Abstract
ADP-ribosylation, the addition of poly-ADP ribose (PAR) onto proteins, is a response signal to cellular challenges, such as excitotoxicity or oxidative stress. This process is catalyzed by a group of enzymes referred to as poly(ADP-ribose) polymerases (PARPs). Because the accumulation of proteins with this modification results in cell death, its negative regulation restores cellular homeostasis: a process mediated by poly-ADP ribose glycohydrolases (PARGs) and ADP-ribosylhydrolase proteins (ARHs). Using linkage analysis and exome or genome sequencing, we identified recessive inactivating mutations in ADPRHL2 in six families. Affected individuals exhibited a pediatric-onset neurodegenerative disorder with progressive brain atrophy, developmental regression, and seizures in association with periods of stress, such as infections. Loss of the Drosophila paralog Parg showed lethality in response to oxidative challenge that was rescued by human ADPRHL2, suggesting functional conservation. Pharmacological inhibition of PARP also rescued the phenotype, suggesting the possibility of postnatal treatment for this genetic condition.
Keywords: ADPRHL2, ARH3, ADP-ribosylation, poly-ADP ribose, oxidative stress, neurodegeneration, epilepsy, ataxia, neuropathy, SUDEP
Main Text
ADP-ribosylation is a tightly regulated posttranslational modification of proteins involved in various essential physiological and pathological processes, including DNA repair, transcription, telomere function, and apoptosis.1, 2, 3 The addition of poly-ADP-ribose (PAR) is mediated by a group of enzymes, referred to as poly(ADP-ribose) polymerases (PARPs), in response to cellular stressors, such as excitotoxicity or reactive oxygen species. PARylated proteins can subsequently initiate cellular stress response pathways. After resolution of the original insult, ADP-ribose polymers are rapidly removed.4, 5 Although PAR modification can protect the cell from death in the setting of cellular stress, excessive PAR accumulation or failure to reverse PAR modification can trigger a cell-death response cascade.6, 7
Humans have two genes encoding specific PAR-degrading enzymes: ADPRHL2 (MIM: 610624; Gene ID: 54936) and PARG (MIM: 603501). Both are capable of hydrolyzing the glycosidic bond between ADP-ribose moieties and are ubiquitously expressed.8, 9 ADPRH (MIM: 603081) and putatively ADPRHL1 (MIM: 610620) encode proteins that can cleave mono-ADP-ribosylated residues and thus are not functionally redundant with ADPRHL2 and PARG.8 Studies of in situ hybridization have shown high Adprhl2 expression in the developing mouse forebrain and that its expression remains high in the cerebellum, cortex, hippocampus, and olfactory bulb in early postnatal ages and persists into adulthood.10 Parg−/− mice die embryonically as a result of PAR accumulation and cellular apoptosis.11 There are no reports of Adprhl2−/− animals, but Adprhl2−/− mouse embryonic fibroblasts (MEFs) engineered to express the catalytic domain of nuclear PARP1 in mitochondria show PAR accumulation, as well as longer mitochondrial PAR polymers.12, 13
Drosophila melanogaster has a single Parg-like gene, and null flies are lethal in the larval stage; however, when grown at a permissive temperature, a few can survive. The surviving flies display PAR accumulation, neurodegeneration, reduced locomotion, and premature death,14 suggesting increased neuronal vulnerability to PAR accumulation. Although mutations in enzymes PARG and PARP have not been reported in human disease, other members of this pathway have been implicated in human phenotypes.15 For example, mutations in XRCC1 (MIM: 194360), encoding a molecular scaffold protein involved in complex assembly during the repair of DNA-strand breaks, lead to PARP-1 overactivation and are associated with cerebellar ataxia, ocular motor apraxia, and axonal neuropathy.16
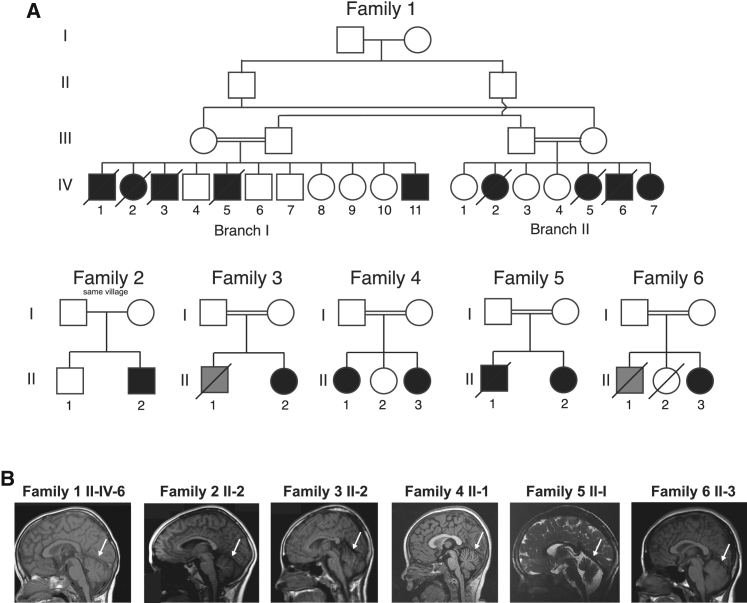
In this study, we show that mutations in ADPRHL2 underlie an age-dependent recessive epilepsy-ataxia syndrome initiating with sudden severe seizures in otherwise healthy individuals followed by progressive loss of milestones, brain atrophy, and death in childhood. We describe six independent families carrying ADPRHL2 mutations leading to a nearly identical epilepsy-ataxia syndrome (Figure 1A). One of the six families (family 2) lacked documentation of parental consanguinity, and the parents from this family were from the same small village. The clinical details of subjects from all included families are shown in Table 1, and detailed clinical history is narrated in the Supplemental Note. The emerging clinical picture is one of a stress-induced neurodegenerative disease of variable progression with developmental delay, intellectual disability, mild cerebellar atrophy (Figure 1B), and recurring seizures.
