In this way, successful knockouts of lncRNA-21A, UCA1 and AK023948 in various human cell lines have been accomplished [80]. Similarly, utilizing a paired-guide RNA library, CRISPR-Cas9-mediated genome-scale deletion of lncRNAs was achieved [81]. The Cas9 system is emerging as a powerful tool for targeting lncRNAs in cancer. Major advantages of the Cas9 system include, but not limited to, programmable targeting effects (e.g. gene knockout, as well as gene mutation and transcriptional activation) and convenience for high-throughput screening of lncRNA knockout phenotypes. Furthermore, a recent development of the Cas9 system was reported by Liu group, which enables conversion of cytidine to uridine, thereby leading to a C -> T (or G -> A) substitution, by fusing the Cas9 protein with a cytidine deaminase enzyme. This DNA base editing system can mediate base conversion in a programmable manner, without requiring double-strand DNA cleavage or DNA repairing templates [82]. Thus this modified Cas9 system bears great potential especially in correcting tumorigenic SNPs on lncRNA genes with higher efficiency and accuracy. In addition, currently the in vivo delivery methods of the Cas9 system are developing very fast. These include viral vectors of lentivirus (LV) and adeno-associated viruses (AAV) for delivering DNAs encoding Cas9 and sgRNA [83–86]; nonviral vectors of lipid-based carriers and polymers for delivering the DNA, mRNA and proteins of the Cas9 system [87–89]; and physical approaches, such as microinjection, electroporation and hydrodynamic injection, for delivering DNA, mRNA and proteins of the Cas9 system [88, 90]. These versatile approaches enable efficient in vivo delivery of the Cas9 system for lncRNA targeting in cancer treatment in the future. However, the Cas9 strategy for lncRNA knockout is genomic context dependent. It is well known that numerous lncRNAs are transcribed from bidirectional promoters, or overlap with promoters or bodies of sense or antisense genes. This unique feature of lncRNAs may expose their neighboring genes to the risk of inadvertent deregulation when using the Cas9 system for lncRNA gene knockout purposes. A study showed that only one third of 15,929 lncRNA loci may be safely targeted for gene knockout via the Cas9 system without perturbation of neighboring genes[91]. Thus, careful study is needed before adopting the Cas9 system for targeting of any lncRNA gene.
In addition to the Cas9 system, novel CRISPR-Cas systems capable of directly targeting RNA, rather than working through DNA recognition, have been established. The class 2 type VI CRISPR-Cas effectors, Cas13a and Cas13b, were recently identified by Zhang group as single-component programmable RNA-guided RNA-targeting RNases that have both RNA processing and RNA cleavage activities [92, 93]. Cas13b from Prevotella sp. P5–125 (PspCas13b) is an RNA-targeted RNase which does not require a PAMmer for target RNA recognition. PspCas13b displays consistent, robust, and specific knockdown of a reporter mRNA in mammalian cells. Thus, it holds great promise as a powerful tool for knocking down lncRNA transcripts in cancer therapy, especially for oncogenic lncRNAs with higher expression levels in cancer cells than in normal tissues and which are unsuitable for knockout by the traditional Cas9 system. Furthermore, the PspCas13b system was modified to an RNA-editing system by fusing the catalytically inactive PspCas13b (dCas13b) with the catalytic domain of adenosine (A) to inosine (I) deaminase 2 (ADAR2), namely RNA Editing for Programmable A to I Replacement (REPAIR) [92, 93]. The REPAIR system can convert A to I in mRNA molecules without disturbing genomic sequences, which avoids unexpected effects such as frameshift and nonsense mutations of the genome. Furthermore, this system is theoretically capable of transformation to other types of base editing platforms, e.g. C-to-U editing, if dCas13b is fused with other RNA base editors. Collectively, the REPAIR system offers advantages in the programmable editing of a specific nucleotide without sequence constraints (no PAM requirement), and independence from endogenous DNA repair pathways, which are usually needed for the Cas9 system. Hopefully, the REPAIR system will function as an accurate tool for editing lncRNAs in cancer. For example, the REPAIR system may reduce cancer risk by altering a single nucleotide in tumorigenic SNPs of lncRNAs [54, 58]. It might also be used to regulate the association of lncRNAs and onco-protein partners by changing specific nucleotides critical for the interaction.
