Abstract
Ketoreductase (KR) domains of modular polyketide synthases (PKSs) control the stereochemistry of C2 methyl and C3 hydroxyl substituents of polyketide intermediates. To understand the molecular basis of stereocontrol exerted by KRs, the crystal structure of a KR from the second module of the amphotericin PKS (AmpKR2) complexed with NADP+ and 2-methyl-3-oxopentanoyl-pantetheine was solved. This first ternary structure provides direct evidence to the hypothesis that a substrate enters into the active site of an A-type KR from the side opposite the coenzyme to generate an L-hydroxyl substituent. A comparison with the ternary complex of a G355T/Q364H mutant sheds light on the structural basis for stereospecificity toward the substrate C2 methyl substituent. Functional assays suggest the pantetheine handle shows obvious influence on the catalytic efficiency and the stereochemical outcome. Together, these findings extend our current understanding of the stereo-chemical control of PKS KR domains.
Keywords: Polyketide, Ketoreductase, Crystal structure, Ternary complex, Stereochemistry, Amphotericin
1. Introduction
Polyketides are a large group of structurally diverse and clinically important natural products synthesized by bacteria, fungi and plants (Cortes et al., 1990). One of the most striking structural features of polyketides is the numerous chiral centers carrying hydroxyl or methyl substituents. Functional characterizations of biosynthetic pathways have revealed that complex polyketides are assembled from short chain carboxylic acids by large multidomain enzymatic machines called modular polyketide synthases (PKSs) in a manner analogous to fatty acid biosynthesis. However, deciphering the molecular basis for the stereochemistry of the chiral centers is still a major challenge to the detailed biochemical understanding and rational engineering of modular PKSs (Reid et al., 2003). A minimal module contains an acyltransferase (AT) to recruit an extender unit, a ketosynthase (KS) to catalyze the decarboxylative condensation of the extender unit onto a growing polyketide intermediate, and an acyl carrier protein (ACP) to carry the growing intermediate via a pantetheine handle. Optional domains of a module that process a nascent polyketide intermediate include a ketoreductase (KR), a dehydratase (DH), and an enoylreductase (ER).
KRs catalyze the reduction of C3 keto groups of polyketide intermediates carried by ACPs to hydroxyl groups utilizing the NADPH coenzyme (Zheng et al., 2013). In hybrid modules generated by domain exchange, KRs usually retain their native stereospecificity, suggesting that stereocontrol of reduced hydroxyl groups is intrinsic to individual KR domains (Kao et al., 1998). Excised KRs are catalytically active toward N-acetyl cysteamine (NAC) thioester derivatives of natural dike-tide intermediates (for example 2-methyl-3-oxopentanoyl-S-NAC, “S” indicates the thioester linkage) (Siskos et al., 2005). The NAC moiety mimics the end of the pantetheine handle of an ACP domain and is therefore widely used as the activating group of PKS substrate surrogates. KRs that naturally process small polyketide intermediates usually retain their intrinsic stereospecificity and reduce NAC substrate surrogates to products with expected stereochemistry; however, KRs that reduce larger polyketides in their native PKSs are usually less active and show aberrant stereochemical control (Piasecki et al., 2011). When 3-ketoacyl-ACPs enzymatically generated in situ are used as the substrates, all KRs maintain their native stereospecificity, highlighting the importance of ACPs in the stereochemistry of KRs (Castonguay et al., 2007). According to the stereochemical outcomes, KRs are classified as A-type KRs that generate L-hydroxyl substituents and B-type KRs that generate D-hydroxyl substituents (Keatinge-Clay, 2007). Distinct sequence motifs correlated with the stereochemical outcome of KR activity have been revealed and used to assign the stereochemistry of uncharacterized polyketides (Caffrey, 2003; Reid et al., 2003). A-type KRs contain a conserved tryptophan (“W” motif) while a leucine-aspartate-aspartate (LDD) motif is observed in B-type KRs.
How modular PKSs control the stereochemistry of C2 methyl substituents is less straightforward. An AT recruits the (2S)-methylmalonyl-CoA extender unit exclusively while the C2 methyl substituent undergoes inversion of configuration in the KS-catalyzed condensation to produce a D-2-methyl substituent (Marsden et al., 1994; Weissman et al., 1997). Compared with modules generating a D-2-methyl substituent, modules that generate an L-2-methyl substituent catalyze an additional epimerization step. Combinations of individually purified KS-AT didomains, KRs and ACPs reveal that KR domains determine the stereochemical outcome of both C2 methyl and C3 hydroxyl substituents in polyketide intermediates (Valenzano et al., 2009). Structural analysis suggests the KR from the first module of the erythromycin PKS (EryKR1) may also catalyze the epimerization of the D-2-methyl to yield the L-methyl (Keatinge-Clay and Stroud, 2006). The proposed mechanism involves enolization of the diketide-ACP thioester followed by an uncatalyzed tautomerization back to the keto-form to generate a mixture of both epimers. When racemic 2-methyl-3-oxopentanoyl-SNAC is used as the substrate surrogate, EryKR1 exclusively prefers the L-epimer to the D-epimer (Siskos et al., 2005). According to this proposal, KRs working on C2 methylated substrates are classified into four distinct categories – KRs catalyzing reduction in the absence of epimerization (A1 and B1) and KRs catalyzing both epimerization and ketoreduction (A2 and B2) (Keatinge-Clay, 2007). A2-type KRs usually contain a histidine three residues N-terminal to the catalytic tyrosine while a conserved proline is usually observed two residues downstream of the catalytic tyrosine in B2-type KRs. However, epimerizing KRs lacking these diagnostic motifs do exist. In equilibrium isotope exchange (EIX) experiments, only epimerizing KRs can catalyze the time-dependent washout of deuterium from the C2 position, providing conclusive evidence that KR domains control the stereochemistry of C2 substituents of polyketide intermediates (Garg et al., 2013).
