Abstract
The emerging importance of taste in medicine and biomedical research, and new knowledge about its genetic underpinnings, has motivated us to supplement classic taste-testing methods in two ways. First, we explain how to do a brief assessment of the mouth, including the tongue, to ensure that taste papillae are present and to note evidence of relevant disease. Second, we draw on genetics to validate taste test data by comparing reports of perceived bitterness intensity and inborn receptor genotypes. Discordance between objective measures of genotype and subjective reports of taste experience can identify data collection errors, distracted subjects or those who have not understood or followed instructions. Our expectation is that fast and valid taste tests may persuade researchers and clinicians to assess taste regularly, making taste testing as common as testing for hearing and vision. Finally, because many tissues of the body express taste receptors, taste responses may provide a proxy for tissue sensitivity elsewhere in the body and, thereby, serve as a rapid, point-of-care test to guide diagnosis and a research tool to evaluate taste receptor protein function.
Keywords: Medicine, Issue 138, psychophysics, human, tongue, taste cells, TAS2R38, phenylthiocarbamide, psychophysical taste testing
Introduction
Measures of human taste perception can be both part of medical care and a target of biomedical research, yet taste has received scant attention compared with hearing and vision (Table 1). From the medical perspective, when clinicians evaluate patients complaining of taste loss, in most cases the actual loss is of smell1, which has led to dismissal of taste loss as an uncommon and often invalid presenting complaint. Taste distortions (dysgeusia) are more common and frequently arise from the secondary effects of medications or peripheral nerve injury2,3, but neither form has an effective treatment (other than stopping the medication). Clinicians have also ignored taste loss because it has hitherto had little diagnostic or prognostic value on its own. However, although the measurement of taste has been a backwater, it may now be entering mainstream medicine with the revival of a historical appreciation that taste may be a diagnostic or prognostic tool4,5. For instance, bitterness perception can predict immune function6 or the willingness of a patient to take medication7. Nonetheless, biomedical researchers have largely neglected taste. This inattention may, in part, reflect the fact that early progress in understanding this sensory system has its roots in experimental psychology8, a field with which those in medicine may be relatively unfamiliar. Moreover, renewed interest in taste has ushered in standardized taste methods9 that build on earlier methods10, which while comprehensive are lengthy and inappropriate for clinical settings. Finally, confidence in taste measures can be weak because subjects report on their own experience and validation of their observations has hitherto been lacking. Our hope is that a simple measure that investigators or clinicians can easily administer will gain in popularity with both constituents. Here we describe a simple taste exam protocol that has three parts: an assessment of the oral cavity, the taste test, and a validation step using inborn genotype. First, we provide biological context for these procedures, which merge simple practices in medicine, sensory measures from experimental psychology and validation of responses using genotype and genetics.
Taste perception starts in the mouth, so an effective taste exam needs to include a brief clinical assessment for obvious oral diseases, redness, swelling and other discoloration. The oral cavity contains seven subsites: the tongue, gingiva, floor of mouth, buccal mucosa, labial mucosa, hard palate, and the retromolar trigone. Past studies of human taste focused on healthy participants or those with well-defined diseases, but as taste testing becomes routine in medical exams, it is important to record the condition of the oral cavity as part of the procedure.
The tongue itself is a muscular structure encased in mucosa; dotting its dorsal surface are papillae, the small raised structures that give the tongue its unique texture and contain taste receptor cells. We classify papillae by their shape: fungiform, filiform, foliate, and circumvallate. Fungiform papillae (FP) are located anterolaterally on the tongue and are round, with a mushroom shape11. Investigators have published several useful methods to quantify FP and we direct readers to these sources for measurement protocols12,13,14,15,16. Foliate papillae, shaped like the pages of a book (folia), are located exclusively on the lateral posterior tongue surface11. Circumvallate papillae, found in the sulcus terminalis of the tongue base, are large dome-shaped structures surrounded by mucosal walls (Latin circum, "surround," + vallum, "wall")11. The most numerous papillae, the filiform, are long and thin and do not contain taste receptors.
People differ in tongue anatomy. While the sources of this anatomic variation are unknown, it is determined in part by inborn genetic variation, with investigators reporting 31% concordance of tongue anatomy among dizygotic twins and 60% concordance among monozygotic twins17. Papillary density also differs among people, and although rare, at least one genetic disease (familial dysautonomia) results in a congenital absence of taste papillae18,19,20. Thus, before performing psychophysical testing, it is helpful to confirm the presence of FP as part of the brief assessment and note the relative size and color of the tongue and evidence of oral disease.