Figure 1.
Pedigrees of Families with Mutations in ADPRHL2 and Their Clinical Presentation
(A) Pedigrees of families 1–6 show consanguineous unions (double bar) and a total of 16 affected individuals. Slashes represent deceased individuals. Black shading indicates affected individuals. Gray shading indicates individuals who passed away from SUDEP; however, no DNA is available.
(B) Panels show midline sagittal MRI for one affected individual from each of the six families. White arrows indicate cerebellar atrophy, evidenced by widely spaced cerebellar folia.
Table 1.
Mutations in ADPRHL2 Cause Various Phenotypes, Including Developmental Delay, Cerebellar Atrophy, Ataxia, and Epilepsy
|
Family 1 |
Family 2 |
Family 3 |
Family 4 |
Family 5 |
Family 6 |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I-IV-1 | I-IV-2 | I-IV-3 | I-IV-5 | I-IV-11 | II-IV-2 | II-IV-5 | II-IV-6 (A1) | II-IV-7 (A2) | II-2 | II-1 | II-1 | II-3 | IV-1 | IV-2 | II-3 | |
| Gender | M | F | M | M | M | F | F | M | F | M | F | F | F | M | F | F |
| Country of origin | UAE | UAE | UAE | UAE | UAE | UAE | UAE | UAE | UAE | Italy | Turkey | Pakistan | Pakistan | Iran | Iran | Turkey |
| Parental consanguinity | + | + | + | + | + | + | + | + | + | same village | + | + | + | + | + | + |
| Current age or age of death | died at 4 years | died at 2 years | died at 7 years | died at 15 years | 4 years | died at 2 years | died at 2 years | died at 9 years | 3 years | 16 years | 15 years | 13 years | 2 years | died at 6 years | 3 years | 10 years |
| Circumstances of death | in sleep | in sleep | seizure | respiratory failure | – | in sleep 1 week after flu-like illness | seizure after playing | respiratory failure after long airplane trip | – | – | – | – | – | in sleep | – | – |
| Mutation | ||||||||||||||||
| Genomic (hg19a) | g.36558895C>T | g.36558895C>T | g.36558895C>T | g.36558895C>T | g.36558895C>T | g.36558895C>T | g.36558895C>T | g.36558895C>T | g.36558895C>T | g.36557226C>T | g.36556868A>C | g.36557324_36557328delTGCCC | g.36557324_36557328delTGCCC | g.36557524C>T | g.36557524C>T | g.36554605G>A |
| cDNA | c.1000C>T | c.1000C>T | c.1000C>T | c.1000C>T | c.1000C>T | c.1000C>T | c.1000C>T | c.1000C>T | c.1000C>T | c.316C>T | c.235A>C | c.414_418delTGCCC | c.414_418delTGCCC | c.530C>T | c.530C>T | c.100G>A |
| Protein | p.Gln334∗ | p.Gln334∗ | p.Gln334∗ | p.Gln334∗ | p.Gln334∗ | p.Gln334∗ | p.Gln334∗ | p.Gln334∗ | p.Gln334∗ | p.Gln106∗ | p.Thr79Pro | p.Ala139Glyfs∗4 | p.Ala139Glyfs∗4 | p.Ser177Leu | p.Ser177Leu | p.Asp34Asn |
| Homozygosity | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Perinatal History | ||||||||||||||||
| Normal birth | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Normal early development | + | + | + | + | + | + | + | + | + | + | + | + | mild developmental delay | + | + | + |
| Psychomotor Development | ||||||||||||||||
| Speech development | spoke in sentences but then deteriorated | few words at 2 years | normal until 2.5 years but then no further development | normal until 3.5 years but then deteriorated | speaks only a few words | normal speech until death | normal speech until death | normal until 25 years but then deteriorated | normal speech but then deteriorated | slow speech | normal | normal | delayed | normal until 1.5 years but then deteriorated with difficulty speaking | speaks only a few words | delayed |
| Motor development | normal but then deteriorated | normal until death | normal but then deteriorated | normal but then deteriorated | normal but then deteriorated | normal but then deteriorated | normal until death | normal but then deteriorated by 2 years | walked normally at 14 months but then had ataxia and poor balance at 19 months | normal but then deteriorated by 2 years | normal | normal but then deteriorated by 2 years | mildly delayed | normal until 1 year but then deteriorated | normal but then deteriorated by 1.5 years | normal |
