Abstract
Oral cavity squamous cell carcinoma is the most common type of head and neck carcinoma. Its incidence is increasing worldwide, and it is associated with major morbidity and mortality. It is often unclear which patients have aggressive, treatment refractory tumors vs those whose tumors will be more responsive to treatment. Better identification of patients with high- vs low-risk cancers could help provide more tailored treatment approaches and could improve survival rates while decreasing treatment-related morbidity. This study investigates computer-extracted image features of nuclear shape and texture on digitized images of H&E-stained tissue sections for risk stratification of oral cavity squamous cell carcinoma patients compared with standard clinical and pathologic parameters. With a tissue microarray cohort of 115 retrospectively identified oral cavity squamous cell carcinoma patients, 50 were randomly chosen as the modeling set, and the remaining 65 constituted the test set. Following nuclear segmentation and feature extraction, the Wilcoxon rank sum test was used to identify the five most prognostic quantitative histomorphometric features from the modeling set. These top ranked features were then combined via a machine learning classifier to construct the oral cavity histomorphometric-based image classifier (OHbIC). The classifier was then validated for its ability to risk stratify patients for disease-specific outcomes on the test set. On the test set, the classifier yielded an area under the receiver operating characteristic curve of 0.72 in distinguishing disease-specific outcomes. In univariate survival analysis, high-risk patients predicted by the classifier had significantly poorer disease-specific survival (P = 0.0335). In multivariate analysis controlling for T/N-stage, resection margins, and smoking status, positive classifier results were independently predictive of poorer disease-specific survival: hazard ratio (95% confidence interval) = 11.023 (2.62–46.38) and P = 0.001. Our results suggest that quantitative histomorpho-metric features of local nuclear architecture derived from digitized H&E slides of oral cavity squamous cell carcinomas are independently predictive of patient survival.
Oral cavity squamous cell carcinoma is the most common type of head and neck carcinoma. Despite decades of study and treatment refinement, it is still a major source of morbidity and mortality.1,2 Over the past few years, there has been an increase in the incidence of oral cavity squamous cell carcinoma, particularly in young adults,3,4 and worldwide incidence rates are on the rise. Oral cavity squamous cell carcinoma is typically treated with primary surgery +/ − adjuvant radiation and/or chemotherapy. Less often, induction or concomitant radiation and chemotherapy are used. However, these aggressive treatments are associated with substantial morbidity and sometimes with mortality.5–7 Further, survival rates in oral cavity squamous cell carcinoma remain rather poor despite significant advances in surgical and adjuvant therapy over the past 50 years. The ability to better predict patients who have lower risk cancers vs those that are higher risk could substantially improve treatment and survival rates and reduce treatment-related morbidity. However, despite years of work and many promising candidates, there are no biomarkers of tumor behavior in oral cavity squamous cell carcinoma that have emerged and that are routinely employed in clinical practice. In theory, such a test could allow a clinician to counsel his or her patient more definitively on their prognosis, and possibly to tailor treatment approaches based on the risk of disease progression or recurrence, rather than just treating them per a standard protocol.
Routine histopathologic examination is still the gold standard for diagnosis of oral cavity squamous cell carcinoma. Tumor differentiation or grade, which essentially represents how much a tumor ‘resembles normal squamous epithelium’, has been assessed in many different ways and is recommended information to provide in pathology reports in routine clinical practice. However, it holds only modest predictive value for tumor behavior in oral cavity squamous cell carcinoma, and primarily in early disease (Stage I or II).8 The growth pattern of the tumor at the leading edge has also been examined extensively and patterns of growth and host response also have been shown to be prognostic, again, mostly in early stage disease.7,9,10 Interobserver variability in these assessments by human reviewers, like most quantitative or semi-quantitative measures, is not well assessed. Most of the studies that assert significant prognostic/predictive ability for these features did not allow for the high numbers of reviewers that would be seen in routine clinical practice.
Recently, there has been interest in using computational image analysis to digitally mine features from digitized histologic images of tumors to predict disease aggressiveness, risk of progression, and outcomes.11–20 Li et al7 reported that computer-extracted measurements relating to nuclear texture and intensity distribution patterns within the cytoplasm were found to be prognostic of disease recurrence in lung cancer patients. Similarly, Beck et al found that computer-extracted image features relating to morphology of stromal nuclei were predictive of survival in breast cancer patients.13
Recent work by Friedl et al13 appears to suggest that more aggressive tumor cells are prone to coordinate as a group and function similar to a cluster of cells. Lewis et al11,14 introduced the concept of cell cluster graphs wherein each cluster of proximally located nuclei in the image was considered as a vertex of a graph. By connecting the graph vertices with edges, one could establish a variety of spatial relationships (eg Voronoi Diagram, Delaunay Triangulation, Minimum Spanning Tree) of nuclear arrangement. Lewis et al used quantitative measurements of nuclear arrangement such inter-vertex distance and nuclear packing which were mined from the cell cluster graphs in conjunction with a machine learning classifier for predicting progression among 160 p16-positive oropharyngeal squamous cell carcinoma patients on a tissue microarray.14 In a 3-fold cross-validation schema, the cell cluster graphs feature based classifier was able to achieve 87.5% accuracy in predicting disease progression in these patients.
In this work, using a tissue microarray cohort of oral cavity squamous cell carcinoma patients, we introduce a new set of quantitative histomorphometric features, called local co-occurrence of morphology, which attempts to capture similarity statistics of nuclear shape, size, and texture within cell clusters. Based on a randomly chosen set of patients for modeling, a binary classifier associated with disease-specific outcomes was developed. This was validated on the remaining patients, who constituted the test set. Results were correlated with clinical and pathologic features and with patient outcomes.
