Abstract
Coronary artery disease (CAD) is the leading cause of death globally. Genome-wide association studies (GWASs) have identified more than 95 independent loci that influence CAD risk, most of which reside in non-coding regions of the genome. To interpret these loci, we generated transcriptome and whole-genome datasets using human coronary artery smooth muscle cells (HCASMCs) from 52 unrelated donors, as well as epigenomic datasets using ATAC-seq on a subset of 8 donors. Through systematic comparison with publicly available datasets from GTEx and ENCODE projects, we identified transcriptomic, epigenetic, and genetic regulatory mechanisms specific to HCASMCs. We assessed the relevance of HCASMCs to CAD risk using transcriptomic and epigenomic level analyses. By jointly modeling eQTL and GWAS datasets, we identified five genes (SIPA1, TCF21, SMAD3, FES, and PDGFRA) that may modulate CAD risk through HCASMCs, all of which have relevant functional roles in vascular remodeling. Comparison with GTEx data suggests that SIPA1 and PDGFRA influence CAD risk predominantly through HCASMCs, while other annotated genes may have multiple cell and tissue targets. Together, these results provide tissue-specific and mechanistic insights into the regulation of a critical vascular cell type associated with CAD in human populations.
Keywords: genetics, genomics, expression quantitative traits, genome-wide association, coronary disease, smooth muscle cells
Introduction
Atherosclerotic coronary artery disease (CAD) is the leading cause of death in both developed and developing countries worldwide, and current estimates predict that more than 1 million individuals will suffer from new and recurrent CAD this year in the U.S. alone.1 Like most polygenic diseases, both genetic and environmental factors influence an individual’s lifetime risk for CAD.2 Early Swedish twin studies and more recent genome-wide association studies (GWASs) have estimated that about 50% of CAD risk is explained by genetic factors.3, 4 To date, GWASs have reported more than 95 replicated independent loci and numerous additional loci that are associated at an FDR < 0.05.5, 6, 7, 8 A majority of these loci reside in non-coding genomic regions and are expected to function through regulatory mechanisms. Also, approximately 75% of CAD loci are not associated with classical risk factors, suggesting that at least part of them function through mechanisms intrinsic to the vessel wall.
Smooth muscle cells (SMCs) constitute the majority of cells in the coronary artery wall. In response to vascular injury (e.g., lipid accumulation, inflammation), SMCs undergo phenotypic switching and ultimately contribute to both atherosclerotic plaque formation and stabilization.9, 10, 11, 12 Recent lineage tracing studies in mice have revealed that although 80% of plaque-derived cells lack traditional SMC markers, roughly half are of SMC origin.13, 14 Thus, genetic studies of human coronary artery smooth muscle cells (HCASMCs) have the potential to shed light on their diverse functions in the vessel wall relevant to human atherosclerosis. In a few cases, the underlying mechanisms have been identified for CAD loci in vascular SMC models.10, 15, 16, 17, 18 Large-scale expression quantitative trait loci (eQTL) mapping efforts such as the Genotype Tissue Expression (GTEx) project have helped refine these mechanisms for multiple traits across human tissues.19 However, due to the lack of HCASMCs in both GTEx and other studies, the overall contribution of this cell type toward heritable CAD risk remains unknown.
Herein, we performed whole-genome sequencing and transcriptomic profiling of 52 HCASMC donors to quantify the effects of cis-acting variation on gene expression and splicing associated with CAD. We evaluated the tissue specificity and disease relevance of our findings in HCASMCs by comparing to publicly available GTEx and ENCODE datasets. We observed significant colocalization of eQTL and GWAS signals for five genes (FES, SMAD3, TCF21, PDGFRA, and SIPA1), which all have the capacity to perform relevant functions in vascular remodeling. Further, comparative analyses with GTEx datasets reveals that SIPA1 and PDGFRA have stronger colocalization signals in HCASMCs than in other tissues. Together, these findings demonstrate the power of leveraging genetics of gene regulation for a critical cell type to generate hypotheses on risk-associated mechanisms for CAD.
Material and Methods
Sample Acquisition and Cell Culture
A total of 62 primary human coronary artery smooth muscle cell (HCASMC) lines collected from donor hearts were purchased, and 52 lines remained after stringent filtering (see Supplemental Material and Methods). These 52 lines were from PromoCell (catalog # C-12511, n = 19), Cell Applications (catalog # 350-05a, n = 25), Lonza (catalog # CC-2583, n = 3), Lifeline Cell Technology (catalog # FC-0031, n = 3), and ATCC (catalog # PCS-100-021, n = 2). All lines were stained with smooth muscle alpha actin to check for smooth muscle content and all lines tested negative for mycoplasma (Table S1). All cell lines were cultured in smooth muscle growth medium (Lonza catalog # CC-3182) supplemented with hEGF, insulin, hFGF-b, and 5% FBS, according to Lonza instructions. All HCASMC lines were expanded to passage 5–6 prior to extraction.
Library Preparation and Sequencing
Whole-Genome Sequencing
Genomic DNA was isolated using QIAGEN DNeasy Blood & Tissue Kit (catalog # 69506) and quantified using NanoDrop 1000 Spectrophotometer (Thermo Fisher). Macrogen performed library preparation using Illumina’s TruSeq DNA PCR-Free Library Preparation Kit and 150 bp paired-end sequencing on Illumina HiSeq X Ten System.
RNA Sequencing
RNA was extracted using QIAGEN miRNeasy Mini Prep Kit (catalog # 74106). Quality of RNA was assessed on the Agilent 2100 Bioanalyzer. Samples with RIN greater than or equal to 8 were sent to the Next-Generation Sequencing Core at the Perelman School of Medicine at the University of Pennsylvania. Libraries were made using Illumina TruSeq Stranded Total RNA Library Prep Kit (catalog # 20020597) and sequenced using 125 bp paired-end on HiSeq 2500 Platform.
ATAC Sequencing
We used ATAC-seq to assess chromatin accessibility with slight modifications to the published protocol.20 Approximately 5 × 104 fresh cells were collected at 500 × g, washed in PBS, and nuclei extracted with cold lysis buffer. Pellets were subjected to transposition containing Tn5 transposases (Illumina) at 37°C for 30 min, followed by purification using the DNA Clean-up and Concentration kit (Zymo). Libraries were PCR amplified using Nextera barcodes, with the total number of cycles empirically determined using SYBR qPCR. Amplified libraries were purified and quantified using bioanalyzer, nanodrop, and qPCR (KAPA) analysis. Libraries were multiplexed and 2 × 75 bp sequencing was performed using an Illumina NextSeq 500.
Alignment and Quantification of Genomic, Transcriptomic, and Epigenomic Features
Whole-genome sequencing data were processed with the GATK best practices pipeline with hg19 as the reference genome,21, 22 and VCF records were phased with Beagle v.4.1.23 Variants with imputation allelic r2 less than 0.8 and Hardy-Weinberg Equilibrium p value less than 1 × 10−6 were filtered out (see Supplemental Material and Methods). De-multiplexed FASTQ files were mapped with STAR version 2.4.0i in 2-pass mode24 over the hg19 reference genome. Prior to expression quantification, we filtered our reads prone to mapping bias using WASP.25 Total read counts and RPKM were calculated with RNA-SeQC v1.1.826 using default parameters with additional flags “-n 1000 -noDoC -strictMode” over GENCODE v.19 reference. Allele-specific read counts were generated with the createASVCF module in RASQUAL.27 We quantified intron excision levels using LeafCutter intron-spanning reads.28 In brief, we converted bam files to splice junction files using the bam2junc.sh script, and defined intron clusters using leafcutter_cluster.py with default parameters, which requires at least 30 reads supporting each intron and allows intron to have a maximum size of 100 kb. We used the ENCODE ATAC-seq pipeline to perform alignment and peak calling (see Web Resources).29 FASTQ files were trimmed with Cutadapt v.1.930 and aligned with Bowtie2 v.2.2.6.31 MACS2 v.2.0.832 was used to call peaks with default parameters. Irreproducible Discovery Rate (IDR)33 analyses were performed based on pseudo-replicates (subsample of reads) with a cutoff of 0.1 to output an IDR call set, which was used for downstream analysis. We used WASP25 to filter out reads that are prone to mapping bias.