| Seizures | ||||||||||||||||
| Seizure onset | 18 months | 19 months | 19 months | 24 months | 15 months | 24 months | 15 months | 18 months | 16 months | – | – | – | 9 months | 24 months | 36 months | – |
| Seizure types | GTCS | GTCS | GTCS | GTCS | absence, GTCS | GTCS | GTCS | absence, GTCS | absence, GTCS | – | – | GTCS with illness | GTCS with illness | multifocal, GTCS | multifocal, GTCS | – |
| Neurological Examination | ||||||||||||||||
| Intellect | normal but then delayed | normal until death | normal but then delayed | normal but then delayed | normal but then delayed | normal until death | normal until death | normal but then delayed | normal but then delayed | normal but then started deteriorating at age 11 yeras | normal | mild ID (IQ 60) | mild global developmental delay | normal but then stagnated | normal but then stagnated | mild ID |
| EEG | – | – | – | – | – | – | – | generalized epileptiform activity, slow background | generalized epileptiform activity, slow background | – | – | mild slowing background activity (3 years) | normal | generalized epileptiform activity, slow background | normal | normal |
| MRI (age performed) | – | – | – | normal (5 years) | – | – | – | mild cerebellar atrophy (7 years) | mild cerebellar atrophy (7 years) | cerebellar vermis atrophy (11 years) | mild cerebellar atrophy, spinal cord atrophy (12 years) | mild cerebellar atrophy (4 years) | normal (11 months) | – | normal (3 years) | mild cerebellar vermis atrophy, spinal cord atrophy (15 years) |
| EMG or biopsy | – | – | – | – | – | – | – | nerve biopsy with severe axonal loss | – | – | axonal polyneuropathy (4 years) | normal muscle biopsy (4 years) | – | normal (4 years) | axonal polyneuropathy (4 years) | axonal polyneuropathy |
| Onset of unsteady gait | – | – | 2.5 years | 3 years | 2.5 years | – | – | 2.5 years | 20 months | 11 years | 4 years | 2.5 years | not yet | 1.5 years | 1.5 years | 10 years |
| Other Clinical Features | ||||||||||||||||
| Exacerbated by illness and/or stress | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Other clinical features | – | – | – | hypotonia with repeated pneumonia, ventilator dependent at time of death | can walk but is very unsteady | progressive weakness | progressive weakness | repeated pneumonia, repeated cardiac arrest, profound type II muscle fiber atrophy | normal hearing but then developed severe SNHL, severe kyphoscoliosis, one episode of cardiac arrest | myopathic changes on muscle biopsy (11 years) | claw hand and pes cavus deformities, scoliosis, SNHL at 10 years, tracheotomy, ventilation | asthma | – | progressive weakness, tremors, frequent falling | progressive weakness, progressive external ophthalmoplegia | distal muscle atrophy, pes cavus deformity, toe abnormality, scoliosis, brisk DTRs, positive Babinski reflex, intentional tremor, ataxia |
Clinical presentation for affected subjects from families 1–6. Abbreviations are as follows: +, yes; –, not available; DTR, deep-tendon reflex; EEG, electroencephalography; EMG, electromyography; F, female; GTCS, generalized tonic-clonic seizure; ID, intellectual disability; M, male; MRI, magnetic resonance imaging; and SNHL, sensorineural hearing loss.
UCSC Genome Browser.
Genome-wide linkage analysis of 14 members of family 1 mapped the disease locus to an 11 Mb locus in chromosomal region 1p36 with a genome-wide-significant multipoint LOD score of 3.4 (Figure S1A). Exome sequencing of individual II-IV-6 at >30× read depth for 96.9% of the exome revealed a single rare (allele frequency < 1:1,000) potentially deleterious variant within the linkage interval: a frameshift ADPRHL2 mutation that segregated with the phenotype according to a recessive mode of inheritance.
