Abstract
A hallmark of breast tumors is its spatial heterogeneity that includes its distribution of cancer stem cells and progenitor cells, but also heterogeneity in the tumor microenvironment. In this study we focus on the contributions of stromal cells, specifically macrophages, fibroblasts, and endothelial cells on tumor progression. We develop a computational model of triple-negative breast cancer based on our previous work and expand it to include macrophage infiltration, fibroblasts, and angiogenesis. In vitro studies have shown that the secretomes of tumor-educated macrophages and fibroblasts increase both the migration and proliferation rates of triple-negative breast cancer cells. In vivo studies also demonstrated that blocking signaling of selected secreted factors inhibits tumor growth and metastasis in mouse xenograft models. We investigate the influences of increased migration and proliferation rates on tumor growth, the effect of the presence on fibroblasts or macrophages on growth and morphology, and the contributions of macrophage infiltration on tumor growth. We find that while the presence of macrophages increases overall tumor growth, the increase in macrophage infiltration does not substantially increase tumor growth and can even stifle tumor growth at excessive rates.
Keywords: tumor, microenvironment, immune cells, angiogenesis, cancer stem cells, hypoxia
Introduction
Tumor-associated macrophages (TAMs) play a significant role in breast cancer growth, invasion, and metastasis1. TAMs are involved in a range of processes that affect cancer growth, such as regulating angiogenesis2–4, inflammation5–7, and therapy resistance8. Macrophage colony-stimulating factor 1 (CSF1) secreted by breast cancer cells is a major factor for macrophage recruitment into the tumor9. Hypoxia was found to stimulate macrophage recruitment in MDA-MB-231 (MB231 for brevity) cells through signaling loops between the MB231 cells and macrophages leading to macrophage recruitment10. Once macrophages are recruited to the breast tumor, they are influenced by breast cancer secreted factors. Triple-negative breast cancer cells secrete factors, such as C-C motif chemokine ligand 2 (CCL2), that differentiate recruited macrophages from immune system enhancing M1-like macrophages11, into immunosuppressive M2-like macrophages12. This conversion from M1-like to M2-like is often mediated by hypoxic cancer cells13. High levels of macrophage recruitment in triple-negative breast cancer are associated with increased metastasis and lower overall survival14. TAM populations are also involved in mediating tumor cell extravasation and intravasation15–19. Due to their many roles in cancer progression and metastasis, several papers have investigated their use as prognostic or diagnostic biomarkers20–22. The influence of macrophages on tumor growth is a complex process that needs to be better understood.
Fibroblasts are part of the connective tissue, secrete extracellular matrix (ECM) and also play a large role in wound healing23. Cancer associated fibroblasts (CAFs) are found in many different types of cancer24, including breast25, pancreatic26, prostate7, and ovarian cancer27 and are associated with poor prognosis28. One of the prominent effects of CAFs in breast cancer is that they cause the epithelial-to-mesenchymal transition to a more invasive type cancer cell through the transforming growth factor beta (TGF-β) pathway29–31. CAFs display higher levels of ECM secretions and are involved in ECM remodeling, leading to increased deposition of ECM32,33. CAFs are activated by breast cancer secreted factors such as interleukin 6 (IL6), TGF-β, and CXC-motif chemokine ligand 12 (CXCL12)34. CAFs can promote tumor growth through stromal-cell derived factor 1 (SDF-1) which binds to CXCR4 receptors and stimulates tumor cell proliferation35. CAFs also secrete several metalloproteases (MMPs) that degrade the extracellular matrix (ECM) and allow for increased cancer cell migration36. There is a complex interplay between heterogenous CAFs and breast cancer cells that have numerous effects on cancer progression37–40. Thus, stromal cells are an important aspect that needs to be included in computational models of tumor growth.
While macrophages and fibroblasts are important aspects of breast tumor growth, there are few computational models that investigate their contribution. One notable model that investigated macrophages was Knutsdottir et al. which examined the contributions of macrophages in a 3D agent-based model of tumor growth41. They incorporated epidermal growth factor (EGF)/CSF-1 paracrine signaling using a system of differential equations; they showed how paracrine signaling is necessary for the co-migration of macrophages and tumor cells41. They found that macrophages contributed to cancer cell motility and invasion. An earlier paper developed a continuous and discrete 2D model of macrophage/tumor signaling42. Another agent-based model examined the transition from immune-enhancing to immuno-suppressive macrophages within the tumor environment43. They included M1 and M2 macrophage states with M1 macrophages secreting tumor lethality signals and converting to an M2 state with exposure to M2 signals (M2S). All macrophages migrate by chemotaxis towards the M2S gradient. They included nonevolving tumor vasculature and proliferating but non-migrating tumor cells. They then predicted engineered cell-based therapies based on their modeling results. A different multiscale model of tumor growth included genetically engineered macrophages as a delivery tool for therapeutics44. Owen and Sharrot modeled the interactions between macrophages and tumors as an immune response45,46. A model based on partial differential equations (PDE) investigated the cell cycle in normal fibroblasts and in cancer cells47. Another PDE-based model investigated fibroblast secretion of EGF and its influence on tumor progression48. An agent-based model investigated the effects of macrophages on angiogenesis using CompuCell3D software platform and found that macrophages facilitated endothelial tip cell bridging49. These important studies provide a background for the present study where we investigate the combined interactions of macrophages, fibroblasts, and angiogenesis on tumor growth and morphology.
Another important aspect of the host stroma is the tumor vasculature. In order for tumors to grow past the diffusion limit of oxygen, they need to recruit new blood vessels in a process called angiogenesis50. Briefly, when tumor cells are in low oxygen environments they become hypoxic and secrete vascular endothelial growth factors (VEGF)51. This initiates sprouting angiogenesis where a branch forms off a mature blood vessel headed by a tip cell, which migrates along the VEGF gradient towards higher VEGF; the tip cell is followed by stalk cells which proliferate to extend the sprout52 followed by quiescent phalanx cells that support the sprout. There has been a large body of mathematical models investigating angiogenesis across different scales, for reviews see53,54. Here we focus on models that combine angiogenesis with tumor development and their interactions, for a review see55. Stephanou et al. developed a 2D on-lattice agent-based model of tumor-induced angiogenesis with a dorsal skinfold chamber and investigated how vascular changes could lead to tumor dormancy56. Another 2D cellular automata model investigated radiation efficacy in a hybrid model combining tumor growth and angiogenesis57. Frieboes et al. have developed models of tumor induced angiogenesis to assess the efficacy of nanoparticle drug delivery on tumor regression58,59. 3D models have investigated the interplay between oxygen and VEGF secretion levels on tumor and vascular growth60. Earlier 3D models have investigated the interplay between tumor growth and angiogenesis using an experimentally derived vasculature47,61 and analyzed the changes in the morphology of the tumor with angiogenesis from spherical to cylindrical to paddle-shaped62. Another 3D mathematical model simulated the effects of an anti-angiogenic agent, endostatin, on tumor growth and angiogenesis63. A hybrid model of cancer cell growth looked at the interplay between stalk cells, tip cells, healthy cells, and cancer cells and the several factors contributing to the cell growth, such as oxygen, glucose, VEGF, and the EGFR and TGF signaling pathways64. These models investigated the interplay between blood vessel growth and tumor expansion and are the basis for our agent-based angiogenesis model which includes the interplay between endothelial cells and macrophages and fibroblasts.
Our laboratory has been building multiscale models of cancer including the interactions between tumor cells, tumor vasculature, fibroblasts and immune cells. Agent-based models were developed examining tumor-associated angiogenesis65, cancer stem cells and CCR5+ cells in triple-negative breast cancer66,67 and immuno-infiltrates in tumors68. Receptor-ligand interactions and intracellular signaling models have been used to study the dynamics of VEGF and thrombospondin (TSP) ligands and their receptors and their effects on angiogenesis69,70. Our laboratory has also found experimentally that macrophages and fibroblasts in the tumor microenvironment (TME) increase triple-negative breast cancer (TNBC) cells proliferation and migration in vitro71.
There is a complex crosstalk between immune cells such a macrophages and tumor vasculature72, that can promote or hinder cancer development. Combination anti-angiogenic therapies and immunotherapies are now in development73. In this work, we develop a computational agent-based model which combines a triple-negative breast cancer model with an angiogenesis model and investigates the contributions of cancer-associated stromal cells to the development of triple-negative breast cancer. The model combines a developing breast tumor, tumor vasculature which responds to hypoxic cancer cells and develops through sprouting angiogenesis, and considers monocyte recruitment through the evolving vasculature that differentiate into macrophages in the tissue, as well as cancer-associated fibroblasts.
Methods
Computational
This study develops an agent-based model of triple-negative breast cancer growth based on our previous studies65–67. It includes a triple-negative breast cancer model with cancer stem cells and CCR5+/− cancer cells67 and it includes a growing vasculature based on our previous angiogenesis study65. We focus on the additional aspects of the model in this methods section.