Previous studies have found that small molecules represent a huge source for drugging proteins. It has always been an interesting question as to whether RNAs can also directly bind to small molecule compounds. Previously, scientists have been pessimistic about the answer until a recent breakthrough in the field [94]. A small molecule LMI070 was found to bind to the pre-mRNA of spinal muscular atrophy-related gene SMN2 which encodes SMN (survival motor neuron). The association boosts the processing of exons and translation of the protein product to antagonize the disease [95]. Later, by screening a library of non-coding RNAs against a library of small molecules to find strong interactions, another small molecule, targaprimir-96, was identified as a binding partner of primary miRNA-96 at a key processing site. This interaction blocks the maturation of mir-96 and induces apoptosis in cancer cells [96]. Using the same strategy, authors also found a small molecule, targaprimir-210, which binds primary miRNA-210 [97]. These inspiring results demonstrate that both coding and non-coding RNAs are druggable just like proteins. Although no relevant studies on drugging lncRNAs have been reported, these studies provide one more prospective strategy for targeting lncRNAs for cancer therapy.
Acknowledgments
We apologize to colleagues whose work was not discussed or cited in this review due to the space limitation. This work was supported, in whole or in part, by the US Department of Defense (PC140683 to C.V. Dang), the National Key Research and Development Program of China 2017YFA0105501 (X. Zhong), the Guangdong Province Science and Technology Project 2015A020212019 (X. Zhong), the US National Institutes of Health (R01CA142776 to L. Zhang, R01CA190415 to L. Zhang, P50CA083638 to L. Zhang, P50CA174523 to L. Zhang, P50CA083639 to A.K. Sood, P50CA098258 to A.K. Sood and R35 CA209904 to A.K. Sood), the Breast Cancer Alliance (L. Zhang and C.V. Dang), the Frank McGraw Memorial Chair in Cancer Research (A.K. Sood), the American Cancer Society Research Professor Award (A.K. Sood), the Marsha Rivkin Center for Ovarian Cancer Research (L. Zhang), the Basser Center for BRCA (L. Zhang), the Harry Fields Professorship (L. Zhang), the Kaleidoscope of Hope Ovarian Cancer Foundation (L. Zhang).
Footnotes
No Conflict of Interest
References
- 1.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H et al. : The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012, 22(9):1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A et al. : Landscape of transcription in human cells. Nature 2012, 489(7414):101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D et al. : Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458(7235):223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee JT: Epigenetic regulation by long noncoding RNAs. Science 2012, 338(6113):1435–1439. [DOI] [PubMed] [Google Scholar]
- 5.Lieberman J, Slack F, Pandolfi PP, Chinnaiyan A, Agami R, Mendell JT: Noncoding RNAs and cancer. Cell 2013, 153(1):9–10. [DOI] [PubMed] [Google Scholar]
- 6.Orom UA, Shiekhattar R: Long noncoding RNAs usher in a new era in the biology of enhancers. Cell 2013, 154(6):1190–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prensner JR, Chinnaiyan AM: The emergence of lncRNAs in cancer biology. Cancer Discov 2011, 1(5):391–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ulitsky I, Bartel DP: lincRNAs: genomics, evolution, and mechanisms. Cell 2013, 154(1):26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R et al. : A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 1991, 349(6304):38–44. [DOI] [PubMed] [Google Scholar]
- 10.Pachnis V, Brannan CI, Tilghman SM: The structure and expression of a novel gene activated in early mouse embryogenesis. The EMBO journal 1988, 7(3):673–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartolomei MS, Zemel S, Tilghman SM: Parental imprinting of the mouse H19 gene. Nature 1991, 351(6322):153–155. [DOI] [PubMed] [Google Scholar]
- 12.Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, Fodor SP et al. : Large-scale transcriptional activity in chromosomes 21 and 22. Science 2002, 296(5569):916–919. [DOI] [PubMed] [Google Scholar]