Using GeneMatcher, this international collaborative group of authors identified further pathogenic alleles in ADPRHL2.17 After obtaining informed consent from all participating individuals in accordance with the ethical standards of the responsible committee on human experimentation at the University of California, San Diego, we identified a total of six distinct mutations in ADP-ribosylhydrolase-like 2 (ADPRHL2 [Gene ID: 54936]) in the six families by whole-exome or genome sequencing (see Supplemental Data). All variants were prioritized by allele frequency, conservation, blocks of homozygosity, and predicted effect on protein function (see Supplemental Data), and in all families the homozygous variant in ADPRHL2 was the top candidate. Variants were confirmed by Sanger sequencing and segregated with the phenotype according to a recessive mode of inheritance. All variants were predicted to be disease causing by MutationTaster.18 These variants were not encountered in dbGaP, the ExAC Browser, 1000 Genomes, genomAD, or the Greater Middle East Variome.
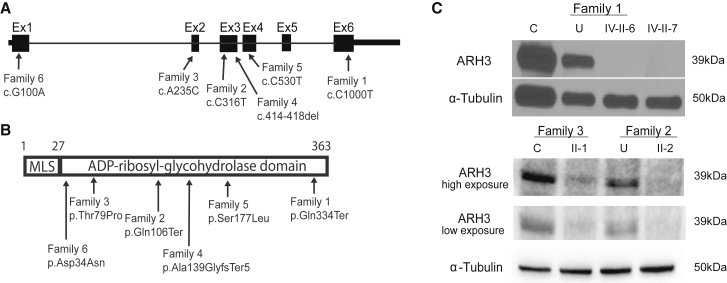
ADPRHL2 contains six coding exons, yielding a single protein-coding transcript, ADP-ribosylhydrolase 3 (ARH3) (Figure 2A). The encoded 363 amino acid ARH3 is predicted to have a mitochondrial localization sequence and single enzymatic ADP-ribosyl-glycohydrolase domain (Figure 2B). Family 1 carried the homozygous exon 6 mutation c.1000C>T (GenBank: NM_017825), which introduces a premature stop codon (p.Gln334Ter) predicted to truncate the highly conserved last 30 amino acids of the protein, including part of the ADP-ribosylhydrolase domain. Family 2 harbored the homozygous exon 3 mutation c.316C>T (GenBank: NM_017825), which also introduces a premature stop codon (p.Gln106Ter) in the ADP-ribosylhydrolase domain. Family 3 revealed the homozygous exon 2 missense mutation c.235A>C (GenBank: NM_017825), which leads to an amino acid change (p.Thr97Pro) in a residue that is highly conserved among vertebrates (Figure S2A). Using a previously published crystal structure of ARH3, we localized this residue to an α-helical loop within the ADP-ribosylhydrolase domain and the substrate binding site, which is defined by the position of two Mg2+ ions located in adjacent binding sites; thus, the residue is predicted to affect protein structure and enzymatic activity (Figure S2B).19 Family 4 carried the homozygous 5 bp, exon 3 deletion c.414_418TGCCC (GenBank: NM_017825), which results in a frameshift (p.Ala139GlyfsTer5) in the ADP-ribosylhydrolase domain. Family 5 carried the homozygous exon 4 missense mutation c.530C>T (GenBank: NM_017825), which leads to an amino acid change (p.Ser177Leu) that is also highly conserved among vertebrates. It is localized in a critical α-helical loop within the ADP-ribosylhydrolase domain, also suggesting an effect on protein structure and activity. Family 6 carried the homozygous exon 1 missense mutation c.100G>A (GenBank: NM_017825), which leads to an amino acid change (p.Asp34Asn) that is highly conserved among vertebrates. This change is also localized in a critical α-helical loop within the ADP-ribosylhydrolase domain, suggesting a potential impact on protein structure and activity.
Figure 2.
Truncating and Missense ADPRHL2 Mutations in Six Independent Families Are Predicted to Be Inactivating
(A) Schematic of ADPRHL2 depicts the coding sequence spanning six exons and the 5′ and 3′ UTRs. Black arrows indicate the positions of the six identified mutations and their coordinates within the cDNA (Gene ID: 54936).
(B) Schematic of ARH3 depicts the mitochondrial localization sequence (MLS) and the ADP-ribosyl-glycohydrolase domain. Black arrows indicate the position and coordinates of the impact of the described mutations.
(C) Western blot of fibroblasts from an unrelated control individual (C), the unaffected carrier father (U), and affected individuals IV-II-6 and IV-II-7 from family 1 shows the absence of ARH3 in affected fibroblasts. α-tubulin was used as the loading control. Western blot of fibroblasts from an unrelated control individual (C) and affected individual II-1 from family 3 and the unaffected carrier mother (U) and affected individual II-3 from family 2 shows significantly reduced amounts of ARH3. α-tubulin was used as the loading control.