Crystal structures of all four types of KR have been solved to elucidate the stereocontrol of KR domains (Keatinge-Clay, 2007; Keatinge-Clay and Stroud, 2006; Zheng et al., 2013; Zheng et al., 2010). KRs belong to the short chain dehydrogenase/reductase (SDR) superfamily. A KR domain contains an N-terminal structural subdomain and a C-terminal catalytic subdomain, each a variation of the Rossmann fold common to SDR enzymes. The structural subdomain lacks the traditional catalytic residues and coenzyme binding site of SDR enzymes, consistent with its role of stabilizing the catalytic subdomain that possesses the active site triad of tyrosine, serine and lysine characteristic of the SDR superfamily. Regardless of the type of KR, the NADPH binds in the catalytic subdomain with the 4′-pro-S hydride facing a catalytic tyrosine. The conserved A-type (“W”) and B-type (LDD) sequence motifs reside on opposite sides of the catalytic grooves and are hypothesized to guide the pantetheine-bound polyketide intermediate to enter the active site from different sides, thereby presenting the alternative face of the C3 keto group to NADPH (Fig. 1) (Caffrey, 2003; Reid et al., 2003). Altering these motifs can indeed switch the stereo-control exerted by KRs, at least in vitro (Baerga-Ortiz et al., 2006). The KR from the second module of amphotericin PKS (AmpKR2) is an A1-type KR reducing the D-methyl epimer of (±)-2-methyl-3-oxopentanoate-S-NAC whereas an AmpKR2 (G355T/Q364H) double mutant behaved as an A2-type KR toward the racemic NAC thioester (Zheng et al., 2013). Since none of the previously reported KR structures has been solved as a ternary complex with both coenzyme and substrate, the detailed roles of these motifs in the stereochemical control of KR-catalyzed reactions remain obscure.
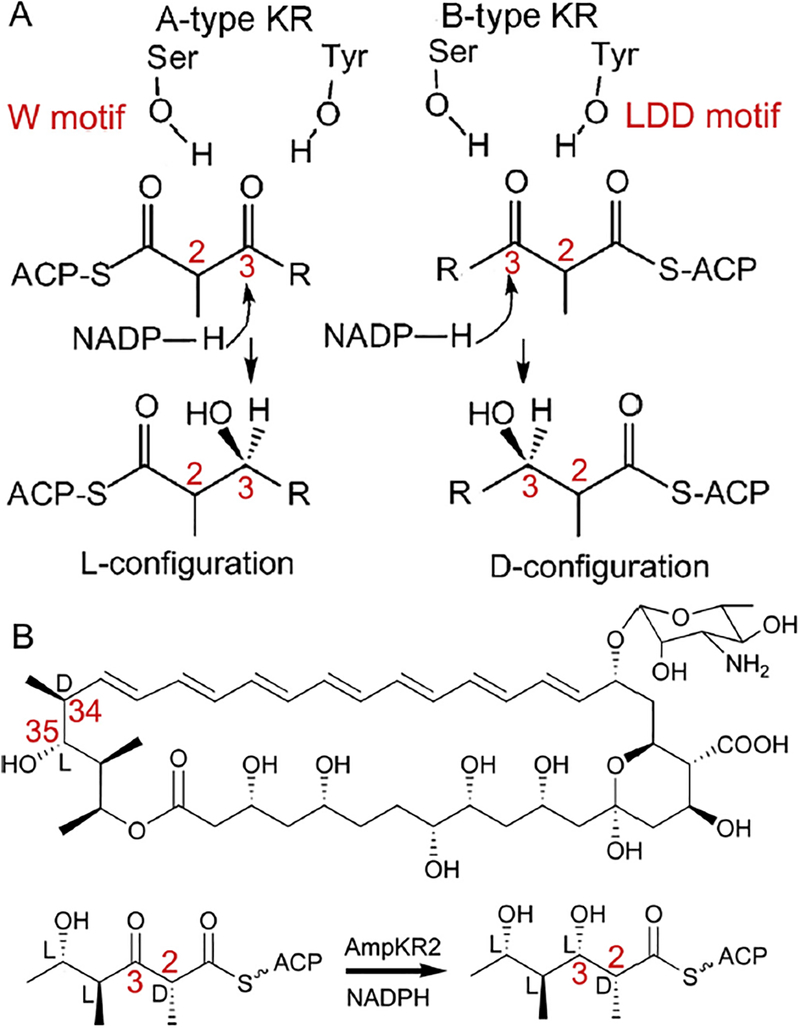
Fig. 1.
The proposed mechanism for control of the stereochemistry of C3 hydroxyl substituents by a KR domain. (A) The W motif of A-type KRs and the LDD motif of B-type KRs are hypothesized to direct the substrate to enter the active site from opposite sides. (B) AmpKR2 in Amphotericin biosynthesis. AmpKR2 reduces the C3 keto group of (2D, 4L, 5L)-5-hydroxy-2,4-dimethyl-3-oxohexanoyl-AmpACP2 to an L-hydroxyl and preserves the D-orientation of the C2-methyl group.
Herein, we report a ternary structure of the A1-type AmpKR2 complexed with NADP+ and 2-methyl-3-oxopentanoyl-pantetheine. The pantetheine handle is oriented in the substrate binding groove along the lid helix and forms a hydrogen bond to the conserved “W” motif of A-type KRs. The 2-methyl-3-oxopentanoyl group is positioned on top of the coenzyme nicotinamide through hydrogen bonding to the catalytic tyrosine and serine. Therefore, the re-side of the C3 keto group faces the pro-4S hydride to generate an L-hydroxyl in the KR-catalyzed reduction. The ternary structure of a G335T/Q364H mutant reveals a slightly larger 2-methyl-3-oxopentanoyl binding pocket compared with the wild type AmpKR2. Functional assays suggest the wild type AmpKR2 exclusively prefers (2D)-2-methyl-3-oxopentanoyl-pantetheine to (2L)-2-methyl-3-oxopentanoyl-pantetheine, while the G355T/Q364H mutant reduces the two epimers to (2D, 3L)- and (2L, 3L)-products almost equally.