- 13.Rinn JL, Euskirchen G, Bertone P, Martone R, Luscombe NM, Hartman S et al. : The transcriptional activity of human Chromosome 22. Genes & development 2003, 17(4):529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA et al. : Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129(7):1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ et al. : Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464(7291):1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D et al. : Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A 2009, 106(28):11667–11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maeda N, Kasukawa T, Oyama R, Gough J, Frith M, Engstrom PG et al. : Transcript annotation in FANTOM3: mouse gene catalog based on physical cDNAs. PLoS genetics 2006, 2(4):e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia H, Osak M, Bogu GK, Stanton LW, Johnson R, Lipovich L: Genome-wide computational identification and manual annotation of human long noncoding RNA genes. Rna 2010, 16(8):1478–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G et al. : Long noncoding RNAs with enhancer-like function in human cells. Cell 2010, 143(1):46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC et al. : Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol 2011, 29(8):742–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A et al. : Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & development 2011, 25(18):1915–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brannan CI, Dees EC, Ingram RS, Tilghman SM: The product of the H19 gene may function as an RNA. Mol Cell Biol 1990, 10(1):28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A: The expanding RNA polymerase III transcriptome. Trends Genet 2007, 23(12):614–622. [DOI] [PubMed] [Google Scholar]
- 24.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT et al. : RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007, 316(5830):1484–1488. [DOI] [PubMed] [Google Scholar]
- 25.Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG et al. : Long noncoding RNAs with snoRNA ends. Mol Cell 2012, 48(2):219–230. [DOI] [PubMed] [Google Scholar]
- 26.Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S et al. : LincRNA-p21 suppresses target mRNA translation. Mol Cell 2012, 47(4):648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F et al. : Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329(5992):689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y et al. : Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 2011, 474(7351):390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L et al. : Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell 2013, 25(1):69–80. [DOI] [PubMed] [Google Scholar]
- 30.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P et al. : A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 2005, 309(5740):1570–1573. [DOI] [PubMed] [Google Scholar]
- 31.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD et al. : Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 2011, 43(7):621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD et al. : ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 2011, 147(4):773–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT et al. : The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 2010, 39(6):925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M et al. : A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011, 147(2):358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP: A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010, 465(7301):1033–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gumireddy K, Li A, Yan J, Setoyama T, Johannes GJ, Orom UA et al. : Identification of a long non-coding RNA-associated RNP complex regulating metastasis at the translational step. EMBO J 2013, 32(20):2672–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES et al. : Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J 2014, 33(9):981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ingolia NT, Lareau LF, Weissman JS: Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 2011, 147(4):789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Juntawong P, Girke T, Bazin J, Bailey-Serres J: Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc Natl Acad Sci U S A 2014, 111(1):E203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruiz-Orera J, Messeguer X, Subirana JA, Alba MM: Long non-coding RNAs as a source of new peptides. Elife 2014, 3:e03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pauli A, Norris ML, Valen E, Chew GL, Gagnon JA, Zimmerman S et al. : Toddler: an embryonic signal that promotes cell movement via Apelin receptors. Science 2014, 343(6172):1248636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES: Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 2013, 154(1):240–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G et al. : lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011, 477(7364):295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D et al. : A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010, 142(3):409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rupaimoole R, Lee J, Haemmerle M, Ling H, Previs RA, Pradeep S et al. : Long Noncoding RNA Ceruloplasmin Promotes Cancer Growth by Altering Glycolysis. Cell reports 2015, 13(11):2395–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T et al. : Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011, 71(20):6320–6326. [DOI] [PubMed] [Google Scholar]