Mapping of cis-Acting Quantitative Trait Loci (QTL)
Prior to QTL mapping, we inferred ancestry principal components (PCs) using the R package SNPRelate34 on a pruned SNP set (Figure S4). We filtered out SNPs based on Hardy-Weinberg equilibrium (HWE < 1 × 10−6), LD (r2 < 0.2), and minor allele frequency (MAF < 0.05).34 To correct for hidden confounders, we extracted 15 covariates using PEER35 on quantile normalized and rank-based inverse normal transformed RPKM values. The number of hidden confounders to be removed was determined by empirically maximizing the power to discover eQTLs on chromosome 20 (for computational speed and to avoid overfitting). We tested combinations of 3 to 5 genotype principal components with 1 to 15 PEER factors. We found that the combination of 4 genotype PCs with 8 PEER factors provides the most power to detect eQTLs. We then used sex, the top four genotype principal components, and the top eight PEER factors in both FastQTL and RASQUAL to map cis-eQTL with a 2 Mb window centered at transcription start sites. Mathematically, the model is the following:
where e stands for gene expression and g stands for the genotype of the test SNP. We used LeafCutter28 to quantify intron excision levels and FastQTL36 to map cis-sQTLs within a 200 kbp window around splice donor sites, controlling for sex, genotype PCs, and splicing PCs. Using a similar approach, we found that 3 genotype PCs and 6 splicing PCs maximized the power to map sQTLs. To control for multiple hypothesis testing, we calculated per-gene eQTL p values using FastQTL with permutation, and controlled transcriptome-wide false discovery rate with the q-value package.37 For RASQUAL, it was not computationally feasible to perform gene-level permutation testing. Instead, we used TreeQTL to simultaneously control for SNP-level FDR and gene-level FDR.38 Note that TreeQTL is more conservative than permutation.
Quantifying Tissue- and Cell Type-Specific Contribution to Coronary Artery Disease (CAD) Risk
We used stratified LD score regression39 to estimate the enrichment of heritability for SNPs around tissue- and cell type-specific genes as described previously.40 We defined tissue-specific genes by first selecting for independent tissues and removing tissues primarily composed of smooth muscle to avoid correlation with HCASMCs (see Supplemental Material and Methods). After filtering, 16 tissues remained: HCASMCs, adipose - subcutaneous, adrenal gland, artery - coronary, brain - caudate (basal ganglia), cells - EBV -transformed lymphocytes, cells - transformed fibroblasts, liver, lung, minor salivary gland, muscle - skeletal, pancreas, pituitary, skin - not sun exposed (suprapubic), testis, and whole blood. We defined tissue-specific genes using gene expression z-score. For each gene, we determined the mean and standard deviation of median RPKM across tissues, from which the z-score is derived:
where et is the RPKM across all individuals in tissue t. We ranked each gene based on the z-scores (a higher z-score indicates more tissue specificity) and defined tissue-specific genes as the top 1,000, 2,000, and 4,000 genes. A given SNP was assigned to a gene if it fell into the union of exon ± 1 kbp of that gene. We estimated the heritability enrichment using stratified LD score regression on a joint SNP annotation across all 16 tissues against the CARDIoGRAMplusC4D GWAS meta-analysis.41 To determine whether CAD risk variants are enriched in the open chromatin regions tissue- and cell type-specific fashion, we used a modified version of GREGOR42 to estimate the likelihood of observing given number of GWAS variants falling into open chromatin regions of each tissue and cell type (see Supplemental Material and Methods). We first defined a GWAS locus as all variants in LD (r2 > 0.7) with the lead variant. Given a set of GWAS loci, we selected 500 background variants matched by (1) number of variants in LD, (2) distance to the nearest gene, (3) minor allele frequency, and (4) gene density in a 1 Mb window. We calculated p values and odds ratios between GWAS variants and background variants across HCASMCs and all ENCODE tissues and primary cell lines.
Colocalization between Molecular QTL and CAD GWASs
We used summary-data-based Mendelian Randomization (SMR)43 to determine GWAS loci that can be explained by cis-acting QTLs. We performed colocalization tests for 3,379 genes with cis-eQTL p value < 5 × 10−5 for the top variant and 2,439 splicing events with cis-sQTL p value < 5 × 10−5 for the top variant in HCASMCs against the latest CARDIoGRAMplusC4D and UK Biobank GWAS meta-analysis.6 We identified genome-wide significant eQTL and sQTL colocalizations based on adjusted SMR p values (Benjamini-Hochberg FDR < 0.05). The equivalent p value was 2.96 × 10−5 and 2.05 × 10−5 for eQTL and sQTL, respectively. SMR uses a reference population to determine linkage between variants; we used genetic data from individuals of European ancestry from 1000 Genomes as the reference population in our analyses. We also used a modified version of eCAVIAR44 to identify colocalized signals (see Supplemental Material and Methods). We calculated colocalization posterior probability (CLPP) using all SNPs within 500 kb of the lead eQTL SNP for all eGenes (FDR < 0.05) against CAD summary statistics from CARDIoGRAMplusC4D and UK Biobank GWAS meta-analysis.6 For computational feasibility, the GWAS and eQTL loci were assumed to have exactly one causal SNP. We defined colocalization events using CLPP > 0.05. Note that this is more conservative than the default eCAVIAR cutoff (CLPP > 0.01). We determined the direction of effect, namely whether gene upregulation increases risk, using the correlation of effect sizes in the GWAS and the eQTL studies. We selected SNPs with p value < 1 × 10−3 in both the GWAS and eQTL datasets (since other SNPs carry mostly noise) and fitted a regression using the GWAS and eQTL effect sizes as the predictor and the response, respectively. We defined the direction of effect as the sign of the regression slope.
Results
HCASMC-Specific Genomic Architecture
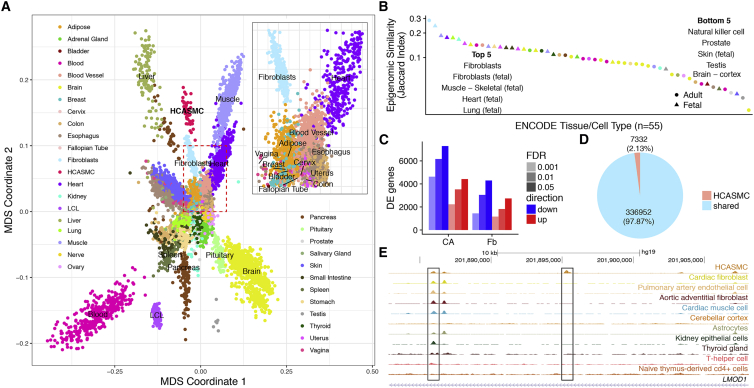
We obtained and cultured 62 primary HCASMC lines, and 52 lines remained for analysis after stringent quality control (Supplemental Material and Methodsand Table S1). We performed whole-genome sequencing to an average depth of 30× and jointly called genotypes using the GATK best practices pipeline,21 producing a total of ∼15.2 million variants after quality control (see Material and Methods). For RNA, we performed 125 bp paired-end sequencing to a median depth of 51.3 million reads, with more than 2.7 billion reads in total. After quantification and quality control, 19,607 genes were expressed in sufficient depth for downstream analysis (Table 1). To confirm that HCASMCs derived from tissue culture reflect in vivo physiology, we first projected their transcriptomes onto the 53 tissues profiled in GTEx19 (Figure 1A). Using multi-dimensional scaling (MDS) to visualize the similarity of HCASMCs to GTEx tissues, we observed that HCASMCs form a distinct cluster and closely neighbors fibroblasts, skeletal muscle, arteries, heart, and various smooth-muscle-enriched tissues (vagina, colon, stomach, uterus, and esophagus). These results were expected given that HCASMCs are predicted to be similar to skeletal muscle, smooth muscle-enriched tissues, as well as tissues representing the same anatomical compartment (e.g., heart and artery).45 In addition, HCASMCs resemble fibroblasts as both can be differentiated from mesenchymal cells from the dorsal mesocardium.46 We also computed the epigenetic similarity between HCASMCs and ENCODE cell types.47 Consistent with the transcriptomic findings, the closest neighbors to HCASMCs using epigenomic data were fibroblasts, heart, lung, and skeletal muscle (Figure 1B).