2. Material and methods
2.1. Protein expression and purification
The expression and purification of AmpKR2 and its mutant have been described previously (Zheng et al., 2013). Briefly, Escherichia coli BL21 (DE3) cells harboring the expression plasmid were grown to OD600 = 0.4 at 37 °C in LB medium containing 50 mg/ml kanamycin. The culture was cooled to 16 °C, supplemented with 0.5 mM IPTG, and incubated for another 12 h. Cells were collected by centrifugation, re-suspended in lysis buffer containing 500 mM NaCl and 50 mM Tris (pH 7.5) and disrupted by sonication. Cell debris was removed by centrifugation at 20,000g for 45 min. The supernatant was loaded onto Nickel-NTA resin equilibrated by lysis buffer followed by washing with lysis buffer containing 15 mM imidazole (pH 7.5). The his-tagged protein was eluted with lysis buffer containing 300 mM imidazole and polished by a Superdex 200 gel filtration column equilibrated with 150 mM NaCl and 10 mM Tris (pH 7.5). The resulting protein was exchanged into a buffer containing 25 mM NaCl, 10 mM Tris (pH 7.5), and 1 mM DTT (pH 7.5), and concentrated to 16 mg/ml. The pH was measured at 20 °C unless mentioned otherwise.
2.2. Synthesis of 2-methyl-3-oxopentanoyl-pantetheine
Propionyl chloride (0.5 mL, 5.7 mmol, 1.0 eq) was added dropwise to a stirred solution of triethylamine (0.8 mL, 5.7 mmol, 1.0 eq.) in 10 mL dichloromethane (DCM). After incubation for an hour at room temperature, the reaction mixture was concentrated in vacuo. The organic residue was chromatographed on silica gel with petroleum ether/ethyl acetate (10:1) to yield the desired 4-ethylidene-3-methyl-2-oxetanone as a clear oil which was stored at −20 °C. D-pantethine (554 mg, 1 mmol, 1.0 eq.) and DTT (154 mg, 1 mmol, 1.0 eq.) were dissolved in 10 mL H2O and incubated at 60 °C for 1 h. The resulting D-pantetheine was concentrated in vacuo and redissolved in 10 mL methanol. Following supplementation with 4-ethylidene-3-methyl-2-oxetanone (2.2 equiv) and triethylamine (2.0 equiv), the stirred reaction mixture was incubated for an additional 1 h at room temperature and then concentrated. The crude product was purified by silica gel column chromatography with ethyl acetate/acetone (3:1) to give 2-methyl-3-oxopentanoyl-pantetheine as a clear oil.
δ1H (600 MHz; CDCl3): 0.92 (s, 3H), 1.01 (s, 3H), 1.06 (t, J = 7.2 Hz, 3H), 1.38 (d, J = 7.1 Hz, 3H), 2.42 (t, J = 5.9 Hz, 2H),2.52–2.63 (m, 2H), 2.98–3.14 (m, 2H), 3.43 (q, J = 6.1 Hz, 1H), 3.47 (s, 1H), 3.48 (s, 2H), 3.52–3.59 (m, 2H), 3.80 (q, J = 7.1 Hz, 1H), 4.00 (d, J = 3.6 Hz, 1H), 6.52 (s, 1H), 7.40 (bt, J = 5.5 Hz, 1H).
2.3. Crystallization and structure determination
For crystallization of AmpKR2 and its mutant, 20% glycerol and 3 mM NADP+ were added to the protein stock solution. Crystals of were grown in sitting drops containing 2 μL protein solution and 1.5 μL precipitant solution (3.4 M ammonium sulfate, 0.1 M HEPES, pH 7.5) at 20 °C. To get the ternary complex, a crystal was soaked in crystallization buffer containing 3 mM 2-methyl-3-oxopantanoate-pantetheine and 3 mM NADP+ for 8–24 h at 20 °C. Data were collected at Shanghai Synchrotron Radiation Facility Beamline BL18U and BL19U and processed with HKL2000. The AmpKR2-NADP+ binary structure (PDB code 3MJE) was used as a search model for molecular replacement in Phaser (McCoy et al., 2007). The model was built in Coot and refined through Refmac (Emsley and Cowtan, 2004; Vagin et al., 2004). Structure coordinates have been deposited in the Protein Data Bank under accession codes 5XWV (AmpKR2 ternary complex) and 5XWW (AmpKR2 G355T/Q364H ternary complex).
2.4. Kinetic assays
The kinetic constants of AmpKR2 and the G355T/Q364H mutant with 2-methyl-3-oxopentanoyl-pantetheine were determined by monitoring the NADPH absorbance at 340 nm over 14 min. All assays were performed at 22 °C in 100 μL of HEPES buffer (50 mM, pH 7.5) containing 0.5–16 mM 2-methyl-3-oxopentanoyl-pantetheine, 1.3 mM NADPH, 6–24 μM enzyme, 10% glycerol, and 100 mM NaCl. Each reaction was performed in triplicate and corrected for the nonenzymatic consumption of NADPH. Initial velocities were plotted against substrate concentration and fit to Michaelis-Menten kinetics to obtain the apparent Km and the apparent kcat.
2.5. Stereochemical assays
Stereochemical assays of KR-catalyzed reduction of 2-methyl-3-oxopentanoyl-pantetheine were performed by a previously reported procedure (Kwan et al., 2011). Reactions were carried out in a 200 μL solution containing 100 mM NaCl, 100 μM NADP+, 500 mM glucose, 50 μM glucose dehydrogenase, 1 mM 2-methyl-3-oxopentanoyl-pantetheine, 6 μM KR, 10% glycerol, and 50 mM HEPES (pH 7.5). After incubation at 30 °C overnight, the products were extracted with 1 mL of ethyl acetate, which was then removed by vacuum evaporation. The residue was redissolved in 200 μL NaHCO3 (50 mM) containing 20 mM NAC and incubated for 12 h at room temperature. The mixture was extracted with 1 mL of ethyl acetate. After evaporation of the organic solvent, the residue was resuspended in 200 μL 93% hexane and 7% ethanol. The products were separated and compared with known standards of the four diastereoisomeric diketides using a ChiralCel OC-H column (250 × 4.6 mm) on an Agilent 1260 HPLC system (Siskos et al., 2005). An isocratic mobile phase of 93% hexane/7% ethanol was run at a flow rate of 0.8 mL/min for 1 h.