Initial Setup
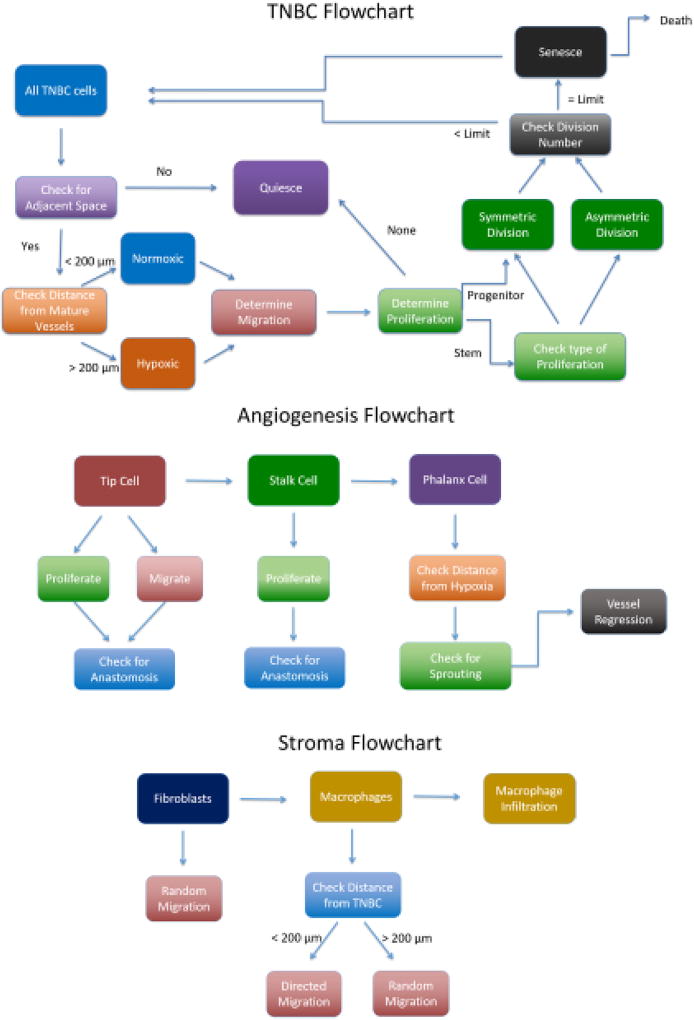
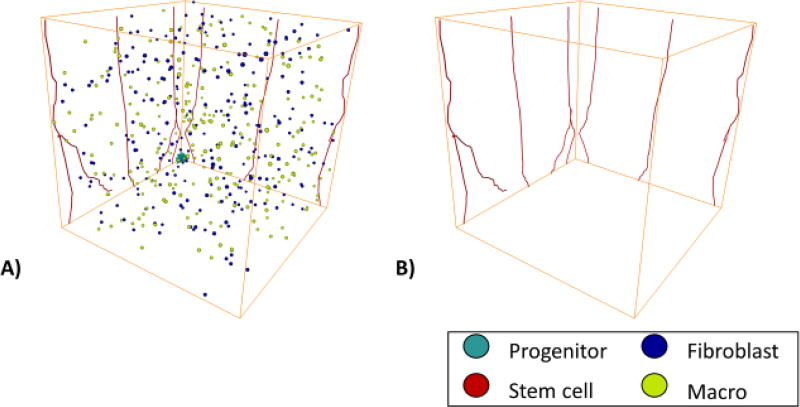
The model consists of two grids, a cellular grid of size 100×100×100 with each voxel having a size of 20 µm3, which is approximately the diameter of a MB231 cell67, and a vascular grid, of size 2000×2000×2000, with each voxel having a size of 1 µm3. The initial tumor is placed in the cellular grid at the origin, occupying the 100×80×100 µm rectangular section of the grid. The initial tumor comprises 100 cells with 20 cancer stem cells, including 1 CCR5+ stem cell, and 80 progenitor cells including 5 CCR5+ progenitor cells. Initially, 250 macrophages and 250 fibroblasts are randomly placed within the cellular grid, Figure 1A. The host vasculature is placed within the vascular grid. The host vasculature consists of 8 vessels with a radius of 5 µm: with 3 vessels on the x-axis spanning the length of the grid on the z axis approximately 900 µm apart, and another set of 3 vessels on the y-axis spanning the length of the grid on the z axis approximately 900 µm apart as shown in Fig. 1B. There are also two vessels sprouted off the outermost vessels on the y-axis, that reach towards the middle vessel on the y-axis. Each of the host vessels consists of 20 endothelial cells that can branch, which are activated at the beginning of the simulation to start sprouting angiogenesis. We assume that the vasculature can overlap with the other cells due to there being interstitial space within the tumor and the fact that vessels often become compressed in tumors. The model consists of three different sections: a TNBC tumor model, an angiogenesis model, and a stroma (macrophages and fibroblasts) model, Figure 2. Briefly, the TNBC cells determine whether they will migrate and their migration speed, then determine whether they will proliferate, and whether they will senesce. The angiogenesis model determines tip cell migration, stalk cell proliferation, and phalanx cell sprouting and regression. Fibroblasts and macrophages migrate, and the macrophage infiltration is determined. We keep track of the number of MB231 progenitor cells, MB231 stem cells, macrophages, fibroblasts, and capillaries each iteration. Each iteration is approximately 6 hours in real time. We ran each simulation in Matlab2016b with the same random seed for comparison unless otherwise stated.
Figure 1.
Initial Setup for the MB231 Tumor. A) This shows the initial setup of the 100 cancer cells and 250 randomly placed fibroblasts and macrophages. B) The initial setup of the mature vasculature in the simulation.
Figure 2.
Flowchart of the Breast Cancer Cells, Stromal Cells, and Angiogenesis Models. MB231 Model: Each cell checks whether there is free space, if so it can migrate, otherwise it becomes quiescent. A cell then decides whether it will migrate, and it migrates based on its CCR5 level, hypoxia, and proximity to stromal cells. A cell decides if it can proliferate based on its stemness, hypoxia, and proximity to stromal cells. A stem cell can proliferate symmetrically or asymmetrically whereas a progenitor cell can only proliferate symmetrically. Each cell then determines if it is hypoxic or normoxic. Senescent cells die probabilistically each iteration. Stroma model: Initially a fixed number of fibroblasts or macrophages are placed randomly in the cellular grid. Macrophages can migrate and infiltrate the tumor from mature vessels probabilistically each iteration. Macrophages migrate towards MB231 cells in proximity to them. Angiogenesis Model: A tip cell checks whether it has a neighboring stalk cell, if not it proliferates to produce a stalk cell. Otherwise it migrates based on the local VEGF concentrations. Stalk cells proliferate if they have reached their cell cycle. Branching can occur if there are hypoxic tumor cells in proximity to the vessel.
MB231 Model
The simulation runs randomly through all the MB231 tumor cells. First, we check whether the cell is hypoxic, based on its distance to the mature vasculature (dead-end capillary sprouts excluded); if the cell is further than 200 µm from the nearest vessel, we assume it is hypoxic, similarly to76. This is a surrogate measure of hypoxia, but it reflects the diffusion distance for oxygen from blood-carrying capillary vessels. Then, we check whether there are any free spaces in the 27-cell neighborhood (Moore neighborhood). If there is free space, we allow the cell to migrate. All parameters with appropriate references are summarized in Table 1.
Table 1.
| Parameter | Value | Reference |
|---|---|---|
| MB231 Stem Cell Symmetric Division Probability | 5% | 74 |
| MB231 CCR5− Migration rate | 0.83 µm/hr | 67 |
| MB231 CCR5+ Migration Rate | 8.3 µm/hr | 67 |
| Senescent Cell Death Probability | 10% | 67 |
| Cell Diameter | 20 µm | 67 |
| MB231 Division Limit | 12 divisions | 67 |
| MB231 Progenitor Division Rate | 0.5/day | 74 |
| MB231 Stem Cell Division Rate | 0.2/day | 74 |
| Stem Cell Initial Percentage | 20% | 67 |
| CCR5+ Cell Initial Percentage | 5% | 67 |
| Stem CCR5+ Initial Percentage | 1% | 67 |
| Endothelial Cell Division Rate | 0.025/hr | 65 |
| Endothelial Cell Migration Rate | 10 µm/hr | 65 |
| Host Vasculature Radius | 5 µm | 75 |
| Chemokine Secretion Threshold | 200 µm | 76 |
| Macrophage CSF1 Migration Speed | 8-fold | 77–82 |
| Macrophage Migration Rate | 3.3 µm/hr | 77–82 |
| Endothelial Segment Max Extension | 1.5*length | 75 |
| Default Migration (noVEGF) | 6.2 µm/hr | 75 |
| VEGF Concentration Per Cell | 30 ng/ml | based on75 |
| Maximum endothelial elongation | 30 µm/hr | 75 |
| Minimum endothelial segment length | 1 µm | 75 |
| Threshold for hypoxia | 200 µm | 76 |
| MB231 stromal educated migration rate | 2.5-fold | 71 |
| MB231 stromal educated proliferation rate | 3.5-fold | 71 |
| Hypoxic MB231 Proliferation Rate | 0.5-fold | 67 |
| Hypoxic MB231 Migration Rate | 3-fold | 67 |
| Macrophage Number | 250 | 41,83 |
| Fibroblast Number | 250 | 41,83 |
| Macrophage Infiltration Probability | 20–80% | 41,83 |
MB231 Migration
First, the proximity of the MB231 cell to a macrophage or fibroblast is checked. If the MB231 cell is within 200 µm of a macrophage or fibroblast, its migration rate is increased by 2.5-fold. When examining the parameter space, we vary the increase in migration rate by 1.5 to 3.5-fold. If the cell is hypoxic, it migrates 3× as far. When examining the parameter space, we vary the increase in migration rate by 1.5 to 3.5-fold based on experimental data71. CCR5+ cells migrate 10× as far as CCR5− cells. Migration is modeled by random motion, such that the MB231 cell checks its 27 neighboring positions in the cellular grid and determines which ones are not occupied. It then chooses one of these positions randomly to move into. This process can occur multiple times each iteration (6 hours) under hypoxic conditions, due to proximity to stromal cells or its CCR5+ state.
MB231 Proliferation
First, the proximity of the MB231 cell to a macrophage or fibroblast is checked. If the MB231 cell is within 200 µm of a macrophage or fibroblast, its proliferation rate is increased by 3.5-fold. When examining the parameter space, we vary the increase in proliferation rate by 1.5 to 3.5-fold. If the cell is hypoxic, the proliferation rate is reduced by half. Cancer stem cells have a base proliferation rate of 0.2 per day whereas progenitor cells have a proliferation rate of 0.5 per day. An MB231 cell can only proliferate if it has an empty space in its neighborhood. The MB231 cell checks its 27 neighboring positions in the cellular grid and determines which ones are not occupied, and if none are open it becomes quiescent. Otherwise the new cell is placed in one of the unoccupied spaces randomly. The cell determines whether it can proliferate this iteration by determining if a random number is lower than its proliferation rate, if so it proliferates. A new cell is created, placed randomly in one of the neighboring positions. The division cycles are updated if the cell is a progenitor cell, and the CCR5 status of the cell is determined. A MB231 cell has a 5% chance of being CCR5+. If a progenitor cell has reached its division limit it becomes senescent and has a 40% chance of dying each day.