Materials and methods
Patient Selection
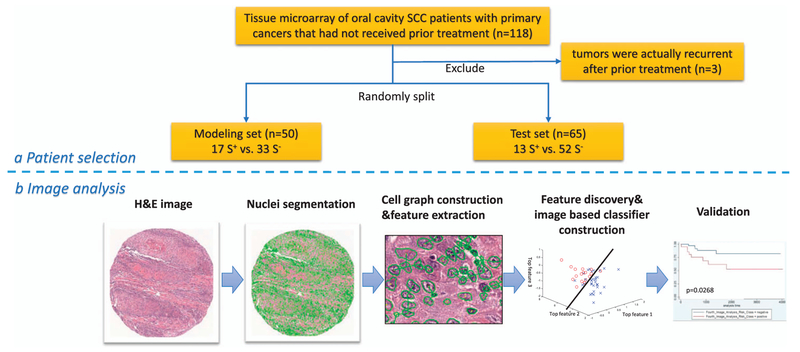
With approval from the Washington University in St. Louis Human Research Protection Office, patients with oral cavity squamous cell carcinoma were identified from Human Research Protection Office-approved clinician databases in Radiation Oncology and Otolaryngology Head and Neck Surgery. The Radiation Oncology database was an approved collection of patients treated by a single radiation oncologist. The Otolaryngology Head and Neck Surgery database was also a Human Research Protection Office-approved collection of patients treated by a single surgeon, all of whom were treated with trans-oral laser surgical resection. Additional cases were gathered by text word searches of the central pathology database. Cases were from 1997 to 2010, and all were primary cancers that had not received prior treatment. Initially, there were 118 patients, but three additional patients were later excluded due to additional follow-up information that the tumors were actually recurrent after prior treatment, thus leaving a final cohort of 115 patients. Slides from each case were reviewed by the study pathologist (JSL) and confirmed to be squamous cell carcinoma. Specific variant types such as basaloid, adenosquamous, verrucous, spindle cell, and papillary were excluded. Two or four 2 mm punches (depending on the amount of tumor in the donor blocks) from each tumor were obtained to construct the array, including leading edge and central tumor, where available. After compilation of the clinical and pathologic information, including American Joint Committee on Cancer (AJCC) 7th edition stage,39 protected health information was then purged from the database. All patients in the study cohort clearly had oral cavity primaries based on clinical and pathologic findings. No oropharyngeal or other primary site tumors were included. We also did not include any patients with primary tumors of the lip.
The slides were digitally scanned on an Aperio Scanscope XT digital scanner at 40 × magnification with a resolution of 0.25 μm per pixel. Each tumor on the array was given a coded number for sharing, with link to actual patient data known only to the study pathologist. For image analysis, one of the central tumor 2 mm punches (ie those punches on the slides that had the most and best represented tumor) was selected by the study pathologist for use.
The whole cohort was then divided into two sets: one for modeling (‘modeling set’) and classifier development and one for validation (‘test set’). For selection of the 50 patients that constituted the modeling set, a random number generator was utilized. The test set then consisted of the remaining 65 patients who were not included in the modeling set. For the modeling set, the tissue microarray tissue punches were designated S+ or S− depending on whether the patient did, or did not, die of recurrent or persistent disease during follow-up, respectively (‘disease-specific survival’). These designations were used in order to provide the basis for feature analysis and classifier development. For the test set, the binary classifier was run without any knowledge of patient mortal or disease status.
Image Analysis
Nuclear detection and segmentation.
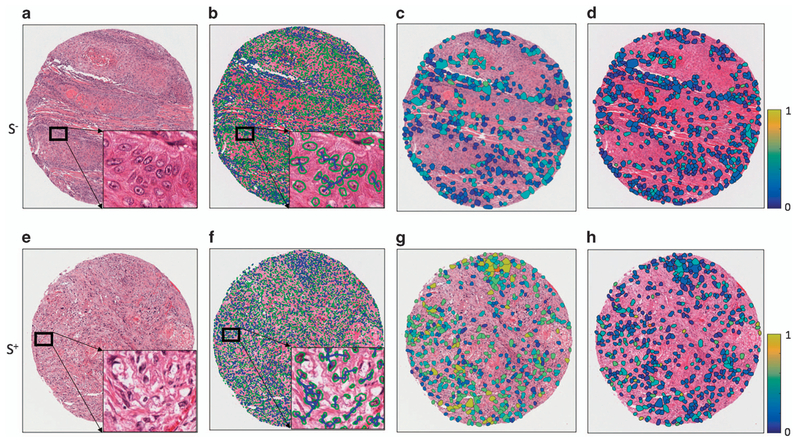
Individual nuclei from both tumor and intervening/surrounding stroma were automatically segmented from the background using a watershed-based nuclear segmentation method21–23 at 40 × magnification (0.25 μm/pixel resolution). The automated segmentation takes into account the intensity/gradient data for each nucleus so that cell boundaries can be extracted for subsequent feature extraction (Figure 2).
Figure 2.
Quantitative histomorphometry feature map illustration. The first column (a, e) shows H&E-stained images of tumors from S− (no disease-specific death) and S+ (disease-specific death) groups. The segmented nuclei contours (shown with green boundaries) are shown in the second column (b, f) with cell cluster graphs. The lines represent connecting edges between nuclei. The third (c, g) and fourth (d, h) columns show quantitative histomorphometric features that capture the variance in nuclear texture and shape, respectively. The blue and yellow colors represent low and high feature values. The S+ case shows a larger range of values than the S− case.
Feature extraction.