3. Results
3.1. Crystallization of ternary complex
During the biosynthesis of amphotericins, AmpKR2 functions as an A1-type KR that reduces the C3 keto group of the (2D, 4L, 5L)-5-hydroxy-2,4-dimethyl-3-oxohexanoate holo-AmpACP2 thioester to an L-hydroxyl group and preserves the C2 D-methyl group generated in the KS-catalyzed condensation step (Fig. 1B) (Caffrey et al., 2001). Previously, we have solved the high-resolution structures of unliganded AmpKR2 (3MJC) and a binary complex with the coenzyme (3MJE) using crystals grown in 3.1 M ammonium sulfate, 200 mM NaCl, and 100 mM sodium cacodylate (pH 6.75). The crystals can also be grown in 2.5 M DL-malic acid (pH 7.0) and two malic acid molecules are observed in the active site of AmpKR2 (3MJS) (Zheng et al., 2010). In the current study, crystals were grown in a slightly different crystallization condition containing 3.4 M ammonium sulfate and 0.1 M HEPES (pH 7.5). Firstly, we tried to obtain a ternary complex with 2-methyl-3-oxopentanoyl-S-NAC which is widely used in the functional assays of KR domains, but the expected electron density was not observed. Steady-state kinetic parameters toward (±)-2-methyl-3-oxopentanoyl-S-NAC have been determined previously for AmpKR2 (kcat/Km = 2.28 ± 0.36 M−1s−1; kcat = 0.28 ± 0.01 s−1 and Km = 120 ± 40 mM) and its G355T/Q364H mutant (kcat/Km = 8.74 ± 1.32 M−1s−1; kcat = 0.52 ± 0.01 s−1 and Km = 60 ± 10 mM) (Zheng et al., 2013). The low affinity may explain why we failed to capture the NAC substrate in the active sites of AmpKR2 and its G355T/Q364H mutant. Thus, we sought to determine the ternary structure complexed with a pantetheine-bound substrate surrogate based on the hypothesis that a pantetheine handle would make more extensive interactions with the AmpKR2 active site. Structures were solved by molecular replacement and well-defined electron density for NADP+ and 2-methyl-3-oxopentanoyl-pantetheine was observed in the active sites of both wild type AmpKR2 and its G355T/Q364H mutant (Figs. 2 and 3, and Table 1). The crystals were also soaked in crystallization buffer containing 3 mM pantetheine, but only very weak electron density was observed suggesting that the 2-methyl-3-oxopentanoyl group promotes binding of the ligand. Two hydrogen bonds are observed between the 2-methyl-3-oxopentanoyl group and the active site residues (Figs. 2 and 3).
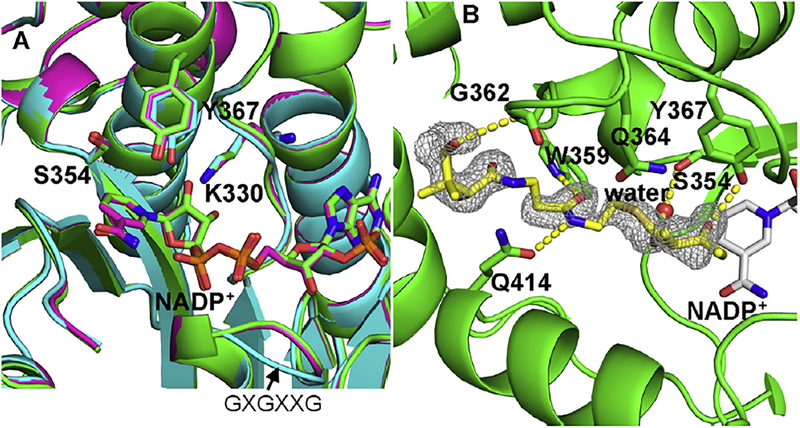
Fig. 2.
The ternary complex of wild type AmpKR2 with NADP+ and 2-methyl-3-oxopentanoyl-pantetheine. (A) Active site comparison of wild type unliganded AmpKR2 (cyan), binary complex (purple), and ternary complex (green). The loop contains GXGXXG motif is labelled. (B) Hydrogen bonds between the substrate and active site of AmpKR2. The |Fo| – |Fc| omit map for the substrate was superimposed on the final model of the wild type AmpKR2 ternary complex and contoured at 2.5 σ.
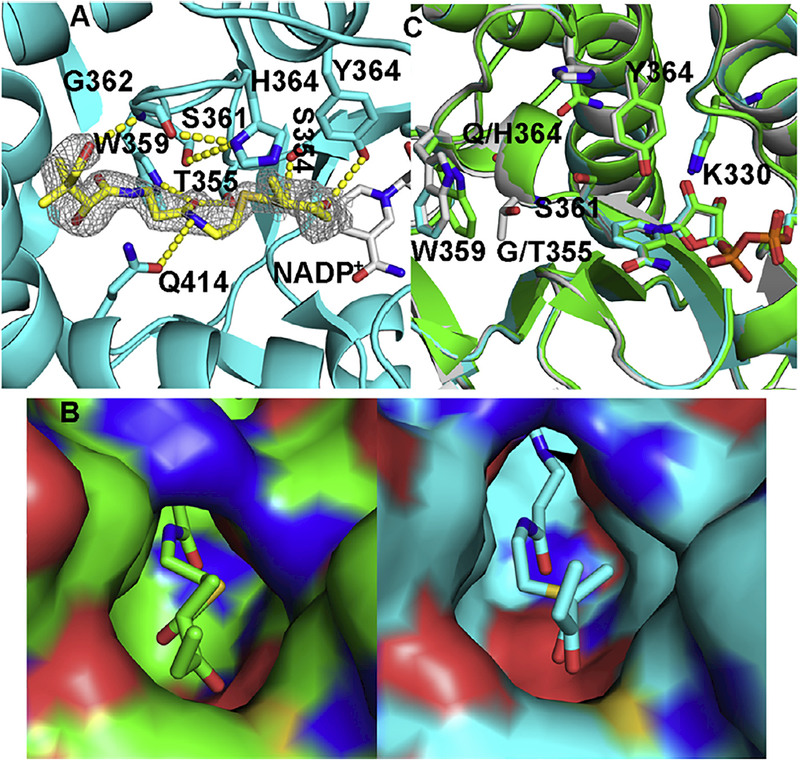
Fig. 3.