- 47.Niinuma T, Suzuki H, Nojima M, Nosho K, Yamamoto H, Takamaru H et al. : Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res 2012, 72(5):1126–1136. [DOI] [PubMed] [Google Scholar]
- 48.Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S et al. : Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 2010, 38(5):662–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M et al. : Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 2011, 30(16):1956–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu X, Feng Y, Zhang D, Zhao SD, Hu Z, Greshock J et al. : A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell 2014, 26(3):344–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu Y, Hu Z, Mangala LS, Stine ZE, Hu X, Jiang D et al. : MYC Targeted Long Noncoding RNA DANCR Promotes Cancer in Part by Reducing p21 Levels. Cancer Res 2018, 78(1):64–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y, He Q, Hu Z, Feng Y, Fan L, Tang Z et al. : Long noncoding RNA LINP1 regulates repair of DNA double-strand breaks in triple-negative breast cancer. Nat Struct Mol Biol 2016, 23(6):522–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM et al. : MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22(39):8031–8041. [DOI] [PubMed] [Google Scholar]
- 54.Zheng J, Huang X, Tan W, Yu D, Du Z, Chang J et al. : Pancreatic cancer risk variant in LINC00673 creates a miR-1231 binding site and interferes with PTPN11 degradation. Nat Genet 2016, 48(7):747–757. [DOI] [PubMed] [Google Scholar]
- 55.Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD et al. : Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell 2015, 28(4):529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leucci E, Vendramin R, Spinazzi M, Laurette P, Fiers M, Wouters J et al. : Melanoma addiction to the long non-coding RNA SAMMSON. Nature 2016, 531(7595):518–522. [DOI] [PubMed] [Google Scholar]
- 57.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y et al. : The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 2015, 47(3):199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pandey GK, Mitra S, Subhash S, Hertwig F, Kanduri M, Mishra K et al. : The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell 2014, 26(5):722–737. [DOI] [PubMed] [Google Scholar]
- 59.Marchese FP, Grossi E, Marin-Bejar O, Bharti SK, Raimondi I, Gonzalez J et al. : A Long Noncoding RNA Regulates Sister Chromatid Cohesion. Mol Cell 2016, 63(3):397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmitt AM, Garcia JT, Hung T, Flynn RA, Shen Y, Qu K et al. : An inducible long noncoding RNA amplifies DNA damage signaling. Nat Genet 2016, 48(11):1370–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang D, Zhang G, Hu X, Wu L, Feng Y, He S et al. : Oncogenic RAS Regulates Long Noncoding RNA Orilnc1 in Human Cancer. Cancer Res 2017, 77(14):3745–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bussemakers MJ, van Bokhoven A, Verhaegh GW, Smit FP, Karthaus HF, Schalken JA et al. : DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res 1999, 59(23):5975–5979. [PubMed] [Google Scholar]
- 63.Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM et al. : Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology 2007, 132(1):330–342. [DOI] [PubMed] [Google Scholar]
- 64.Shao Y, Ye M, Jiang X, Sun W, Ding X, Liu Z et al. : Gastric juice long noncoding RNA used as a tumor marker for screening gastric cancer. Cancer 2014, 120(21):3320–3328. [DOI] [PubMed] [Google Scholar]
- 65.Cai B, Wu Z, Liao K, Zhang S: Long noncoding RNA HOTAIR can serve as a common molecular marker for lymph node metastasis: a meta-analysis. Tumour Biol 2014, 35(9):8445–8450. [DOI] [PubMed] [Google Scholar]
- 66.Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M et al. : Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics 2013, 14:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Q, Shao Y, Zhang X, Zheng T, Miao M, Qin L et al. : Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol 2015, 36(3):2007–2012. [DOI] [PubMed] [Google Scholar]
- 68.Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y et al. : Exosome-Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell 2016, 29(5):653–668. [DOI] [PubMed] [Google Scholar]
