SUMMARY:
In the last few decades, we have seen significant advances in brain imaging, which have resulted in more detailed anatomic and functional localization of gliomas in relation to the eloquent cortex, as well as improvements in microsurgical techniques and enhanced delivery of adjuvant stereotactic radiation. While these advancements have led to a relatively modest improvement in clinical outcomes for patients with malignant gliomas, much more work remains to be done. As with other types of cancer, we are now rapidly moving past the era of histopathology dictating treatment for brain tumors and into the realm of molecular diagnostics and associated targeted therapies, specifically based on the genomic architecture of individual gliomas. In this review, we discuss the current era of molecular glioma characterization and how these profiles will allow for individualized, patient-specific targeted treatments.
KEYWORDS : 1p/19q, anaplastic glioma, Avastin, BRAF inhibitors, convection based therapy, cyclin inhibitors, EGRF, EGRFvIII, glioblastoma, glioma, glioma vaccines, oligodendroglioma, p53, PCV, personalized medicine, temozolamide
Practice Points.
High-grade gliomas (HGG) are historically classified by the histopathological WHO system as either anaplastic astrocytoma (grade III) or glioblastoma (GBM, grade IV); however, this system does not take into account underlying genetic alterations driving the disease and in turn differing outcomes.
Despite an aggressive standard of care, including surgical resection followed by radiotherapy with temozolomide, median survival for GBM remains poor.
Recent efforts aimed at dissecting the genomic architecture of glial tumors has led to significant advancements in our understanding of the molecular pathways important in gliomagenesis, revealing several genes recurrently mutated during tumor formation.
Similarly, gene expression profiling of GBMs has identified at least four different genetic subtypes, yielding specific prognostic expectations.
The use of next generation genomic technologies, such as massively parallel sequencing, now allows for identification of somatic driver mutations and genomic events in individual tumors, marking the start of personalized oncologic care for patients with HGG.
Agents currently being employed or in development to target HGGs include receptor tyrosine kinase inhibitors, PI3K/Akt inhibitors, mTOR inhibitors, BRAF inhibitors, HDAC inhibitors and proteasome inhibitors, as well as VEGF inhibitors.
Other strategies include targeting glioma cells and developing vaccines against an individual’s specific tumor.
Institutions with genomic capabilities should consider sequencing their patient’s glioma samples in an effort to forge the beginnings of personalized medicine by selecting treatment targets based on genomic signatures of individual gliomas.
Background
Malignant gliomas are the most frequently occurring primary brain tumor [1], with an annual incidence of 6–7 cases per 100,000 and a median age of onset in the 5th and 6th decades of life [1,2]; they pose a significant challenge for care practitioners despite multimodal treatment strategies. Malignant gliomas are derived from glial cells and are heterogeneous in appearance, typically with a central region of necrosis surrounded by contrast-enhancing proliferative glioma cells [3]. These tumors are highly infiltrative and extend beyond the areas of contrast enhancement [3]. The WHO classifies gliomas into four grades based on histology: grade 1 (pilocytic astrocytoma), grade II (diffuse astrocytoma), grade III (anaplastic astrocytoma or AA), and grade IV (glioblastoma or GBM) [3], with the latter two considered malignant or high-grade (HGG) and accounting for approximately 75% of cases [1,3]. This histopathological classification system has served as the cornerstone that guides management and predicts prognosis [4]; yet despite the standard of care, including maximal safe surgical resection followed by radiotherapy with temozolomide [5], and a variety of salvage therapies at recurrence, median survival for GBM remains less than 15–20 months [6,7]. Furthermore, patients with histologically identical tumors may have very different outcomes, underscoring the heterogeneity of underlying molecular derangements in these tumors. Thus, in an effort to increase meaningful survival, the focus has shifted away from the general histopathological characterization towards understanding the distinct molecular and genetic alterations in gliomas, with the goal of developing more rational therapies. In this review, we will discuss the current known molecular alterations in malignant gliomas and offer insight into the potential for targeted, patient-specific therapies.
Classification schemes & survival
Gliomas are classified into four pathological grades based on tumor cell density, presence of nuclear atypia and necrosis, as well as the mitotic index [3]. In addition, gliomas can be categorized based on histological subtypes as determined by their cell of origin, including astrocytic, oligodendroglial, or mixed [3]. The genomic architecture of a glioma varies not only according to the cell of origin and pathological grade, but also the patient’s age [1,2]. Indeed, there is a significant difference in the genetic makeup of gliomas in the pediatric versus adult age. For example, WHO grade I gliomas, which are mainly observed in children, mostly harbor the recurrent activating BRAF mutations or the BRAF–KIAA1549 fusion [8]. By contrast, most of the grade II low-grade gliomas (LGGs) are typically observed in young adults [9]. Genetically, the majority of these LGGs harbor a recurrent mutation affecting the R132 residue of the IDH1 gene [10]. The mutated IDH1 enzyme gains the catalytic ability to produce an oncometabolite, 2-hydroxyglutarate, affecting epigenetic regulations and establishing a stereotypic CpG island hypermethylator phenotype in these tumors [10]. Interestingly, this hypermethylator phenotype has been associated with a better outcome, being observed in a subset of long-term GBM survivors (>3 years) [11]. Importantly, the recurrent IDH1 R132H mutation co-exists either with TP53 and ATRX mutations along with chromosome 17 loss in tumors of astrocytic origin or with CIC and FUBP1 mutations [12], as well as chromosome 1p and 19q loss in oligodendroglial tumors [13]. Mixed tumors contain a combination of the above genomic alterations [12,13].
Although these tumors pathologically appear low grade and carry an indolent clinical course in the beginning, they do not necessarily carry a benign long-term prognosis. Median overall survival (OS ) following surgical resection, chemotherapy, and radiation depends on the histological subtype: 4–10 years for grade II astrocytoma [1,14], 2–5 years for grade III astrocytoma [15] and 11.6 years for grade II oligodendroglioma [16]. Similarly, the rate of secondary malignant transformation into anaplastic glioma or GBM is high at 74% for astrocytoma versus 45% for oligodendroglioma, with this transformation associated with OS of less than 14 months for GBM [17]. Although the molecular mechanisms responsible for the transformation of LGGs into these secondary HGGs are poorly understood, based on the efforts of The Cancer Genome Atlas (TCGA) research network, the pathways underlying formation of primary HGGs, specifically GBMs, are described in detail in the Molecular Signaling Pathways Section (Figure 1) [18].
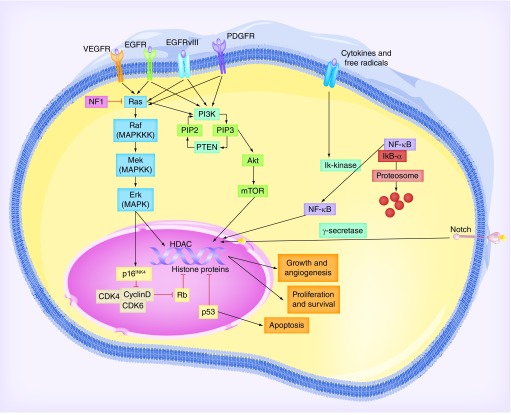
Figure 1. The main signaling pathways affected in high-grade gliomas.