The local co-occurrence of morphology feature extraction process comprised the following steps. (1) We identified the local cell clusters based on a distance threshold, where all nuclei within a fixed distance of each other were assigned to the same cluster (additional details provided in the Supplementary Materials Section). (2) We extracted a total of seven size, shape, and texture features for each nucleus within each of the cell clusters. The two nuclear size features were area and major axis length of best fit ellipse for that nucleus.24 The three nuclear shape features were nuclear eccentricity, solidity, and equivalent diameter.15 The two texture descriptors were mean intensity within and immediately outside the nuclear border.19 (3) In order to identify feature similarities between co-clustered nuclei we computed the co-occurrence matrix for each of the 7 extracted nuclear features. The co-occurrence matrix enables the capture of frequency of co-occurrence of diverse features for different nuclei within the cell cluster graphs. Intuitively, if all of the nuclei are identical in appearance, the co-occurrence matrix is 1 × 1. On the other hand, the greater the diversity of attributes, the larger the co-occurrence matrix. We discretized the nuclear morphological features along each feature dimension and created the co-occurrence matrix for each cell cluster graphs. Intuitively, the discretization operation categorizes the nuclei into sub-classes. (4) We then extracted second order statistics, eg, entropy, from each co-occurrence matrix. (5) First order statistics (mean, median, standard derivation, skewness, and kurtosis) were then used to summarize the underlying feature distribution of all cell cluster graphs for each tissue microarray spot image (additional details provided in the Supplementary Materials Section). Intuitively, then, if higher entropy values were observed, this suggests greater diversity in the morphologic features of co-clustered nuclei, with the converse true for lower entropy values.
Figure 1 illustrates the workflow for the construction and evaluation of the classifier. The local co-occurrence of morphology feature maps for two representative S− and S+ cases are shown in Figure 2. Each of the color patches superimposed on the original tissue microarray spot image represents a local cell cluster, in which the patch color represents the normalized value for the feature being analyzed. The size of each patch reflects the size of the local cell cluster. The third and fourth columns show quantitative histomorphometry features that capture the variance of nuclear texture and shape, respectively. The blue and yellow colors represent the lower and higher bounds of a given feature value.
Figure 1.
(a) Illustration of procedure for patient selection for the modeling and test sets. (b) Image analysis flowchart of the tissue microarray spots.
Feature selection.
Three different feature selection methods were implemented to select the local cooccurrence of morphology features most correlated with survival outcomes within the modeling set. Three feature selection methods were considered: (1) Minimum redundancy maximum relevance25 (2) Wilcoxon rank sum test, and (3) Random Forest.26 Each method was employed in conjunction with a three-fold cross-validation scheme and run over 100 iterations within the modeling set to identify the local co-occurrence of morphology features that maximally distinguished the two classes (S+ and S−), while minimizing intra-feature correlation. The top five most frequently selected features were identified and were displayed using box and whisker plots to compare feature expression between the S+ and S− patients. We limited the number of features to five in order to avoid model overfitting and the curse of dimensionality problem, an issue relating to too many features with respect to the number of training examplars.
Classifier Construction
Four different machine learning classifiers, Linear Discriminant Analysis, Quadratic Discriminant Analysis, Support Vector Machine, and Random Forest were implemented in conjunction with the top five features identified using the different feature selection schemes. The machine learning classifiers were evaluated across 100 iterations of three-fold cross-validation within the modeling set. We then identified the top performing classifier feature selection scheme combination based on the area under the receiver operating characteristic (ROC) curve (AUC) values obtained on the learning set. This best performing feature selection-classification combination was designated as the oral cavity squamous cell carcinoma histomorphometric-based image classifier (OHbIC or ‘classifier’).
For each image corresponding to the test set, each machine classifier assigned a probability of recurrence. This probability was then thresholded at different levels from 0 to 1, thereby assigning each test instance to either the S+ or S− categories for each threshold. The predicted labels for each classifier were then compared with the ground truth labels (true patient outcomes) to determine accuracy and AUC performance metrics.
Survival Analysis
Associations between the binary classifier results and the other categorical clinical and pathologic variables were determined by two-sided Fisher’s exact test. Welch’s unequal variance t-test was used for age. Overall survival was defined as the time interval between the start of treatment (either the date of surgical resection or, if treated only non-surgically, the date of the start of radiation and/or chemotherapy) and the date of death from any cause. Disease-specific survival was defined as the time interval between the start of treatment and the date of death in patients with known recurrent tumor in their bodies at the time of death. The Kaplan–Meier (KM) product limit method was used to estimate empirical survival probabilities as illustrated by KM curves. Log-rank tests were applied to examine survival differences, indicating the significance of a categorical variable being prognostic for a survival endpoint. Multivariate Cox proportional hazard models were also used to investigate the independent prognostic ability of variables after accounting for the other clinical and pathologic variables. Hazard ratios, associated 95% confidence intervals, and P-values from Wald tests were reported. All tests were 2 sided, with the significance level set at 0.05. All analyses were performed using Stata v14 statistical software (http://www.stata.com/).
Results
Clinicopathologic Features of the Patient Cohorts
Clinical and pathologic features of the modeling and test sets are provided in Table 1. Patients were primarily in their 60 s, about 40% of whom were current or former smokers. Approximately 50% of patients were white (Caucasian) in both cohorts. More than 60% patients were treated with postoperative radiation and more than 50% of patients’ tumors were T1 or T2. Approximately 75% of the tumors were well or moderately differentiated. They arose predominantly in the oral tongue and less often in the floor of mouth, alveolar ridges, retromolar trigone, buccal mucosa, and hard palate. These features match well with contemporary oral cavity squamous cell carcinoma patient cohorts in the United States27,28 and worldwide. Other than patient sex, there were no significant differences between the modeling and test sets. In the modeling set, 25 patients (50%) suffered disease recurrence vs 21 patients (32%) in the test set.
Table 1.