The emerging phenotype of recessive ADPRHL2 mutations is a degenerative pediatric-onset stress-induced epileptic-ataxia syndrome. Individuals with mutations in this gene are asymptomatic early after birth but gradually develop a cyclic pattern of illness-related spontaneous epileptic seizures or present with a neurodegenerative course including weakness, ataxia, and loss of milestones followed by clinical deterioration that ultimately leads to premature death. Most of the subjects succumbed to sudden unexpected death in epilepsy (SUDEP) or an apnoic-attack-like clinical presentation, suggesting a hyperacute presentation prior to the family’s recognition of a predisposition. We could not establish an obvious genotype-phenotype correlation given that we show below that the missense mutation also leads to a severe loss of function. Thus, the clinical variability in the age of onset might occur because the genetic background or environmental challenges lead to variable susceptibility to illness-related cellular stress.
The differential diagnosis for this condition was based upon the presentation of a recessive condition with recurrent exacerbations and predominant features of global developmental delay, intellectual disability, seizures, neurogenic changes on electromyography, hearing impairment, regression, and mild cerebellar atrophy but not microcephaly or cataracts. The differential diagnosis in our families included mitochondrial disorders, spastic ataxia, and peripheral neuropathy.
To determine the impact of these mutations on protein folding and binding activity, we generated recombinant proteins in E.coli and purified them by His-tag affinity chromatography. Our results showed that the p.Gln334Ter protein was not evident in the soluble fraction, whereas the wild-type (WT) was recovered with good purity (Figure S3A). The p.Thr79Pro protein was expressed and soluble, although possibly recovered with slightly less purity than WT ARH3. We studied the deleterious impact of p.Thr79Pro by using circular dichroism spectroscopy (Figures S3B and S3C). Compared with the WT, this mutant exhibited reduced α-helical content and an altered secondary structure, in agreement with the fact that p.Thr79Pro occurred within an α-helical domain. Further, the melting temperature (Tm) of p.Thr79Pro was reduced by more than 10°C, confirming destabilization of the mutant (Figures S3D–S3F). We also found that in contrast to WT ARH3, the p.Thr79Pro protein was not stabilized by ligands such as adenosine diphosphate ribose (ADPr) (Figures S3G–S3I). We confirmed the specificity of this assay by using adenosine triphosphate (ATP) and ribose-5-phosphate as negative controls, which were not predicted to bind or stabilize ARH3. Together, these data suggest that both disease-causing, truncating mutants and amino acid substitutions should be destabilized when expressed in cells.
Because the c.1000C>T (p.Gln334ter) mutation of family 1 was in the last exon, we first excluded nonsense-mediated decay (NMD) of the mutant mRNA. We collected skin biopsies from the father (III-II) and two affected individuals (II-IV-6 and II-IV-7), generated primary fibroblasts, and then performed RT-PCR by using primers designed to amplify the last three exons of ADPRHL2 (Figure S1B). The father’s and affected individuals’ cells revealed a band of the expected size and of similar intensity to that of a healthy control individual, arguing against NMD. However, when we used an antibody recognizing amino acids 231–245, lysates derived from the affected individuals showed no detectable ARH3 (Figure 2C; Supplemental Data), consistent with a null effect of the truncating mutation. Further, western blot analysis of individual II-2 from family 2 showed an absence of the protein, as predicted by the early stop codon. Fibroblasts from individual II-1 from family 3 showed a severely reduced amount of ARH3 (Figure 2C), consistent with the thermal instability of this mutant protein (Figures S3D–S3F) and the severe alteration of its secondary structure (Figures S3B and S3C).
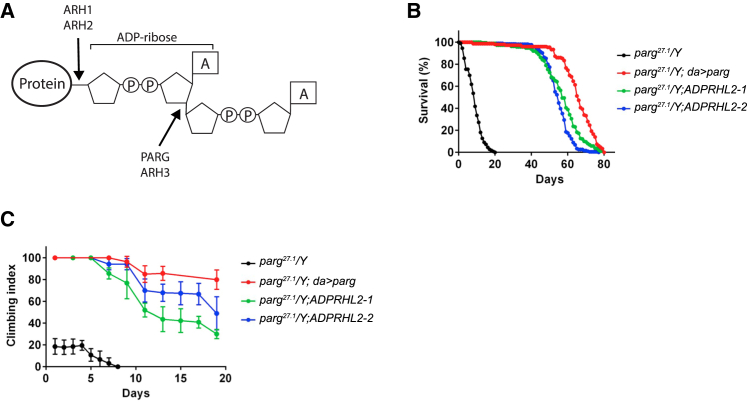
Whereas humans have two known genes with specific PARG activity (PARG and ADPRHL2; Figure 3A), Drosophila have a single gene that regulates this process: Parg. Using the Gal4-UAS system to drive RNAi expression, we found that Parg knockdown led to a 60% decrease in total Parg mRNA for flies with the ubiquitous da promoter and a 50% decrease with the neuron-specific promoter, elav (embryonic lethal abnormal visual system) (Figure S4A). Whereas the da-Gal4 and PargRNAi lines showed normal survival, crossing the two together led to daughterless (da)-mediated expression of PargRNAi, which reduced survival substantially (Figure S4B). Ubiquitous knockdown of Parg also led to decreased survival when animals were exposed to stress with either hydrogen peroxide (H2O2) in their water or environmental hypoxia (2% O2) (Figures S4C and S4D). Furthermore, knockdown of Parg specifically in neurons largely recapitulated this phenotype by using the same two environmental stressors (Figures S4E and S4F). These data provide evidence that stress leads to premature death in the absence of Parg and that neurons play an important role in this phenotype.