Receptor tyrosine kinases (i.e., EGFR, VEGFR, PDGFR) signal through a MAPK cascade to promote cell proliferation, survival, angiogenesis, and differentiation. In gliomas, this pathway is mutated such that it is deregulated and overactivated, leaving DNA transcription unchecked. NF-1 is normally a brake that inhibits Ras, but is often mutated in gliomas and thus is nonfunctional. The tyrosine kinases also signal through PI3K, which phosphorylates PIP2 to the active PIP3, which goes on to activate nuclear transcription through mTOR. This pathway is kept in check by PTEN, but mutations in gliomas result in constitutively active PI3K or an inactive PTEN. NF-κB is normally found in the cytoplasm bound with an inhibitor-α, but when activated the inhibitor-α and the free NF-kB can then translocate to the nucleus to regulate transcription. Notch pathway activation results in the cleavage of the cytoplasmic domain of the transmembrane receptor by γ-secretase, which then translocates to the nucleus to affect transcription. In terms of direct nuclear regulation, p53 is mutated in a majority of gliomas and DNA damage goes unregulated allowing continuously new mutations to occur. Lastly, Rb is a brake that keeps transcription off and is normally inhibited by cyclins that promote transcription. p16 normally inhibits the cyclin proteins to keep the cycle in check, but p16 is often mutated in gliomas, thereby leading to deregulated proliferation. In the end, all the various treatment strategies discussed go after one of these cascades either directly or indirectly.
EGFR: EGF receptor; PDGFR: PDGF receptor; PIP2: Phosphatidylinositol 4,5-bisphosphate; PIP3: Phosphatidylinositol 3,4,5-bisphosphate; PTEN: Phosphate and tensin homolog; VEGFR: VEGF receptor.
• Pediatric GBMs
As mentioned above, the genomic architecture of gliomas differ significantly based on the age of the patients. Although malignant gliomas are rare in children, recent reports focusing on the genomic architecture of pediatric GBMs failed to identify mutations in the aforementioned molecules. Instead, in pediatric cases, driver mutations in histone and chromatin were identified, with 44% of cases harboring mutations in the H3.3–ATRX–DAXX chromatin remodeling pathway [19]. In addition, recurrent mutations in H3F3A, which encodes the replication-independent histone 3 variant H3.3, were found in 31% of pediatric GBMs [19]. Mutations in ATRX and DAXX, which encode subunits of the chromatin-remodeling complex, were identified in 31% of all patients, but remarkably in all of the patients with histone mutations [19]. Further supporting a central role for epigenetic regulation in gliomagenesis in pediatric cases, mutations in the genes encoding H3.3 core histone proteins, H3F3A and HIST1H3B, were recently identified in 78 and 22% of pediatric diffuse intrinsic pontine glioma and nonbrainstem pediatric glioblastomas, respectively [20]. It is important to emphasize that adult GBMs are much less likely to harbor mutations in these genes, once again reflecting the differences in genomic architecture of pediatric versus adult gliomas.
• Primary versus secondary GBMs
Although primary GBMs that form de novo and secondary GBMs that form due to malignant transformation of LGGs appear histologically identical, their genomic architecture differ quite significantly [21]. Primary GBMs are typically observed in patients older than 50 years of age, and are commonly associated with EGFR amplifications and/or activating mutations, loss of chromosomes 10q (PTEN), as well as 9p21 (CDKN2A locus, encoding for the p16Ink4A) [15,21]. Secondary GBMs, on the other hand, are much less common and result from the sequential accumulation of somatic mutations as well as chromosomal aberrations of LGGs [21]. These tumors usually occur in younger individuals, harbor p53 tumor suppressor and IDH gene mutations, loss of heterozygosity (LOH) of chromosome 10q, as well as abnormalities in p16 and Rb [15,21]. Specifically, IDH1 mutations are a definitive diagnostic molecular marker of secondary GBMs [15,21]. Generally, based on gene expression profiles, GBMs have been divided into four subtypes, including proneural, neural, classical and mesenchymal [22]. Among these, aberrations and gene expression of EGFR, NF1 and PDGFRA/IDH1 have been shown to define the classical, mesenchymal and proneural subtypes [22], respectively, with response to therapy differing by subtype, with the greatest seen in the classical subtype and no benefit seen in proneural subtypes [22]. Overall, these classification systems have laid the foundation to catalog the genetic landscape of gliomas, providing prognostic information and potential targets for therapy.
Molecular signaling pathways & pathogenesis
Malignant gliomas can arise either de novo or secondarily from lower grade tumors through the acquisition of additional genetic alterations [1]. Over the last two decades, the advancements in genomic technologies, particularly with the introduction of next generation mass sequencing, has allowed characterization of these tumors at a genomic level. Various genomic alterations have been cataloged, and distinct patterns in molecular pathways have emerged, which are discussed below.
• RTK/Ras/PI3K/AKT1 pathway
Alterations in receptor tyrosine kinase (RTK) signaling have long been associated with gliomagenesis. RTKs transduce extracellular growth factors into intracellular cascades through the MAPK pathway, inducing cell proliferation. Amplification of the EGFR gene is the most frequent RTK affected in gliomas, seen in 40% of astrocytic tumors, and/or a constitutively active variant EGFRvIII seen in 20–30% of astrocytic tumors, both of which enhance tumor growth, survival, progression and resistance to therapy [23]. The EGFRvIII mutation is characterized by a deletion of 267 amino acids in the extracellular domain, leading to a constitutively active receptor that is unable to bind ligand [24]. This continuously active receptor has impaired internalization and degradation, thus leading to enhanced tumorigenic potential by activating and maintaining mitosis pathways, anti-apoptotic pathways, as well as invasive signaling pathways [24]. Given that EGFRvIII is not found in normal tissues, targeted therapy has been actively sought, which will be discussed later. Similarly, the PDGFR gene is mutated and constitutively active in oligodendroglial tumors, with high-level amplification of the PDGFRA gene seen in approximately 13% of adult GBMs, and it appears to be a commonly affected RTK in pediatric GBMs and diffuse pontine gliomas [25]. Activation of these and other RTKs, such as the Met oncogene, leads to increased Ras–Raf–MEK–ERK pathway signaling, resulting in cell division and malignant transformation [18]. Recent studies have established that more than one RTK is affected in a substantial portion of GBMs, which could explain the limited efficacy of drugs targeting a single RTK pathway in gliomas [25].
Even though RTK amplifications are rare in LGG, increased PDGF signaling has been noted in these tumors, which could be accounted for by ligand-driven tumorigenesis [25]. Downstream components of the RTK pathways are also commonly affected, with 88% of GBMs having been reported as having significant genomic alterations in the PI3K–AKT–mTOR and RAS–MAPK molecular pathways [25]. PI3K has a regulatory (PIK3CA) and a catalytic (PIK3R1) unit, with mutations in either subunit having been noted in 15% of adult GBMs, while 36% of tumors have been seen to have silencing mutations or deletions affecting the PTEN gene, the primary negative regulator of the PI3K–AKT–mTOR pathway [26]. Phosphate and tensin homolog (PTEN) is normally a regulator that keeps the tyrosine kinase pathways in check by removing phosphates that are placed by kinases. Loss of PTEN, along with activated RTK signaling, results in increased PI3K/AKT1 pathway activity, leading to inhibition of apoptosis and increased survival, as a main regulator has been lost [26]. Additionally, frequent epigenomic repression of the PTEN gene, located on chromosome 10q, is observed in the majority of the LGGs [27]. Loss of PTEN, along with activated RTK signaling, results in increased PI3K–AKT1 pathway activity, leading to inhibition of apoptosis and increased survival [26]
Additional alterations, such as NF1 mutations in glioblastoma, a negative regulator of Ras, contribute further to increased cell proliferation by allowing the Ras signaling pathway to run unchecked [28]. Overall, the RTK–Ras–PI3K–AKT1 pathway is altered in nearly 90% of GBMs, significantly contributing to GBM formation (Figure 1) [18].