Ternary complex of AmpKR2 G355T/Q364H mutant. (A) Hydrogen bonds between the substrate and the active site of the AmpKR2 G355T/Q364H mutant. The |Fo| – |Fc| omit map of the substrate was superimposed on the final model of the ternary complex of AmpKR2 G355T/Q364H mutant and contoured at 2.5 σ. (B) The 2-methyl-3-oxopentanoyl binding pocket of wild type AmpKR2 (green) and the G355T/Q364H mutant (cyan). (C) Structural comparison of ternary structure of wild type AmpKR2 (green), the ternary structure of the G335T/Q364H mutant (cyan), and the binary structure of the G335T/Q364H mutant (grey).
Table 1.
Data collection and refinement statistics (molecular replacement).
| AmpKR2 | AmpKR2 G355T/Q364H | |
|---|---|---|
| Data collection | ||
| Space group | P1 | P1 |
| a, b, c (Å) | 61.319 63.564 71.886 | 61.066 63.627 71.586 |
| α,β,γ (°) | 73.094 67.196 89.759 | 72.670 67.211 89.879 |
| Resolution (Å) | 50–1.80 | 50–1.96 |
| Rmerge | 0.065 (0.265) | 0.10 (0.368) |
| I/σI | 9.8 (2.0) | 8.6 (2.0) |
| Completeness (%) | 91.6 (89.5) | 94.6 (90.6) |
| Redundancy | 1.9 (1.8) | 1.8 (1.6) |
| Refinement | ||
| Resolution (Å) | 50–1.8 | 50–1.96 |
| No. reflections | 76,537 | 60,085 |
| Rwork/Rfree | 0.18/0.22 | 0.18/0.22 |
| No. atoms | ||
| Protein | 6938 | 6946 |
| NADP+ | 96 | 96 |
| 8H6a | 52 | 52 |
| Water | 266 | 102 |
| B-factors | ||
| Protein | 24 | 32 |
| NADP+ | 19 | 25 |
| 8H6a | 43 | 51 |
| Water | 24 | 27 |
| Rmsd | ||
| Bond lengths (Å) | 0.009 | 0.007 |
| Bond angles (°) | 0.943 | 1.088 |
(2D)-2-methyl-3-oxopentanoyl-pantetheine.
3.2. Substrate binding in the active site of wild type AmpKR2
The ternary structure is very similar to the binary complex with NADP+ (PDB 3MJE, 0.16 Å rmsd) and the unliganded structure (PDB 3MJC, 0.20 Å rmsd) (Fig. 2) (Zheng et al., 2010). The catalytic triad residues (K330, S354, and Y367) are well ordered and organized as observed in the binary structure. The phenolic hydroxyl group of the catalytic Y367 in the unliganded structure (cyan) is shifted ~1 Å away from the position observed in binary and ternary structures (Fig. 2A). The slightly displacement of Y367 is likely induced by the binding of NADP+ since the catalytic residues in binary and ternary complex exactly match each other as shown in Fig. 3A. The binding of NADP+ also results in slightly conformation of a loop containing the conserved GXGXXG dinucleotide binding motif (Fig. 2A). It has been widely accepted that the hydroxyl group of the catalytic tyrosine functions as a proton donor following a hydride transfer from NADPH to the electrophilic carbon of the substrate (Kavanagh et al., 2008).
The 2-methyl-3-oxopentanoyl group is positioned appropriately for reduction by the SDR catalytic machinery (Fig. 2B). Y367 forms a hydrogen bond with the C3 keto group of the substrate and S354 forms a water mediated hydrogen bond with the thioester carbonyl group. Consequently, the C3 of the substrate is positioned 4.3 Å from NADP+ with the re-face of the keto toward the pro-4S hydride of the coenzyme nicotinamide group, consistent with the observation of an L-hydroxyl on C35 of amphotericin. The conserved W motif of A-type KRs has been hypothesized to guide the substrate into the catalytic groove. As expected, a hydrogen bond is observed between the side-chain indole NH of W359 and the C4′ carbonyl group of the pantetheine handle (Figs. 2B and4). The amide NH forms another hydrogen bond with the side chain of Q414 located on the C-terminal end of the lid helix. Q414 is not conserved in A-type KRs, suggesting that this hydrogen bond is not obligatory for orientation of the substrate. The C11′ hydroxyl group of the pantetheine handle forms a hydrogen bond with the backbone NH of G362, whereas this hydrogen bond could not form for ACP-linked substrates. A large part of the substrate binding groove is formed by the nicotinamide ring so that the substrate can only bind effectively after the formation of a binary complex with the coenzyme. In contrast, the reduced product has to depart first before NADP+ can vacate the active site. Such an ordered, sequential mechanism with the coenzyme binding first and leaving last has been proposed by kinetic studies of SDR enzymes (Kavanagh et al., 2008).
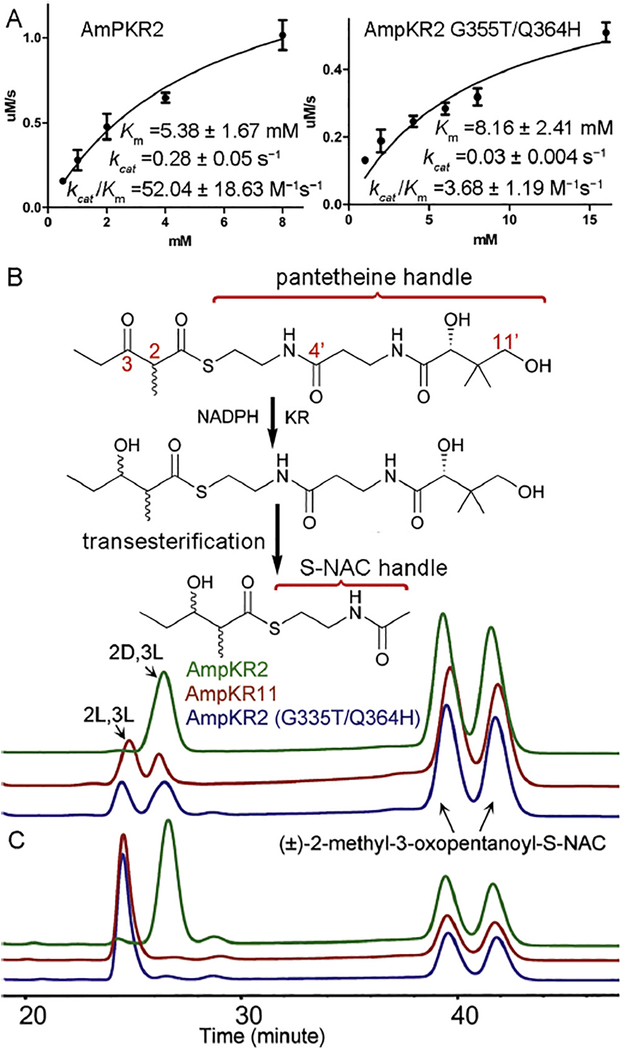
Fig. 4.