Hypoxia
We determine whether a MB231 cell is hypoxic based on its distance from mature vessels within the vasculature. The initial vessels within the simulation are all mature, and thus we assume they have blood flow through them. Vessels that have sprouted and anastomosed are also mature and would have blood flow. Sprouts that have not anastomosed are considered immature and would not have blood flow. Any tumor cell that is farther than 200 µm away from a mature vessel is considered hypoxic, as justified in76.
Apoptosis
Forty percent of the senescent cells die each iteration. Thus, we randomly choose 40% of the senescent cells to be eliminated each iteration. These cells are removed from the simulation and all tallies are updated.
Macrophages and Fibroblasts
In the simulations, macrophages and fibroblasts are randomly placed within the cellular grid of 100×100×100 voxels. Macrophages and fibroblasts move randomly throughout the grid, with fibroblasts moving first. Fibroblasts randomly migrate one voxel per iteration, at 3.33 µm/hr. A macrophage within 200 µm of a tumor cell will migrate towards the tumor 8 times as fast, at 26.67 µm/hr, due to the CSF-1 factor that tumor cells release77–82. Macrophages check whether they can move towards the nearest MB231 cell, otherwise they move randomly84. Fibroblasts and macrophages migrate until they reach the edge of the grid.
Macrophage Infiltration
Monocytes are recruited to the tumor through the vasculature by secreted factors released by tumor cells, such as CSF-1 and then differentiate into tissue macrophages; for brevity, we refer to macrophage recruitment. Thus, macrophages can infiltrate the tumor through mature vessels in the simulation. If a mature vessel is within 200 µm of a tumor cell, we assume it receives a signal through tumor cell secreted factors to recruit macrophages. Any vessel within 200 µm of a tumor cell has a probability of recruiting a macrophage. If the vessel can recruit a macrophage, it checks whether there is an unoccupied space in the 27 neighbors of the cellular grid. If there is an unoccupied space, it randomly chooses one of those spaces for the new macrophage to move into.
MB231 interaction with Stromal Cells
Proximity to macrophages and fibroblasts increases the tumor cell’s proliferation and migration rates. A tumor cell within 200 µm of a macrophage increases its proliferation rate by 3.5-fold and its migration rate by 2.5-fold. A tumor cell within 200 µm of a fibroblast increases its proliferation rate by 3.5-fold and its migration rate by 1.7-fold. In the simulations where the influence of macrophages and fibroblasts are varied, the influence on proliferation rate varies between 1.5-fold and 3.5-fold and the influence on migration rate varies between 1.5-fold and 3.5-fold for both macrophages and fibroblasts.
Angiogenesis Model
The model is a modification to that presented in65.Sprouting angiogenesis is initiated by the release of VEGF from hypoxic tumor cells and thus in the model it is initiated once a hypoxic cell appears in the simulation. The simulation starts with a migration parameter which is multiplied by the elongation of the tip cell during the tip cell migration step and a proliferation value which governs the cell cycle length of a stalk cell endothelial cell. The rest of the sprout is made up of quiescent phalanx cells that are not affected by these parameters and support the rest of the cells.
Tip Cell Actions
The simulation loops through all the sprouted vessels and finds the tip cell in each capillary. If the tip cell is still active and not mature, it can migrate or proliferate. A tip cell only proliferates to produce a stalk cell, after that it no longer proliferates. The tip cell proliferates once its cell clock has reached its cell cycle, otherwise its cell clock is incremented. If the cell clock reaches its cell cycle, a new stalk cell replaces the old tip cell and a new tip cell is created extending in the direction of the old tip cell with a length of one µm since that is the minimum length possible for a sprout, for more details see65. The tip cell then checks whether it has hit another vessel and if so it anastomoses. If the tip cell leaves the grid, the capillary becomes mature and can no longer migrate or proliferate.
The main function for a tip cell is to migrate in the direction of VEGF gradients. The tip cell searches the 27 grid spaces near it on the cell grid to find whether there are cells occupying any of those grid spaces. We assume that all cells produce approximately the same amount of VEGF, resulting in a concentration 30 ng/ml within their cellular grid space. This is based on the constant VEGF value of 20 ng/ml in65 considering tumors cell death is around 40% of senescent cells, thus 30 ng/ml per cell would approximately result in between 12–30 ng/ml in a packed tumor. The tip cell finds the positions with cells near it and randomly chooses to migrate towards one of these cells, such that is does not move backwards. The formula for elongation distance is M = T2[VEGF] + migNoVegfMatrix, where T2 and migNoVegfMatrix are constants75. VEGF concentration is calculated by searching the 27 grid spaces near the end of the tip cell on the cell grid to find how many cells are occupying any of those grid spaces. We multiply the number of cells occupying the cell grid by 30 ng/ml to find VEGF concentration. The cell cannot elongate greater than 1.5× of its current length nor can it migrate farther than 60 µm75. The cell does not migrate if migration would cause it to leave the vascular grid. After migration, the cell checks whether it anastomoses.
Stalk Cell Actions
After tip cell actions, we find the adjacent stalk cell and determine whether it proliferates. The stalk cell proliferates once its cell clock has reached its cell cycle, otherwise its cell clock is incremented. Only immature stalk cells proliferate, and they do not proliferate if proliferating would make the tip cell leave the grid. Proliferation causes a new stalk cell to be produced, replacing the previous tip cell location and a new tip cell is created extending in the direction of the old tip cell with a length of one µm to initialize the start of the sprout. The cell clock is reset after proliferation and the new cell is added to the capillary. The new tip cell then checks whether stalk cell proliferation has caused it to hit another vessel and if so it anastomoses.
Branching
Only phalanx cells, those following the stalk or tip cell are allowed to branch. Branching is triggered by hypoxic cells, so a phalanx cell can only branch if one of its nodes is within 250 µm of a hypoxic MB231 cell76. If these conditions are met, it has a 20% probability to branch, see65. The phalanx cell finds the closest hypoxic MB231 cell and branches in that direction, specifically the new sprout’s second node is equal to the second node of the phalanx cell plus 3*(normalized vector of the phalanx cell’s second node to the nearest hypoxic cell). Branching only occurs if the resulting branch is within the vascular grid boundary. We include vascular cell regression if the new branch is within 50 µm of a mature vessel, not including the parent vessels, for more information see65. Once branching occurs, a new tip cell is created, its nodes are updated, and it is given a new cycle length based on the simulation proliferation value with random noise.
Experimental
Mouse Xenografts
Animal protocols described in this study were approved by the Institutional Care and Use Committee at the Johns Hopkins Medical Institutions. Before tumor cell inoculation, athymic nude mice (female, 5–6 weeks, 18–20 g) were pre-treated by injecting 50 µl tumor-conditioned media (TCM) of MDA-MB-231 cells subcutaneously for two weeks daily. After two weeks of induction, 2 ×106 MDA-MB-231 cells were injected into the upper inguinal mammary fat pad of the animals with 100 ul of 1:1 PBS/Matrigel (BD Biosciences, San Jose, CA). When tumors grow to approximately 1.5 cm diameter, they will be excised for immunohistochemistry (IHC) analysis71.).
Immunohistochemistry analysis
Immunohistochemical analysis of Iba-1 was performed using monoclonal antibodies against Iba-1 (Ionized calcium binding adaptor molecule 1), a microglia/macrophage-specific calcium-binding protein (Wako). For the IHC staining, after blocking with 5% goat serum in PBST for 1 hour at room temperature, the sections were treated with the Iba-1 antibody overnight at 4°C, then the peroxidase-conjugated streptavidin complex method was performed, followed by the 3, 3' diaminobenzidine (DAB) procedure according to manufacturer’s protocols (VECTASTAIN Elite ABC Kit, Vector Lab, Burlingame, CA)71.
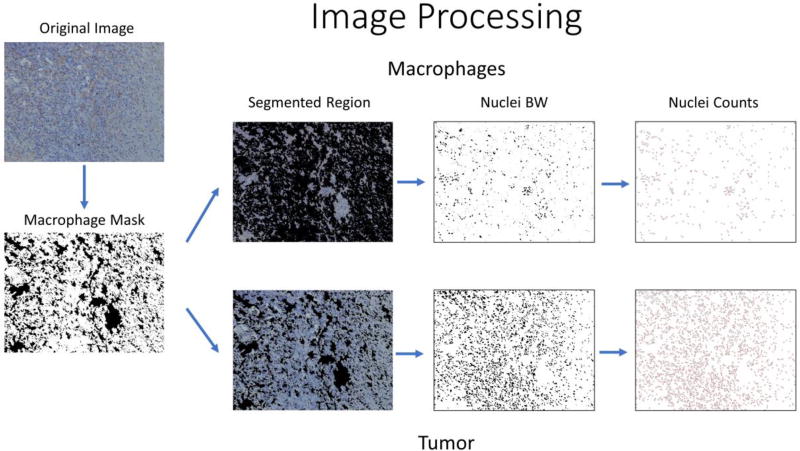
Image Processing
To estimate the number of macrophages in the xenograft tumor sample, standard image processing in ImageJ85 was used. First, we binarized the image in the RGB channel, using threshold ranges of 104–154 in the red channel, 91–141 in the green channel, and 95–150 in the blue channel to segment out the areas stained brown, i.e. the macrophages. We then performed the following morphological functions on the binarized image: “dilate” which increases the region to fill in any gaps due to inconsistent staining, “fill holes” which fills in any holes in the region, “erode” which decreases the region back to its original size, and “convert to mask” which creates a mask which will be applied to the original image to segment out the macrophages or tumor. To count the number of macrophages, we used the “mask” to remove the tumor region and binarized the masked image using ranges of 0–103 in the red channel, 0–122 in the green channel, and 55–193 in the blue channel to segment out the nuclei. We then dilated the binarized image to seal any holes, used the watershed algorithm to find individual nuclei and erode the binarized image to bring the image back to its original size. Lastly, we analyzed particles to count the number of nuclei that are greater than 100 pixels in size. The same algorithm was used on the masked tumor image to calculate the number of tumor nuclei in the image. We then calculated the number of macrophages/ (macrophages + tumor cells) to determine the ratio of macrophages to total cells within the tumor.