• p53 & Rb pathways
Disruption of the retinoblastoma (RB1) and p53 tumor suppressor pathways has been shown to be a frequent event in GBM formation, with p53 (TP53 gene) signaling altered in up to 87% of patients [18]. TP53, located on chromosome 17, normally causes cell-cycle arrest in the presence of DNA damage by halting the growth phase or by inducing cellular apoptosis [18]. The p53 protein is stabilized by stress-sensing agents within the cell that respond to genotoxic and cytotoxic environments, and functions predominantly as a transcription factor, by regulating the promoter of thousands of potential effector genes [29]. Thus, dysfunction of p53 not only allows for unchecked growth and provides glioma cells with a growth advantage [29], but also leads to genomic instability secondary to a lack of proper DNA repair checkpoints [30].
The Rb protein is considered to be the brake of the cell cycle, keeping it in check until it is phosphorylated by the cyclins, including cyclin D1, CDK4 and CDK6 [31]. The Rb protein functions by sequestering the E2F family of transcription factors, which are necessary to progress through the cell cycle [31]. Once the MAPK cascade is activated, Rb is phosphorylated allowing the transcription of E2F targets and thus the entry into the S phase [31]. Mutations in RB1 are infrequent in GBM, but its upstream regulators are frequently altered. The RB1 gene, located on chromosome 13q14, is mutated in approximately 25% of high-grade astrocytomas and loss of 13q is seen in tumors that have progressed from low grade to intermediate-grade gliomas, allowing for loss of cell cycle control and continued growth potential for gliomas [18]. CDK4 amplification, CNKN2A deletion (normally an activator of RB1 and TP53), and loss of p16INK4a (a CDK4 suppressor) are more common, and result in functional inactivation of Rb; specifically, CDK4 amplification is seen in up to 15% of HGGs [18]. p16 INK4a, generated as one of two transcripts at the CDKN2A locus on chromosome 9p21, is inactivated by allelic loss or hypermethylations in 50–70% of HGGs (Figure 1) [18].
• NF-κB signaling
Heterozygous deletion of the NF-κB inhibitor-α (NFKBIA), an inhibitor of EGF receptor (EGFR) signaling, has been recently described in 25% of GBMs, and has an effect similar to EGFR amplification (Figure 1) [32]. NFKBIA deletion and EGFR amplification have been shown to be mutually exclusive, strongly suggesting that the two genetic events converge on the same pathway [32]. As expected, both genetic events are associated with similar prognostic outcomes, which is inferior to that of patients with normal expression levels of these two genes [32]. However, the detailed molecular mechanism for the role of NF-κB in glioma development and progression, in association with EGFR signaling, remains to be investigated.
• Angiogenic pathways
VEGF promotes cell proliferation and survival by binding to its receptor (VEGFR), resulting in subsequent activation of the MAPK–Ras–PI3K pathway [33,34] and resultant proliferation. Tumor vessels are destabilized by angiopoietin-2, thereby promoting angiogenesis [35], with additional mediators including the Notch signaling pathway that stimulates transcription and in turn promotes angiogenesis (Figure 1) [36]. Under physiologic conditions, these pathways are regulated by antiangiogenic factors, including angiostatin [35,36], which are less effective in tumors [35].
Biomarkers
Biomarkers can be divided into three types based on utility: diagnostic, prognostic and predictive [37]. While each of these entities may have overlap, strictly speaking, diagnostic markers allow for a more specific diagnosis, whereas prognostic markers provide further expectations regarding natural history and predictive markers demonstrate the probability of responsiveness to a particular treatment. Using the current knowledge regarding malignant gliomas, a paradigm for potential individualized treatment based on an individual tumor’s genomic profile can be created (Figure 2). Individual biomarkers including MGMT promoter methylation, LOH at chromosome 1p/19q, IDH, EGFR, p53, PTEN, cyclins, mitotic markers, BRAF, VEGF, cytochrome c oxidase and miRNAs, among others, are discussed in detail below.
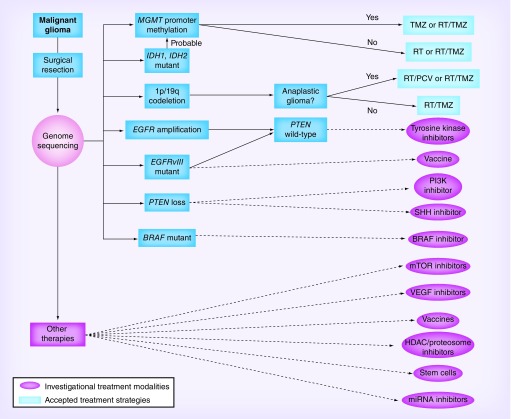
Figure 2. Targeted high-grade glioma therapy based upon surgical resection or biopsy that allows genomic sequencing of an individual’s tumor in order to target therapy based on specific mutations.
This is in addition to the standard alkylating agents and radiotherapy already used today. The figure is conceptual and it would be imagined that in personalized medicine, multiple agents would be used simultaneously based on the genetic profile.
PCV: Procarbazine, lomustine and vincristine chemotherapy; RT: Radiotherapy; TMZ: Temozolomide.
• MGMT promoter methylation
The MGMT gene encodes for the DNA repair enzyme O(6)-methylguanine-DNA methyltransferase [38]. It functions in removing the alkyl groups from the O6 position of guanine, commonly produced by alkylating drugs, such as temozolomide [38]. Silencing of the MGMT gene, through methylation of its promoter, has been shown to result in increased response to temozolomide and is associated with a favorable prognosis in gliomas [5,38]. Furthermore, irrespective of treatment, MGMT promoter methylation is an independent prognostic factor predicting responsiveness and survival in patients, with the highest OS noted in patients treated with temozolomide and radiotherapy (23.4 months) compared with radiotherapy alone (15.3 months) [5].
• Chromosome 1p/19q LOH
Initial studies have identified 1p/19q codeletions to serve not only as diagnostic markers, being observed in oligodendroglial as opposed to astrocytic tumors, but also as powerful prognostic markers for chemotherapeutic and radiotherapeutic response [39]. Later studies identified FUBP1 (encoding far-upstream element binding protein) and CIC (homolog of the Drosophila gene capicua) genes to be the tumor suppressor genes somatically mutated and deleted on chromosomes 1p and 19q, respectively [12]. In pure anaplastic oligodendrogliomas, polychemotherapy with procarbacine, lomustin, and vincristine (PCV) either before or immediately after radiotherapy, has been shown to have a survival advantage [40]. Importantly, in GBMs, the presence of an oligodendroglioma-like component carries no prognostic significance and these tumors behave more like astrocytomas, carrying a worse prognosis [41]. However, this study is controversial because all of the GBMs with an oligodendroglioma component had EGFR amplication. Regardless, in a more recent study, 1p/19q was found to correlated with alpha-thalassemia/mental retardation syndrome X-linked (ATRX status), with a loss seen in 27% in anaplastic oligoastrocytomas compared with 10% of anaplastic oligodendrogliomas [42]. Given that ATRX is mutually exclusive to 1p/19q, anaplastic oligoastrocytomas had a similar clinical course with anaplastic astrocytoma, whereas anaplastic oligoastrocytomas carrying 1p/19q codeletion shared a similar course with anaplastic oligodendrogliomas [42]. This means that because ATRX loss is a hallmark of astrocytic tumors, mixed tumors with ATRX loss behave like astrocytomas [42].
• IDH
As mentioned above, the recurrent IDH1 or IDH2 mutations indicate a survival benefit among patients with HGGs [43]. For instance, patients with GBMs with IDH1 mutations have a better prognosis than patients with AA or GBMs without the IDH1 mutation, with a survival advantage of 31 months versus 15 months, respectively [43]. Furthermore, IDH1 mutations inversely correlate with grade, such that 75% of grade II gliomas, 50% of grade III, and only 5% of primary GBMs harbor the mutations, compared with 80% of secondary GBMs carrying the mutation [44]. There also appears to be a relation with MGMT promoter methylation, with the presence of an IDH mutation rendering the chance of methylation as practically 100% [5,45].