Table 1.
Molecular Quantitative Trait Loci Discoveries
| Molecular Phenotype | Trait Type | # of Traits Tested |
# of Traits with at Least One QTL |
||
|---|---|---|---|---|---|
| FDR = 0.05 | FDR = 0.01 | FDR = 0.001 | |||
| Gene expression | protein coding | 13,624 | 1,048 (7.69%) | 841 (6.17%) | 636 (4.67%) |
| lincRNA | 1,266 | 51 (4.03%) | 41 (3.24%) | 33 (2.61%) | |
| pseudogene | 2,616 | 50 (1.91%) | 34 (1.3%) | 25 (0.96%) | |
| other | 2,101 | 71 (3.38%) | 56 (2.67%) | 44 (2.09%) | |
| Total | 19,607 | 1,220 (6.22%) | 972 (4.96%) | 738 (3.76%) | |
| Splicing | protein coding | 24,461 | 519 (2.12%) | 349 (1.43%) | 245 (1%) |
| lincRNA | 300 | 11 (3.67%) | 7 (2.33%) | 5 (1.67%) | |
| pseudogene | 376 | 22 (5.85%) | 15 (3.99%) | 12 (3.19%) | |
| other | 541 | 29 (5.36%) | 19 (3.51%) | 17 (3.14%) | |
| Total | 25,678 | 581 (2.96%) | 390 (1.99%) | 279 (1.42%) | |
We report the number of tests performed and the number of significant loci at FDR < 0.05, 0.01, and 0.001 for eQTL and sQTL stratified by molecular trait type. We used permutation and the Benjamini-Hochberg adjustment for sQTL discovery, and a multi-level FDR correction procedure (TreeQTL38) for eQTL discovery, where permutation was not computationally feasible (see Material and Methods).
Figure 1.
The Relationship between HCASMCs and GTEx and ENCODE Cell and Tissue Types
(A) The multidimensional scaling plot of gene expression shows that HCASMCs form a distinct cluster, which neighbors fibroblast, skeletal muscle, heart, blood vessel, and various types of smooth muscle tissues such as esophagus and vagina (inset).
(B) Jaccard similarity index between HCASMCs and ENCODE cell and tissue types reveals that fibroblast, skeletal muscle, heart, and lung are most closely related to HCASMCs.
(C) Thousands of genes are differentially expressed between HCASMCs and its close neighbors, fibroblast, as well as the tissue of origin, coronary artery.
(D) A total of 344,284 open chromatin peaks are found in HCASMCs, of which 7,332 (2.1%) are HCASMC specific.
(E) An example of a HCASMC-specific peak located within the intron of LMOD1, which is an HCASMC-specific gene.
Next, we determined the pathways that may be selectively upregulated in HCASMCs compared to closely related tissues. We performed differential expression analysis of HCASMCs against fibroblasts and coronary artery in GTEx after correcting for batch effects and other hidden confounders (see Supplemental Material and Methods). Overall, 2,610 and 6,864 genes were found to be differentially expressed, respectively (FDR < 1 × 10−3, Figures 1C and S1), affecting pathways involved in cellular proliferation, epithelial-mesenchymal transition (EMT), and extracellular matrix (ECM) secretion (Table S2). Additionally, we determined the cellular content in human coronary artery48 and found that smooth muscle cells are the most abundant, followed by endothelial cells (Figure S16). Next, we sought to identify HCASMC-specific epigenomic signatures by comparing HCASMC open chromatin profiles, as determined with ATAC-seq, against DNaseI hypersensitivity (DHS) sites across all ENCODE primary cell types and tissues (Table S3). We processed HCASMC ATAC-seq data with the ENCODE pipeline and standardized peaks as 75 bp around the peak summit for all tissues and cell lines to mitigate batch effect (see Material and Methods). A total of 7,332 peaks (2.1%) were not previously identified in ENCODE and represent HCASMC-specific sites (Figure 1D). For example, an intronic peak within LMOD1 was found to be restricted to HCASMCs (Figure 1E). This gene is expressed primarily in vascular and visceral smooth muscle cells where it is involved in actin polymerization and has been mapped as a candidate causal CAD gene.11 We then sought to identify transcription factor binding sites overrepresented in HCASMC-specific peaks. Motif enrichment analyses indicated that HCASMC-specific open chromatin sites are enriched with binding sites for members of the forkhead box (FOX) transcription factor family (see Material and Methods). We performed motif enrichment analysis using 50-, 200-, and 1,000-bp regions flanking HCASMC-specific peaks and found that the enrichment was robust to selection of window size, indicating the result is not simply due to selection bias (Figure S2). The FOX transcription factors are known to regulate tissue- and cell type-specific gene transcription,49 and a subgroup of this family includes those with the ability to serve as pioneer factors.50 To validate that FOX motif enrichment is specific to HCASMCs, we performed similar analyses for brain-, heart-, and fibroblast-specific open chromatin regions and observed a depletion of FOX motifs (Figure S3). Together these results suggest that HCASMC-specific transcriptomic and epigenomic profiles identify regulatory mechanisms not previously established with large publicly available datasets.
Expression and Splicing Quantitative Trait Locus Discovery
In order to investigate the genetic regulatory mechanisms of gene expression in HCASMCs, we conducted genome-wide mapping of eQTLs using both FastQTL36 and RASQUAL27 on the 52 donor samples from diverse ethnic backgrounds (Table S1 and Figure S4). RASQUAL has been previously shown to increase the cis-eQTL discovery power in small sample sizes by leveraging allele-specific information.27 Indeed, using a threshold of FDR < 0.05, RASQUAL increased the number of eQTLs discovered approximately 7-fold as compared to FastQTL (RASQUAL:1220 versus FastQTL:167, Table 1). We next evaluated whether these eQTLs were enriched in regions of open chromatin using data from a subset of individuals with ATAC-seq profiles. We observed that eQTLs within HCASMC open chromatin regions had more significant p values compared to all eQTLs (Figure S5, two-sided rank-sum test p value < 9.2 × 10−5). This is consistent with putative effects of cis-acting variation, potentially functioning through altered TF binding around these accessible regions. Next, using a Bayesian meta-analytic approach,51 we sought to identify HCASMC-specific eQTLs using GTEx tissues as a reference. Under the most stringent criteria (eQTL posterior probability > 0.9 for HCASMCs and < 0.1 for all GTEx tissues, see Material and Methods), we identified four HCASMC-specific eQTLs (Figure S6). For example, rs1048709 is the top eQTL-SNP and confers HCASMC-specific regulatory effects on Complement Factor B (Figure S6B), a gene that has been previously implicated in atherosclerosis and other inflammatory diseases.52 In addition to regulatory effects on gene expression, previous studies have identified splicing as a major source of regulatory impact of genetic variation on complex diseases.53 Therefore, we mapped splicing QTLs (sQTLs) using LeafCutter28 and identified 581 sQTLs associated at FDR < 0.05 (Table 1). As a quality control, we estimated the enrichment of sQTLs and eQTLs against a matched set of background variants. As expected, eQTLs were enriched around the 5′ UTR (Figure S7A), whereas sQTLs were enriched in splicing regions, particularly splice donor and acceptor sites (Figure S7B).