Results
An agent-based model of triple-negative breast cancer stromal interactions
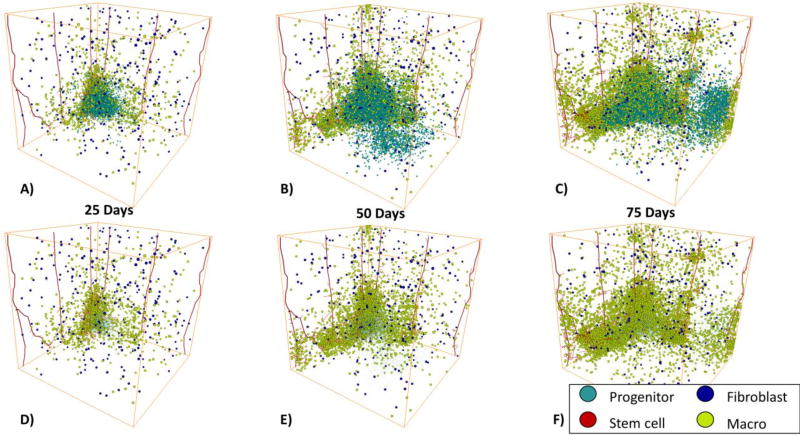
In Figure 3, we show an example of an in silico tumor progression from 25 days to 75 days. The top row shows the entire tumor progression with progenitor cells, stem cells, fibroblasts, and macrophages. The middle row shows the progression with a reduced tumor cell size to show the interior of the tumor. The bottom row shows tumor vasculature. In panel A the tumor has started to grow but remains fairly solid with few extensions off the tumor. There were many cells shed from the tumor but overall it did not demonstrate any clear finger-like projections. In panel D, it is clear that macrophages have started to infiltrate out of the vasculature close to the tumor; more accurately, monocytes extravasate from the vasculature and differentiate into macrophages. On the right side of the tumor, we observe tens of progenitor cells that seem to have migrated off the bulk tumor with tens of macrophages in close proximity. In panel B at day 50, these tens of progenitor cells and macrophages have expanded into larger extensions off the tumor with increasing numbers of macrophage recruitment into the extensions, as shown in panel E. On the bottom part of panel B there is also a clear invasion of hundreds of progenitor cells off the tumor with seemingly few macrophages or fibroblasts. By day 75, in panel C, both the tumor and the macrophage recruitment substantially increased. The extension on the right side of the tumor expanded even farther and continues to be composed of both progenitor cells and macrophages as shown in panel C. In addition, even farther to the right of the bulk tumor another extension of progenitor cells has evolved. This second extension has also established new macrophage recruitment into this part of the tumor. Interestingly, the hundreds of progenitor cells that could be seen extending from the tumor in panel B have largely disappeared. This leads to the hypothesis that macrophages may contribute to the lifespan of a finger/extension off the tumor due to their influence on proliferation. In panels C and F, the macrophages are starting to encase parts of the tumor. In panels G though I, we show the progression of angiogenesis over time. As the tumor grows larger it recruits new vasculature which eventually leads to the reduction of its hypoxic regions.
Figure 3.
3D Complete Tumor Progression. A) Tumor progression on day 25 with the full tumor. B) Day 50 with the full tumor. C) Day 75 with the full tumor. D) Day 25 with reduced tumor cell size for visualization purpose to show the stroma. E) Day 50 with reduced tumor cell size to show the stroma. F) Day 75 with reduced tumor cell size to show the stroma. G) Day 25 with just the tumor vasculature. H) Day 50 with just the tumor vasculature. I) Day 75 with just the tumor vasculature. MB231 progenitor cells are shown in cyan, cancer stem cells are shown in red, macrophages are shown in yellow, and fibroblasts are shown in blue. The vasculature is shown in red.
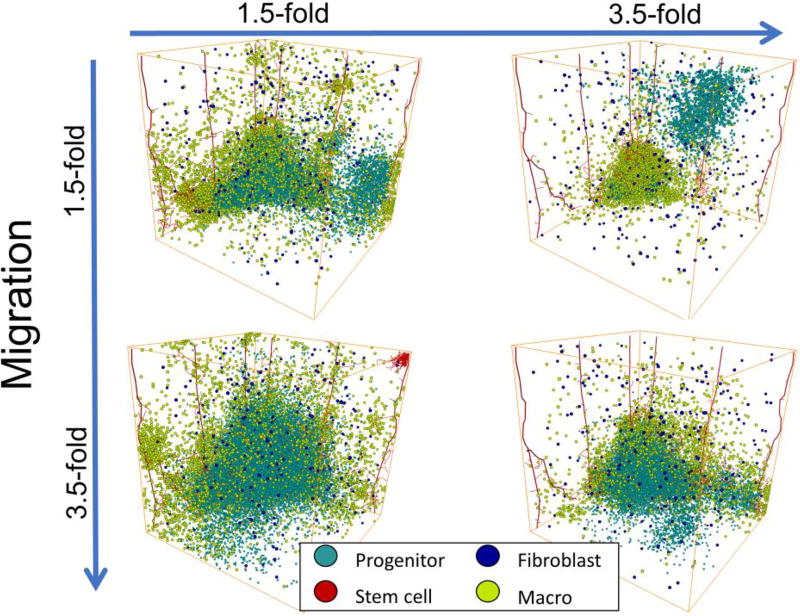
Increasing stromal influence on MB231 proliferation decreases tumor growth whereas increasing stromal influence on MB231 migration increases tumor growth
To investigate the specific effects of the influence of stromal cells on MB231 cell migration and proliferation, the MB231 proliferation rate was varied between 1.5-fold and 3.5-fold and the MB231 migration rate was varied between 1.5-fold and 3.5-fold when a cell was in proximity to either a macrophage or a fibroblast on day 75, Figure 4. We find that increasing stromal effects on migration rate, increases the size of the tumor. In contrast, increasing stromal effects on proliferation rate, decreases the size of the tumor. The tumor with 3.5-fold proliferation rate and 1.5-fold migration rate resulted in a total of 4,272 MB231 cells, whereas the tumor with 1.5-fold proliferation rate and 3.5-fold migration rate resulted in a total of 35,583 MB231 cells. The other two tumors resulted in approximately 17,000 MB231 cells. The growth of the tumor over time was fastest for a 3.5-fold migration rate and a 1.5-fold proliferation rate and slowest for a tumor with a 1.5-fold migration rate and a 3.5-fold proliferation rate, as shown in Supplemental Figure 1. Increasing stromal influence on migration also seems to decrease the fingers and results in a more disperse tumor.
Figure 4.
3D Tumor Parameter Space with Differences in Stromal Contributions to Migration and Proliferation Rates on day 75. The x-axis shows increasing proliferation rates from 1.5-fold to 3.5-fold and the y-axis shows increasing migration rates from 1.5-fold to 3.5-fold. We show the changes in the size and morphology of the tumor with changes in stromal influence on proliferation and migration rates.
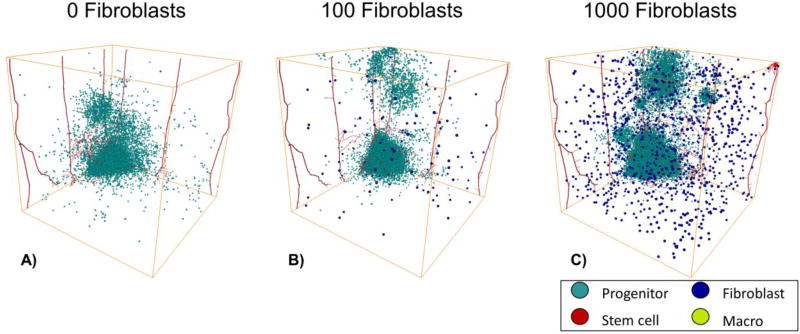
Increasing the number of fibroblasts increases invasion and growth
To determine the individual effects of fibroblasts on tumor growth and invasion, we varied the number of fibroblasts between 0 and 1,000 on day 75, Figure 5. We found that with increasing numbers of fibroblasts, there were increases in “self-metastases”86–89, which are invading tumor cells that have separated from the bulk tumor. Originally, these self-metastases start out as invasive fingers90 but then they continue to migrate off the tumor and become separate entities. We also find that with large numbers of fibroblasts the tumor growth increases. The tumor with 1,000 fibroblasts had a quick increase in growth and then the cell number fluctuate as the self-metastases grow and shrink, Supplemental Figure 2. Thus, fibroblasts contribute to cancer cell invasion and metastasis
Figure 5.
3D Tumor Progression with just Fibroblasts on day 75. A) 0 initial fibroblasts. B) 100 initial fibroblasts. C) 1000 initial fibroblasts. MB231 cells are shown in cyan, stem cells are shown in red, and fibroblasts are shown in blue. The vasculature is shown in red.