Summary of clinical and pathological features of the studied oral cavity squamous cell carcinoma cohorts
| Entire cohort N (%) | Modeling set N (%) | Test set N (%) | P-value | |
|---|---|---|---|---|
| No. of patients | 115 | 50 | 65 | |
| Age | 61.0 ± 12.0 | 62.9 ± 13.3 | 59.5 ± 10.7 | 0.1545 |
| Sex | 0.0006 | |||
| Female | 63 (54.8) | 18 (36.0) | 45 (69.2) | |
| male | 52 (45.2) | 32 (64.0) | 20 (30.7) | |
| Race | 0.8530 | |||
| White | 54 (47.0) | 24 (48.0) | 30 (46.2) | |
| Other | 61 (53.0) | 26 (52.0) | 35 (53.8) | |
| Smoking | 1 | |||
| Ever | 29 (90.6) | 16 (94.1) | 13 (86.7) | |
| Never | 3 (9.4) | 1 (5.9) | 2 (13.3) | |
| Comorbidity score | 0.8263 | |||
| None/mild | 59 (64.8) | 26 (52.0) | 33 (50.8) | |
| Moderate/severe | 32 (35.2) | 13 (26.0) | 19 (29.2) | |
| Treatment | 0.1494 | |||
| Surgery only | 33 (29.7) | 11 (22.0) | 22 (33.9) | |
| Postoperative or definitive | 78 (70.3) | 38 (76.0) | 40 (61.5) | |
| Pathology histology | 0.8262 | |||
| Well/moderately differentiated | 88 (76.5) | 39 (78.0) | 49 (75.4) | |
| Poorly differentiated | 27 (23.5) | 11 (22.0) | 16 (24.6) | |
| N-stage | 0.5492 | |||
| N0/N1 | 75 (66.4) | 35 (70.0) | 40 (61.5) | |
| N2 | 38 (33.6) | 15 (30.0) | 23 (35.4) | |
| N-stage | 0.7055 | |||
| N0 | 55 (48.7) | 23 (46.0) | 32 (50.8) | |
| N1/N2 | 58 (51.3) | 27 (54.0) | 31 (49.2) | |
| T-stage | 0.3433 | |||
| T1/T2 | 64 (57.1) | 26 (52.0) | 38 (58.5) | |
| T3/T4 | 48 (42.9) | 24 (48.0) | 24 (36.9) | |
| Margin | 0.4762 | |||
| Negative | 91 (80.5) | 37 (74.0) | 54 (83.1) | |
| Positive | 22 (19.5) | 11 (22.0) | 11 (16.9) | |
| Chemotherapy | 0.2321 | |||
| No | 78(67.0) | 34 (68.0) | 44 (67.7) | |
| Yes | 22 (33.0) | 13 (26.0) | 9 (13.9) | |
| Patient status | 0.2577 | |||
| Alive | 69 (60.0) | 27 (54.0) | 42 (64.6) | |
| Dead | 46 (40.0) | 23 (46.0) | 23 (35.4) | |
| Disease recurrence | 0.1330 | |||
| No | 85 (73.9) | 33 (66.0) | 52 (80.0) | |
| Yes | 30 (26.1) | 17 (34.0) | 52 (80.0) |
Value in bold is statistically significantly different between the Modeling and Test sets, P < 0.05.
Consensus Clustering with Local Co-occurrence of Morphology Features
The top most predictive local co-occurrence of morphology features identified on the modeling set were contrast entropy of nuclear mean intensity, intensity average of nuclear shape (equivalent diameter), intensity average of nuclear border mean intensity, entropy of nuclear mean intensity immediately outside the border, and entropy of nuclear size. These are best described in visual terms as follows:
-
(1)
Contrast entropy of nuclear mean intensity: reflecting the cellular diversity by quantifying the relative difference between cells in terms of the nuclear appearance. Higher values indicate a higher likelihood that nuclei that are extremely dark and extremely bright in appearance present as local cell clusters. Implicitly, this may reflect nuclear pleomorphism on a local scale.
-
(2)
Intensity average of nuclear shape (equivalent diameter): quantifying the cellular diversity in terms of the nuclear shape, higher values suggest a higher likelihood of the presence of extremely large cells within local cell clusters.
-
(3)
Intensity average of nuclear border mean intensity: quantifying the cellular diversity in terms of the nuclear texture, lower values suggest a higher likelihood that nuclei that are darker are present within local cell clusters.
-
(4)
Entropy of nuclear mean intensity immediately outside the border: quantifying the disorder of the inter-cellular regions in terms of image intensity, high values indicate that large variation is present, whereas lower values reflect homogenous inter-cellular appearances in local cell clusters.
-
(5)
Entropy of nuclear size: quantifying the variations of cells in terms of nuclear size, high values indicate large variation between nuclei, whereas lower values reflect relatively uniform sizes of the nuclei present in local cell clusters.
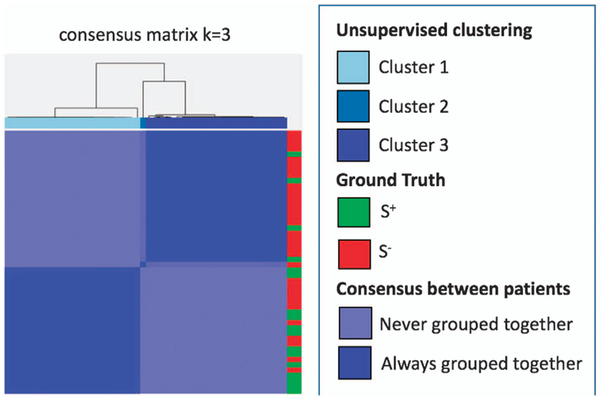
Figure 3 shows consensus clustering plots utilizing local co-occurrence of morphology features on the modeling set. The presence of distinct chusters in the clustergram, which in turn correlate with patient outcome appear, suggest that the local co-occurrence of morphology features appear to be similar for patients in the S+ and S− categories and substantially different across the two outcome categories.
Figure 3.
Consensus clustering utilizing local co-occurrence of morphology features. The local co-occurrence of morphology feature yielded clear clusters which were strongly correlated to outcome (ie S+ and S−).
Classifier Evaluation
The performance of the 12 combinations of feature selection and classifier schemes in terms of AUC on the modeling set are summarized in Table 2. Note that since the combination of Wilcoxon rank sum test and Quadratic Discriminant Analysis yielded the best performance in distinguishing two patient groups on the modeling set, we settled on for a combination of these two analytical tools for constructing OHbIC. Table 3 shows the performance characteristics of the classifier on the test set.