Figure 3.
Premature Death and Locomotor Defects in Drosophila Parg Mutants Are Rescued by Human ADPRHL2
(A) Schematic of a poly-ADP-ribosylated protein and the location of cleavage. PARG and ADPRHL2 both remove poly-ADP-ribose (PAR) from proteins and cleave the same site. Drosophila melanogaster has one PAR-removing enzyme, Parg.
(B) Parg27.1 mutant flies (black) show a severe climbing defect, which was rescued by ubiquitous forced expression of Parg (red) or mis-expression of human ADPRHL2 in two different transgenic lines (green and blue).
(C) Parg27.1 mutant flies (black) displayed decreased survival, which was rescued with ubiquitous forced expression of Parg (red) and two different transgenic lines expressing human ADPRHL2 (green and blue). Data represent the mean ± SEM of eight experiments.
However, lethality of these flies was not as severe as in the Parg27.1 line, which carries a p-element insertion that deletes two-thirds of the open reading frame (nucelotides 34,622–36,079 of GenBank: Z98254),14 suggesting that PargRNAi is partially inactivating. These mutant flies with Parg loss of function lack the protein Parg and show elevated amounts of PAR, especially in nervous tissue.14 Mutant flies die in larval stages, but 25% of the animals survive when grown at the permissive 29°C temperature. These adult flies display progressive neurodegeneration, reduced locomotion, and reduced lifespan,14 consistent with the individuals’ phenotypes in our families. We confirmed lethality of the Parg27.1 line and found that forced expression of Drosophila Parg under the ubiquitous da promoter in the mutant background increased both survival and motor activity as measured by an established “climbing index” (Figures 3B and 3C).20 Likewise, expression of the human ADPRHL2 under the same da promoter showed a nearly identical degree of rescue of both survival and locomotor activity (Figures 3B and 3C). These results suggest that human ADPRHL2 is a functional paralog of Drosophila Parg.
We next tested whether this phenotype might be ameliorated by inhibition of protein PARylation. We reasoned that the requirement for dePARylation should be reduced by the blockage of stress-induced PARylation. Minocycline displays PARP inhibitory activity with an IC50 of 42 nM in humans21 and is well tolerated in flies.22 We fed flies with a range of concentrations from 0 to 1 mg/mL minocycline for 24 hr before stress and measured survival rates 96 hr after stress induction. Drug treatment of flies with ubiquitous knockdown of Parg revealed a dose-dependent partial rescue of the lethality (Figure S4G). This rescue was also seen when the drug was given to flies with neuron-specific knockdown of Parg (Figure S4H), providing evidence that PARP inhibition can rescue lethality in vivo. Although we expect that the effect of minocycline on survival in this assay was due to its effect on PARP, we cannot exclude off-target or non-specific effects.22
Given that PARP inhibitors are currently in trials for various types of cancer, it is possible that these drugs could be tested for clinical effectiveness in this orphan disease, where they could have a positive effect. Potentially clinically relevant PARP inhibitors include (1) minocycline, an FDA-approved tetracycline derivative that displays PARP inhibitory activity; (2) dihydroisoquinoline (DPQ), a non-FDA-approved potent PARP-1 inhibitor used in experimental research; and (3) veliparib (ABT: 888), a potent PARP-1 and PARP-2 inhibitor currently in clinical trials for the treatment of various type of cancers (IC50 = 42, 37, and 4.4 nM, respectively).21, 23
The extent to which ADPRHL2 and PARG functionally diverge or converge is not well understood, partly because of a lack of detailed comparative expression analysis and biochemical function. PARG demonstrates greater specific activity than ARH3 for removing PAR from proteins,8 and loss of Parg in mice is embryonically lethal.13 Together, these data suggest that PARG is likely to be the major contributor to PAR removal in cells that express both genes under basal conditions. One possibility is that ADPRHL2 acts as a backup for PARG to remove excessive PAR moieties under stress conditions. This would be consistent with the clinical pesentation of individuals with loss of ADPRHL2, where phenotypes appear to be induced by environmental stress. Recent studies have shown that ARH3 acts on a recently discovered form of Ser-ADP ribosylation.24 For example, studies have illustrated an excessive accumulation of Ser-poly-ADP-ribosylated enzymes in ADPRHL2−/− cell lines and that ARH3 acts mainly on Ser-ADPr removal.25 This would be consistent with the phenotype we see in subjects with loss of ARH3, where phenotypes do not emerge until environmental stress insults are encountered. Finally, ARH3 contains a mitochondrial localization signal, and thus another possibility is that ARH3 functions as a mitochondrial-specific glycohydrolase that is required after the induction of oxidative stress.13