The taste papillae contain the sensory cells that when stimulated initiate taste sensation. Humans are capable of sensing at least five classes of taste: salty, sour, bitter, sweet, and umami. While salty, sweet, and umami tastes signal the presence of valuable food sources containing sodium chloride, glucose, and amino acids, respectively, bitterness and sourness signal the presence of potential toxins and acids from the bacterial decomposition of food, respectively, and induce aversive behavior21. Salty and sour tastes are transduced through the activation of ion channels found in some types of taste cells, though the understanding of salt transduction is evolving and it may require type I cells as well22,23. Bitter, sweet, and umami arise from activation of G-protein-coupled receptors on type II taste cells, each attuned to a particular taste. Heterodimers of subunits of three particular receptors transduce sweet and umami while bitter compounds activate a group of 25 different bitter receptors24. These bitter receptors can respond to multiple bitter compounds, and a single bitter compound often stimulates more than one receptor25. Despite the recent expansion of knowledge about the molecular basis of taste, novel pathways26 and new discoveries beyond the traditional five taste qualities (e.g., calcium27 or fatty acid28 perception) may lie ahead.
There are at least two surprising aspects of the taste families of receptors: genes that code for these receptors can differ markedly in DNA sequence and hence function among people, and many tissues of the body express these genes21,29,30,31. These extraoral sites include the brain, thyroid, upper and lower respiratory tract, and the gastrointestinal tract, among many others21,29,30,31. While the taste receptors at these locations do not participate in taste perception in the traditional sense, they likely sense the local chemical environment29,32. For example, the ciliated epithelium of the upper respiratory tract expresses the bitter receptor T2R38 (Bitter Taste Receptor 38), which responds to chemical compounds produced by bacteria and influences the innate immune response32, such as increasing mucociliary clearance and levels of anti-microbial peptides and nitric oxide. This finding has medical implications for chronic rhinosinusitis, a disease of chronic bacterial infection and inflammation of the upper respiratory tract and paranasal sinuses.
Of particular relevance to the taste exam we describe here is that the T2R38 bitter taste receptor, encoded by the TAS2R38 gene, exhibits genetic variability and therefore variable taste sensitivity. Perceptual differences for the bitter compound phenylthiocarbamide (PTC) were first described by the chemist Arthur Fox33; later this compound was identified as an agonist of the T2R38 receptor34. Individual differences arise from the DNA sequence of the TAS2R38 gene, which has three single-nucleotide polymorphisms, each yielding amino acid substitutions (A49P, A262V, and I296V; A: Alanine, P: Proline, V: Valine, I: Isoleucine). Two common haplotypes result, PAV and AVI, with PAV/PAV individuals being highly sensitive to PTC ("tasters"), AVI/AVI individuals being relatively insensitive ("non-tasters"), and heterozygous AVI/PAV individuals being more variable in their sensitivity35. There are more examples of genetic variation affecting bitter perception, e.g., taste receptor T2R19, encoded by the TAS2R19 gene, similarly exhibits genetic variability and differing taste sensitivity to the bitter compound quinine36. Likewise, variation in TAS2R31 affects the perceived bitterness of one of the high-potency sweeteners37,38,39.
Here we describe a rapid method to characterize a patient's sense of taste that draws on high-yield protocols in clinical medicine, experimental psychology and genetics.
Protocol
The University of Pennsylvania Institutional Review board approved this protocol. We excluded subjects if they were under 18 years of age or were pregnant.
1. Oral Cavity Evaluation: Disease Assessment and Papilla Identification
Instruct the subject to open the mouth.
- Using a light source such as a penlight or headlamp, illuminate the oral cavity and examine the seven subsites of the area (tongue, floor of mouth, buccal mucosa, labial mucosa, gingiva, hard palate, and retromolar trigone).
- Visualize the dorsal surface of the tongue. Instruct the subject to lift up the tongue, and examine the ventral tongue surface and floor of mouth, making sure to extend the examination posteriorly to the molars.
- Using a tongue depressor, lateralize the subject's cheek to visualize the buccal mucosa, as well as the lateral gingiva bilaterally surrounding the upper and lower teeth.
- Extend the examination anteriorly by lifting the upper and lower lips to visualize the surfaces of the labial mucosal and anterior gingiva.