Furthermore, IDH mutations are responsible for CpG island gene silencing in gliomas and thus widespread epigenetic changes, including MGMT promoter methylation [10]. Not surprisingly, IDH1 and IDH2 mutations can aid in the diagnosis of diffuse glioma versus similar looking radiographic entities such as pilocytic astrocytoma or glioneuronal tumor [46].
• EGFR, p53 & PTEN
As discussed above, EGFR alterations are quite common in primary GBMs, especially amplification and the EGFRvIII variant, but the usefulness of inhibitors has remained controversial [47]. However, patients treated with temozolomide in the setting of EGFR amplification, with retained PTEN and p53 strongly predicts better survival [48]. Finally, as mentioned above, loss of PTEN due to chromosome 10q deletions, which are frequent in primary GBMs, are associated with diminished survival [21,28,49].
• Cyclin & mitotic markers
Deregulation of the p16INK4a-cyclin pathway is commonly found in patients with GBM [18]. Normally p16 binds to cyclin kinases to promote antiproliferative signaling via means of Rb [18]. Loss of p16 occurs in up to 57% of patients with GBM, but predictions in terms of prognosis have been inconsistent [50]. Checkpoint kinases in the mitotic cycle can also serve as biomarkers: specifically, monopolar spindle 1, which positively correlates with grade and negatively with patient survival [51].
• BRAF
Part of the MAP kinase cascade, RAF kinases regulate transcription factors and protein kinases that control cell proliferation, differentiation, and apoptosis (Figure 1) [52]. Several different types of mutations have been identified in gliomas, but the most common is a single point mutation of BRAF (V600E) [53]. These mutations are more common in children and are beginning to serve as a biomarker with therapeutic promise given success in other neoplasms, such as melanoma [52].
• VEGF
Given the strong implication of neovascularization in gliomas, VEGF is thought to be the driving force for angiogenesis and increased expression is observed in 61% of glioblastomas [28]. There appears to be a strong correlation between VEGF expression and survival [28].
• miRNAs
miRNAs are noncoding RNA molecules that can have oncogenic or tumor suppressor activities [54]. They have been implicated in temozolomide resistance, as well as glioma stem cell resistance [28]. Several studies have demonstrated their ability to predict OS and progression-free survival (PFS) and thus demonstrate a promising avenue for both future biomarkers and future personalized targets [28].
• Other biomarkers
ELTD1 is a new biomarker that is intimately involved with angiogenesis, and has significantly higher expression in high-grade gliomas compared with LGG [55]. In addition to its association with grade, it relates to survival and the mesenchymal subtype, meaning that it may be able to serve as a future biomarker [55]. Other investigators have started looking at the phospholipid metabolic environment and telomerase activity, which have been shown to correlate with survival and disease-free survival [28].
Targeted therapies
The current standard of care for newly diagnosed HGGs is maximal safe surgical resection, followed by adjuvant chemotherapy (temozolomide) and radiotherapy [5]. Gross total resection is virtually impossible due to the infiltrative nature of these tumors, but resecting as much of the contrast-enhancing tumor as safely possible improves symptoms and quality of life, prolongs survival, and provides tissue for histologic and molecular diagnosis [56]. Given the increased understanding of the multitude of signaling pathways involved in malignant gliomas, a rudimentary theoretical treatment paradigm is offered that will continually change as new knowledge is gathered (Figure 2).
• Tyrosine kinase inhibitors
Given the high frequency of deregulation of the EGFR pathway in HGGs, this pathway would seem promising for targeted therapy. However, the first-generation EGFR inhibitors, erlotinib and gefitinib, have not been effective in GBM as seen in preclinical trials [57,58] and only had a modest effect in a Phase II trial [59]. A similar lack of response was seen with a monoclonal antibody against EGFR known as cetuximab [60], as well as with a more recent EGFR inhibitor known as lapatinib [61]. However, patients treated with temozolomide in the setting of EGFR amplification, with retained PTEN and p53 strongly predicts improved survival [48]. Activation of multiple downstream signaling pathways has been implicated as a potential explanation of why single EGFR inhibitors have failed [62]. However, these drugs are re-emerging given new insights into the refinement of such molecules including escape mechanisms, resistance, immunogenicity and conformational binding with some proposing protein–protein interactions as the most important in the era of effective targeted therapies, ushering in a potential new realm of proteonomics [63].
• PI3K/Akt inhibitors
PI3K signaling is usually activated by PTEN loss in malignant gliomas, as well as SHH, thereby synergizing to promote tumor growth and viability [64]. Targeting of both pathways results in apoptosis and reduces growth of PTEN-deficient GBM in vitro and in vivo [64]. The PI3K inhibitor PX-866 prohibits glioma cell proliferation and migration, and prolongs cell survival [65]. Given these effects, it is currently under clinical trial for progressive GBM [65]. Several other agents are under investigation and development, all having been shown to reduce in vivo tumor growth, vascularity and angiogenesis, with ongoing clinical trials [65].
• mTOR target inhibitors
First-generation mTOR inhibitors, such as temsirolimus and everolimus, have been used to treat several types of solid tumors, including renal cell carcinoma, subependymal giant cell astrocytoma and progressive neuroendocrine tumors of pancreatic origin. Yet GBM clinical trials for the use mTOR inhibitors have not shown changes in outcomes. The lack of efficacy of these drugs on GBMs is though to be due to resistance from negative feedback loops (i.e., activation of Akt) [66], parallel signaling pathways, and lack of target specificity [67,68]. To circumvent this problem, combined agents inhibiting PI3K and mTOR have been shown to block GBM growth and are currently in clinical trials [50]. Similarly, targeting of the Notch pathway by a γ-secretase inhibitor potentially targets tumor-initiating cells and is currently in clinical trial as monotherapy, in combination with temozolomide and radiotherapy, as well as with bevacizumab [50].
• BRAF inhibitors
BRAF inhibitors, such as vemurafenib, have outstanding clinical activity in patients with melanomas harboring the BRAF (V600E) mutation [69]. BRAF inhibitors for HGG have shown in vitro promise against specific cells with driver mutations [53], but are unlikely to work on BRAF fusion proteins, as they respond to MAPK inhibition instead [69]. Regardless, our institution is trialing these inhibitors in patients with appropriately confirmed mutations.
• HDAC & proteasome inhibitors
Inhibition of HDACs, which serve as regulators of the chromatic structure for gene expression, is another strategy currently being explored. A Phase I trial of the HDAC inhibitor panobinostat in combination with bevacizumab was well tolerated [70], with Phase II trials now needed. Vorinostat, another HDAC inhibitor, is well tolerated as a monotherapy and has a modest effect with PFS of 15.2% at 6 months [71]. When combined with the proteasome inhibitor, bortezomib, there were no patients who experienced PFS and thus the study was terminated [72]. However, bortezomib induces cell death in GBM cell lines and temozolomide-resistant gliomas [73] and thus requires further study. Histone dysregulation is given further strength by evidence that pediatric high-grade pontine gliomas require it for pathogenesis [20]. Interestingly, valproic acid, an anti-epileptic, has HDAC properties and a survival benefit in GBM in those patients also treated with temozolomide and radiation [40]. This was confirmed in a more recent study of 544 patients, with a median OS of 16.9 months in those taking valproic acid, compared with 13.6 months in those taking a different agent [74]. In terms of seizure prophylaxis, the American Academy of Neurology’s practice guideline states that there is no evidence for prophylactic antiepileptic drugs and advises against the routine use in those patients without seizures [75,76]. However, this evidence makes one consider using valproic acid in most patients with HGG, or at least as a first-line agent in those patients who do have seizures, given this survival benefit. Many other agents are being explored in Phase I/II trials, both as single agents and as combination therapies including vorinostat and panobinostat, with efficacy trials pending [77].