PAR signaling has been shown to play a role in a number of cellular processes—including the regulation of transcription, telomere function, mitotic spindle formation, intracellular trafficking, and energy metabolism—in addition to apoptosis-inducing-factor (AIF)-mediated apoptosis.2, 3 Although we hypothesize that the disease mechanism is through cell death, it is possible that PAR accumulation could affect other cellular processes before this. Further work is needed to characterize these effects in the context of this disease.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
The authors thank the subjects and their families for participating in this study. S.G. was sponsored by the Ruth L. Kirschstein Institutional National Research Service Award (T32 GM008666) from the National Institute on Deafness and Other Communication Disorders and by award F31HD095602 from the NIH Eunice Kennedy Shriver National Institute of Child Health and Human Development. We thank the Broad Institute (U54HG003067 to E. Lander and UM1HG008900 to D. MacArthur) and the Yale Center for Mendelian Disorders (U54HG006504 to R. Lifton and M. Gunel). We also thank Lisa Weixler for technical assistance and all the staff of the Cologne Center for Genomics for next-generation sequencing. The sequencing data were analyzed with the CHEOPS high-performace computer cluster of the Regional Computing Center of the University of Cologne. We thank DeCode for whole-genome sequencing. This work was supported by NIH grants R01NS048453 and R01NS052455, the Simons Foundation Autism Research Initiative, the Howard Hughes Medical Institute (to J.G.G.), the Deutsche Forschungsgemeinschaft Emmy Noether Programme (grant CI 218/1-1 to S.C.), the Wellcome Trust (WT093205MA and WT104033AIA), Ataxia UK, the UCL/UCLH NIHR Biomedical Research Centre, the Medical Research Council (to H.H. and M.H), EU Horizon 2020 Solve-RD, and the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant 2012-305121 for the “Integrated European -omics research project for diagnosis and therapy in rare neuromuscular and neurodegenerative diseases (NEUROMICS)” to B.W.
Published: August 9, 2018
Footnotes
Supplemental Data include a Supplemental Note, Supplemental Material and Methods, four figures, and one table and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.07.010.
Contributor Information
Sebahattin Cirak, Email: sebahattin.cirak@uk-koeln.de.
Joseph G. Gleeson, Email: jogleeson@ucsd.edu.
Accession Numbers
The exome sequencing data from individuals from the University of California, San Diego, study site have been deposited in the Database of Genotypes and Phenotypes under accession number dbGaP: phs000288.v1.
Web Resources
1000 Genomes, http://browser.1000genomes.org
Exome Aggregation Consortium (ExAC) Browser, http://exac.broadinstitute.org/
FlyBase, http://flybase.org
GeneMatcher, https://genematcher.org
HaplotypeCaller and GATK, https://www.broadinstitute.org/gatk/
Iranome, http://www.iranome.ir/
Mutation Assessor, http://mutationassessor.org/
MutationTaster, htp://mutationtaster.org/
NHLBI Exome Sequencing Project Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
Provean, http://provean.jcvi.org
SIFT, http://sift.jcvi.org/
UniProt, http://www.uniprot.org
Supplemental Data
References
- 1.Hassa P.O., Haenni S.S., Elser M., Hottiger M.O. Nuclear ADP-ribosylation reactions in mammalian cells: Where are we today and where are we going? Microbiol. Mol. Biol. Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luo X., Kraus W.L. On PAR with PARP: Cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;26:417–432. doi: 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schreiber V., Dantzer F., Ame J.C., de Murcia G. Poly(ADP-ribose): Novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 4.De Vos M., Schreiber V., Dantzer F. The diverse roles and clinical relevance of PARPs in DNA damage repair: Current state of the art. Biochem. Pharmacol. 2012;84:137–146. doi: 10.1016/j.bcp.2012.03.018. [DOI] [PubMed] [Google Scholar]
- 5.Wang Z., Wang F., Tang T., Guo C. The role of PARP1 in the DNA damage response and its application in tumor therapy. Front. Med. 2012;6:156–164. doi: 10.1007/s11684-012-0197-3. [DOI] [PubMed] [Google Scholar]
- 6.Andrabi S.A., Kim N.S., Yu S.W., Wang H., Koh D.W., Sasaki M., Klaus J.A., Otsuka T., Zhang Z., Koehler R.C. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc. Natl. Acad. Sci. USA. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y., Dawson V.L., Dawson T.M. Poly(ADP-ribose) signals to mitochondrial AIF: A key event in parthanatos. Exp. Neurol. 2009;218:193–202. doi: 10.1016/j.expneurol.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oka S., Kato J., Moss J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 2006;281:705–713. doi: 10.1074/jbc.M510290200. [DOI] [PubMed] [Google Scholar]