- Finally, visualize the hard palate and retromolar trigone.
- Note lesions, abrasions, and masses or signs of inflammation.
- Again, ask the subject to open the mouth and extend the tongue.
- Use a light source to visualize the dorsal surface of the tongue.
- Identify the presence or absence of FP, e.g., a smooth tongue surface
Note the results of the oral cavity examination before proceeding with taste testing. If investigators conduct this taste test in a medical context, unexpected findings should prompt further work-up.
2. Psychophysical Taste Testing
NOTE: Resources and descriptions for the psychophysical taste testing that follow are also available from the following web page: https://osf.io/hn87s/.
- Tastant preparation
- Prepare solutions as directed below. Make each solution using a volumetric flask to ensure precision of concentrations to ± 0.0002 M. Dissolve samples using ultrapure water. Tailor the choice of compounds to the research goals. The compounds included here are meant as one example.
- Denatonium benzoate (bitter): prepare a stock solution of 4.99 x 10-3 M denatonium benzoate by dissolving 2.228 g of denatonium benzoate in 1 L of water. Add 180 µL of this stock solution to a 500 mL volumetric flask. Add water to bring the volume to 500 mL, producing a solution with a final concentration of 1.8 µM.
- PTC (bitter): Place 0.0135 g of PTC in a 500 mL volumetric flask. Add water to bring the volume to 500 mL. PTC is difficult to dissolve, so place a stir bar in the flask and heat the solution to 70 °C on a hot plate. Use the stir bar to mix the solution until all solute has dissolved (~15 min). This produces a solution with a final concentration of 180 µM.
- Quinine (bitter): Place 0.011 g of quinine HCl dihydrate in a 500 mL volumetric flask. Add water to bring the volume to 500 mL, producing a solution with a final concentration of 56 µM.
- Sodium chloride (salty): Place 7.5 g of sodium chloride in a 500 mL volumetric flask. Add water to bring the volume to 500 mL, producing a solution with a final concentration of 0.25 M.
- Sucrose (sweet): Place 60 g of sucrose in a 500 mL volumetric flask. Add water to bring the volume to 500 mL, producing a solution with a final concentration of 0.35 M.
- Store taste solutions at 4 °C. Some commonly used taste compounds are light-sensitive, and investigators should wrap them in foil or other materials to reduce their exposure to light.
- To identify common errors in solution preparation, fill one tasting cup with the old solution and one with the new solution. Taste each solution to verify they are identical in strength.
- Include water as a control solution, presented in the first position to verify subjects understand the testing procedure. Present subjects with each tastant and the control tastant twice, taking care to avoid presenting the same tastant consecutively. For example, to test five unique tastants, feature twelve samples in the questionnaire (see 2.2): the five tastants and water, each presented twice. Aliquot 5 mL of water into individual glass scintillation vials. Label the vial caps with a dark blue sticker bearing the number 1.
- Repeat this process for each tastant. Label vial caps with a circular sticker according to the order of presentation and some color code, example detailed below; match the vial labels with the labels on the taste questionnaire (see 2.2). Water (dark blue) Quinine (light blue) NaCl (green) PTC (yellow) Sucrose (orange) Denatonium benzoate (red) NaCl (dark blue) Denatonium benzoate (light blue) Water (green) Quinine (yellow) Sucrose (orange) PTC (red)
- Package samples by placing them into two boxes, samples 1–6 and 7–12 in boxes labeled "box A" and "box B," respectively (Figure 1); other packaging strategies are possible.
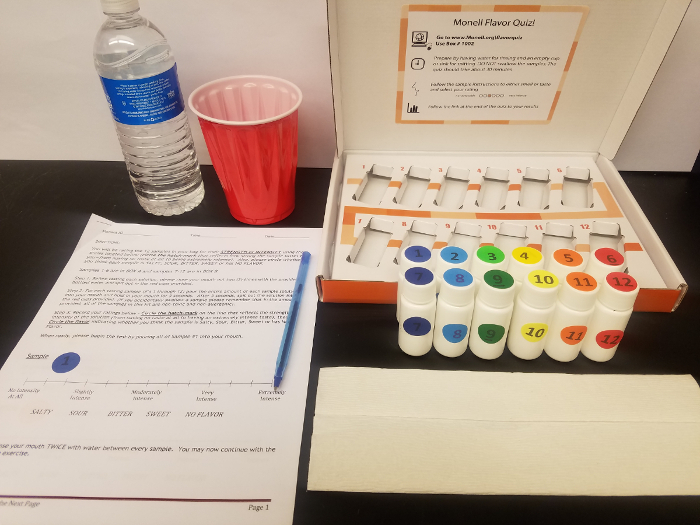
Figure 1: Taste kit. Subjects use the kit to rate taste intensity and quality of various color-coded tastants. Box A contains samples 1–6, box B contains 7–12. Please click here to view a larger version of this figure.