• miRNAs
miRNAs represent a promising therapeutic agent for gliomas, with the current constraint being delivery past the blood–brain barrier, but nanoparticle delivery and/or convection based delivery systems hold promise in circumventing this obstacle [50]. The therapeutic strategy involves substituting miRNA with tumor suppressor functions and inhibiting miRNAs that have oncogenic properties, with significantly positive results in animal models [78]. Human trials should be forthcoming in the near future.
• VEGF inhibitor: bevacizumab
Antioangiogenic therapy with bevacizumab (Avastin®, Genentech, CA, USA), a humanized monoclonal antibody directed against the VEGF-A ligand [79], is the most extensively tested of the antiangiogenic agents and has received approval in the USA as monotherapy for the treatment of recurrent GBM [80]. It likely inhibits angiogenesis through several mechanisms, including direct inhibition of tumor associated angiogenesis, a direct anti-GBM effect on VEGFR-expressing GBM cells, disruption of the glioma stem cell microvascular niche and improved vascular function and normalization [81,82]. Bevacizumab received accelerated US FDA approval in 2009 [80] for use as monotherapy in progressive GBM based on improved radiologic response rates seen in two Phase II trials [80,83]. These trials demonstrated improved PFS, at 6 months, for recurrent GBM. Additionally, the Dutch BELOB randomized Phase II trial demonstrated increased survival when bevacizumab was combined with lomustine [84]. Specifically, 41% of those receiving bevacizumab and lomustine had PFS at 6 months, compared with 18 and 11%, for respective use of bevacizumab and lomustine alone. The ongoing EORTC 26101 trial is exploring this concept in more detail and it would appear that certain chemotherapeutic combinations provide even better survival [85]. Given these results, two multicenter Phase III, randomized-controlled trials were started including the RTOG 0825 trial [86] and the AVAglio trials [87], where patients with newly diagnosed GBM were randomly assigned to receive standard therapy (radiation and temozolomide) or standard therapy plus bevacizumab. While OS was not improved, PFS was prolonged, but this has unclear clinical significance. Thus, while the role of bevacizumab for newly diagnosed GBM remains obscure, there remains convincing evidence for continuing its use for recurrent GBM. In terms of AAs, there is only a modest benefit, which requires further study [88].
• Other antiangiogenic therapies
Given the results of bevacizumab, there is a strong interest in effective antiangiogenic agents. Aflibercept, a VEGFR fusion protein, failed a Phase II trial as a single agent in recurrent malignant gliomas [89]. Enzastaurin, another anti-angiogenesis inhibitor, failed a Phase III trial [90]. However, combination trials are ongoing, including bevacizumab with another antiangiogenesis molecule, cediranib, and the results are pending [90]. Many other molecules are being explored in early Phase I/II trials, with results eagerly anticipated.
• Glioma stem cells & inhibitors
Stem cells have been implicated in the resistance of gliomas to cytotoxic therapies, including radiotherapy and chemotherapy, thereby providing a mechanism for recurrence [91]. Direct targeting has remained challenging and investigators have recommended combined strategies. Interestingly, given that glioma stem cells (GCSs) have higher Notch signaling, when exposed to γ-secretase they have inhibited proliferation, increased differentiation and reduced tumorigenicity [50]. A similar effect is seen with hedgehog inhibitors [50]. Modification of the Akt pathway can occur via TGF-β, which in itself is a modifier of radiation responses, as in vitro evidence demonstrates increased radiosensitivity of GSCs given TGF-β inhibitors [92]. More specifically, selective inhibitors of the TGF-β receptor kinase potentiate radiation responses in GBM by increasing apoptosis, blocking DNA damage repair and blocking invasion and mesenchymal transition, as well as angiogenesis [92]. Sonic hedgehog inhibitors are antiproliferative and reduce tumor volume, especially after temozolomide in vitro [93], and can deplete gliomas stem cells [94]. Other strategies for targeting GSCs include targeting the tumor microenvironment and modifying the immune system to prevent evasion [50].
• Immunotherapies & vaccines
Given that gliomas cause immunosuppression of the host against the tumor, gliomas are further able to escape immune detection and thus survive [95]. Positive preclinical results have led to several Phase II and III vaccine trials, based on the premise that when tumor epitopes are presented to MHC molecules, peptides can be purified and then employed as vaccines. In a Phase II trial for an EGFRvIII vaccine in patients with newly diagnosed GBM, the OS was significantly increased at 26 months, compared with 15 months for nonvaccinated controls [96]. Even more interesting is that at recurrence, there was loss of EGFRvIII in all patients, demonstrating that recurrence occurred through other mechanisms and strengthened the utility of such a vaccine. A Phase III trial (ACT IV) is currently ongoing [97].
Given that the EGFRvIII vaccine is monovalent and the gliomas cells were able to escape, another strategy centers on using polyvalent vaccines to target multiple epitopes in order to prevent escape mechanisms. To potentiate a greater response, investigators have conjugated heat shock proteins (HSPs), which chaperone peptides to APCs, with glioma antigens [98]. A recent Phase II trial of recurrent malignant gliomas injected with heat shock protein–peptide complexes reported a 93% PFS rate, and now a Phase II randomized trial in combination with bevacizumab is underway [98]. An additional strategy is to load ex vivo dendritic cells with antigens derived from an individual patient’s tumor [99]. DCVax-L is such a vaccine composed of dendritic cells that are charged with tumor lysate [99]. Phase I and II trials have documented a 25% 6-year survival rate [99], with a Phase III trial ongoing [100].
Conclusion & future perspective
OS in patients with malignant glioma remains poor despite the current treatment strategies. Genetic and epigenetic alterations in these tumors allow for disease resistance and progression through proliferation of transformed cells with selected driver mutations. MGMT promoter methylation, 1p/19q codeletion, and IDH1 mutations all are favorable prognostic markers that can help guide patient expectations. Secondary mutations result from selection pressures during treatment in addition to an altered epigenetic profile and can change depending on treatment paradigm. Now in the era of molecular diagnostics, genomic sequencing for detection of tumor-specific mutations will become the standard and introduction of tumor-specific therapies will be the key to altering the course of malignant gliomas. The ability to identify additional biomarkers will not only predict the benefits of targeted treatments, but will also allow practitioners to provide explicit patient stratification and follow response to treatment. However, it must be emphasized that new, effective agents will need to be added to the armamentarium before personalized medicine becomes a reality for patients with gliomas. In the end, effective therapy will likely require both targeted drugs based on a tumor’s specific genetic profile to block cells with driver mutations, as well as less specific therapies to fight against cells that develop secondary mutations. Patient referral to centers that have the capability of sequencing gliomas should be pursued.
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.CBTRUS. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2004–2008. CBTRUS; Hinsdale, IL, USA: 2012. [Google Scholar]
- 2.Fisher JL, Schwartzbaum JA, Wrensch M, Wiemels JL. Epidemiology of brain tumors. Neurol Clin. 2007;25:867–890. doi: 10.1016/j.ncl.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Rousseau A, Mokhtari K, Duyckaerts C. The 2007 WHO classification of tumors of the central nervous system – what has changed? Curr. Opin. Neurol. 2008;21:720–727. doi: 10.1097/WCO.0b013e328312c3a7. [DOI] [PubMed] [Google Scholar]
- 4.Fuller GN, Scheithauer BW. The 2007 Revised World Health Organization (WHO) Classification of Tumours of the Central Nervous System: newly codified entities. Brain Pathol. 2007;17:304–307. doi: 10.1111/j.1750-3639.2007.00084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 6.Westphal M, Lamszus K. The neurobiology of gliomas: from cell biology to the development of therapeutic approaches. Nat. Rev. Neurosci. 2011;12:495–508. doi: 10.1038/nrn3060. [DOI] [PubMed] [Google Scholar]; • Good complimentary review detailing molecular pathways in gliomas and potential therapeutic targets.