Table 2.
Performance of four different classifiers with three different feature selection methods in the modeling set
| Classifier | Feature selection | AUC | Accuracy | Specificity | Sensitivity |
|---|---|---|---|---|---|
| LDA | WRST | 0.74 ± 0.07 | 0.83 ± 0.03 | 0.91 ± 0.10 | 0.67 ± 0.04 |
| MRMR | 0.83 ± 0.05 | 0.79 ± 0.07 | 0.87 ± 0.08 | 0.65 ± 0.09 | |
| RF | 0.77 ± 0.05 | 0.81 ± 0.03 | 0.91 ± 0.10 | 0.62 ± 0.06 | |
| QDA | WRST | 0.87 ± 0.02 | 0.88 ± 0.02 | 0.93 ± 0.10 | 0.78 ± 0.04 |
| MRMR | 0.81 ± 0.04 | 0.84 ± 0.05 | 0.88 ± 0.14 | 0.76 ± 0.04 | |
| RF | 0.83 ± 0.06 | 0.85 ± 0.3 | 0.91 ± 0.15 | 0.72 ± 0.06 | |
| RF | WRST | 0.81 ± 0.05 | 0.77 ±0.04 | 0.87 ± 0.06 | 0.59 ± 0.02 |
| MRMR | 0.84 ± 0.03 | 0.81 ±0.04 | 0.87 ± 0.06 | 0.68 ± 0.06 | |
| RF | 0.78 ± 0.04 | 0.74 ± 0.04 | 0.83 ± 0.13 | 0.58 ± 0.05 | |
| SVM | WRST | 0.86 ± 0.02 | 0.82 ± 0.03 | 0.93 ± 0.06 | 0.62 ± 0.04 |
| MRMR | 0.79 ± 0.07 | 0.72 ± 0.04 | 0.90 ± 0.15 | 0.35 ± 0.06 | |
| RF | 0.84 ± 0.02 | 0.79 ± 0.02 | 0.92 ± 0.08 | 0.53 ± 0.03 |
Abbreviations: AUC, area under curve; LDA/QDA, linear/quadratic discriminant analysis; MRMR, minimum redundancy, maximum relevance feature selection method; RF, random forest; SVM, support vector machine; WRST, Wilcoxon rank sum test.
The best performance in each metric/column is shown in bold.
Table 3.
Performance of OHbIC classifier on the test set
| AUC | Accuracy | Specificity | Sensitivity |
|---|---|---|---|
| 0.72 | 70.77 | 73.08 | 61.54 |
Abbreviations: AUC, area under curve; OHbIC, oral cavity squamous cell carcinoma quantitative histomorphometric-based image classifier.
Survival Analysis
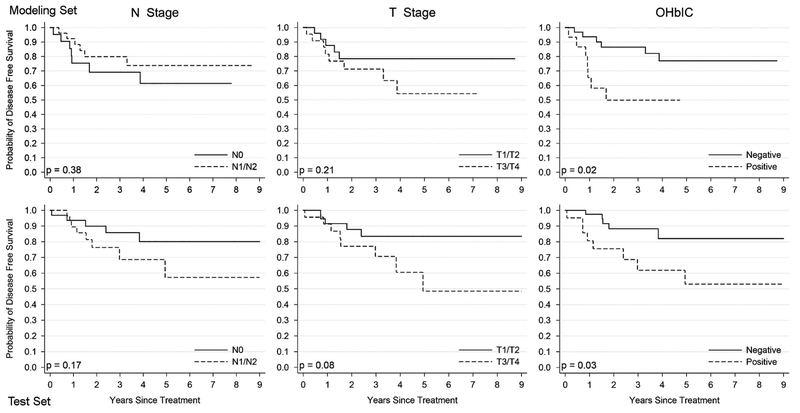
Patients were followed for an average of 38 months (range 1–130 months). Table 4 provides results of the univariate log-rank survival analysis for the major clinical and pathologic variables, and for the classifier on the test set. On the test set, classifier-positive patients (ie patients identified by the classifier to be high-risk) had statistically significantly worse disease-specific survival in univariate analysis (hazard ratio = 3.31, 95% confidence interval = 1.08–10.14 P = 0.0335). Kaplan–Meier survival curves for the classifier, T-stage, and N-stage for the modeling and test sets are presented in Figure 4. Multivariate survival analysis controlling for the other major prognostic variables is presented in Table 5. The classifier results correlated with worse disease-specific survival independent of the other variables (hazard ratio = 11.02, 95% confidence interval = 2.62–46.38, P = 0.001). Thus, patients with positive results were 11 times more likely to develop disease recurrence and die from it.
Table 4.
Univariate log-rank analysis for disease-specific survival on the test set including OHbIC (n = 65 patients)
| Variable | Disease-specific survival HR (95% CI) | P-value |
|---|---|---|
| Age ≤60 vs >60 | 0.39 (0.12–1.26) | 0.103 |
| Race Nonwhite vs white | 0.80 (0.20–3.26) | 0.7435 |
| PaQi histology Well/moderately differentiated vs poorly differentiated | 1.93 (0.59–6.29) | 0.3006 |
| T-stage T1/T2 vs T3/T4 | 2.62 (0.86–8.03) | 0.0855 |
| N-stage N0 vs N1/N2 | 1.66 (0.89–3.08) | 0.1088 |
| N-stage N0/N1 vs N2 | 2.14 (0.69–6.61) | 0.1771 |
| Comorbidity score Non/mild vs moderate/severe | 3.02 (0.80–11.45) | 0.1047 |
| Overall stage I/II vs III/IV | 3.71 (0.82–16.80) | 0.0519 |
| Chemotherapy Yes vs no | 1.96 (0.40–9.66) | 0.4359 |
| Treatment Surgery only vs postoperative/definitive | 1.75 (0.47–6.48) | 0.3830 |
| OHbIC Positive vs neeative | 3.31 (1.08–10.14) | 0.0335 |
Abbreviations: CI, confidence interval; HR, hazard ratio; OHbIC, oral cavity squamous cell carcinoma quantitative histomorphometric-based image classifier.