- Taste questionnaire
- Prepare the taste questionnaire using a category scale for rating taste intensity and a forced choice for identifying the taste quality of each tastant (Figure 2).
- Place circular labels of the appropriate color and number next to the appropriate sample in the taste questionnaire (see 2.1.6).
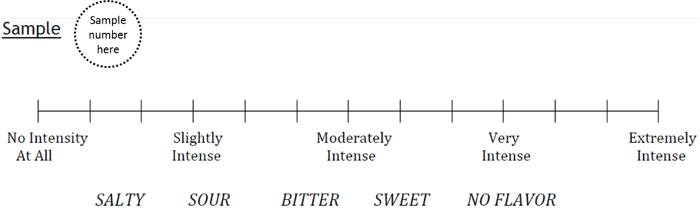
Figure 2: Taste questionnaire entry, comprising a category scale for intensity rating and forced choice response for tastant quality. The taste questionnaire will include one entry for each of the color-coded tastants tested. Please click here to view a larger version of this figure.
- Taste Test Administration
- Provide subjects with box A, box B, a bottle of water, empty cup, pen, and pen-and-paper taste questionnaire containing entries for 12 samples. Use the same brand of bottled water throughout the duration of any given study. As an alternative to a paper questionnaire, port the material into online survey software and administered via tablet or desktop/Laptop. A sample template is available at https://osf.io/hn87s/.
- Instruct subjects that they will be asked to rate both the intensity and quality (e.g., salty, sour, bitter, sweet, or no flavor) of each tastant. Also, inform subjects they may not experience all qualities.
- Explain the testing procedure, as follows: Rinse your mouth twice with water and spit it out in the cup provided. Pour all of sample 1 into your mouth and hold it there for 5 seconds before spitting the solution into the cup. Do not gargle or swallow the solution. Circle one of the 13 vertical lines corresponding with the sample's intensity, on a scale of 0 to 12, from "no intensity at all" to "extremely intense" and choose a single quality to describe the taste. Afterward, rinse your mouth with water twice before proceeding to the next sample.
- Observe the subject tasting and rating sample 1 (water). Should the rating deviate from "no intensity at all" and "no flavor," reiterate the questionnaire instructions before allowing the test to proceed.
- Review the finished questionnaire for completeness.
- Score intensity ratings on a scale of 0 to 12 from the vertical lines that the subjects circled. Average the two intensity ratings for each tastant; this value will be used for analysis.
3. Genotype
Collect saliva from each subject using a saliva DNA collection kit.
Purify genomic DNA from the sample by following manufacturer instructions.
Determine TAS2R38 genotype using SNP genotyping assays (rs713598,rs1726866, rs10246939)40.
Determine TAS2R19 genotype using SNP genotyping assays (rs10772420)36.
4. Genotype-phenotype Validation
Review the available pooled control data from over 800 subjects genotyped for TAS2R38 (rs713598, rs1726866, rs10246939) and TAS2R19 (rs10772420) at the following web page: https://carayata.shinyapps.io/TasteBoxplots/.
Based on a subject's TAS2R38 genotype, compare his or her psychophysical taste response for the bitter compound PTC with the norms for individuals of the same genotype. Responses should match; however, in rare cases, TAS2R38 genotype does not perfectly predict PTC sensory results35.
Should responses show significant divergence, compare the subject's taste response for quinine with the norms for individuals of the same TAS2R19 genotype. Responses for quinine intensity and genotype should match36. Should all taste results fail to correspond with genotype, it is possible that the subject (a) does not understand the task (b) is providing spurious ratings or malingering or (c) there has been an data collection error on the part of the investigator.
Identify data point outliers, and perhaps exclude them from analysis (Figure 3). Correspondence of sensory results with objective genotyping validates the reliability of the psychophysical testing procedure.