- 9.Poitras M.F., Koh D.W., Yu S.W., Andrabi S.A., Mandir A.S., Poirier G.G., Dawson V.L., Dawson T.M. Spatial and functional relationship between poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in the brain. Neuroscience. 2007;148:198–211. doi: 10.1016/j.neuroscience.2007.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magdaleno S., Jensen P., Brumwell C.L., Seal A., Lehman K., Asbury A., Cheung T., Cornelius T., Batten D.M., Eden C. BGEM: An in situ hybridization database of gene expression in the embryonic and adult mouse nervous system. PLoS Biol. 2006;4:e86. doi: 10.1371/journal.pbio.0040086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koh D.W., Lawler A.M., Poitras M.F., Sasaki M., Wattler S., Nehls M.C., Stöger T., Poirier G.G., Dawson V.L., Dawson T.M. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc. Natl. Acad. Sci. USA. 2004;101:17699–17704. doi: 10.1073/pnas.0406182101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niere M., Kernstock S., Koch-Nolte F., Ziegler M. Functional localization of two poly(ADP-ribose)-degrading enzymes to the mitochondrial matrix. Mol. Cell. Biol. 2008;28:814–824. doi: 10.1128/MCB.01766-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niere M., Mashimo M., Agledal L., Dölle C., Kasamatsu A., Kato J., Moss J., Ziegler M. ADP-ribosylhydrolase 3 (ARH3), not poly(ADP-ribose) glycohydrolase (PARG) isoforms, is responsible for degradation of mitochondrial matrix-associated poly(ADP-ribose) J. Biol. Chem. 2012;287:16088–16102. doi: 10.1074/jbc.M112.349183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanai S., Kanai M., Ohashi S., Okamoto K., Yamada M., Takahashi H., Miwa M. Loss of poly(ADP-ribose) glycohydrolase causes progressive neurodegeneration in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2004;101:82–86. doi: 10.1073/pnas.2237114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bütepage M., Eckei L., Verheugd P., Lüscher B. Intracellular mono-ADP- ribosylation in signaling and disease. Cells. 2015;4:569–595. doi: 10.3390/cells4040569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoch N.C., Hanzlikova H., Rulten S.L., Tétreault M., Komulainen E., Ju L., Hornyak P., Zeng Z., Gittens W., Rey S.A., Care4Rare Canada Consortium XRCC1 mutation is associated with PARP1 hyperactivation and cerebellar ataxia. Nature. 2017;541:87–91. doi: 10.1038/nature20790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: A matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarz J.M., Rödelsperger C., Schuelke M., Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 19.Mueller-Dieckmann C., Kernstock S., Lisurek M., von Kries J.P., Haag F., Weiss M.S., Koch-Nolte F. The structure of human ADP-ribosylhydrolase 3 (ARH3) provides insights into the reversibility of protein ADP-ribosylation. Proc. Natl. Acad. Sci. USA. 2006;103:15026–15031. doi: 10.1073/pnas.0606762103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madabattula S.T., Strautman J.C., Bysice A.M., O’Sullivan J.A., Androschuk A., Rosenfelt C., Doucet K., Rouleau G., Bolduc F. Quantitative analysis of climbing defects in a Drosophila model of neurodegenerative disorders. J. Vis. Exp. 2015;100:e52741. doi: 10.3791/52741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alano C.C., Kauppinen T.M., Valls A.V., Swanson R.A. Minocycline inhibits poly(ADP-ribose) polymerase-1 at nanomolar concentrations. Proc. Natl. Acad. Sci. USA. 2006;103:9685–9690. doi: 10.1073/pnas.0600554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee G.J., Lim J.J., Hyun S. Minocycline treatment increases resistance to oxidative stress and extends lifespan in Drosophila via FOXO. Oncotarget. 2017;8:87878–87890. doi: 10.18632/oncotarget.21224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donawho C.K., Luo Y., Luo Y., Penning T.D., Bauch J.L., Bouska J.J., Bontcheva-Diaz V.D., Cox B.F., DeWeese T.L., Dillehay L.E. ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models. Clin. Cancer Res. 2007;13:2728–2737. doi: 10.1158/1078-0432.CCR-06-3039. [DOI] [PubMed] [Google Scholar]
- 24.Fontana P., Bonfiglio J.J., Palazzo L., Bartlett E., Matic I., Ahel I. Serine ADP-ribosylation reversal by the hydrolase ARH3. eLife. 2017;6:e28533. doi: 10.7554/eLife.28533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palazzo L., Leidecker O., Prokhorova E., Dauben H., Matic I., Ahel I. Serine is the major residue for ADP-ribosylation upon DNA damage. eLife. 2018;7:e34334. doi: 10.7554/eLife.34334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.