- 69.Ren S, Peng Z, Mao JH, Yu Y, Yin C, Gao X et al. : RNA-seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer-associated long noncoding RNAs and aberrant alternative splicings. Cell Res 2012, 22(5):806–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Arun G, Diermeier S, Akerman M, Chang KC, Wilkinson JE, Hearn S et al. : Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev 2016, 30(1):34–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G et al. : The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res 2013, 73(3):1180–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu SY, Lopez-Berestein G, Calin GA, Sood AK: RNAi therapies: drugging the undruggable. Science translational medicine 2014, 6(240):240ps247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu SY, Yang X, Gharpure KM, Hatakeyama H, Egli M, McGuire MH et al. : 2’-OMe-phosphorodithioate-modified siRNAs show increased loading into the RISC complex and enhanced anti-tumour activity. Nat Commun 2014, 5:3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nishimura M, Jung EJ, Shah MY, Lu C, Spizzo R, Shimizu M et al. : Therapeutic synergy between microRNA and siRNA in ovarian cancer treatment. Cancer Discov 2013, 3(11):1302–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ma L, Reinhardt F, Pan E, Soutschek J, Bhat B, Marcusson EG et al. : Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol 2010, 28(4):341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N et al. : Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339(6121):819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hsu PD, Lander ES, Zhang F: Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157(6):1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Komor AC, Badran AH, Liu DR: CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell 2017, 168(1–2):20–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE et al. : RNA-guided human genome engineering via Cas9. Science 2013, 339(6121):823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ho TT, Zhou N, Huang J, Koirala P, Xu M, Fung R et al. : Targeting non-coding RNAs with the CRISPR/Cas9 system in human cell lines. Nucleic acids research 2015, 43(3):e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhu S, Li W, Liu J, Chen CH, Liao Q, Xu P et al. : Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat Biotechnol 2016, 34(12):1279–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR: Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533(7603):420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera Mdel C, Yusa K: Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol 2014, 32(3):267–273. [DOI] [PubMed] [Google Scholar]
- 84.Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR et al. : CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 2014, 159(2):440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T et al. : Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014, 343(6166):84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang T, Wei JJ, Sabatini DM, Lander ES: Genetic screens in human cells using the CRISPR-Cas9 system. Science 2014, 343(6166):80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liang X, Potter J, Kumar S, Zou Y, Quintanilla R, Sridharan M et al. : Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J Biotechnol 2015, 208:44–53. [DOI] [PubMed] [Google Scholar]
- 88.Zuckermann M, Hovestadt V, Knobbe-Thomsen CB, Zapatka M, Northcott PA, Schramm K et al. : Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat Commun 2015, 6:7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH et al. : Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol 2015, 33(1):73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Han X, Liu Z, Jo MC, Zhang K, Li Y, Zeng Z et al. : CRISPR-Cas9 delivery to hard-to-transfect cells via membrane deformation. Sci Adv 2015, 1(7):e1500454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Goyal A, Myacheva K, Gross M, Klingenberg M, Duran Arque B, Diederichs S: Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic acids research 2017, 45(3):e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J et al. : RNA editing with CRISPR-Cas13. Science 2017, 358(6366):1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang L, Chen LL: Enhancing the RNA engineering toolkit. Science 2017, 358(6366):996–997. [DOI] [PubMed] [Google Scholar]
- 94.Mullard A: Small molecules against RNA targets attract big backers. Nat Rev Drug Discov 2017, 16(12):813–815. [DOI] [PubMed] [Google Scholar]
- 95.Palacino J, Swalley SE, Song C, Cheung AK, Shu L, Zhang X et al. : SMN2 splice modulators enhance U1-pre-mRNA association and rescue SMA mice. Nat Chem Biol 2015, 11(7):511–517. [DOI] [PubMed] [Google Scholar]
- 96.Velagapudi SP, Cameron MD, Haga CL, Rosenberg LH, Lafitte M, Duckett DR et al. : Design of a small molecule against an oncogenic noncoding RNA. Proc Natl Acad Sci U S A 2016, 113(21):5898–5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Costales MG, Haga CL, Velagapudi SP, Childs-Disney JL, Phinney DG, Disney MD: Small Molecule Inhibition of microRNA-210 Reprograms an Oncogenic Hypoxic Circuit. J Am Chem Soc 2017, 139(9):3446–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]