Overall CAD Genetic Risk Mediated by HCASMCs
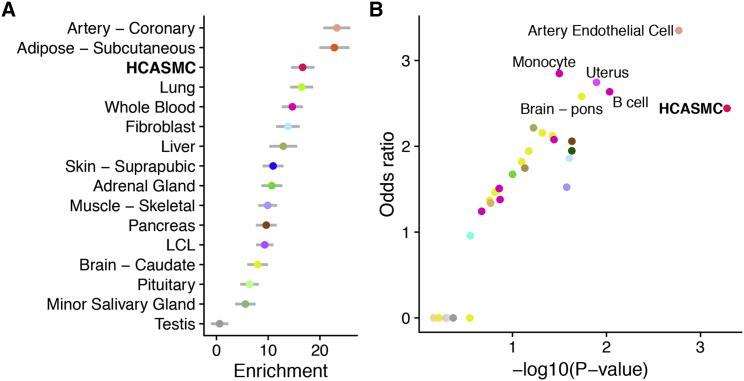
We next examined the heritable contribution of HCASMCs toward the risk of CAD. Previous reports have suggested that disease-associated SNPs are often enriched in genes expressed in the relevant tissue types.40 Thus, we estimated the contribution to CAD risk from SNPs in or near genes showing patterns of tissue-specific expression and identified the top 2,000 tissue-specific genes for HCASMCs and GTEx tissues (see Material and Methods). We then applied stratified LD score regression39 to estimate CAD heritability explained by SNPs within 1 kb of tissue-specific genes. We found that HCASMCs, along with coronary artery and adipose tissues, contribute substantially toward CAD heritability (Figure 2A). These enrichment results were robust to the tissue-specificity cutoff (top 1,000, 2,000, or 4,000 genes), suggesting that they were not simply due to selection bias (Figure S8). Complementary epigenomic evidence previously demonstrated that risk variants for complex diseases are often enriched in open chromatin regions in relevant tissue types.39, 42, 47 Thus, we estimated the degree of overlap between CAD variants and open chromatin in HCASMCs and ENCODE cell types using a modified version of GREGOR42 (see Material and Methods). We observed that open chromatin regions in HCASMCs, as well as vascular endothelial cells, monocytes, uterus (smooth muscle), and B cells, are enriched for CAD risk variants (Figure 2B). These findings support the role of HCASMCs as an appropriate cellular model to map the genetic basis of CAD, which may be supplemented by the contribution of other vessel wall cell types.
Figure 2.
Tissue- and Cell Type-Specific Contribution to CAD Risk
(A) Tissue-specific enrichment of CAD heritability. We used stratified LD score regression to estimate the CAD risk explained by SNPs close to tissue-specific genes, defined as the 2,000 genes with highest expression z-scores (see Material and Methods). Genes whose expression is specific to coronary artery, adipose, and HCASMCs harbor SNPs with large effects on CAD. Error bars indicate standard error of the enrichments.
(B) Overlap between CAD risk variants and tissue- and cell type-specific open chromatin regions. We used a modified version of GREGOR (see Material and Methods) to estimate the probability and odds ratio (compared with matched background SNPs) of overlap between CAD risk variants and open chromatin regions in HCASMCs and across ENCODE tissues. HCASMCs, arterial endothelial cells, monocytes, B cell, uterus (composed primarily of smooth muscle), and pons (possibly through regulation of blood pressure) showed the highest degrees of overlap.
Fine-Mapping CAD Risk Variants
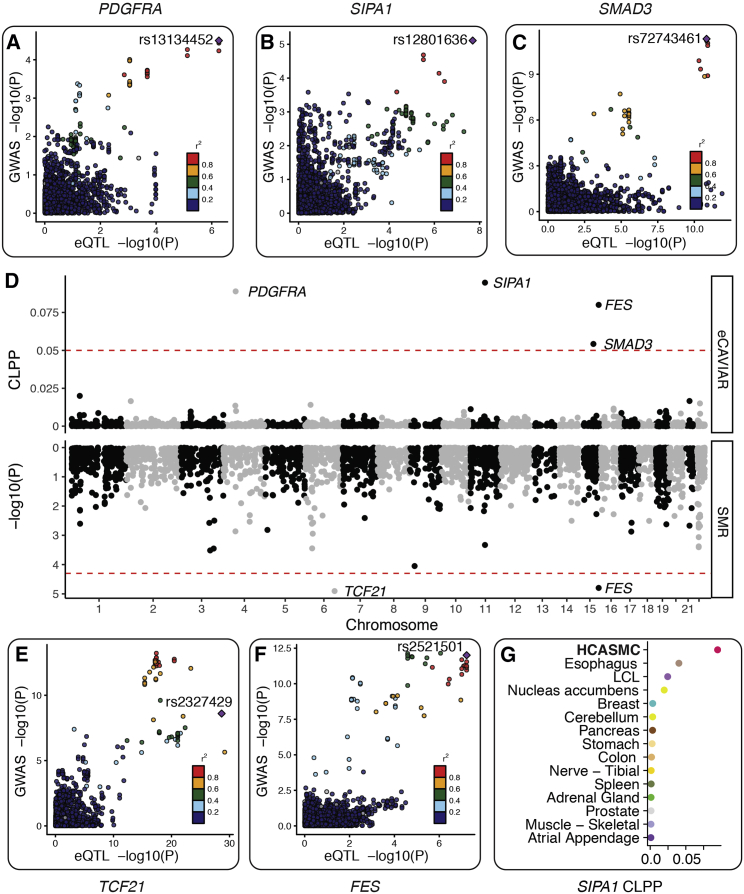
Whole-genome sequencing of our HCASMC population sample provides the opportunity to fine-map CAD risk loci. Several studies have used colocalization between GWAS and eQTL signals as a fine-mapping approach to identify candidate causal regulatory variants,43, 44, 54, 55 and in several cases pinpointing single causal variants.56, 57 Given the global overlap between CAD risk variants and genetic regulation in HCASMCs, we sought to find evidence for colocalization between GWAS and eQTL signals. We thus compiled publicly available genome-wide summary statistics from the latest meta-analysis.6 We then applied two methods with different statistical assumptions, eQTL and GWAS Causal Variants Identification in Associated Regions (eCAVIAR)44 and Summary-data-based Mendelian Randomization (SMR)43 to identify colocalizing variants and genes across all CAD loci, and we focused on the union of results from the two independent methods. We used FDR < 0.05 and colocalization posterior probability (CLPP) > 0.05 as cutoffs for SMR and eCAVIAR, respectively (note that CLPP > 0.05 is more conservative than the CLPP > 0.01 recommended in the publication of the eCAVIAR method). From this approach, we identified five genes that showed statistically significant colocalization, namely FES, SMAD3, TCF21, PDGFRA, and SIPA1 (Figure 3). Although the top genes found by two methods differed, we observed that the SMR p values and eCAVIAR CLPPs positively correlate (Figure S9) and that two of the three genes found by eCAVIAR achieved nominal significance in SMR (Table S4). We then investigated whether these colocalizations were restricted to HCASMCs by conducting colocalization tests across all GTEx tissues. For SIPA1 and PDGFRA, colocalization appears to be HCASMC-specific (Figures 3G, S10A, and S10D). For SMAD3, both HCASMCs and thyroid have strong colocalization signals (Figure S10B). TCF21 and FES colocalization were found to be shared across multiple tissues (Figures S10C and S11D). Next, we conducted colocalization analysis between sQTL and GWAS summary statistics with both eCAVIAR and SMR. We identified colocalization with four genes (Table S4 and Figure S12). The most significant colocalization event is at the SMG9 locus. Interestingly, the top sQTL variant, rs4760, is a coding variant located in the exon of the PLAUR (plasminogen activator urokinase receptor) gene and is also a GWAS variant for circulating cytokines and multiple immune cell traits.58, 59 However, experimental validation is required to confirm these candidate genes. By correlating eQTL and GWAS effect sizes, we observed that increased TCF21 and FES expression levels are associated with reduced CAD risk, while increased PDGFRA, SIPA1, and SMAD3 expression levels are associated with increased CAD risk (Figure S17). These results provide genetic evidence that pathways promoting SMC phenotypic transition during atherosclerosis can be both protective and detrimental depending on the genes implicated (Figure 4).
Figure 3.