Representative Results
Results from the taste test have been pooled for all subjects evaluated (n = 840) and are presented after segregation by genotype. The full data set is accessible at https://carayata.shinyapps.io/TasteBoxplots/ and can be reviewed for each tastant assessed and for TAS2R38 and TAS2R19 genotypes. Results confirm the existence of perceptual taste differences for PTC among subjects grouped by TAS2R38 receptor genotype (Figure 3). Ratings of PTC intensity are significantly different across TAS2R38 genotypes (AVI/AVI, 0.86; AVI/PAV, 6.95; PAV/PAV, 8.18; one-way ANOVA,p <0.0001). Results of quinine intensity are also significantly different across TAS2R19 genotype (A:A, 3.77; A:G, 3.08; G:G, 2.26; one-way ANOVA,p <0.0001).
Figure 3: PTC taste questionnaire results by TAS2R38 genotype. The taste questionnaire can be used to segregate individuals by TAS2R38 genotype based on PTC bitterness intensity ratings on a category scale (*p <0.0001). Here we see a few outliers, data points located outside the fences ("whiskers") of this boxplot (e.g., outside 1.5 times the interquartile range above the upper quartile and bellow the lower quartile) Please click here to view a larger version of this figure.
| Metric | Taste | Hearing | Vision | Hearing/Taste | Vision/Taste |
| PubMed | 36,302 | 123,101 | 171,522 | 3.39 | 4.72 |
| NIH Reporter | 6,144 | 23,873 | 54,858 | 3.89 | 8.93 |
| We conducted a text search of PubMed and NIH Reporter using a particular sense as a key word. PubMed is an online database of published experimental results and NIH Reporter is an online database that lists research projects funded through the National Institutes of Health. Values in the 'Hearing/Taste' column show a ratio, i.e., there are over three times as many publications referencing hearing compared with taste. We accessed the URLs below on January 31, 2018 at 10 am EST. | |||||
| https://www.ncbi.nlm.nih.gov/pubmed/ | |||||
| https://projectreporter.nih.gov/reporter.cfm |
Table 1: Records identified using 'Taste' versus 'Hearing' and 'Vision' as keywords.
Discussion
The significance of this method is that it uses a multidisciplinary approach with features from medicine (the oral exam), experimental psychology (the taste test) and genetics (a validation step). Taste information is likely to develop as a diagnostic and prognostic tool because taste provides a window into the function of proteins elsewhere in the body. From an experimental psychology viewpoint, the addition of a simple exam can identify subjects who are not appropriate for the study of normative taste function. From a genetics point of view, these procedures provide a simple way to study easily reproducible genotype-phenotype relationships.
Measuring human taste has several critical features that are intangible but important, including helping the subject feel at ease and oriented to the task, and especially in medical settings, keeping the procedure short, so the attention of the subject does not waver. It is also important to intervene if subjects appear uncomfortable, and trouble-shoot their concerns, such as regarding the nature of the testing stimuli. Subjects are often reassured to learn that most of the testing stimuli are in foods, e.g., salt and sugar. This procedure, while simple, has significant limitations. While the oral exam is routine for investigators with medical training, those with experimental psychology or genetics training are likely to be less facile at recognizing oral disease. Another limitation is the rating scale that while easily understood by subjects with no prior training, may obscure differences between individuals, or groups of individuals41. Finally, the guidance on how to treat subjects with mismatches between genotype and phenotype in the statistical processing of the data is not yet codified into simple rules, e.g., dropping subjects who fail to meet certain criterion.
Looking ahead to future applications, taste exams may become routine parts of medicine like vision and hearing tests, which would increase our understanding of how taste relates to human disease and well-being and will allow us to refine this simple test.
Disclosures
NAC and DRR are co-inventors on a patent under review (Therapy and Diagnostics for Respiratory Infection 61/697,652, WO2013112865).
Acknowledgments
Awards from the National Institutes of Health supported this research (R01DC013588 to NAC, R21DC013886 to NAC and DRR, and NIDCD Administrative Research Supplement to Promote Emergence of Clinician-Scientists in Chemosensory Research to JED). We collected genotype data from equipment purchased in part with NIH funds from OD018125.