- 7.Grossman SA, Ye X, Piantadosi S, et al. Survival of patients with newly diagnosed glioblastoma treated with radiation and temozolomide in research studies in the United States. Clin. Cancer Res. 2010;16:2443–2449. doi: 10.1158/1078-0432.CCR-09-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, Wu G, Miller CP, et al. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat. Genet. 2013;45:602–612. doi: 10.1038/ng.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Landmark paper identifying multiple new genetic alterations in low-grade gliomas, including BRAF, RAF1, FGFR1, MYB, MYBL1 and genes with histone-related functions, including H3F3A and ATRX.
- 9.van den Bent MJ, Snijders TJ, Bromberg JE. Current treatment of low grade gliomas. Memo. 2012;5:223–227. doi: 10.1007/s12254-012-0014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]; • IDH1 mutation is responsible for epigenetic methylation.
- 11.Shinawi T, Hill VK, Krex D, et al. DNA methylation profiles of long- and short-term glioblastoma survivors. Epigenetics. 2013;8:149–156. doi: 10.4161/epi.23398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bettegowda C, Agrawal N, Jiao Y, et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science. 2011;333:1453–1455. doi: 10.1126/science.1210557. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Specific genes mutated in oligodendrogliomas besides the well-known 1p/19q mutation.
- 13.Zhang C, Moore LM, Li X, Yung WK, Zhang W. IDH1/2 mutations target a key hallmark of cancer by deregulating cellular metabolism in glioma. Neuro Oncol. 2013;15:1114–1126. doi: 10.1093/neuonc/not087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smoll NR, Gautschi OP, Schatlo B, Schaller K, Weber DC. Relative survival of patients with supratentorial low-grade gliomas. Neuro Oncol. 2012;14:1062–1069. doi: 10.1093/neuonc/nos144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 16.Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J. Neuropathol. Exp. Neurol. 2005;64:479–489. doi: 10.1093/jnen/64.6.479. [DOI] [PubMed] [Google Scholar]
- 17.Dolecek TA, Propp JM, Stroup NE, Kruchko C. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro Oncol. 2012;14(Suppl. 5):v1–v49. doi: 10.1093/neuonc/nos218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Glioblastoma has key mutations in MGMT, TP53, ERB and PI3K pathways.
- 19.Schwartzentruber J, Korshunov A, Liu XY. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]; •• New mutations identified in histone and epigenetic remodeling systems in glioblastoma (GBM). This paper helped pave the way that mutations in GBM go beyond just mutations in the genetic structure and instead stretch into the epigenetic realm.
- 20.Wu G, Broniscer A, McEachron TA, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat. Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am. J. Pathol. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao Q, Lei T, Ye F. Therapeutic targeting of EGFR-activated metabolic pathways in glioblastoma. Expert Opin. Investig. Drugs. 2013;22:1023–1040. doi: 10.1517/13543784.2013.806484. [DOI] [PubMed] [Google Scholar]
- 24.Gan HK, Kaye AH, Luwor RB. The EGFRvIII variant in glioblastoma multiforme. J. Clin. Neurosci. 2009;16:748–754. doi: 10.1016/j.jocn.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 25.Joensuu H, Puputti M, Sihto H, Tynninen O, Nupponen NN. Amplification of genes encoding KIT, PDGFRalpha and VEGFR2 receptor tyrosine kinases is frequent in glioblastoma multiforme. J. Pathol. 2005;207:224–231. doi: 10.1002/path.1823. [DOI] [PubMed] [Google Scholar]
- 26.Quayle SN, Lee JY, Cheung LW, et al. Somatic mutations of PIK3R1 promote gliomagenesis. PLoS ONE. 2012;7:e49466. doi: 10.1371/journal.pone.0049466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 28.McNamara MG, Sahebjam S, Mason WP. Emerging biomarkers in glioblastoma. Cancers (Basel) 2013;5:1103–1119. doi: 10.3390/cancers5031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sidransky D, Mikkelsen T, Schwechheimer K, Rosenblum ML, Cavanee W, Vogelstein B. Clonal expansion of p53 mutant cells is associated with brain tumour progression. Nature. 1992;355:846–847. doi: 10.1038/355846a0. [DOI] [PubMed] [Google Scholar]
- 30.Bogler O, Huang HJ, Cavenee WK. Loss of wild-type p53 bestows a growth advantage on primary cortical astrocytes and facilitates their in vitro transformation. Cancer Res. 1995;55:2746–2751. [PubMed] [Google Scholar]
- 31.Tanaka S, Louis DN, Curry WT, Batchelor TT, Dietrich J. Diagnostic and therapeutic avenues for glioblastoma: no longer a dead end? Nat. Rev. Clin. Oncol. 2013;10:14–26. doi: 10.1038/nrclinonc.2012.204. [DOI] [PubMed] [Google Scholar]; • Discusses additional potential therapeutic targets in glioma.
- 32.Bredel M, Scholtens DM, Yadav AK, et al. NFKBIA deletion in glioblastomas. N. Engl. J. Med. 2011;364:627–637. doi: 10.1056/NEJMoa1006312. [DOI] [PMC free article] [PubMed] [Google Scholar]; • New pathway mutation identifed in GBM allows for constitutively active NF-κB.
- 33.Chowdhary S, Chamberlain M. Bevacizumab for the treatment of glioblastoma. Expert Rev. Neurother. 2013;13:937–949. doi: 10.1586/14737175.2013.827414. [DOI] [PubMed] [Google Scholar]
- 34.Zerbini G, Lorenzi M, Palini A. Tumor angiogenesis. N. Engl. J. Med. 2008;359:763. doi: 10.1056/NEJMc081278. [DOI] [PubMed] [Google Scholar]
- 35.Jain RK, di Tomaso E, Duda DG, Loeffler JS, Sorensen AG, Batchelor TT. Angiogenesis in brain tumours. Nat. Rev. Neurosci. 2007;8:610–622. doi: 10.1038/nrn2175. [DOI] [PubMed] [Google Scholar]
- 36.Ie M Shih, Wang TL. Notch signaling, gamma-secretase inhibitors, and cancer therapy. Cancer Res. 2007;67:1879–1882. doi: 10.1158/0008-5472.CAN-06-3958. [DOI] [PubMed] [Google Scholar]
- 37.Jansen M, Yip S, Louis DN. Molecular pathology in adult gliomas: diagnostic, prognostic, and predictive markers. Lancet Neurol. 2010;9:717–726. doi: 10.1016/S1474-4422(10)70105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]; Landmark paper demonstrating the MGMT gene silencing predicts chemotherapy response.