Colocalization between HCASMC eQTL and Coronary Artery Disease GWASs
(A–C) Three candidate genes identified by eCAVIAR.
(A) Platelet-derived growth factor alpha (PDFGRA) eQTL signal colocalized with the KDR GWAS locus, which has p value < 3.16 × 10−5 (FDR < 0.05) in the latest CARDIoGRAMplusC4D and UK Biobank GWAS meta-analysis.6
(B) Signal-Induced Proliferation-Associated 1 (SIPA1) eQTL signal colocalized with the PCNX3 locus, which has p value < 7.75 × 10−6 in the UK Biobank meta-analysis, and reached genome-wide significance (p value < 9.71 × 10−9) in Howson et al.5 Note that the latter study has a larger sample size than the UK Biobank study.
(C) SMAD3 eQTL signal colocalized with the SMAD3 locus, which was identified in the UK Biobank meta-analysis.6
(D) Transcriptome-wide colocalization signals between HCASMC eQTL and CAD GWAS. We used eCAVIAR (top) and SMR (bottom) to fine-map GWAS causal variants and to identify eQTL signals that can explain CAD risk variants (see Material and Methods). We found five genes whose eQTL signals show significant colocalization with CAD GWAS signal (SMR FDR < 0.05 or eCAVIAR colocalization posterior probability > 0.05).
(E and F) Two candidate genes identified by SMR.
(E) Transcription factor 21 (TCF21) eQTL signal colocalized with the TCF21 locus, which was identified by Schunkert et al.74 and replicated in the UK Biobank meta-analysis.
(F) FES eQTL signal colocalized with the FURIN-FES locus, which was identified by Deloukas et al.75 and replicated in the UK Biobank meta-analysis.
(G) SIPA1 colocalization is strongest in HCASMCs, suggesting that this gene may influence CAD risk through this specialized cell type.
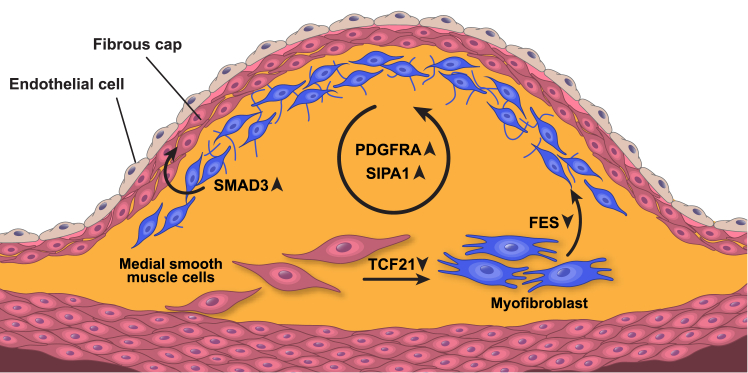
Figure 4.
Candidate Genes Are Involved in HCASMC-Related Vascular Remodeling
Hypothetical functions of five candidate genes. Upregulation of TCF21 facilitates the transition of smooth muscle cells from a contractile to a synthetic state.76 Upon phenotypic transition, FES assists in smooth muscle cell migration to the neo-intima.77 Both SIPA1 and PDGFRA promote HCASMC proliferation.67, 78SMAD3 induces synthetic smooth muscle re-differentiation into the synthetic phenotype for vessel wall repair.79 Upward arrows indicate genetic upregulation increases CAD risk, and downward arrows indicate genetic upregulation is protective against CAD risk.
Discussion
In this study, we have integrated genomic, transcriptomic, and epigenetic datasets to create the first map of genetic regulation of gene expression in human coronary artery smooth muscle cells. Comparison with publicly available transcriptomic and epigenomic datasets in GTEx and ENCODE revealed regulatory patterns specific to HCASMCs. By comparing against neighboring tissues in GTEx, we found thousands of differentially expressed genes, which were enriched in pathways such as EMT, protein secretion, and cellular proliferation, consistent with our current understanding of HCASMC physiology in vivo. In comparison with ENCODE, we found 7,332 (∼2.1%) specific open chromatin peaks in HCASMCs, and we showed that these peaks are enriched with binding motifs for Forkhead box family proteins, which are known to regulate cell-type-specific gene expression.60 FOXP1 in particular has been shown to increase collagen production in smooth muscle cells,61 supporting a potential role in extracellular matrix remodeling in the vessel wall.
Using both transcriptomic and epigenomic profiles, we established that HCASMCs represent an important cell type for coronary artery disease. On a tissue level, we demonstrated that genes highly expressed in HCASMCs, coronary artery and adipose tissue are enriched for SNPs associated with CAD risk. While the proximal aortic wall is also susceptible to atherosclerosis, the coronary arteries represent the primary origin of ischemic coronary artery disease in humans.9 Given that the majority of coronary arteries in the epicardium are encapsulated by perivascular adipose tissue in individuals with disease, one would expect these tissues to share gene responses involved in both vascular inflammation and lipid homeostasis.62 Further, we demonstrated that HCASMCs, endothelial cells, and immune cells also contribute toward the genetic risk of coronary artery disease. Recent -omic profiling of human aortic endothelial cells (HAECs) isolated from various donors identified a number of genetic variants and transcriptional networks mediating responses to oxidized phospholipids and pro-inflammatory stimuli.63 Likewise, systems approaches investigating resident macrophages and other immune cells involved in vessel inflammation have provided additional insights into context-specific disease mechanisms.64, 65
Our integrative analyses identified a number of CAD-associated genes that may offer clues into potentially targetable HCASMC-mediated disease mechanisms. Although two of these associated genes, TCF21 and SMAD3, have established roles in regulating vascular remodeling and inflammation during disease,12, 16, 66 the other identified genes, PDGFRA, FES, and SIPA1, appear to also be SMC-associated genes. While the role for PDGFRB-mediated signaling has been well documented in atherosclerosis and modulation of SMC phenotype, the possible involvement of PDGFRA has not been investigated in detail.67, 68 It is worth noting that the GWAS signal for PDGFRA reached FDR < 0.05 and not genome-wide significance. In the latest meta-analysis using an interim release of UKBB data,6 12 of the 13 loci identified at genome-wide significance were on the previous list of loci meeting the FDR < 0.05 threshold, and the study argued that most remaining loci at the FDR < 0.05 threshold likely represent genuine signals. Similarly, we chose to include PDGFRA on the reasonable expectation that it may become genome-wide significant in the next release of GWAS integrating full UKBB data. Interestingly, FES and SIPA1 were found to harbor CpGs identified in current smokers in the Rotterdam Study, based on targeted methylation profiling of CAD loci in whole blood.69 The two identified CpGs in FES were located near the transcription start site, while the one CpG identified in SIPA1 was located in the 5′ UTR, suggesting potential environmental influences on gene expression levels. SIPA1 encodes a mitogen-induced GTPase activating protein (GAP), specifically activating Ras and Rap GTPases.70 SIPA1 may be a specific mitogen response signal in HCASMCs undergoing phenotypic transition in the injured vessel wall; however, these hypotheses should be explored in relevant functional models. Another HCASMC eQTL variant, rs2327429, located in the TCF21 promoter region, was also the lead SNP in this locus in a recent CAD meta-analysis and has been identified as an mQTL for TCF21 expression in two separate studies.71, 72 These data suggest that regulation of methylation is a molecular trait that may mediate risk for CAD. Splicing QTL colocalization analysis reveals that alternative splicing in SMG9 also influences CAD risk. SMG9 has been shown to regulate the nonsense-mediated decay (NMD) pathway in human cells and has been implicated in several developmental disorders such as brain malformations and congenital heart disease.73 It is worth noting that TCF21, which was the top hit for SMR, received low CLPP from eCAVIAR. This is because SMR uses the top eQTL SNP as the instrumental variable. In this case, the SNP rs2327429 is genome-wide significant for both eQTL and GWAS (eQTL p value < 2.3 × 10−29 and GWAS p value < 2.5 × 10−09), and thus SMR returned a significant causal probability. On the other hand, eCAVIAR first assigns causal posterior probability independently for GWAS and eQTL. Because the GWAS and eQTL does not share a lead variant (rs2327429 for eQTL and rs12202017 for GWAS) for TCF21, eCAVIAR assigns high posterior to rs2327429 and low posterior to rs12202017 in eQTL and vice versa in GWAS. As a result, the product of the causal posterior probability (i.e., colocalization posterior probability, CLPP) was low. Due to these differences, we argue that a systematic comparison across colocalization methods is needed in the future. In addition, our power to detect causal genes is limited by the modest sample size, and an increase in the number of sample will aid in identifying weaker eQTLs and colocalization events.