References
- Cowart BJ, Young IM, Feldman RS, Lowry LD. Clinical disorders of smell and taste. Occupational Medicine. 1997;12(3):465–483. [PubMed] [Google Scholar]
- Ackerman BH, Kasbekar N. Disturbances of taste and smell induced by drugs. Pharmacotherapy. 1997;17(3):482–496. [PubMed] [Google Scholar]
- Kveton JF, Bartoshuk LM. The effect of unilateral chorda tympani damage on taste. Laryngoscope. 1994;104:25–29. doi: 10.1288/00005537-199401000-00006. [DOI] [PubMed] [Google Scholar]
- Fischer RA, Griffin F. Pharmacogenetic aspects of gustation. Drug Research. 1964;14(14):673–686. [PubMed] [Google Scholar]
- Joyce CR, Pan L, Varonos DD. Taste sensitivity may be used to predict pharmacological effects. Life Science. 1968;7(9):533–537. doi: 10.1016/0024-3205(68)90058-1. [DOI] [PubMed] [Google Scholar]
- Adappa ND, et al. Genetics of the taste receptor T2R38 correlates with chronic rhinosinusitis necessitating surgical intervention. International Forum of Allergy & Rhinology. 2013. [DOI] [PubMed]
- Lipchock SV, Reed DR, Mennella JA. Relationship between bitter-taste receptor genotype and solid medication formulation usage among young children: a retrospective analysis. Clinical Therapeutics. 2012;34(3):728–733. doi: 10.1016/j.clinthera.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoshuk LM. In: Handbook of perception: Tasting and smelling. Carterette EC, Friedman MP, editors. Academic Press; 1978. pp. 2–18. Vol. VIA. [Google Scholar]
- Coldwell SE, et al. Gustation assessment using the NIH Toolbox. Neurology. 2013;80(11 Suppl 3):S20–S24. doi: 10.1212/WNL.0b013e3182872e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller C, et al. Quantitative assessment of gustatory function in a clinical context using impregnated "taste strips". Rhinology. 2003;41(1):2–6. [PubMed] [Google Scholar]
- Reed DR, Tanaka T, McDaniel AH. Diverse tastes: Genetics of sweet and bitter perception. Physiological Behavior. 2006;88(3):215–226. doi: 10.1016/j.physbeh.2006.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller IJ, Jr, Reedy FE., Jr Variations in human taste bud density and taste intensity perception. Physiological Behavior. 1990;47(6):1213–1219. doi: 10.1016/0031-9384(90)90374-d. [DOI] [PubMed] [Google Scholar]
- Shahbake M, Hutchinson I, Laing DG, Jinks AL. Rapid quantitative assessment of fungiform papillae density in the human tongue. Brain Research. 2005;1052(2):196–201. doi: 10.1016/j.brainres.2005.06.031. [DOI] [PubMed] [Google Scholar]
- Spielman AI, Pepino MY, Feldman R, Brand JG. Technique to collect fungiform (taste) papillae from human tongue. Journal of Visualized Experiments. 2010;18(42):2201. doi: 10.3791/2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuessle TM, Garneau NL, Sloan MM, Santorico SA. Denver papillae protocol for objective analysis of fungiform papillae. Journal of Visualized Experiments. 2015. p. e52860. [DOI] [PMC free article] [PubMed]
- Sanyal S, O'Brien SM, Hayes JE, Feeney EL. TongueSim: development of an automated method for rapid assessment of fungiform papillae density for taste research. Chemical Senses. 2016;41(4):357–365. doi: 10.1093/chemse/bjw008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman AI, Brand JG, Buischi Y, Bretz WA. Resemblance of tongue anatomy in twins. Twin Research and Human Genetics. 2011;14(3):277–282. doi: 10.1375/twin.14.3.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmus H, Smith SM. The antimode and lines of optimal separation in a genetically determined bimodal distribution, with particular reference to phenylthiocarbamide sensitivity. Annals of Human Genetics. 1965;29(2):127–138. doi: 10.1111/j.1469-1809.1965.tb00508.x. [DOI] [PubMed] [Google Scholar]
- Pearson J, Finegold MJ, Budzilovich G. The tongue and taste in familial dysautonomia. Pediatrics. 1970;45(5):739–745. [PubMed] [Google Scholar]
- Fukutake T, et al. Late-onset hereditary ataxia with global thermoanalgesia and absence of fungiform papillae on the tongue in a Japanese family. Brain. 1996;119(Pt 3):1011–1021. doi: 10.1093/brain/119.3.1011. [DOI] [PubMed] [Google Scholar]