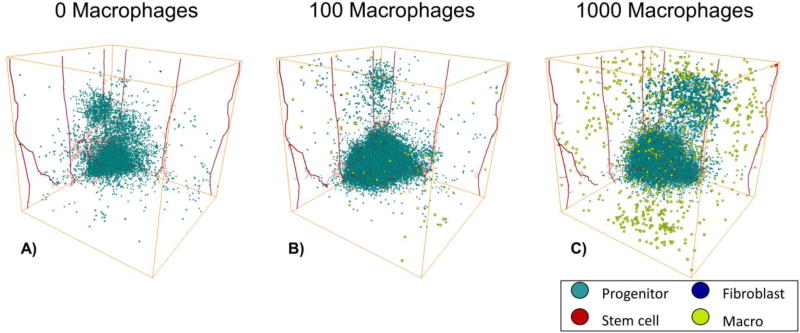
Small numbers of macrophages increase the size of the tumor
To determine the individual effects of macrophages on tumor growth and invasion, we varied the number of macrophages between 0 and 1,000, Figure 6. We find that including 100 macrophages more than doubles the size of the tumor, while including 1,000 macrophages has less of an effect on the tumor growth, Supplemental Figure 3. This is due to two factors, one is that having large numbers of macrophages can slow tumor growth due to space limitations and the other is that if there are large clumps of macrophages then the number of cells they actually affect remains similar as the cytokines and other factors they release only affect cells within approximately 200 µm of them. We also see “self-metastases” forming with macrophages but they are more spread out and less compact than those formed with fibroblasts only. This is probably due to the fact that the macrophages tend to migrate with the invading cancer cells, since they migrate toward cancer cells within 200 µm of them, whereas fibroblasts migrate randomly. Macrophages are continually influencing the cancer cell’s migration and proliferation rates, whereas a fibroblast may just influence that cell for a brief period of time.
Figure 6.
3D Tumor Progression with just Macrophages on day 75. A) 0 initial macrophages. B) 100 initial macrophages. C) 1000 initial macrophages. MB231 cells are shown in cyan, cancer stem cells are shown in red, and macrophages are shown in yellow. The vasculature is shown in red.
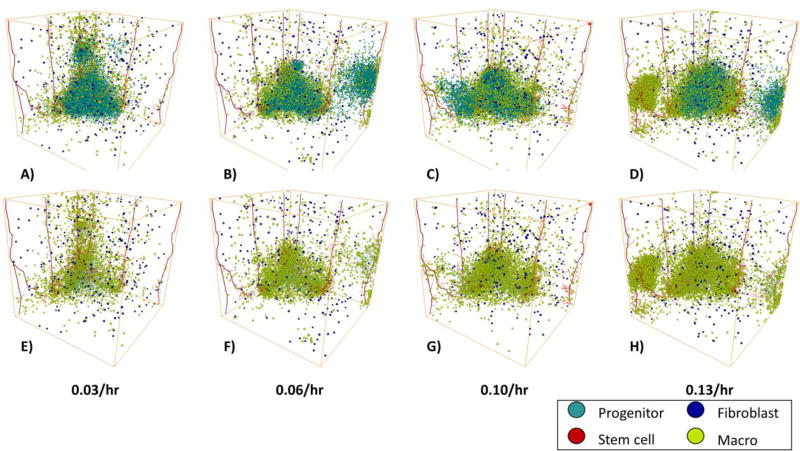
Increasing macrophage infiltration rates does not increase overall tumor size
We investigated the effect of increasing macrophage infiltration rates on tumor growth by varying the rate of macrophage infiltration from mature vessels from 0.033 to 0.133 per hour Figure 7. We find that while increasing the macrophage infiltration rate increases the number of macrophages it does not increase the number of MB231 cells, Supplemental Figure 4. In fact, at very high macrophage infiltration rates panel D, it can actually slow the tumor growth by spatially inhibiting the tumor cells. The tumors all exhibit invasive fingers and “self-metastases” as shown in panels A–D, which shows all cells within the tumor. In addition, except at very high macrophage infiltration rates, the macrophages are mostly confined to the tumor E-G, which shows the macrophages and reduced size tumor cells, so the interior of the tumor is viewable. At an infiltration rate of 0.133 per hour shown in panel D and H, a large cluster of macrophages form around the vasculature on the left without many tumor cells within the cluster.
Figure 7.
MB231 Tumor Growth vs Time with Increasing Macrophage Infiltration on day 75. AD) The number of MB231 cells over time with a 0.3–0.9/hr probability of infiltration of macrophages. The tumor increases more quickly with increasing macrophage numbers. E–H) The same tumors as in A–D) but with reduced size tumor cells to show the stroma. MB231 cells are shown in cyan, cancer stem cells are in red, macrophages in yellow, and fibroblasts in blue. The vasculature is shown in red.
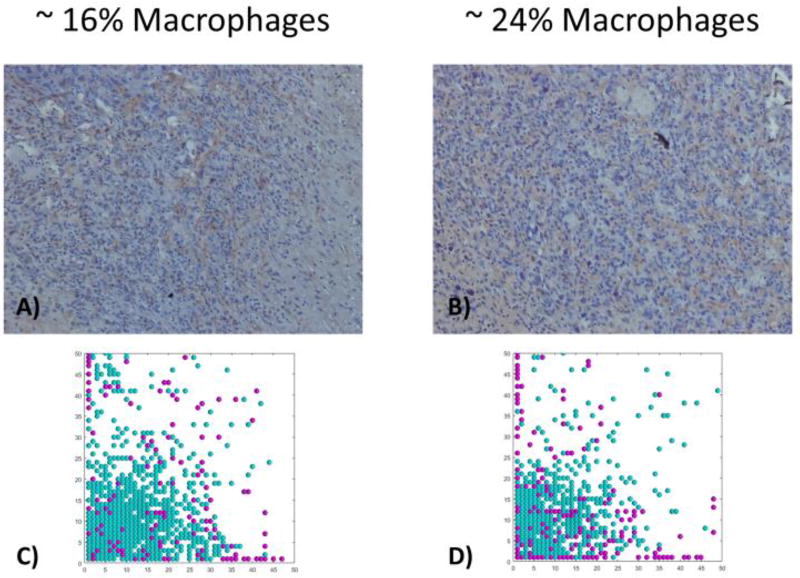
Both in vivo and in silico tumors exhibit varying percentages of macrophages
We used image processing to calculate the percentages of macrophages in xenograft mouse models and in the bulk tumor of the in silico simulations, Figure 8. The slices on xenograft tumors had 16% and 24% of macrophages, Figure 9 panel A and B. We also found comparable macrophage percentages within slices of the in silico tumor with different infiltration rates. The tumor with an infiltration rate of 0.067 per hour had an overall macrophage percentage of 17% within the bulk tumor and the tumor with an infiltration probability of 0.1 per hour, had an overall macrophage percentage of 20% within the bulk tumor, Figure 9 panel C and D. Although the overall macrophage percentage within the tumor with an infiltration rate of 0.1 per hour ranged from 8% to 47% within the different slices. We show example slices that had a macrophage percentage of 16% in panel C and a macrophage percentage of 24% in panel D. While separate slices ranged from between 0–47%, the macrophage percentages of the entire tumors ranged from 0.02 – 33.7%, comparable to experimental results ranging from 1% to 25%41,83. This illustrates that choosing a representative slice is critical for understanding the actual macrophage percentage within the tumor.
Figure 8.
Flowchart of the Stromal Image Processing. First, we binarize the image to segment out only the stromal region using RGB color segmentation on the brown stained cells. We then create a mask to remove either the tumor cells or the stromal cells. Then we use thresholding to segment out the nuclei. Lastly, we use a watershed algorithm to identify and count each individual nuclei.
Figure 9.
Comparison of 2D in silico Tumor Slices with in vivo Samples with Macrophage Infiltration. The top panel shows two in vivo samples with increasing amounts of macrophage infiltration. The bottom panel shows two in silico tumor slices with comparable amounts of macrophage infiltration, around 14–16% for the left column and between 19–25% for the right column.
Comparison of Spatial Metrics for in vivo and in silico data
We compared an average of 10 in silico simulation results at day 17, 33, and 50 to in vivo from our tumor xenograft experiments and literature results for several spatial statistical measures Shannon’s index, Simpson’s index, and Morista-Horn91. We found that all metrics were comparable between the in silico and in vivo results. Shannon’s Index and Simpson’s Index were the most robust across days, with very little difference between them. The Morista-Horn score decreased when compared across the first 5 slices as the days increased. While the tumor is still small, the Morista-Horn score has not stabilized across slice 21–25 but once the tumor has grown the score stays similar.
Discussion
We have developed an agent-based model, based on our previous models65–67 of triple-negative breast cancer cells interaction with their microenvironment, including breast cancer stem cells, cancer progenitor cells, endothelial cells, macrophages, and fibroblasts. We specifically model tumor hypoxia, and angiogenesis in response to hypoxia, as well as macrophage recruitment.
Breast cancer cells in the model invade by forming “invasive fingers” or “self-metastases” and we find that these are accompanied by macrophages. Initially, the cancer cells migrate with only a few macrophages but eventually the infiltrating macrophages get recruited to the invasive self-metastases. This is corroborated by in vivo evidence showing that macrophages are involved in tumor cell invasion and metastasis and are frequently present at the invasive fronts of a tumor1. This coupling of macrophages and tumor cells supports metastasis as macrophages have been shown to migrate together along collagen fibers and intravasate into nearby blood vessels95. In addition, tumor cells and macrophages have been shown to migrate together and intravasate specifically at the junction where the endothelial cell, tumor cell and macrophage meet96. In support of this, another mathematical model of cancer immunotherapy, showed that more migratory cells resulted in tumor growth, escape, and relapse97. Fibroblasts also contribute to invasion and spread of tumor cells, but they tend to form less compact “self-metastases” and more single-cell invasion events. This is supported by previous adhesive particle mechanistic modeling, in which clusters with both tumor cells and stromal cells showed increased invasiveness and had increased displacement98. This supports the role of macrophages and fibroblasts in the invasiveness of the tumor.