Functional assays of A1-type AmpKR2, the AmpKR G335T/Q364H mutant, and A2-type AmpKR11. (A) Kinetic assays of A1-type AmpKR2 and the AmpKR G335T/Q364H mutant. (B) Reduction of (±)-2-methyl-3-oxopentanoyl-pantetheine. The reduced diketide- pantetheine was converted to the corresponding diketide-S-NAC through a thiol-thioester exchange reaction. Additional (±)-2-methyl-3-oxopentanoyl-S-NAC was supplemented as a reference in the chiral HPLC analysis. (C) Reduction of (±)-2-methyl-3-oxopentanoyl-S-NAC. The HPLC traces are colored as in panel A.
3.3. Substrate binding in the active site of a G355T/Q364H mutant
AmpKR2 is an A1-type KR and reduces (±)-2-methyl-3-oxopentanoyl-S-NAC almost exclusively to (2D, 3L)-2-methyl-3-hydro-xypentanoyl-S-NAC (Zheng et al., 2010). The side chain of Q364 is 2.8 Å from the thioester sulfur and 3.2 Å from the C2 methyl substituent (Fig. 2B). The AmpKR2 G355T/Q364H mutant almost exclusively prefers (2L)-2-methyl-3-oxopentanoyl-S-NAC and reduces it to a (2L, 3L)-product (Zheng et al., 2013). To better understand the molecular basis for this reversed stereospecificity, the structure of the G355T/Q364H mutant complexed with pantetheine-bound substrate was solved (Fig. 3). The ternary structure of the G355T/Q364H mutant is very similar to that of the wild type AmpKR2 (0.22 Å rmsd) except the displacement of W359 that has already been observed in the binary structure of the G355T/Q364H mutant (Fig. 3C) (Zheng et al., 2013). The catalytic residues in the ternary structure of the G355T/Q364H mutant exactly match these in the binary structure without the substrate (Fig. 3C). H364 is within hydrogen-bond distance of the G362 carbonyl and the S361 side chain. Compared with the Q364 residue in the wild type enzyme, the H364 side chain of the mutant swings away from the substrate binding groove (Fig. 3C). Consequently, the 2-methyl-3-oxopentanoyl binding pocket of the G335T/Q364H mutant is slightly larger than that of the wild type AmpKR2 (Fig. 3B).
3.4. Functional assays with pantetheine-bound substrate
Since the ternary structures reported here suggest that the pantetheine handle contributes to the enzyme-substrate interactions, NADPH-dependent kinetic assays were carried out using the 2-methyl-3-oxopentanoyl-pantetheine substrate. As expected, both AmpKR2 (kcat/Km = 52.04 ± 18.63 M−1s−1, kcat = 0.28 ± 0.05 s−1 and Km = 5.38 ± 1.67 mM) and its G355T/Q364H mutant (k/K 1 cat m = 3.68 ± 1.19 M− s−1, kcat = 0.03 ± 0.004 s−1 and Km = 8.16 ± 2.41 mM) exhibit lower Km values toward the pantetheine-bound thioester compared to the NAC substrate (Fig. 4A). We next investigated the stereospecificity of AmpKR2 and its G355T/Q364H mutant towards the pantetheine-bound substrate. By a thiol-thioester exchange reaction, the reduced pantetheinyl-diketide was converted to the corresponding diketide-S-NAC (Kwan et al., 2011), of which the stereo-chemistry could be easily assigned by chiral HPLC analysis, as reported previously (Siskos et al., 2005). Wild type AmpKR2 reduced the pantetheine-bound substrate exclusively to a (2D, 3L)-product as expected, whereas both a (2D, 3L)-product (~57%) and a (2L, 3L)-product (~43%) were generated by the G355T/Q364H mutant (Fig. 4B). The influence of the pantetheine handle on KR stereospecificity was further confirmed by the observation that AmpKR11, an A2-type KR that converts (±)-2-methyl-3-oxopentanoate-SNAC to a (2L, 3L)-product (~100%) (Zheng et al., 2013), also reduced the pantetheine-bound substrate to ~37% of the (2D, 3L)-product and ~63% of the (2L, 3L)-product (Fig. 4C).
4. Discussion
A-type and B-type KRs generate C2 hydroxyl substituents of opposite stereochemistry in the biosynthesis of polyketides, despite sharing similarly organized active sites and binding NADPH with the 4′-pro-S hydride facing the catalytic tyrosine. Sequence motifs highly correlated with the stereochemical outcomes of KR-catalyzed reactions have been revealed for both A-type KRs (the W motif) and B-type KRs (the LDD motif) (Keatinge-Clay, 2007). These motifs are located on opposite sides of the catalytic tyrosine and are hypothesized to control the directions of entry of the pantetheine-bound substrates into the active site (Fig. 1) (Caffrey, 2003; Reid et al., 2003). Therefore, different face of the C3 keto group can be presented to NADPH for hydride transfer. In agreement with this hypothesis, the substrate enters the active site of AmpKR2 from the side opposite the coenzyme nicotinamide group with the conserved W359 forming a hydrogen bond with pantetheine handle (Fig. 2B). Mutation to W359 of AmpKR2 and the corresponding residue in EryKR1 does affect the stereospecificity of KR domains (Baerga-Ortiz et al., 2006; Zheng et al., 2010). The “W” motifs of PlmKR1 and AmpKR11 are buried more deeply and may not form hydrogen bonds with the pantetheine handles directly (Bonnett et al., 2013; Zheng et al., 2013). The ternary structure of a B-type KR complexed with a substrate is still unavailable. However, the structures of a high-molecular weight FabG complexed with hexanoyl-CoA (PDB: 3V1U) (Dutta et al., 2013) and a substrate-bound acetoacetyl-CoA reductase (PDB: 4N5M) (Kim et al., 2014) reveal that an aspartate equivalent to the third residue of the LDD motif of a B-type KR helps orientate the pantetheine handle by forming a hydrogen bond with the C4′ carbonyl group (Fig. 4B). Inspired by these two ternary structures, a substrate has been modeled into the active site of a B-type KR from the side closest to the coenzyme nicotinamide group (Keatinge-Clay, 2007; Keatinge-Clay and Stroud, 2006). Functional assays revealed significant influence of the pantetheine handle on the catalytic efficiency and stereochemistry of the A-type AmpKR2 domain (Fig. 4B and C). In contrast, the B-type EryKR1 domain shows almost identical kinetic parameters and stereospecificity toward NAC- and pantetheine-bound thioesters (Kwan et al., 2011).