Value in bold is statistically significant, P < 0.05..
Figure 4.
The first row shows Kaplan–Meier disease-specific survival curves for American Joint Committee on Cancer 7th edition T-stage, N-stage, and classifier results in the modeling set. The second row shows corresponding survival curves for the test set.
Table 5.
Multivariate disease-specific survival analysis on the test set including OHbIC (n = 65 patients)
| Variable | P-value | HR (95% CI) |
|---|---|---|
| T-stage T1/T2 vs T3/T4 | 0.47 | 1.59 (0.45–5.61) |
| N-stage N0 vs N1/N2 | 0.026 | 4.97 (1.21–20.39) |
| OHbIC Positive vs negative | 0.001 | 11.02 (2.62–46.38) |
Abbreviations: CI, confidence interval; HR, hazard ratio; OHbIC, oral cavity squamous cell carcinoma quantitative histomorphometric-based image classifier. Values in bold are statistically significant at P < 0.05.
Discussion
Multiple recent studies have found a causal link between intra-tumoral molecular heterogeneity and worse disease prognosis.29–32 It has also been shown that molecular heterogeneity of tumors is reflected in corresponding morphologic heterogeneity on tissue slides.33–35 The ability to ‘unlock’ subtle, sub-visual attributes of tumors from tissue slides via computerized image analysis offers the opportunity to glean a wealth of information regarding morphologic heterogeneity compared with what may be discerned via traditional visual (or manual) inspection of slides alone.
In this study, we generated an image analysis based classifier, which utilizes quantitative histomorphometric features relating to diversity in nuclear shape, size, and texture within cell clusters from digitized H&E slides of 2 mm oral cavity squamous cell carcinoma microarray tumor sections. Our aim was to quantify and associate local nuclear morphologic heterogeneity with long term outcomes in these patients based on the concept that the greater the local variation (or diversity) in the nuclear features, the more heterogeneous the tumor and hence the worse the outcome.
While there have been previous attempts at using quantitative image analysis for characterizing oral cavity squamous cell carcinoma morphology and predicting behavior, these approaches have not explicitly focused on capturing morphologic heterogeneity. Bose et al18 showed that a combination of measurements including nuclear fractal dimension from DAPI images, Ki67 expression within the tumor from immune-fluorescence images, and lymphocytic infiltration on H&E images measured by a patholo-gist, were significantly associated with disease-specific survival. Other studies have primarily focused on capturing nuclear density or nuclear shape. Vedam et al assessed nuclear diameter, area, perimeter, and shape in the invasive front of 32 oral cavity squamous cell carcinomas and found a correlation of these features with disease recurrence.36 While our approach did evaluate features relating to nuclear shape, texture and appearance, it is different from the approaches in18 and36 in at least three important ways. Firstly, the goal of the new local co-occurrence of morphology feature approach was to evaluate and correlate diversity in nuclear morphologic features with disease recurrence, as opposed to correlating the absolute feature values with outcomes.36 Secondly, cellular diversity was captured via measurement of co-occurring second order statistics (eg entropy) of nuclear size, shape, and texture within local nuclear clusters. By first constructing local cell graphs, we were able to define local neighborhoods of proximal nuclei within which we were then able to compute entropy and diversity of cellular morphologic features. Thus, our approach to capturing intra-tumoral heterogeneity involved quantifying cellular diversity at a local as opposed to a global scale. Finally, and most critically, our approach involved distinct modeling and test sets for evaluating the association between the local co-occurrence of morphology features with disease recurrence. Despite somewhat small numbers, the use of an independent validation set is good evidence of the robustness of this approach.
As molecular studies have begun to show that malignant tumors consist of large numbers of clonal and sub-clonal tumor cell populations, it is possible that these local co-occurrence of morphology features are a surrogate of similarity (or lack thereof) of certain subpopulations of tumor cells, perhaps reflecting the degree of heterogeneity of the tumors. It is becoming clear that markedly heterogeneous tumors with many sub-clones (at least to a point), are clinically more aggressive.36,37 The five most discriminating local cooccurrence of morphology features were identified as (1) contrast entropy of nuclear mean intensity, (2) intensity average of nuclear shape (equivalent diameter) (3) nuclear mean intensity immediately outside the boundary (4) entropy of nuclear mean intensity immediately outside the boundary, and (5) nuclear size. The local co-occurrence of morphology features were found to have higher entropy values in worse prognosis tumors compared with the ones with better prognosis. This is intuitive since higher entropy values are reflective of greater diversity in local nuclear morphologic features while lower entropy values would reflect the converse. The third and fourth columns of Figure 2 illustrate the differences in nuclear morphologic diversity within local nuclear clusters between patients in the S+ and S− groups. For the patients in the S+ group, there is a greater variation in nuclear shape, as evidenced by the brighter colors in (Figure 2g and h). By contrast, for the patients in the S− group, nuclear texture appears to be more uniform, at least locally (Figure 2c and d).