We find that macrophage recruitment to the invasive “self-metastases” contributes to the compactness and long-term survival of the “self-metastases.” Without macrophage recruitment the fingers that form tend to spread out or disappear. From previous work, we have found that invasive fingers form due to migrating cancer stem cells as compared to migrating progenitor cells67. Thus, the increased migration rate due to the stromal cells contributes to stem cell migration and “self-metastases” formation. In addition, migrating cancer stem cells have a higher chance of proliferating symmetrically to create an additional cancer stem cell due to the lack of spatial inhibition, increasing the growth and survival of the “self-metastasis” and the overall tumor. This is supported by in vivo work showing that the depletion of tumor-associated macrophages in a mouse xenograft model led to a decrease in cancer stem cells99 and in vitro studies that showed co-culture of breast cancer cell lines and M2-type macrophages increased mammosphere production100. MCF-7 cancer cells co-cultured with M2-type macrophages were also found to prevent hypoxia-induced cell death101. The increased survival of tumor cells due to the compact, surviving invasive fingers are a result of migrating stem cells as compared to migrating progenitor cells. Migrating progenitor cells might cause a transient finger to form but the finger will eventually senesce and die due to the limited number of times the cells can divide. By increasing the migration rate of the stem cells, macrophages are contributing both to the invasive morphology of the tumor but also its growth and survival.
There is a trade-off between the effects of increasing proliferation and increasing migration of the macrophage secretome on MB231 cells. Increasing migration benefits tumor growth while increasing proliferation reduces tumor growth. This is caused by spatial inhibition, where tumor cells with higher proliferation rates and lower migration rates tend to become overcrowded and stop proliferating. This is especially important for stem cells because stem cell proliferation is the only way a tumor can expand in a non-transient way since progenitor cells eventually stop proliferating and die. Increased migration leads to a more disperse tumor with less contact inhibition which promotes tumor growth. These results are supported by previous work showing that increasing progenitor cell division led to inhibited tumor growth66,87 due to spatial inhibition and that increased cancer cell migration leads to increased tumor growth66. Small numbers of macrophages seem to be enough to increase the tumor growth, whereas large numbers can inhibit growth due to overcrowding. A computational model of colorectal cancer also predicted that high numbers of immune cells contributed to reduced tumor growth and improved treatment102. Inhibiting macrophage recruitment has shown variable success, anti-CSF-1 treatment early in the primary tumor growth (before macrophage infiltration) decreases primary tumor growth but the effects was modest103 while others failed104. Other immunotherapies, specifically monoclonal antibodies targeting checkpoint inhibitors on T-cells, have been tested in a variety of cancers105. Normally, the adaptive immune system targets cancer cells by releasing cytotoxic T-cells that recognize and eliminates cancer cells as threats due to mutations in their genes68. Unfortunately, some cancers evade the immune system by tricking them into recognizing them as self through checkpoint inhibitors such as programmed cell death protein-1 (PD-1) and its ligand (PD-L1)68. Therapies targeting PD-1/PD-L1 and cytotoxic T lymphocyte antigen-4 (CTLA-4) have been shown to be effective in melanoma, bladder cancer, and non-small cell lung cancer and are in clinical trials for breast cancer, especially triple-negative breast cancer106.
There are several limitations and future directions that could be taken from this work. It has been shown that macrophages promote stem cell expansion107,108. While we focused on the effects on macrophages and fibroblasts on overall proliferation rate, which includes stem cell proliferation, we did not specifically incorporate the effects on symmetric stem cell division. This can be examined in further research. We also did not consider an immune response to the tumor and the effects on cytotoxic T cells on tumor death. In future studies it would be of interest to investigate the effects on therapies targeting the MB231 and stromal secretomes to determine how these would affect overall tumor growth and macrophage recruitment.
Supplementary Material
Table 2.
A Comparison of Spatial Metrics between simulations and in vivo results (mean +− standard deviation)
| Spatial Metric | in silico Day 17 | in silico Day 33 | in silico Day 50 | literature | Reference |
|---|---|---|---|---|---|
| Shannon's Index | 0.63 +−(0.06) | 0.64 +−0.05 | 0.64 +−(0.05) | 0–1.6 | 92 |
| Simpson's Index | 1.60 +−(0.13) | 1.67 +−0.11 | 1.65 +−(0.12) | 1.13–2.88 | 93 |
| Morista-Horn (1–5 slice) | 0.71 +−(0.05) | 0.60 +−0.09 | 0.52 +−(0.47) | 0.68–0.9 | 94 |
| Morista-Horn (21–25 slice) | 0.57 +−(0.27) | 0.94 +−0.08 | 0.96 +−(0.02) |
Highlights.
Modeled the complex interplay between breast cancer stem cells, cancer progenitor cells, macrophages, fibroblasts, and endothelial cells.
Increasing stromal influence on individual cancer cell proliferation decreases overall tumor growth.
Even small numbers of macrophages increase overall tumor growth.
Increasing macrophage infiltration rate has varying effects on tumor growth.
Acknowledgments
The authors would like to acknowledge Dr. Chang Gong for his help in visualizations of the in silico tumors and Dr. Adam Mirando for his help in analyzing the macrophage percentages using ImageJ. This work was supported by the National Institutes of Health grants R01 CA138264 and R01 CA196701 (ASP) and American Cancer Society postdoctoral fellowship PF-13-174-01-CSM (KAN).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests:
The authors declare no competing interests.
References
- 1.Condeelis J, Pollard JW. Macrophages: Obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501 doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 3.Lewis CE, Leek R, Harris A, McGee JO. Cytokine regulation of angiogenesis in breast cancer: the role of tumor-associated macrophages. J. Leukoc. Biol. 1995;57:747–51. doi: 10.1002/jlb.57.5.747. [DOI] [PubMed] [Google Scholar]
- 4.Leek RD, et al. Association of macrophage infiltration with angiogenesis and prognosis in invasive breast carcinoma. Cancer Res. 1996;56:4625–9. [PubMed] [Google Scholar]
- 5.Solinas G, Germano G, Mantovani A, Allavena P. Tumor-associated macrophages (TAM) as major players of the cancer-related inflammation. J. Leukoc. Biol. 2009;86:1065–73. doi: 10.1189/jlb.0609385. [DOI] [PubMed] [Google Scholar]
- 6.Qian B-Z, et al. FLT1 signaling in metastasis-associated macrophages activates an inflammatory signature that promotes breast cancer metastasis. J. Exp. Med. 2015;212:1433–48. doi: 10.1084/jem.20141555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Comito G, et al. Cancer-associated fibroblasts and M2-polarized macrophages synergize during prostate carcinoma progression. Oncogene. 2014;33:2423–2431. doi: 10.1038/onc.2013.191. [DOI] [PubMed] [Google Scholar]
- 8.Straussman R, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487 doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaturvedi P, Gilkes DM, Takano N, Semenza GL. Hypoxia-inducible factor-dependent signaling between triple-negative breast cancer cells and mesenchymal stem cells promotes macrophage recruitment. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E2120–9. doi: 10.1073/pnas.1406655111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Semenza GL. The hypoxic tumor microenvironment: A driving force for breast cancer progression. Biochimica et Biophysica Acta - Molecular Cell Research. 2016;1863:382–391. doi: 10.1016/j.bbamcr.2015.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hollmén M, Roudnicky F, Karaman S, Detmar M. Characterization of macrophage--cancer cell crosstalk in estrogen receptor positive and triple-negative breast cancer. Sci. Rep. 2015;5:9188. doi: 10.1038/srep09188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sousa S, et al. Human breast cancer cells educate macrophages toward the M2 activation status. Breast Cancer Res. 2015;17:101. doi: 10.1186/s13058-015-0621-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tripathi C, et al. Macrophages are recruited to hypoxic tumor areas and acquire a proangiogenic M2-polarized phenotype via hypoxic cancer cell derived cytokines Oncostatin M and Eotaxin. Oncotarget. 2014;5:5350–68. doi: 10.18632/oncotarget.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan Z-Y, Luo R-Z, Peng R-J, Wang S-S, Xue C. High infiltration of tumor-associated macrophages in triple-negative breast cancer is associated with a higher risk of distant metastasis. Onco. Targets. Ther. 2014;7:1475–80. doi: 10.2147/OTT.S61838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qian B, et al. A distinct macrophage population mediates metastatic breast cancer cell extravasation, establishment and growth. PLoS One. 2009;4:e6562. doi: 10.1371/journal.pone.0006562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su S, et al. A Positive Feedback Loop between Mesenchymal-like Cancer Cells and Macrophages Is Essential to Breast Cancer Metastasis. Cancer Cell. 2014;25:605–620. doi: 10.1016/j.ccr.2014.03.021. [DOI] [PubMed] [Google Scholar]
- 17.Chen J, et al. CCL18 from tumor-associated macrophages promotes breast cancer metastasis via PITPNM3. Cancer Cell. 2011;19:541–555. doi: 10.1016/j.ccr.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dovas A, Patsialou A, Harney AS, Condeelis J, Cox D. Imaging interactions between macrophages and tumour cells that are involved in metastasis in vivo and in vitro. J. Microsc. 2013;251:261–269. doi: 10.1111/j.1365-2818.2012.03667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pignatelli J, et al. Macrophage-dependent tumor cell transendothelial migration is mediated by Notch1/MenaINV-initiated invadopodium formation. Sci. Rep. 2016;6:37874. doi: 10.1038/srep37874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang X. Tumor-associated macrophages as potential diagnostic and prognostic biomarkers in breast cancer. Cancer Lett. 2013;332:3–10. doi: 10.1016/j.canlet.2013.01.024. [DOI] [PubMed] [Google Scholar]