- 39.Nutt CL. Molecular genetics of oligodendrogliomas: a model for improved clinical management in the field of neurooncology. Neurosurg. Focus. 2005;19:E2. doi: 10.3171/foc.2005.19.5.3. [DOI] [PubMed] [Google Scholar]
- 40.Weller M, Gorlia T, Cairncross JG, et al. Prolonged survival with valproic acid use in the EORTC/NCIC temozolomide trial for glioblastoma. Neurology. 2011;77:1156–1164. doi: 10.1212/WNL.0b013e31822f02e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hegi ME, Janzer RC, Lambiv WL, et al. Presence of an oligodendroglioma-like component in newly diagnosed glioblastoma identifies a pathogenetically heterogeneous subgroup and lacks prognostic value: central pathology review of the EORTC_26981/NCIC_CE.3 trial. Acta Neuropathol. 2012;123:841–852. doi: 10.1007/s00401-011-0938-4. [DOI] [PubMed] [Google Scholar]
- 42.Wiestler B, Capper D, Holland-Letz T, et al. ATRX loss refines the classification of anaplastic gliomas and identifies a subgroup of IDH mutant astrocytic tumors with better prognosis. Acta Neuropathol. 2013;126:443–451. doi: 10.1007/s00401-013-1156-z. [DOI] [PubMed] [Google Scholar]
- 43.Hartmann C, Hentschel B, Wick W, et al. Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta Neuropathol. 2010;120:707–718. doi: 10.1007/s00401-010-0781-z. [DOI] [PubMed] [Google Scholar]
- 44.Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J. Clin. Oncol. 2009;27:4150–4154. doi: 10.1200/JCO.2009.21.9832. [DOI] [PubMed] [Google Scholar]
- 45.Mulholland S, Pearson DM, Hamoudi RA. MGMT CpG island is invariably methylated in adult astrocytic and oligodendroglial tumors with IDH1 or IDH2 mutations. Int. J. Cancer. 2012;131:1104–1113. doi: 10.1002/ijc.26499. [DOI] [PubMed] [Google Scholar]
- 46.Horbinski C, Kofler J, Yeaney G, et al. Isocitrate dehydrogenase 1 analysis differentiates gangliogliomas from infiltrative gliomas. Brain Pathol. 2011;21:564–574. doi: 10.1111/j.1750-3639.2011.00480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patel M, Vogelbaum MA, Barnett GH, Jalali R, Ahluwalia MS. Molecular targeted therapy in recurrent glioblastoma: current challenges and future directions. Expert Opin. Investig. Drugs. 2012;21:1247–1266. doi: 10.1517/13543784.2012.703177. [DOI] [PubMed] [Google Scholar]
- 48.Ang C, Guiot MC, Ramanakumar AV, Roberge D, Kavan P. Clinical significance of molecular biomarkers in glioblastoma. Can. J. Neurol. Sci. 2010;37:625–630. doi: 10.1017/s0317167100010805. [DOI] [PubMed] [Google Scholar]
- 49.Ohgaki H, Dessen P, Jourde B, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004;64:6892–6899. doi: 10.1158/0008-5472.CAN-04-1337. [DOI] [PubMed] [Google Scholar]
- 50.Tanase CP, Enciu AM, Mihai S, Neagu AI, Calenic B, Cruceru ML. Anti-cancer therapies in high grade gliomas. Curr. Proteomics. 2013;10:246–260. doi: 10.2174/1570164611310030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tannous BA, Kerami M, Van der Stoop PM. Effects of the selective MPS1 inhibitor MPS1-IN-3 on glioblastoma sensitivity to antimitotic drugs. J. Natl Cancer Inst. 2013;105:1322–1331. doi: 10.1093/jnci/djt168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sievert AJ, Lang SS, Boucher KL, et al. Paradoxical activation and RAF inhibitor resistance of BRAF protein kinase fusions characterizing pediatric astrocytomas. Proc. Natl Acad. Sci. USA. 2013;110:5957–5962. doi: 10.1073/pnas.1219232110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nicolaides TP, Li H, Solomon D, et al. Targeted therapy for BRAFV600E malignant astrocytoma. Clin. Cancer Res. 2011;17:7595–7604. doi: 10.1158/1078-0432.CCR-11-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hummel R, Maurer J, Haier J. MicroRNAs in brain tumors : a new diagnostic and therapeutic perspective? Mol. Neurobiol. 2011;44:223–234. doi: 10.1007/s12035-011-8197-x. [DOI] [PubMed] [Google Scholar]
- 55.Towner RA, Jensen RL, Colman H, et al. ELTD1, a potential new biomarker for gliomas. Neurosurgery. 2013;72:77–90. doi: 10.1227/NEU.0b013e318276b29d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lacroix M, Abi-Said D, Fourney DR, et al. A multivariate analysis of 416 patients with glioblastoma multiforme: prognosis, extent of resection, and survival. J. Neurosurg. 2001;95:190–198. doi: 10.3171/jns.2001.95.2.0190. [DOI] [PubMed] [Google Scholar]
- 57.Mellinghoff IK, Wang MY, Vivanco I, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N. Engl. J. Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 58.Brown PD, Krishnan S, Sarkaria JN, et al. Phase I/II trial of erlotinib and temozolomide with radiation therapy in the treatment of newly diagnosed glioblastoma multiforme: North Central Cancer Treatment Group Study N0177. J. Clin. Oncol. 2008;26:5603–5609. doi: 10.1200/JCO.2008.18.0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raizer JJ, Abrey LE, Lassman AB, et al. A Phase II trial of erlotinib in patients with recurrent malignant gliomas and nonprogressive glioblastoma multiforme postradiation therapy. Neuro Oncol. 2010;12:95–103. doi: 10.1093/neuonc/nop015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Neyns B, Sadones J, Joosens E, et al. Stratified Phase II trial of cetuximab in patients with recurrent high-grade glioma. Ann. Oncol. 2009;20:1596–1603. doi: 10.1093/annonc/mdp032. [DOI] [PubMed] [Google Scholar]
- 61.Thiessen B, Stewart C, Tsao M, et al. A Phase I/II trial of GW572016 (lapatinib) in recurrent glioblastoma multiforme: clinical outcomes, pharmacokinetics and molecular correlation. Cancer Chemother. Pharmacol. 2010;65:353–361. doi: 10.1007/s00280-009-1041-6. [DOI] [PubMed] [Google Scholar]
- 62.Takeuchi K, Ito F. Receptor tyrosine kinases and targeted cancer therapeutics. Biol. Pharm. Bull. 2011;34:1774–1780. doi: 10.1248/bpb.34.1774. [DOI] [PubMed] [Google Scholar]
- 63.Hegi ME, Rajakannu P, Weller M. Epidermal growth factor receptor: a re-emerging target in glioblastoma. Curr. Opin. Neurol. 2012;25:774–779. doi: 10.1097/WCO.0b013e328359b0bc. [DOI] [PubMed] [Google Scholar]
- 64.Filbin M Gruber, Dabral SK, Pazyra-Murphy MF, et al. Coordinate activation of Shh and PI3K signaling in PTEN-deficient glioblastoma: new therapeutic opportunities. Nat. Med. 2013;19:1518–1523. doi: 10.1038/nm.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sami A, Karsy M. Targeting the PI3K/AKT/mTOR signaling pathway in glioblastoma: novel therapeutic agents and advances in understanding. Tumour Biol. 2013;34:1991–2002. doi: 10.1007/s13277-013-0800-5. [DOI] [PubMed] [Google Scholar]
- 66.Hu X, Pandolfi PP, Li Y, Koutcher JA, Rosenblum M, Holland EC. mTOR promotes survival and astrocytic characteristics induced by Pten/AKT signaling in glioblastoma. Neoplasia. 2005;7:356–368. doi: 10.1593/neo.04595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Faivre S, Kroemer G, Raymond E. Current development of mTOR inhibitors as anticancer agents. Nat. Rev. Drug Discov. 2006;5:671–688. doi: 10.1038/nrd2062. [DOI] [PubMed] [Google Scholar]
- 68.Thoreen CC, Kang SA, Chang JW, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Poulikakos PI, Persaud Y, Janakiraman M. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Drappatz J, Lee EQ, Hammond S, et al. Phase I study of panobinostat in combination with bevacizumab for recurrent high-grade glioma. J. Neurooncol. 2012;107:133–138. doi: 10.1007/s11060-011-0717-z. [DOI] [PubMed] [Google Scholar]