- Kinnamon SC. Acta physiologica. 2. Vol. 204. Oxford, England: 2012. Taste receptor signalling - from tongues to lungs; pp. 158–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowski BC, Sukumaran SK, Margolskee RF, Bachmanov AA. Amiloride-insensitive salt taste is mediated by two populations of type iii taste cells with distinct transduction mechanisms. Journal of Neuroscience. 2016;36(6):1942–1953. doi: 10.1523/JNEUROSCI.2947-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbeuch A, Clapp TR, Kinnamon SC. Amiloride-sensitive channels in type I fungiform taste cells in mouse. BMC Neuroscience. 2008;9:1. doi: 10.1186/1471-2202-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolskee RF. The biochemistry and molecular biology of taste transduction. Current Opinions in Neurobiology. 1993;3(4):526–531. doi: 10.1016/0959-4388(93)90051-y. [DOI] [PubMed] [Google Scholar]
- Meyerhof W, et al. The molecular receptive ranges of human TAS2R bitter taste receptors. Chemical Senses. 2010;35(2):157–170. doi: 10.1093/chemse/bjp092. [DOI] [PubMed] [Google Scholar]
- Yee KK, Sukumaran SK, Kotha R, Gilbertson TA, Margolskee RF. Glucose transporters and ATP-gated K+ (KATP) metabolic sensors are present in type 1 taste receptor 3 (T1r3)-expressing taste cells. Proceedings of the National Academy of Sciences USA. 2011. [DOI] [PMC free article] [PubMed]
- Tordoff MG. Calcium: taste, intake and appetite. Physiological Review. 2001;81:1567–1597. doi: 10.1152/physrev.2001.81.4.1567. [DOI] [PubMed] [Google Scholar]
- Reed DR, Xia MB. Recent advances in fatty acid perception and genetics. Advances in Nutrition. 2015;6(3):353S–360S. doi: 10.3945/an.114.007005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blekhman R, et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biology. 2015;16:191. doi: 10.1186/s13059-015-0759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoon MA, et al. Putative mammalian taste receptors: a class of taste-specific GPCRs with distinct topographic selectivity. Cell. 1999;96(4):541–551. doi: 10.1016/s0092-8674(00)80658-3. [DOI] [PubMed] [Google Scholar]
- Laffitte A, Neiers F, Briand L. Functional roles of the sweet taste receptor in oral and extraoral tissues. Current Opinion in Clinical Nutrition and Metabolic. 2014;17(4):379–385. doi: 10.1097/MCO.0000000000000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An SS, et al. Tas2r activation promotes airway smooth muscle relaxation despite beta2-adrenergic receptor tachyphylaxis. American Journal of Physiology-Lung Cellular and Molecular. 2012. [DOI] [PMC free article] [PubMed]
- Fox AL. The relationship between chemical composition and taste. Science. 1931;74:607. [Google Scholar]
- Fox AL. The relationship between chemical constitution and taste. Proceedings of the National Academy of Sciences USA. 1932;18:115–120. doi: 10.1073/pnas.18.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bufe B, et al. The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception. Current Biol.ogy. 2005;15(4):322–327. doi: 10.1016/j.cub.2005.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed DR, et al. The perception of quinine taste intensity is associated with common genetic variants in a bitter receptor cluster on chromosome 12. Human Molecular Genetics. 2010;19(21):4278–4285. doi: 10.1093/hmg/ddq324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobowski N, Reed DR, Mennella JA. Variation in the TAS2R31 bitter taste receptor gene relates to liking for the nonnutritive sweetener Acesulfame-K among children and adults. Science Reports. 2016;6:39135. doi: 10.1038/srep39135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen AL, McGeary JE, Knopik VS, Hayes JE. Bitterness of the non-nutritive sweetener acesulfame potassium varies with polymorphisms in TAS2R9 and TAS2R31. Chemical Senses. 2013;38(5):379–389. doi: 10.1093/chemse/bjt017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudnitzky N, et al. Genomic, genetic, and functional dissection of bitter taste responses to artificial sweeteners. Human Molecular Genetics. 2011;20(17):3437–3449. doi: 10.1093/hmg/ddr252. [DOI] [PubMed] [Google Scholar]
- Guo SW, Reed DR. The genetics of phenylthiocarbamide perception. Annals in Human Biology. 2001;28(2):111–142. doi: 10.1080/03014460151056310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoshuk LM, et al. Labeled scales (e.g., category, Likert, VAS) and invalid across-group comparisons: what we have learned from genetic variation in taste. Food Quality Prefererences. 2003;14(2):125–138. [Google Scholar]