- 21.Mahmoud SMA, et al. Tumour-infiltrating macrophages and clinical outcome in breast cancer. J. Clin. Pathol. 2012;65:159–63. doi: 10.1136/jclinpath-2011-200355. [DOI] [PubMed] [Google Scholar]
- 22.Sparano JA, et al. Molecular Pathology of Breast Cancer. Springer International Publishing; 2016. The tumor microenvironment as a metastasis biomarker in breast cancer; pp. 153–165. [DOI] [Google Scholar]
- 23.Darby IA, Laverdet B, Bonté F, Desmoulière A. Fibroblasts and myofibroblasts in wound healing. Clin. Cosmet. Investig. Dermatol. 2014;7:301–11. doi: 10.2147/CCID.S50046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rønnov-Jessen L, Petersen OW, Koteliansky VE, Bissell MJ. The origin of the myofibroblasts in breast cancer. Recapitulation of tumor environment in culture unravels diversity and implicates converted fibroblasts and recruited smooth muscle cells. J. Clin. Invest. 1995;95:859–73. doi: 10.1172/JCI117736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takai K, Le A, Weaver VM, Werb Z. Targeting the cancer-associated fibroblasts as a treatment in triple-negative breast cancer. Oncotarget. 2016;7:82889–82901. doi: 10.18632/oncotarget.12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugimoto H, Mundel TM, Kieran MW, Kalluri R. Identification of fibroblast heterogeneity in the tumor microenvironment. Cancer Biol. Ther. 2006;5:1640–1646. doi: 10.4161/cbt.5.12.3354. [DOI] [PubMed] [Google Scholar]
- 27.Corvigno S, et al. Markers of fibroblast-rich tumor stroma and perivascular cells in serous ovarian cancer: Inter- and intra-patient heterogeneity and impact on survival. Oncotarget. 2016;7:18573–84. doi: 10.18632/oncotarget.7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korkaya H, Liu S, Wicha MS. Breast cancer stem cells, cytokine networks, and the tumor microenvironment. J Clin Invest. 2011;121:3804–3809. doi: 10.1172/JCI57099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhowmick NA, et al. TGF-beta signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science. 2004;303:848–51. doi: 10.1126/science.1090922. [DOI] [PubMed] [Google Scholar]
- 30.Yu Y, et al. Cancer-associated fibroblasts induce epithelial-mesenchymal transition of breast cancer cells through paracrine TGF-β signalling. Br. J. Cancer. 2014;110:724–32. doi: 10.1038/bjc.2013.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeisberg EM, Potenta S, Xie L, Zeisberg M, Kalluri R. Discovery of endothelial to mesenchymal transition as a source for carcinoma-associated fibroblasts. Cancer Res. 2007;67:10123–8. doi: 10.1158/0008-5472.CAN-07-3127. [DOI] [PubMed] [Google Scholar]
- 32.Bochet L, et al. Adipocyte-derived fibroblasts promote tumor progression and contribute to the desmoplastic reaction in breast cancer. Cancer Res. 2013;73:5657–68. doi: 10.1158/0008-5472.CAN-13-0530. [DOI] [PubMed] [Google Scholar]
- 33.Gilkes DM, Semenza GL, Wirtz D. Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat. Rev. Cancer. 2014;14:430–439. doi: 10.1038/nrc3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao Y, Keller ET, Garfield DH, Shen K, Wang J. Stromal cells in tumor microenvironment and breast cancer. Cancer Metastasis Rev. 2013;32:303–315. doi: 10.1007/s10555-012-9415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diao X-W, Feng J-Y, Wang Q-W, Sun J-G, Chen Z-T. SDF-1/CXCR4 axis promotes prostate cancer cell invasion and bone metastasis through p38, NF-κB and HIF-1α pathways. Int J Clin Exp Pathol. 2016;9:2706–2717. [Google Scholar]
- 36.Luo H, Tu G, Liu Z, Liu M. Tumor-Associated Macrophages: From Mechanisms to Therapy. Immunity. 2014;41:49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bauer AL, Jackson TL, Jiang Y, Rohlf T. Receptor cross-talk in angiogenesis: mapping environmental cues to cell phenotype using a stochastic, Boolean signaling network model. J Theor Biol. 2010;264:838–846. doi: 10.1016/j.jtbi.2010.03.025. [DOI] [PubMed] [Google Scholar]
- 38.Peng Q, et al. Biological Characteristics and Genetic Heterogeneity between Carcinoma- Associated Fibroblasts and Their Paired Normal Fibroblasts in Human Breast Cancer. PLoS One. 2013;8:e60321. doi: 10.1371/journal.pone.0060321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orimo A, Weinberg RA. Heterogeneity of stromal fibroblasts in tumor. Cancer Biol. Ther. 2007;6:618–619. doi: 10.4161/cbt.6.4.4255. [DOI] [PubMed] [Google Scholar]
- 40.Ishii G, Ochiai A, Neri S. Phenotypic and functional heterogeneity of cancer-associated fibroblast within the tumor microenvironment. Advanced Drug Delivery Reviews. 2016;99:186–196. doi: 10.1016/j.addr.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 41.Knutsdottir H, Condeelis JS, Palsson E. 3-D individual cell based computational modeling of tumor cell-macrophage paracrine signaling mediated by EGF and CSF-1 gradients. Integr. Biol. (Camb) 2016;8:104–19. doi: 10.1039/c5ib00201j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knútsdóttir H, Pálsson E, Edelstein-Keshet L. Mathematical model of macrophage-facilitated breast cancer cells invasion. J. Theor. Biol. 2014;357:184–199. doi: 10.1016/j.jtbi.2014.04.031. [DOI] [PubMed] [Google Scholar]
- 43.Wells DK, et al. Spatial and Functional Heterogeneities Shape Collective Behavior of Tumor-Immune Networks. PLOS Comput. Biol. 2015;11:e1004181. doi: 10.1371/journal.pcbi.1004181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Owen MR, et al. Mathematical modeling predicts synergistic antitumor effects of combining a macrophage-based, hypoxia-targeted gene therapy with chemotherapy. Cancer Res. 2011;71:2826–37. doi: 10.1158/0008-5472.CAN-10-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Owen MR, Sherratt JA. Modelling the macrophage invasion of tumours: effects on growth and composition. IMA J. Math. Appl. Med. Biol. 1998;15:165–85. [PubMed] [Google Scholar]
- 46.Owen MR, Sherratt JA. Pattern formation and spatiotemporal irregularity in a model for macrophage-tumour interactions. J. Theor. Biol. 1997;189:63–80. doi: 10.1006/jtbi.1997.0494. [DOI] [PubMed] [Google Scholar]
- 47.Alarcón T, Byrne HM, Maini PK. A mathematical model of the effects of hypoxia on the cell-cycle of normal and cancer cells. J. Theor. Biol. 2004;229:395–411. doi: 10.1016/j.jtbi.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 48.Kim Y, Friedman A. Interaction of tumor with its micro-environment: a mathematical model. Bull. Math. Biol. 2010;72:1029–1068. doi: 10.1007/s11538-009-9481-z. [DOI] [PubMed] [Google Scholar]
- 49.Kleinstreuer N, et al. A Computational Model Predicting Disruption of Blood Vessel Development. PLoS Comput. Biol. 2013;9:e1002996. doi: 10.1371/journal.pcbi.1002996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Folkman J. Fighting cancer by attacking its blood supply. Sci. Am. 1996;275:150–154. doi: 10.1038/scientificamerican0996-150. [DOI] [PubMed] [Google Scholar]
- 51.Ferrara N. VEGF and the quest for tumour angiogenesis factors. Nat. Rev. Cancer. 2002;2:795–803. doi: 10.1038/nrc909. [DOI] [PubMed] [Google Scholar]
- 52.Hellström M, Phng LK, Gerhardt H. VEGF and Notch signaling: the yin and yang of angiogenic sprouting. Cell adhesion & migration. 2007;1:133–136. doi: 10.4161/cam.1.3.4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scianna M, Bell CG, Preziosi L. A review of mathematical models for the formation of vascular networks. J. Theor. Biol. 2013;333:174–209. doi: 10.1016/j.jtbi.2013.04.037. [DOI] [PubMed] [Google Scholar]
- 54.Perfahl H, et al. 3D hybrid modelling of vascular network formation. J. Theor. Biol. 2017;414:254–268. doi: 10.1016/j.jtbi.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 55.Oden JT, et al. Toward predictive multiscale modeling of vascular tumor growth: computational and experimental oncology for tumor prediction. Arch. Comput. Methods Eng. 2016;23:735–779. [Google Scholar]
- 56.Stéphanou A, et al. How tumour-induced vascular changes alter angiogenesis: Insights from a computational model. J. Theor. Biol. 2017;419:211–226. doi: 10.1016/j.jtbi.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 57.Scott JG, Fletcher AG, Anderson ARA, Maini PK. Spatial metrics of tumour vascular organisation predict radiation efficacy in a computational model. PLoS Comput. Biol. 2016;12:e1004712. doi: 10.1371/journal.pcbi.1004712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Curtis LT, Frieboes HB. The tumormicroenvironment as a barrier to cancer nanotherapy. Advances in Experimental Medicine and Biology. 2016;936:165–190. doi: 10.1007/978-3-319-42023-3_9. [DOI] [PubMed] [Google Scholar]