- 71.Galanis E, Jaeckle KA, Maurer MJ, et al. Phase II trial of vorinostat in recurrent glioblastoma multiforme: a north central cancer treatment group study. J. Clin. Oncol. 2009;27:2052–2058. doi: 10.1200/JCO.2008.19.0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Friday BB, Anderson SK, Buckner J, et al. Phase II trial of vorinostat in combination with bortezomib in recurrent glioblastoma: a north central cancer treatment group study. Neuro Oncol. 2012;14:215–221. doi: 10.1093/neuonc/nor198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bota DA, Alexandru D, Keir ST, Bigner D, Vredenburgh J, Friedman HS. Proteasome inhibition with bortezomib induces cell death in GBM stem-like cells and temozolomide-resistant glioma cell lines, but stimulates GBM stem-like cells’ VEGF production and angiogenesis. J. Neurosurg. 2013;119:1415–1423. doi: 10.3171/2013.7.JNS1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barker CA, Bishop AJ, Chang M, Beal K, Chan TA. Valproic acid use during radiation therapy for glioblastoma associated with improved survival. Int. J. Radiat. Oncol. Biol. Phys. 2013;86:504–509. doi: 10.1016/j.ijrobp.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bergen DC. Prophylactic antiepileptic drugs in patients with brain tumors. Epilepsy Curr. 2005;5:182–183. doi: 10.1111/j.1535-7511.2005.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Perry J, Zinman L, Chambers A, Spithoff K, Lloyd N, Laperriere N. The use of prophylactic anticonvulsants in patients with brain tumours-a systematic review. Curr. Oncol. 2006;13:222–229. [PMC free article] [PubMed] [Google Scholar]
- 77.Lee EQ, Puduvalli VK, Reid JM, et al. Phase I study of vorinostat in combination with temozolomide in patients with high-grade gliomas: North American Brain Tumor Consortium Study 04–03. Clin. Cancer Res. 2012;18:6032–6039. doi: 10.1158/1078-0432.CCR-12-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mizoguchi M, Guan Y, Yoshimoto K, et al. Clinical implications of microRNAs in human glioblastoma. Front. Oncol. 2013;3:19. doi: 10.3389/fonc.2013.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Presta LG, Chen H, O’Connor SJ, et al. Humanization of an anti-vascular endothelial growth factor monoclonal antibody for the therapy of solid tumors and other disorders. Cancer Res. 1997;57:4593–4599. [PubMed] [Google Scholar]
- 80.Kreisl TN, Kim L, Moore K, et al. Phase II trial of single-agent bevacizumab followed by bevacizumab plus irinotecan at tumor progression in recurrent glioblastoma. J. Clin. Oncol. 2009;27:740–745. doi: 10.1200/JCO.2008.16.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Willett CG, Boucher Y, di Tomaso E, et al. Direct evidence that the VEGF-specific antibody bevacizumab has antivascular effects in human rectal cancer. Nat. Med. 2004;10:145–147. doi: 10.1038/nm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Borgstrom P, Hillan KJ, Sriramarao P, Ferrara N. Complete inhibition of angiogenesis and growth of microtumors by anti-vascular endothelial growth factor neutralizing antibody: novel concepts of angiostatic therapy from intravital videomicroscopy. Cancer Res. 1996;56:4032–4039. [PubMed] [Google Scholar]
- 83.Friedman HS, Prados MD, Wen PY, et al. Bevacizumab alone and in combination with irinotecan in recurrent glioblastoma. J. Clin. Oncol. 2009;27:4733–4740. doi: 10.1200/JCO.2008.19.8721. [DOI] [PubMed] [Google Scholar]
- 84.Taal W, Oosterkamp HM, Walenkamp AME, et al. A randomized Phase II study of bevacizumab versus bevacizumab plus lomustine versus lomustine single agent in recurrent glioblastoma: the Dutch BELOB study. J. Clin. Oncol. 2013:31. [Google Scholar]
- 85.http://clinicaltrials.gov/show/NCTM01290939 Clinicaltrials.gov: US National Library of Medicine 2013.
- 86.Gilbert MR, Dignam J, Won M, et al. RTOG 0825: Phase III double-blind placebo-controlled trial evaluating bevacizumab (Bev) in patients (Pts) with newly diagnosed glioblastoma (GBM) J. Clin. Oncol. 2013:31. [Google Scholar]
- 87.Chinot O, Wick W, Mason W, et al. OT-03. Phase III trial of bevacizumab added to standard radiotherapy and temozolomide for newly-diagnosed glioblastoma: mature progression-free survival and preliminary overall survival results in AVAglio. Neuro. Oncol. 2012;14(Suppl.) Abstract OT-03. [Google Scholar]
- 88.Reardon DA, Herndon JE, 2nd, Peters K, et al. Outcome after bevacizumab clinical trial therapy among recurrent grade III malignant glioma patients. J. Neurooncol. 2012;107:213–221. doi: 10.1007/s11060-011-0740-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.de Groot JF, Lamborn KR, Chang SM, et al. Phase II study of aflibercept in recurrent malignant glioma: a North American Brain Tumor Consortium study. J. Clin. Oncol. 2011;29:2689–2695. doi: 10.1200/JCO.2010.34.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Seystahl K, Weller M. Is there a world beyond bevacizumab in targeting angiogenesis in glioblastoma? Expert Opin. Investig. Drugs. 2012;21:605–617. doi: 10.1517/13543784.2012.670219. [DOI] [PubMed] [Google Scholar]
- 91.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 92.Zhang M, Kleber S, Rohrich M, et al. Blockade of TGF-beta signaling by the TGFbetaR-I kinase inhibitor LY2109761 enhances radiation response and prolongs survival in glioblastoma. Cancer Res. 2011;71:7155–7167. doi: 10.1158/0008-5472.CAN-11-1212. [DOI] [PubMed] [Google Scholar]
- 93.Ferruzzi P, Mennillo F, De Rosa A, et al. In vitro and in vivo characterization of a novel Hedgehog signaling antagonist in human glioblastoma cell lines. Int. J. Cancer. 2012;131:e33–e44. doi: 10.1002/ijc.27349. [DOI] [PubMed] [Google Scholar]
- 94.Bar EE, Chaudhry A, Lin A, et al. Cyclopamine-mediated hedgehog pathway inhibition depletes stem-like cancer cells in glioblastoma. Stem Cells. 2007;25:2524–2533. doi: 10.1634/stemcells.2007-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dunn GP, Fecci PE, Curry WT. Cancer immunoediting in malignant glioma. Neurosurgery. 2012;71:201–222. doi: 10.1227/NEU.0b013e31824f840d. [DOI] [PubMed] [Google Scholar]
- 96.Sampson JH, Heimberger AB, Archer GE. Immunologic escape after prolonged progression-free survival with epidermal growth factor receptor variant III peptide vaccination in patients with newly diagnosed glioblastoma. J. Clin. Oncol. 2010;28:4722–4729. doi: 10.1200/JCO.2010.28.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.2013. http://clinicaltrials.gov/ct2/show/NCT01480479 Clinicaltrials.gov. US National Library of Medicine. Phase III Study of Rindopepimut/GM-CSF in Patients With Newly Diagnosed Glioblastoma (ACT IV)
- 98.Parsa AT, Crane C, Han S, et al. A Phase 2 multicenter trial of autologous heat shock protein-peptide vaccine (HSPPC-96) for recurrent glioblastoma multiforme (GBM) patients shows improved survival compared with a contemporary cohort controlled for age, KPS and extent of resection. AANS. 2012 [Google Scholar]
- 99.Wheeler CJ, Black KL. DCVax-Brain and DC vaccines in the treatment of GBM. Expert Opin. Investig. Drugs. 2009;18:509–519. doi: 10.1517/13543780902841951. [DOI] [PubMed] [Google Scholar]
- 100.2013. http://clinicaltrials.gov/show/NCTM01290939 Clinicaltrials.gov. US National Library of Medicine. Study of a Drug [DCVax®-L] to Treat Newly Diagnosed GBM Brain Cancer.