AmpKR2 is stereospecific toward the pantetheine-bound surrogate and reduces the 2D epimer out of a racemic mixture to the expected (2D, 3L)-product. Consequently, (2D)-2-methyl-3-oxopentanoyl-pantetheine was modeled in the AmpKR2 active site (Fig. 2B). MD simulations of AmpKR2 with NAC thioesters suggest the 2D epimer is retained in a conformation close to the reactive configuration, whereas Q364 can form a hydrogen bond to the 2L epimer to trap it in an un-reactive conformation (Mugnai et al., 2015). Based on the structural comparison between A1- and A2-type KRs, a G355T/Q364H mutant of AmpKR2 was engineered which exhibits reversed stereospecificity toward the C2 methyl group of the racemic NAC thioester, reducing the 2L epimer to a (2L, 3L)-product (Zheng et al., 2013). Surprisingly, when pantetheine thioester was used as the substrate, both (2D, 3L)-product (~43%) and a (2L, 3L)-product (~57%) were produced by the G355T/Q364H mutant (Fig. 4B). The Felkin-Ahn rule predicts the reactions catalyzed by A2-type KRs are more favorable compared with the reactions catalyzed byA1-type KRs (Bailey et al., 2016; Mengel and Reiser, 1999). Therefore, the generation of a (2L, 3L)-product by the G355T/Q364H mutant is likely due to a decreased capability for stereocontrol. The A2-type AmpKR11 reduces both epimers of the pantetheine thioester to 3L-products (Fig. 4C), suggesting that proper protein-protein interactions with the ACP domain are necessary for AmpKR11 to maintain its intrinsic stereoselectivity.
Deciphering the mechanism for the epimerization of C2 methyl substituents has been much more complicated. Complex polyketides harbor numerous methyl groups possessing L stereochemistry, whereas the KSs of modular PKSs catalyze the generation of (2D)-2-methyl-3-ketoacyl-ACPs exclusively during the elongation of polyketide intermediates (Castonguay et al., 2007; Weissman et al., 1997; Xie et al., 2017). In addition, the binding of the acyl group to a patch on the surface of the ACP prevents the water-catalyzed epimerization observed for the C2 methyl group of the NAC- or pantetheine-bound thioesters (Castonguay et al., 2007; Vance et al., 2016). The EIX assays provide direct evidence that A2- and B2-type KRs have intrinsic epimerase activities. Disappointingly, the AmpKR2 G355T/Q364H mutant failed to exhibit any detectable epimerase activity in the EIX assay, suggesting that the introduced double mutations only alter the diastereoselectivity of AmpKR2 (Xie et al., 2017). Site-directed mutagenesis of epimerase-active KRs suggests the epimerization reaction utilizes the same catalytic residues as the ketoreduction reaction. The proposed mechanism involves the generation of an enolate by removal of the H2 proton followed by reprotonation at C2 on the opposite face of the transient intermediate to form an epimerized (2L)-2-methyl-3-ketoacyl-ACP (Xie et al., 2016). A ternary structure of an epimerizing KR would help decode the mechanism of this intriguing epimerase activity.
5. Conclusions
We report here the first crystal structure of a PKS KR domain complexed with both NADP+ and a 2-methyl-3-oxopentanoyl-pantetheine substrate surrogate. This ternary structure shows that an A-type KR binds its substrate in a groove that runs along the lid helix. The conserved “W” motif located on the side opposite the coenzyme nicotinamide group helps orient the substrate by forming a hydrogen bond with the pantetheine handle. Functional assays reveal the pantetheine handle has significant influence on the catalytic efficiency and stereo-chemical outcome of KR-catalyzed reactions. In summary, the ternary structures presented here provide the first snapshots of how a polyke-tide intermediate binds at the active site of a modular PKS KR.
Acknowledgments
This work was supported in part by National Natural Science Foundation of China (31370101, 31570056, 31770068), National Program on Key Basic Research Project (973 program, 2013CB734002), State Key Laboratory of Pathogen and Biosecurity (Academy of Military Medical Science SKLPBS1526), Recruitment Program of Global Experts of China, and Fundamental Research Funds for the Central Universities.