Oral cavity squamous cell carcinoma prognosis can be stratified in many ways, most notably by TNM classification using combinations of tumor size, invasiveness, and nodal and distant metastases.7,10,38 None of these features are based on the actual morphologic attributes of the tumors. Patients that were identified with poor prognosis by the classifier were 12 times more likely to suffer from disease recurrence, and six times more likely to develop distant metastases. Based on the binary output generated by the classifier, the log-rank test showed a strong association between classifier scores and disease-specific survival (P = 0.03) in the test set. In multivariate analysis, controlling for AJCC 7th edition39 T and N classification, classifier results were still strongly associated with worse disease-specific survival (P = 0.001). Unfortunately, we do not have extra nodal extension or depth of invasion data to analyze outcomes with the very recently published 8th edition AJCC staging system.40
The classifier utilizes H&E-stained images and was modeled on just single 2 mm tissue microarray spots for each tumor. This suggests that very small samples of tumor may be sufficient for this testing to be prognostic, meaning that it should work for very small tumors and also for small biopsy specimens. One can imagine a potential clinical management strategy in which the classifier is applied alongside routine pathologic evaluation of patient specimens and then results are combined with (or utilized in specific subgroups from) other major clinical and pathologic variables, such as TNM stage, to more effectively stratify patients into low-risk and high-risk groups. Patients with different group labels could have tailored treatment approaches developed.
Limitations of this study are that the image analysis was performed on tissue microarrays, which are only a small portion of the whole tumors. Morphological variation is known to exist within the same tumor, which may not be captured in small tissue microarray punches. Whole slide images may provide a more comprehensive picture of the tumors. Even though we had an independent validation cohort, the overall numbers of patients in the study are small and some of the other prognostic features in oral cavity squamous cell carcinoma patients, such as depth of invasion and nodal extracapsular extension, were not controlled for. Future studies will be needed to analyze the classifier on whole slide images and in a statistically-powered retrospective cohort of patients, one in which we control for all of the well-established clinical and pathologic features and in well-established outcome subgroups of patients.
In summary, this study models and validates the independent prognostic value of a computer-assisted histomorphometric classifier of nuclear shape and texture in a cohort of oral cavity squamous cell carcinoma patients. It shows independent prognostic ability for these morphologic features, providing strong evidence for their utility. These results pave the way for future studies to further refine the classifier and independently validate it in prospective patient cohorts.
Supplementary Material
Acknowledgments
We thank Autumn Watson B.A. for her expert technical assistance with the tissue microarray construction. We also thank Donna M. Posey for her wonderful assistance with clerical support and spreadsheet data management for the various aspects of this study. We also thank Tyler Smallman for his assistance with the digital image scanning and file management. Dr. Lu is partially supported by the National Natural Science Foundation of China (Grant Numbers 61401263, 61672333, 61672334, 61673251, 61502290, 61573232, 61501287). Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award numbers 1U24CA199374–01, R01CA20 2752–01A1, R01CA208236–01A1, R21CA179327–01, R21CA195152–01, the National Institute of Diabetes and Digestive and Kidney Diseases under award number R01DK098503–02, National Center for Research Resources under award number 1 C06 RR12463–01, the DOD Prostate Cancer Synergistic Idea Development Award (PC120857); the DOD Lung Cancer Idea Development New Investigator Award (LC130463), the DOD Prostate Cancer Idea Development Award; the DOD Peer Reviewed Cancer Research Program W81XWH-16–1-0329, the Case Comprehensive Cancer Center Pilot Grant, VelaSano Grant from the Cleveland Clinic, the Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering at Case Western Reserve University. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Disclosure/conflict of interest
Dr. Madabhushi is an equity holder in Elucid Bioimaging and in Inspirata Inc. He is also a scientific advisory consultant for Inspirata Inc., sits on its scientific advisory board, and is an equity holder in the company. Additionally, his technology has been licensed to Elucid Bioimaging and Inspirata Inc. He is also involved in a NIH U24 grant with PathCore Inc. The remaining authors declare no conflict of interest.
Supplementary Information accompanies the paper on Modern Pathology website (http://www.nature.com/modpathol)
References
- 1.Chaturvedi AK, Anderson WF, Lortet-Tieulent J, et al. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J Clin Oncol Off J Am Soc Clin Oncol 2013;31:4550–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta B, Johnson NW, Kumar N. Global epidemiology of head and neck cancers: a continuing challenge. Oncology 2016;91:13–23. [DOI] [PubMed] [Google Scholar]
- 3.D’Souza G, Kreimer AR, Viscidi R, et al. Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med 2007;356:1944–1956. [DOI] [PubMed] [Google Scholar]
- 4.Sturgis EM, Cinciripini PM. Trends in head and neck cancer incidence in relation to smoking prevalence: an emerging epidemic of human papillomavirus-associated cancers? Cancer 2007;110:1429–1435. [DOI] [PubMed] [Google Scholar]
- 5.Hutcheson KA, Lewin JS. Functional outcomes after chemoradiotherapy of laryngeal and pharyngeal cancers. Curr Oncol Rep 2012;14:158–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Group, E. B. C. T. C.. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet 2005;365:1687–1717. [DOI] [PubMed] [Google Scholar]
- 7.Li Y, Bai S, Carroll W, et al. Validation of the risk model: high-risk classification and tumor pattern of invasion predict outcome for patients with low-stage oral cavity squamous cell carcinoma. Head Neck Pathol 2013;7:211–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas B, Stedman M, Davies L. Grade as a prognostic factor in oral squamous cell carcinoma: a population-based analysis of the data: early oral cancer-grade matters. Laryngoscope 2014;124:688–694. [DOI] [PubMed] [Google Scholar]