- 59.Curtis LT, Wu M, Lowengrub J, Decuzzi P, Frieboes HB. Computational modeling of tumor response to drug release from vasculature-bound nanoparticles. PLoS One. 2015;10:e0144888. doi: 10.1371/journal.pone.0144888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bloch N, Harel D. The tumor as an organ: comprehensive spatial and temporal modeling of the tumor and its microenvironment. BMC Bioinformatics. 2016;17:317. doi: 10.1186/s12859-016-1168-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perfahl H, et al. Multiscale modelling of vascular tumour growth in 3D: the roles of domain size and boundary conditions. PLoS One. 2011;6:e14790. doi: 10.1371/journal.pone.0014790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shirinifard A, et al. 3D multi-cell simulation of tumor growth and angiogenesis. PLoS One. 2009;4:e7190. doi: 10.1371/journal.pone.0007190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cai Y, Zhang J, Li Z. Multi-scale mathematical modelling of tumour growth and microenvironments in anti-angiogenic therapy. Biomed. Eng. Online. 2016;15:155. doi: 10.1186/s12938-016-0275-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zangooei MH, Habibi J. Hybrid multiscale modeling and prediction of cancer cell behavior. PLoS One. 2017;12:e0183810. doi: 10.1371/journal.pone.0183810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Norton K-A, Popel AS. Effects of endothelial cell proliferation and migration rates in a computational model of sprouting angiogenesis. Sci. Rep. 2016;6:36992. doi: 10.1038/srep36992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Norton K-A, Popel AS. An agent-based model of cancer stem cell initiated avascular tumour growth and metastasis: The effect of seeding frequency and location. J. R. Soc. Interface. 2014;11 doi: 10.1098/rsif.2014.0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Norton K-A, Wallace T, Pandey NB, Popel AS. An agent-based model of triple-negative breast cancer: The interplay between chemokine receptor CCR5 expression, cancer stem cells, and hypoxia. BMC Syst. Biol. 2017;11 doi: 10.1186/s12918-017-0445-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gong C, et al. A computational multiscale agent-based model for simulating spatio-temporal tumour immune response to PD1 and PDL1 inhibition. J. R. Soc. Interface. 2017;14:20170320. doi: 10.1098/rsif.2017.0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bazzazi H, Isenberg JS, Popel AS. Inhibition of VEGFR2 activation and its downstream signaling to erk1/2 and calcium by thrombospondin-1 (tsp1): In silico investigation. Frontiers in Physiology. 2017;8:48. doi: 10.3389/fphys.2017.00048. [Front. Physiol. 8, 48(2017)] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bazzazi H, Popel AS. Computational investigation of sphingosine kinase 1 (SphK1) and calcium dependent ERK1/2 activation downstream of VEGFR2 in endothelial cells. PLOS Comput. Biol. 2017;13:e1005332. doi: 10.1371/journal.pcbi.1005332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jin K, Pandey NB, Popel AS. Crosstalk between stromal components and tumor cells of TNBC via secreted factors enhances tumor growth and metastasis. Oncotarget. 2017;8:60210–60222. doi: 10.18632/oncotarget.19417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang Y, Snuderl M, Jain RK. Polarization of tumor-associated macrophages: a novel strategy for vascular normalization and antitumor immunity. Cancer Cell. 2011;19:1–2. doi: 10.1016/j.ccr.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ramjiawan RR, Griffioen AW, Duda DG. Anti-angiogenesis for cancer revisited: Is there a role for combinations with immunotherapy? Angiogenesis. 2017;20:185–204. doi: 10.1007/s10456-017-9552-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Norton KA, Popel AS. An agent-based model of cancer stem cell initiated avascular tumour growth and metastasis: the effect of seeding frequency and location. J R Soc Interface. 2014;11:20140640. doi: 10.1098/rsif.2014.0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qutub AA, Popel AS. Elongation, proliferation & migration differentiate endothelial cell phenotypes and determine capillary sprouting. BMC Syst. Biol. 2009;3:13. doi: 10.1186/1752-0509-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gillies RJ, Gatenby RA. Adaptive landscapes and emergent phenotypes: why do cancers have high glycolysis? J Bioenerg Biomembr. 2007;39:251–257. doi: 10.1007/s10863-007-9085-y. [DOI] [PubMed] [Google Scholar]
- 77.Allen WE, Zicha D, Ridley AJ, Jones GE. A role for Cdc42 in macrophage chemotaxis. J. Cell Biol. 1998;141:1147–57. doi: 10.1083/jcb.141.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vanhaesebroeck B, et al. Distinct PI(3)Ks mediate mitogenic signalling and cell migration in macrophages. Nat. Cell Biol. 1999;1:69–71. doi: 10.1038/9045. [DOI] [PubMed] [Google Scholar]
- 79.Chaubey S, Jones GE, Shah AM, Cave AC, Wells CM. Nox2 Is Required for Macrophage Chemotaxis towards CSF-1. PLoS One. 2013;8:e54869. doi: 10.1371/journal.pone.0054869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zicha, et al. Chemotaxis of macrophages is abolished in the Wiskott-Aldrich syndrome. Br. J. Haematol. 1998;101:659–665. doi: 10.1046/j.1365-2141.1998.00767.x. [DOI] [PubMed] [Google Scholar]
- 81.Sturge J, Todd SK, Kogianni G, McCarthy A, Isacke CM. Mannose receptor regulation of macrophage cell migration. J. Leukoc. Biol. 2007;82:585–93. doi: 10.1189/jlb.0107053. [DOI] [PubMed] [Google Scholar]
- 82.Jones GE, et al. Requirement for PI 3-kinase γ in macrophage migration to MCP-1 and CSF-1. Exp. Cell Res. 2003;290:120–131. doi: 10.1016/s0014-4827(03)00318-5. [DOI] [PubMed] [Google Scholar]
- 83.Colegio OR, et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014;513:559–63. doi: 10.1038/nature13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Leung E, et al. Blood vessel endothelium-directed tumor cell streaming in breast tumors requires the HGF/C-Met signaling pathway. Oncogene. 2017;36:2680–2692. doi: 10.1038/onc.2016.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Enderling H, Hlatky L, Hahnfeldt P. The promoting role of a tumour-secreted chemorepellent in self-metastatic tumour progression. Math. Med. Biol. 2012;29:21–29. doi: 10.1093/imammb/dqq015. [DOI] [PubMed] [Google Scholar]
- 87.Enderling H, Hlatky L, Hahnfeldt P. Migration rules: tumours are conglomerates of self-metastases. Br. J. Cancer. 2009;100:1917–25. doi: 10.1038/sj.bjc.6605071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Enderling H, Hlatky L, Hahnfeldt P. Immunoediting: evidence of the multifaceted role of the immune system in self-metastatic tumor growth. Theor. Biol. Med. Model. 2012;9:31. doi: 10.1186/1742-4682-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vermeulen PB, Van Laere SJ, Dirix LY. Inflammatory breast carcinoma as a model of accelerated self-metastatic expansion by intravascular growth. Br. J. Cancer. 2009;101:1028–9. doi: 10.1038/sj.bjc.6605251. author reply 1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Eddy RJ, Weidmann MD, Sharma VP, Condeelis JS. Tumor cell invadopodia: invasive protrusions that orchestrate metastasis. Trends Cell Biol. 2017;27:595–607. doi: 10.1016/j.tcb.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maley CC, et al. Classifying the evolutionary and ecological features of neoplasms. Nat. Rev. Cancer. 2017;17:605–619. doi: 10.1038/nrc.2017.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Maley CC, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat. Genet. 2006;38:468–473. doi: 10.1038/ng1768. [DOI] [PubMed] [Google Scholar]
- 93.Merlo LMF, et al. A comprehensive survey of clonal diversity measures in Barrett’s esophagus as biomarkers of progression to esophageal adenocarcinoma. Cancer Prev. Res. 2010;3:1388–1397. doi: 10.1158/1940-6207.CAPR-10-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maley CC, Koelble K, Natrajan R, Aktipis A, Yuan Y. An ecological measure of immune-cancer colocalization as a prognostic factor for breast cancer. Breast Cancer Res. 2015;17:131. doi: 10.1186/s13058-015-0638-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41:49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Harney AS, et al. Real-time imaging reveals local, transient vascular permeability, and tumor cell intravasation stimulated by TIE2hi macrophage–derived VEGFA. Cancer Discov. 2015;5:932–943. doi: 10.1158/2159-8290.CD-15-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Frascoli F, Flood E, Kim PS. A model of the effects of cancer cell motility and cellular adhesion properties on tumour-immune dynamics. Math. Med. Biol. 2016;34:dqw004. doi: 10.1093/imammb/dqw004. [DOI] [PubMed] [Google Scholar]
- 98.Chang WK, Carmona-Fontaine C, Xavier JB. Tumour-stromal interactions generate emergent persistence in collective cancer cell migration. Interface Focus. 2013;3:20130017–20130017. doi: 10.1098/rsfs.2013.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mitchem JB, et al. Targeting tumor-infiltrating macrophages decreases tumor-initiating cells, relieves immunosuppression, and improves chemotherapeutic responses. Cancer Res. 2013;73:1128–41. doi: 10.1158/0008-5472.CAN-12-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ward R, et al. Monocytes and macrophages, implications for breast cancer migration and stem cell-like activity and treatment. Oncotarget. 2015;6:14687–99. doi: 10.18632/oncotarget.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jeong SK, et al. Tumor associated macrophages provide the survival resistance of tumor cells to hypoxic microenvironmental condition through IL-6 receptor-mediated signals. Immunobiology. 2017;222:55–65. doi: 10.1016/j.imbio.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 102.Kather JN, et al. In Silico Modeling of Immunotherapy and Stroma-Targeting Therapies in Human Colorectal Cancer. Cancer Res. 2017;77:6442–6452. doi: 10.1158/0008-5472.CAN-17-2006. [DOI] [PubMed] [Google Scholar]
- 103.Lohela M, et al. Intravital imaging reveals distinct responses of depleting dynamic tumor-associated macrophage and dendritic cell subpopulations. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E5086–95. doi: 10.1073/pnas.1419899111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.MacDonald KPA, et al. An antibody against the colony-stimulating factor 1 receptor depletes the resident subset of monocytes and tissue- and tumor-associated macrophages but does not inhibit inflammation. Blood. 2010;116:3955–63. doi: 10.1182/blood-2010-02-266296. [DOI] [PubMed] [Google Scholar]
- 105.Ott PA, Hodi FS, Kaufman HL, Wigginton JM, Wolchok JD. Combination immunotherapy: a road map. J. Immunother. Cancer. 2017;5:16. doi: 10.1186/s40425-017-0218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Emens LA, et al. Cancer immunotherapy: Opportunities and challenges in the rapidly evolving clinical landscape. European Journal of Cancer. 2017;81:116–129. doi: 10.1016/j.ejca.2017.01.035. [DOI] [PubMed] [Google Scholar]
- 107.Wan S, et al. Tumor-associated macrophages produce interleukin 6 and signal via STAT3 to promote expansion of human hepatocellular carcinoma stem cells. Gastroenterology. 2014;147:1393–1404. doi: 10.1053/j.gastro.2014.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Williams CB, Yeh ES, Soloff AC. Tumor-associated macrophages: unwitting accomplices in breast cancer malignancy. NPJ breast cancer. 2016;2:15025. doi: 10.1038/npjbcancer.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.