In summary, the current study confirms the value of detailed genomic and genetic analyses of disease-related tissues and cell types, which when analyzed in the context of publicly available data can provide deep insights into the physiology of human traits and pathophysiology of complex human disease. We expect that these findings will provide a rich resource for the community and prompt detailed functional investigations of candidate loci for preclinical development.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank Professor Nicolas Mermod for providing biological material and Normal Cyr for making illustrations. B.L. is supported in part by the Stanford Center for Computational, Evolutionary and Human Genomics Fellowship. T.Q. is supported by NIH grants R01HL109512, R01HL134817, R33HL120757, R01DK107437, and R01HL139478. C.L.M. is supported by R00HL125912 (NIH). S.B.M. is supported by R33HL120757 (NHLBI), U01HG009431 (NHGRI; ENCODE4), R01MH101814 (NIH Common Fund; GTEx Program), R01HG008150 (NHGRI; Non-Coding Variants Program), and the Edward Mallinckrodt, Jr. Foundation.
Published: August 23, 2018
Footnotes
Supplemental Data include 17 figures, 4 tables, and Supplemental Material and Methods and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.08.001.
Accession Numbers
RNA sequencing data has been deposited at Gene Expression Omnibus (GEO), accession number GSE113348. All eQTL and sQTL summary statistics are accessible through the Montgomery lab website (see Web Resources). All code used to perform analyses and generate figures are in the GitHub repository (hcasmc_eqtl in Web Resources).
Web Resources
1000 Genomes, http://www.internationalgenome.org/
ATAC-seq and DNase-seq processing pipeline, https://github.com/kundajelab/atac_dnase_pipelines
BEAGLE, http://faculty.washington.edu/browning/beagle/beagle.html
Bowtie2, http://bowtie-bio.sourceforge.net/bowtie2/index.shtml
DESeq2, https://bioconductor.org/packages/release/bioc/html/DESeq2.html
ENCODE, https://www.encodeproject.org/
ENCODE ATAC-seq/DNase-seq pipeline, https://github.com/kundajelab/atac_dnase_pipelines
FastQC, https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
FastQTL, http://fastqtl.sourceforge.net/
FINEMAP, http://www.christianbenner.com
Gencode v.17, https://www.gencodegenes.org
hcasmc_eqtl, https://github.com/boxiangliu/hcasmc_eqtl
JASPAR, http://jaspar.genereg.net/
LD score regression, https://github.com/bulik/ldsc
LeafCutter, https://github.com/davidaknowles/leafcutter
METASOFT, http://genetics.cs.ucla.edu/meta/
Montgomery lab, http://montgomerylab.stanford.edu/resources.html
NOISeq, https://bioconductor.org/packages/release/bioc/html/NOISeq.html
PLINK 1.9, https://www.cog-genomics.org/plink2/
RASQUAL, https://github.com/natsuhiko/rasqual
RNA-SeQC, https://software.broadinstitute.org/cancer/cga/rna-seqc
sva, https://bioconductor.org/packages/release/bioc/html/sva.html
VerifyBamID, https://genome.sph.umich.edu/wiki/VerifyBamID
Supplemental Data
References
- 1.Benjamin E.J., Blaha M.J., Chiuve S.E., Cushman M., Das S.R., Deo R., de Ferranti S.D., Floyd J., Fornage M., Gillespie C., American Heart Association Statistics Committee and Stroke Statistics Subcommittee Heart disease and stroke statistics-2017 update: a report from the American Heart Association. Circulation. 2017;135:e146–e603. doi: 10.1161/CIR.0000000000000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khera A.V., Emdin C.A., Drake I., Natarajan P., Bick A.G., Cook N.R., Chasman D.I., Baber U., Mehran R., Rader D.J. Genetic risk, adherence to a healthy lifestyle, and coronary disease. N. Engl. J. Med. 2016;375:2349–2358. doi: 10.1056/NEJMoa1605086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Won H.-H., Natarajan P., Dobbyn A., Jordan D.M., Roussos P., Lage K., Raychaudhuri S., Stahl E., Do R. Disproportionate contributions of select genomic compartments and cell types to genetic risk for coronary artery disease. PLoS Genet. 2015;11:e1005622. doi: 10.1371/journal.pgen.1005622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zdravkovic S., Wienke A., Pedersen N.L., Marenberg M.E., Yashin A.I., De Faire U. Heritability of death from coronary heart disease: a 36-year follow-up of 20 966 Swedish twins. J. Intern. Med. 2002;252:247–254. doi: 10.1046/j.1365-2796.2002.01029.x. [DOI] [PubMed] [Google Scholar]
- 5.Howson J.M.M., Zhao W., Barnes D.R., Ho W.-K., Young R., Paul D.S., Waite L.L., Freitag D.F., Fauman E.B., Salfati E.L., CARDIoGRAMplusC4D. EPIC-CVD Fifteen new risk loci for coronary artery disease highlight arterial-wall-specific mechanisms. Nat. Genet. 2017;49:1113–1119. doi: 10.1038/ng.3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson C.P., Goel A., Butterworth A.S., Kanoni S., Webb T.R., Marouli E., Zeng L., Ntalla I., Lai F.Y., Hopewell J.C., EPIC-CVD Consortium. CARDIoGRAMplusC4D. UK Biobank CardioMetabolic Consortium CHD working group Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat. Genet. 2017;49:1385–1391. doi: 10.1038/ng.3913. [DOI] [PubMed] [Google Scholar]
- 7.Klarin D., Zhu Q.M., Emdin C.A., Chaffin M., Horner S., McMillan B.J., Leed A., Weale M.E., Spencer C.C.A., Aguet F., CARDIoGRAMplusC4D Consortium Genetic analysis in UK Biobank links insulin resistance and transendothelial migration pathways to coronary artery disease. Nat. Genet. 2017;49:1392–1397. doi: 10.1038/ng.3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Harst P., Verweij N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ. Res. 2018;122:433–443. doi: 10.1161/CIRCRESAHA.117.312086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khera A.V., Kathiresan S. Genetics of coronary artery disease: discovery, biology and clinical translation. Nat. Rev. Genet. 2017;18:331–344. doi: 10.1038/nrg.2016.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pu X., Xiao Q., Kiechl S., Chan K., Ng F.L., Gor S., Poston R.N., Fang C., Patel A., Senver E.C. ADAMTS7 cleavage and vascular smooth muscle cell migration is affected by a coronary-artery-disease-associated variant. Am. J. Hum. Genet. 2013;92:366–374. doi: 10.1016/j.ajhg.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller C.L., Pjanic M., Wang T., Nguyen T., Cohain A., Lee J.D., Perisic L., Hedin U., Kundu R.K., Majmudar D. Integrative functional genomics identifies regulatory mechanisms at coronary artery disease loci. Nat. Commun. 2016;7:12092. doi: 10.1038/ncomms12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braitsch C.M., Combs M.D., Quaggin S.E., Yutzey K.E. Pod1/Tcf21 is regulated by retinoic acid signaling and inhibits differentiation of epicardium-derived cells into smooth muscle in the developing heart. Dev. Biol. 2012;368:345–357. doi: 10.1016/j.ydbio.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shankman L.S., Gomez D., Cherepanova O.A., Salmon M., Alencar G.F., Haskins R.M., Swiatlowska P., Newman A.A.C., Greene E.S., Straub A.C. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat. Med. 2015;21:628–637. doi: 10.1038/nm.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cherepanova O.A., Gomez D., Shankman L.S., Swiatlowska P., Williams J., Sarmento O.F., Alencar G.F., Hess D.L., Bevard M.H., Greene E.S. Activation of the pluripotency factor OCT4 in smooth muscle cells is atheroprotective. Nat. Med. 2016;22:657–665. doi: 10.1038/nm.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nurnberg S.T., Cheng K., Raiesdana A., Kundu R., Miller C.L., Kim J.B., Arora K., Carcamo-Oribe I., Xiong Y., Tellakula N. Coronary artery disease associated transcription factor TCF21 regulates smooth muscle precursor cells that contribute to the fibrous cap. PLoS Genet. 