- 9.Brandwein-Gensler M, Smith RV, Wang B, et al. Validation of the histologic risk model in a new cohort of patients with head and neck squamous cell carcinoma. Am J Surg Pathol 2010;34:676–688. [DOI] [PubMed] [Google Scholar]
- 10.Sinha P, Mehrad M, Chernock RD, et al. Histologic and systemic prognosticators for local control and survival in margin-negative transoral laser microsurgery treated oral cavity squamous cell carcinoma: systemic and histologic prognosticators in transoral laser microsurgery-treated oral cavity cancers. Head Neck 2015;37:52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ali S, Lewis J, Madabhushi A. Spatially aware cell cluster(spACC1) graphs: predicting outcome in oropharyngeal pl6+ tumors. Med Image Comput Comput-Assist Interv 2013;16:412–419. [DOI] [PubMed] [Google Scholar]
- 12.Doyle S, Hwang M, Shah K et al. Automated grading of prostate cancer using architectural and textural image features. In Proceedings of the 4th IEEE International Symposium on Biomedical Imaging 2007; 1284–1287. [Google Scholar]
- 13.Beck AH, Sangoi AR, Leung S, et al. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med 2011;3:108ra113. [DOI] [PubMed] [Google Scholar]
- 14.Lewis JSJ, Ali S, Luo J, et al. A quantitative histomorphometric classifier (QuHbIC) identifies aggressive versus indolent p16-positive oropharyngeal squamous cell carcinoma. Am J Surg Pathol 2014;38:128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Basavanhally A, Ganesan S, Feldman MD, et al. Multi-field-of-view framework for distinguishing tumor grade in ER+ breast cancer from entire histopathology slides. IEEE Trans Biomed Eng 2013;60:2089–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu K-H, Zhang C, Berry GJ, et al. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun 2016;7: 12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romo-Bucheli D, Janowczyk A, Romero A, et al. Automated tubule nuclei quantification and correlation with oncotype DX risk categories in ER+ breast cancer whole slide images. Sci Rep 2016;6:32706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bose P, Brockton NT, Guggisberg K, et al. Fractal analysis of nuclear histology integrates tumor and stromal features into a single prognostic factor of the oral cancer microenvironment. BMC Cancer 2015;15: 409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madabhushi A, Lee G. Image analysis and machine learning in digital pathology: challenges and opportunities. Med Image Anal 2016;33:170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhargava R, Madabhushi A. Emerging themes in image informatics and molecular analysis for digital pathology. Annu Rev Biomed Eng 2016;18:387–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu H, Lu C, Berendt R, et al. Automatic nuclei detection based on generalized Laplacian of Gaussian filters. IEEE J Biomed Health Inform 2016;21:826–837. [DOI] [PubMed] [Google Scholar]
- 22.Veta M, van Diest PJ, Kornegoor R, et al. Automatic nuclei segmentation in H&E stained breast cancer histopathology images. PloS One 2013;8:e70221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu C, Mahmood M, Jha N, et al. A robust automatic nuclei segmentation technique for quantitative histo-pathological image analysis. Anal Quant Cytopathol Histopathol 2012;34:296–308. [PubMed] [Google Scholar]
- 24.Lu Cheng, Mandal M. Toward automatic mitotic cell detection and segmentation in multispectral histopathological images. IEEE J Biomed Health Inform 2014;18:594–605. [DOI] [PubMed] [Google Scholar]
- 25.Peng H, Long F, Ding C. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Anal Mach Intell 2005;27:1226–1238. [DOI] [PubMed] [Google Scholar]
- 26.Breiman L Random forests. Mach Learn 2001;45:5–32. [Google Scholar]
- 27.Bobdey S, Balasubramaniam G, Mishra P. Nomogram prediction for survival of patients with oral cavity squamous cell carcinoma: nomogram prediction for survival of patients with oral cancer. Head Neck 2016;38:1826–1831. [DOI] [PubMed] [Google Scholar]
- 28.Marchiano E, Patel TD, Eloy JA, et al. Impact of nodal level distribution on survival in oral cavity squamous cell carcinoma: a population-based study. Otolaryngol-Head Neck Surg 2016;155:99–105. [DOI] [PubMed] [Google Scholar]
- 29.Zhang XC, Xu C, Mitchell RM, et al. Tumor evolution and intratumor heterogeneity of an oropharyngeal squamous cell carcinoma revealed by whole-genome sequencing. Neoplasia N Y N 2013;15:1371–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mroz EA, Rocco JW. MATH, a novel measure of intratumor genetic heterogeneity, is high in poor-outcome classes of head and neck squamous cell carcinoma. Oral Oncol 2013;49:211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mroz EA, Tward AD, Pickering CR, et al. High intratumor genetic heterogeneity is related to worse outcome in patients with head and neck squamous cell carcinoma. Cancer 2013;119:3034–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multi-region sequencing. N Engl J Med 2012;366:883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh RR, Murugan P, Patel LR, et al. Intratumoral morphologic and molecular heterogeneity of rhabdoid renal cell carcinoma: challenges for personalized therapy. Mod Pathol 2015;28:1225–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sciallis AP, Bedroske PP, Schoolmeester JK, et al. High-grade endometrial stromal sarcomas: a clinicopathologic study of a group of tumors with heterogenous morphologic and genetic features. Am J Surg Pathol 2014;38:1161–1172. [DOI] [PubMed] [Google Scholar]
- 35.Mafficini A, Amato E, Fassan M, et al. Reporting tumor molecular heterogeneity in histopathological diagnosis. PloS One 2014;9:e104979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vedam VKV, Boaz K, Natarajan S. Prognostic efficacy of nuclear morphometry at invasive front of oral squamous cell carcinoma: an image analysis microscopic study. Anal Cell Pathol 2014;1:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mroz EA, Tward AM, Hammon RJ, et al. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from The Cancer Genome Atlas. PLOS Med 2015;12:e1001786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang Y-C, Nieh S, Chen SF, et al. Invasive pattern grading score designed as an independent prognostic indicator in oral squamous cell carcinoma: Invasive pattern grading score in OSCC. Histopathology 2010;57:295–303. [DOI] [PubMed] [Google Scholar]
- 39.Chapter 3: lip and oral cavity In: Edge S, Byrd DR, Compton CC, et al. (eds). AJCC Cancer Staging Manual, 7th edn Springer: New York, 2010, pp 29–40. [Google Scholar]
- 40.Ridge JA, Lydiatt WM, Patel SG, et al. Chapter 7: lip and oral cavity In: Amin MB, Edge S, Greene F, et al. (eds). AJCC Cancer Staging Manual, 8th edn Springer: New York, 2016, pp 113–121. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.