References
- Baerga-Ortiz A, Popovic B, Siskos AP, O’Hare HM, Spiteller D, Williams MG, Campillo N, Spencer JB, Leadlay PF, 2006. Directed mutagenesis alters the stereochemistry of catalysis by isolated ketoreductase domains from the erythromycin polyketide synthase. Chem. Biol 13, 277–285. [DOI] [PubMed] [Google Scholar]
- Bailey CB, Pasman ME, Keatinge-Clay AT, 2016. Substrate structure-activity relationships guide rational engineering of modular polyketide synthase ketoreductases. Chem. Commun. (Camb.) 52, 792–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnett SA, Whicher JR, Papireddy K, Florova G, Smith JL, Reynolds KA, 2013. Structural and stereochemical analysis of a modular polyketide synthase ketoreductase domain required for the generation of a cis-alkene. Chem. Biol 20, 772–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffrey P, 2003. Conserved amino acid residues correlating with ketoreductase stereo-specificity in modular polyketide synthases. Chembiochem 4, 654–657. [DOI] [PubMed] [Google Scholar]
- Caffrey P, Lynch S, Flood E, Finnan S, Oliynyk M, 2001. Amphotericin biosynthesis in Streptomyces nodosus: deductions from analysis of polyketide synthase and late genes. Chem. Biol 8, 713–723. [DOI] [PubMed] [Google Scholar]
- Castonguay R, He W, Chen AY, Khosla C, Cane DE, 2007. Stereospecificity of ketoreductase domains of the 6-deoxyerythronolide B synthase. J. Am. Chem. Soc 129, 13758–13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes J, Haydock SF, Roberts GA, Bevitt DJ, Leadlay PF, 1990. An unusually large multifunctional polypeptide in the erythromycin-producing polyketide synthase of Saccharopolyspora erythraea. Nature 348, 176–178. [DOI] [PubMed] [Google Scholar]
- Dutta D, Bhattacharyya S, Roychowdhury A, Biswas R, Das AK, 2013. Crystal structure of hexanoyl-CoA bound to beta-ketoacyl reductase FabG4 of Mycobacterium tuberculosis. Biochem. J 450, 127–139. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K, 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Garg A, Khosla C, Cane DE, 2013. Coupled methyl group epimerization and reduction by polyketide synthase ketoreductase domains. Ketoreductase-Catalyzed equilibrium isotope exchange. J. Am. Chem. Soc 135, 16324–16327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CM, McPherson M, McDaniel RN, Fu H, Cane DE, Khosla C, 1998. Alcohol stereochemistry in polyketide backbones is controlled by the β-Ketoreductase domains of modular polyketide synthases. J. Am. Chem. Soc 120, 2478–2479. [Google Scholar]
- Kavanagh KL, Jornvall H, Persson B, Oppermann U, 2008. Medium- and short-chain dehydrogenase/reductase gene and protein families: the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell. Mol. Life Sci 65, 3895–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keatinge-Clay AT, 2007. A tylosin ketoreductase reveals how chirality is determined in polyketides. Chem. Biol 14, 898–908. [DOI] [PubMed] [Google Scholar]
- Keatinge-Clay AT, Stroud RM, 2006. The structure of a ketoreductase determines the organization of the beta-carbon processing enzymes of modular polyketide synthases. Structure 14, 737–748. [DOI] [PubMed] [Google Scholar]
- Kim J, Chang JH, Kim EJ, Kim KJ, 2014. Crystal structure of (R)-3-hydroxybutyryl-CoA dehydrogenase PhaB from Ralstonia eutropha. Biochem. Biophys. Res. Commun 443, 783–788. [DOI] [PubMed] [Google Scholar]
- Kwan DH, Tosin M, Schlager N, Schulz F, Leadlay PF, 2011. Insights into the stereospecificity of ketoreduction in a modular polyketide synthase. Org. Biomol. Chem 9, 2053–2056. [DOI] [PubMed] [Google Scholar]
- Marsden AF, Caffrey P, Aparicio JF, Loughran MS, Staunton J, Leadlay PF, 1994. Stereospecific acyl transfers on the erythromycin-producing polyketide synthase. Science 263, 378–380. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ, 2007. Phaser crystallographic software. Appl. Crystallogr 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengel A, Reiser O, 1999. Around and beyond Cram’s Rule. Chem. Rev 99, 1191–1224. [DOI] [PubMed] [Google Scholar]
- Mugnai ML, Shi Y, Keatinge-Clay AT, Elber R, 2015. Molecular dynamics studies of modular polyketide synthase ketoreductase stereospecificity. Biochemistry 54, 2346–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piasecki SK, Taylor CA, Detelich JF, Liu J, Zheng J, Komsoukaniants A, Siegel DR, Keatinge-Clay AT, 2011. Employing modular polyketide synthase ketoreductases as biocatalysts in the preparative chemoenzymatic syntheses of diketide chiral building blocks. Chem. Biol 18, 1331–1340. [DOI] [PubMed] [Google Scholar]
- Reid R, Piagentini M, Rodriguez E, Ashley G, Viswanathan N, Carney J, Santi DV, Hutchinson CR, McDaniel R, 2003. A model of structure and catalysis for ketoreductase domains in modular polyketide synthases. Biochemistry 42, 72–79. [DOI] [PubMed] [Google Scholar]
- Siskos AP, Baerga-Ortiz A, Bali S, Stein V, Mamdani H, Spiteller D, Popovic B,Spencer JB, Staunton J, Weissman KJ, Leadlay PF, 2005. Molecular basis of Celmer’s rules: stereochemistry of catalysis by isolated ketoreductase domains from modular polyketide synthases. Chem. Biol 12, 1145–1153. [DOI] [PubMed] [Google Scholar]
- Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN, 2004. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr 60, 2184–2195. [DOI] [PubMed] [Google Scholar]
- Valenzano CR, Lawson RJ, Chen AY, Khosla C, Cane DE, 2009. The biochemical basis for stereochemical control in polyketide biosynthesis. J. Am. Chem. Soc 131, 18501–18511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance S, Tkachenko O, Thomas B, Bassuni M, Hong H, Nietlispach D, Broadhurst W, 2016. Sticky swinging arm dynamics: studies of an acyl carrier protein domain from the mycolactone polyketide synthase. Biochem. J 473, 1097–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman KJ, Timoney M, Bycroft M, Grice P, Hanefeld U, Staunton J, Leadlay PF, 1997. The molecular basis of Celmer’s rules: the stereochemistry of the condensation step in chain extension on the erythromycin polyketide synthase. Biochemistry 36, 13849–13855. [DOI] [PubMed] [Google Scholar]
- Xie X, Garg A, Khosla C, Cane DE, 2017. Mechanism and stereochemistry of polyketide chain elongation and methyl group epimerization in polyether biosynthesis. J. Am. Chem. Soc 139, 3283–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Garg A, Keatinge-Clay AT, Khosla C, Cane DE, 2016. Epimerase and reductase activities of polyketide synthase ketoreductase domains utilize the same conserved tyrosine and serine residues. Biochemistry 55, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Piasecki SK, Keatinge-Clay AT, 2013. Structural studies of an A2-type modular polyketide synthase ketoreductase reveal features controlling alpha-substituent stereochemistry. ACS Chem. Biol 8, 1964–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Taylor CA, Piasecki SK, Keatinge-Clay AT, 2010. Structural and functional analysis of a-type ketoreductases from the amphotericin modular polyketide synthase. Structure 18, 913–922. [DOI] [PubMed] [Google Scholar]