2015;11:e1005155. doi: 10.1371/journal.pgen.1005155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller C.L., Haas U., Diaz R., Leeper N.J., Kundu R.K., Patlolla B., Assimes T.L., Kaiser F.J., Perisic L., Hedin U. Coronary heart disease-associated variation in TCF21 disrupts a miR-224 binding site and miRNA-mediated regulation. PLoS Genet. 2014;10:e1004263. doi: 10.1371/journal.pgen.1004263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srivastava R., Zhang J., Go G.-W., Narayanan A., Nottoli T.P., Mani A. Impaired LRP6-TCF7L2 activity enhances smooth muscle cell plasticity and causes coronary artery disease. Cell Rep. 2015;13:746–759. doi: 10.1016/j.celrep.2015.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J.B., Pjanic M., Nguyen T., Miller C.L., Iyer D., Liu B., Wang T., Sazonova O., Carcamo-Orive I., Matic L.P. TCF21 and the environmental sensor aryl-hydrocarbon receptor cooperate to activate a pro-inflammatory gene expression program in coronary artery smooth muscle cells. PLoS Genet. 2017;13:e1006750. doi: 10.1371/journal.pgen.1006750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Battle A., Brown C.D., Engelhardt B.E., Montgomery S.B., GTEx Consortium. Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group. Statistical Methods groups—Analysis Working Group. Enhancing GTEx (eGTEx) groups. NIH Common Fund. NIH/NCI. NIH/NHGRI. NIH/NIMH. NIH/NIDA. Biospecimen Collection Source Site—NDRI. Biospecimen Collection Source Site—RPCI. Biospecimen Core Resource—VARI. Brain Bank Repository—University of Miami Brain Endowment Bank. Leidos Biomedical—Project Management. ELSI Study. Genome Browser Data Integration &Visualization—EBI. Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz. Lead analysts. Laboratory, Data Analysis &Coordinating Center (LDACC) NIH program management. Biospecimen collection. Pathology. eQTL manuscript working group Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. [Google Scholar]
- 20.Buenrostro J.D., Wu B., Chang H.Y., Greenleaf W.J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 2015;109:1–9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2002. From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Browning B.L., Yu Z. Simultaneous genotype calling and haplotype phasing improves genotype accuracy and reduces false-positive associations for genome-wide association studies. Am. J. Hum. Genet. 2009;85:847–861. doi: 10.1016/j.ajhg.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van de Geijn B., McVicker G., Gilad Y., Pritchard J.K. WASP: allele-specific software for robust molecular quantitative trait locus discovery. Nat. Methods. 2015;12:1061–1063. doi: 10.1038/nmeth.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeLuca D.S., Levin J.Z., Sivachenko A., Fennell T., Nazaire M.-D., Williams C., Reich M., Winckler W., Getz G. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28:1530–1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumasaka N., Knights A.J., Gaffney D.J. Fine-mapping cellular QTLs with RASQUAL and ATAC-seq. Nat. Genet. 2016;48:206–213. doi: 10.1038/ng.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y.I., Knowles D.A., Humphrey J., Barbeira A.N., Dickinson S.P., Im H.K., Pritchard J.K. Annotation-free quantification of RNA splicing using LeafCutter. Nat. Genet. 2018;50:151–158. doi: 10.1038/s41588-017-0004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sloan C.A., Chan E.T., Davidson J.M., Malladi V.S., Strattan J.S., Hitz B.C., Gabdank I., Narayanan A.K., Ho M., Lee B.T. ENCODE data at the ENCODE portal. Nucleic Acids Res. 2016;44(D1):D726–D732. doi: 10.1093/nar/gkv1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal. 2011;17:10–12. [Google Scholar]
- 31.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Q., Brown J.B., Huang H., Bickel P.J. Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat. 2011;5:1752–1779. [Google Scholar]
- 34.Zheng X., Levine D., Shen J., Gogarten S.M., Laurie C., Weir B.S. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28:3326–3328. doi: 10.1093/bioinformatics/bts606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stegle O., Parts L., Piipari M., Winn J., Durbin R. Using probabilistic estimation of expression residuals (PEER) to obtain increased power and interpretability of gene expression analyses. Nat. Protoc. 2012;7:500–507. doi: 10.1038/nprot.2011.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ongen H., Buil A., Brown A.A., Dermitzakis E.T., Delaneau O. Fast and efficient QTL mapper for thousands of molecular phenotypes. Bioinformatics. 2016;32:1479–1485. doi: 10.1093/bioinformatics/btv722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peterson C.B., Bogomolov M., Benjamini Y., Sabatti C. TreeQTL: hierarchical error control for eQTL findings. Bioinformatics. 2016;32:2556–2558. doi: 10.1093/bioinformatics/btw198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finucane H.K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P.-R., Anttila V., Xu H., Zang C., Farh K., ReproGen Consortium. Schizophrenia Working Group of the Psychiatric Genomics Consortium. RACI Consortium Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boyle E.A., Li Y.I., Pritchard J.K. An expanded view of complex traits: from polygenic to omnigenic. Cell. 2017;169:1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nikpay M., Goel A., Won H.-H., Hall L.M., Willenborg C., Kanoni S., Saleheen D., Kyriakou T., Nelson C.P., Hopewell J.C. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt E.M., Zhang J., Zhou W., Chen J., Mohlke K.L., Chen Y.E., Willer C.J. GREGOR: evaluating global enrichment of trait-associated variants in epigenomic features using a systematic, data-driven approach. Bioinformatics. 2015;31:2601–2606. doi: 10.1093/bioinformatics/btv201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu Z., Zhang F., Hu H., Bakshi A., Robinson M.R., Powell J.E., Montgomery G.W., Goddard M.E., Wray N.R., Visscher P.M., Yang J. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 44.Hormozdiari F., van de Bunt M., Segrè A.V., Li X., Joo J.W.J., Bilow M., Sul J.H., Sankararaman S., Pasaniuc B., Eskin E. Colocalization of GWAS and eQTL signals detects target genes. Am. J. Hum. Genet. 2016;99:1245–1260. doi: 10.1016/j.ajhg.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang G., Jacquet L., Karamariti E., Xu Q. Origin and differentiation of vascular smooth muscle cells. J. Physiol. 2015;593:3013–3030. doi: 10.1113/JP270033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vrancken Peeters M.P., Gittenberger-de Groot A.C., Mentink M.M., Poelmann R.E. Smooth muscle cells and fibroblasts of the coronary arteries derive from epithelial-mesenchymal transformation of the epicardium. Anat. Embryol. (Berl.) 1999;199:367–378. doi: 10.1007/s004290050235. [DOI] [PubMed] [Google Scholar]
- 47.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aran D., Hu Z., Butte A.J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li S., Weidenfeld J., Morrisey E.E. Transcriptional and DNA binding activity of the Foxp1/2/4 family is modulated by heterotypic and homotypic protein interactions. Mol. Cell. Biol. 2004;24:809–822. doi: 10.1128/MCB.24.2.809-822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iwafuchi-Doi M., Donahue G., Kakumanu A., Watts J.A., Mahony S., Pugh B.F., Lee D., Kaestner K.H., Zaret K.S. The pioneer transcription factor FoxA maintains an accessible nucleosome configuration at enhancers for tissue-specific gene activation. Mol. Cell. 2016;62:79–91. doi: 10.1016/j.molcel.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han B., Eskin E. Interpreting meta-analyses of genome-wide association studies. PLoS Genet. 2012;8:e1002555. doi: 10.1371/journal.pgen.1002555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hovland A., Jonasson L., Garred P., Yndestad A., Aukrust P., Lappegård K.T., Espevik T., Mollnes T.E. The complement system and toll-like receptors as integrated players in the pathophysiology of atherosclerosis. Atherosclerosis. 2015;241:480–494. doi: 10.1016/j.atherosclerosis.2015.05.038. [DOI] [PubMed] [Google Scholar]
- 53.Li Y.I., van de Geijn B., Raj A., Knowles D.A., Petti A.A., Golan D., Gilad Y., Pritchard J.K. RNA splicing is a primary link between genetic variation and disease. Science. 2016;352:600–604. doi: 10.1126/science.aad9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., Plagnol V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nica A.C., Montgomery S.B., Dimas A.S., Stranger B.E., Beazley C., Barroso I., Dermitzakis E.T. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 2010;6:e1000895. doi: 10.1371/journal.pgen.1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Claussnitzer M., Dankel S.N., Kim K.-H., Quon G., Meuleman W., Haugen C., Glunk V., Sousa I.S., Beaudry J.L., Puviindran V. FTO obesity variant circuitry and adipocyte browning in humans. N. Engl. J. Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Musunuru K., Strong A., Frank-Kamenetsky M., Lee N.E., Ahfeldt T., Sachs K.V., Li X., Li H., Kuperwasser N., Ruda V.M. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ahola-Olli A.V., Würtz P., Havulinna A.S., Aalto K., Pitkänen N., Lehtimäki T., Kähönen M., Lyytikäinen L.-P., Raitoharju E., Seppälä I. Genome-wide association study identifies 27 loci influencing concentrations of circulating cytokines and growth factors. Am. J. Hum. Genet. 2017;100:40–50. doi: 10.1016/j.ajhg.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Astle W.J., Elding H., Jiang T., Allen D., Ruklisa D., Mann A.L., Mead D., Bouman H., Riveros-Mckay F., Kostadima M.A. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. 2016;167:1415–1429.e19. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Golson M.L., Kaestner K.H. Fox transcription factors: from development to disease. Development. 2016;143:4558–4570. doi: 10.1242/dev.112672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bot P.T., Grundmann S., Goumans M.J., de Kleijn D., Moll F., de Boer O., van der Wal A.C., van Soest A., de Vries J.P., van Royen N. Forkhead box protein P1 as a downstream target of transforming growth factor-β induces collagen synthesis and correlates with a more stable plaque phenotype. Atherosclerosis. 2011;218:33–43. doi: 10.1016/j.atherosclerosis.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 62.Berg A.H., Scherer P.E. Adipose tissue, inflammation, and cardiovascular disease. Circ. Res. 2005;96:939–949. doi: 10.1161/01.RES.0000163635.62927.34. [DOI] [PubMed] [Google Scholar]
- 63.Hogan N.T., Whalen M.B., Stolze L.K., Hadeli N.K., Lam M.T., Springstead J.R., Glass C.K., Romanoski C.E. Transcriptional networks specifying homeostatic and inflammatory programs of gene expression in human aortic endothelial cells. eLife. 2017;6:e22536. doi: 10.7554/eLife.22536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ghattas A., Griffiths H.R., Devitt A., Lip G.Y.H., Shantsila E. Monocytes in coronary artery disease and atherosclerosis: where are we now? J. Am. Coll. Cardiol. 2013;62:1541–1551. doi: 10.1016/j.jacc.2013.07.043. [DOI] [PubMed] [Google Scholar]
- 65.Kinlay S., Libby P., Ganz P. Endothelial function and coronary artery disease. Curr. Opin. Lipidol. 2001;12:383–389. doi: 10.1097/00041433-200108000-00003. [DOI] [PubMed] [Google Scholar]
- 66.Turner A.W., Martinuk A., Silva A., Lau P., Nikpay M., Eriksson P., Folkersen L., Perisic L., Hedin U., Soubeyrand S., McPherson R. Functional analysis of a novel genome-wide association study signal in SMAD3 that confers protection from coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 2016;36:972–983. doi: 10.1161/ATVBAHA.116.307294. [DOI] [PubMed] [Google Scholar]
- 67.Andrae J., Gallini R., Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008;22:1276–1312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.He C., Medley S.C., Hu T., Hinsdale M.E., Lupu F., Virmani R., Olson L.E. PDGFRβ signalling regulates local inflammation and synergizes with hypercholesterolaemia to promote atherosclerosis. Nat. Commun. 2015;6:7770. doi: 10.1038/ncomms8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Steenaard R.V., Ligthart S., Stolk L., Peters M.J., van Meurs J.B., Uitterlinden A.G., Hofman A., Franco O.H., Dehghan A. Tobacco smoking is associated with methylation of genes related to coronary artery disease. Clin. Epigenetics. 2015;7:54. doi: 10.1186/s13148-015-0088-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kurachi H., Wada Y., Tsukamoto N., Maeda M., Kubota H., Hattori M., Iwai K., Minato N. Human SPA-1 gene product selectively expressed in lymphoid tissues is a specific GTPase-activating protein for Rap1 and Rap2. Segregate expression profiles from a rap1GAP gene product. J. Biol. Chem. 1997;272:28081–28088. doi: 10.1074/jbc.272.44.28081. [DOI] [PubMed] [Google Scholar]
- 71.Gutierrez-Arcelus M., Lappalainen T., Montgomery S.B., Buil A., Ongen H., Yurovsky A., Bryois J., Giger T., Romano L., Planchon A. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. eLife. 2013;2:e00523. doi: 10.7554/eLife.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gibbs J.R., van der Brug M.P., Hernandez D.G., Traynor B.J., Nalls M.A., Lai S.-L., Arepalli S., Dillman A., Rafferty I.P., Troncoso J. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shaheen R., Anazi S., Ben-Omran T., Seidahmed M.Z., Caddle L.B., Palmer K., Ali R., Alshidi T., Hagos S., Goodwin L. Mutations in SMG9, encoding an essential component of nonsense-mediated decay machinery, cause a multiple congenital anomaly syndrome in humans and mice. Am. J. Hum. Genet. 2016;98:643–652. doi: 10.1016/j.ajhg.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schunkert H., König I.R., Kathiresan S., Reilly M.P., Assimes T.L., Holm H., Preuss M., Stewart A.F.R., Barbalic M., Gieger C., Cardiogenics. CARDIoGRAM Consortium Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Deloukas P., Kanoni S., Willenborg C., Farrall M., Assimes T.L., Thompson J.R., Ingelsson E., Saleheen D., Erdmann J., Goldstein B.A., CARDIoGRAMplusC4D Consortium. DIAGRAM Consortium. CARDIOGENICS Consortium. MuTHER Consortium. Wellcome Trust Case Control Consortium Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Acharya A., Baek S.T., Huang G., Eskiocak B., Goetsch S., Sung C.Y., Banfi S., Sauer M.F., Olsen G.S., Duffield J.S. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development. 2012;139:2139–2149. doi: 10.1242/dev.079970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Greer P. Closing in on the biological functions of Fps/Fes and Fer. Nat. Rev. Mol. Cell Biol. 2002;3:278–289. doi: 10.1038/nrm783. [DOI] [PubMed] [Google Scholar]
- 78.Hattori M., Tsukamoto N., Nur-e-Kamal M.S., Rubinfeld B., Iwai K., Kubota H., Maruta H., Minato N. Molecular cloning of a novel mitogen-inducible nuclear protein with a Ran GTPase-activating domain that affects cell cycle progression. Mol. Cell. Biol. 1995;15:552–560. doi: 10.1128/mcb.15.1.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Verrecchia F., Mauviel A. Transforming growth factor-beta signaling through the Smad pathway: role in extracellular matrix gene expression and regulation. J. Invest. Dermatol. 2002;118:211–215. doi: 10.1046/j.1523-1747.2002.01641.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.