Abstract
Inherited bone-marrow-failure syndromes (IBMFSs) include heterogeneous genetic disorders characterized by bone-marrow failure, congenital anomalies, and an increased risk of malignancy. Many lines of evidence have suggested that p53 activation might be central to the pathogenesis of IBMFSs, including Diamond-Blackfan anemia (DBA) and dyskeratosis congenita (DC). However, the exact role of p53 activation in each clinical feature remains unknown. Here, we report unique de novo TP53 germline variants found in two individuals with an IBMFS accompanied by hypogammaglobulinemia, growth retardation, and microcephaly mimicking DBA and DC. TP53 is a tumor-suppressor gene most frequently mutated in human cancers, and occasional germline variants occur in Li-Fraumeni cancer-predisposition syndrome. Most of these mutations affect the core DNA-binding domain, leading to compromised transcriptional activities. In contrast, the variants found in the two individuals studied here caused the same truncation of the protein, resulting in the loss of 32 residues from the C-terminal domain (CTD). Unexpectedly, the p53 mutant had augmented transcriptional activities, an observation not previously described in humans. When we expressed this mutant in zebrafish and human-induced pluripotent stem cells, we observed impaired erythrocyte production. These findings together with close similarities to published knock-in mouse models of TP53 lacking the CTD demonstrate that the CTD-truncation mutations of TP53 cause IBMFS, providing important insights into the previously postulated connection between p53 and IBMFSs.
Keywords: TP53, p53, the C-terminal domain, inherited bone marrow failure syndrome, Diamond-Blackfan anemia, dyskeratosis congenita, zebrafish, human-induced pluripotent stem cell, iPSC, gene editing
Main Text
Inherited bone-marrow-failure syndromes (IBMFSs) include a heterogeneous group of genetic disorders characterized by bone-marrow failure, congenital anomalies, and an increased risk of malignacy.1 Many lines of evidence suggested that p53 activation may be central to the pathogenesis of IBMFSs, including Diamond-Blackfan anemia (DBA [MIM: 105650]) and dyskeratosis congenita (DC [MIM: 305000]).2, 3
DBA is characterized by macrocytic pure red-cell aplasia involving a decreased number of erythroid progenitors in the bone marrow and various constitutive abnormalities.4 Except for rare germline GATA1 (MIN: 305371) mutations,5 all known causative mutations involve ribosomal protein (RP) genes.6 It is widely accepted that the pathogenesis of DBA involves elevated p53 accumulation caused by haploinsufficiency of a ribosomal component; this haploinsufficiency leads to an increased stability of the protein by an MDM2-mediated mechanism, thereby inducing cell-cycle arrest and apoptosis of erythroid precursors.2 DC is caused by germline mutations in the telomerase complex or the shelterin telomere protection complex.7 An individual with DC typically presents with a classic triad of phenotypes, including nail dystrophy, oral leukopathies, and skin hyperpigmentation. However, these phenotypes are usually not evident until the second decade of life. In most DC cases, the initial hematologic abnormalities include thrombocytopenia or macrocytic anemia followed by pancytopenia.1 Hypogammaglobulinemia is also common.8, 9 It is widely accepted that telomere or telomerase dysfunction in DC leads to uncapped chromosomes, resulting in p53 activation and induction of apoptosis.3
Although the p53 pathway appears to be a critical mediator in both diseases, the exact role of p53 activation in each clinical feature remains unknown. In this report, we describe two individuals with a unique form of IBMFS caused by de novo germline mutation of TP53 (MIM: 191170), unequivocally suggesting a role of augmented p53 functions in the pathogenesis of bone-marrow failure.
The two individuals in our DBA cohort had similar clinical phenotypes that partially mimicked DBA (e.g., pure red-cell aplasia [Figure 1A] and growth retardation [Figure 1B]).4 Other features, such as hypogammaglobulinemia and microcephaly (Figure 1C), are also seen in DBA4, 10 but are more common in DC.8 In fact, the older case (individual 1) presented typical DC-like features, including reticular skin pigmentation (Figure 1C), hypogonadism, and tooth anomalies (Table 1) (see Supplemental Note).9 Their anemia did not respond to steroid therapy. They required a regular blood transfusion from early infancy. However, anemia of individual 1 showed spontaneous remission at 13 years of age, and he became transfusion independent. Both individuals needed regular IgG replacement therapy for persistent hypogammaglobulinemia. No cancers were identified in either individual. No mutations or deletions were detected in the known DBA-associated genes. To identify causative mutations, we first performed whole-exome sequencing (WES) of germline DNA obtained from the peripheral blood of the two individuals and their parents and/or siblings as previously described (Supplemental Material and Methods).12 Gene mutations were confirmed by Sanger sequencing, performed with their hair follicles and/or buccal tissues. Written informed consent was obtained from their parents. The ethics committees at Hirosaki University and the University of Tokyo approved this study. In WES and copy-number analysis, we did not find mutations or deletions in the genes associated with IBMFSs (Tables S1 and S2). Intriguingly, however, in both individuals we identified heterozygous frameshift variants commonly affecting TP53 (c.1083delG in individual 1 [II-3] and c.1077delA in individual 2 [II-3]: RefSeq accession: NM_001126112.2) (Figure 2A). These TP53 mutations were not found in their parents (Figure 2A and 2B), indicating that both variants represented de novo germline mutations. These frameshift mutations led to identical C-terminal truncations (p.Ser362Alafs∗8) (Figure 1C). Both variants were located in exon 10, resulting in the introduction of premature terminal codons in the last coding exon (exon 11) (Figure 1D). Therefore, mutant transcripts most likely escaped from the nonsense-mediated mRNA decay pathway.13 Indeed, Sanger sequencing detected the mutant transcripts in amounts comparable to those of normal transcripts in peripheral blood mononuclear cells (PBMCs) from individual 1 (Figure S1). A spontaneous remission of the IBMF phenotype within the second decade of life in individual 1 suggests the possibility of a clonal genetic reversion event. However, deep sequencing showed comparable variant allele frequency (VAF) of the TP53 mutation in PBMC in remission (VAF: 0.49).
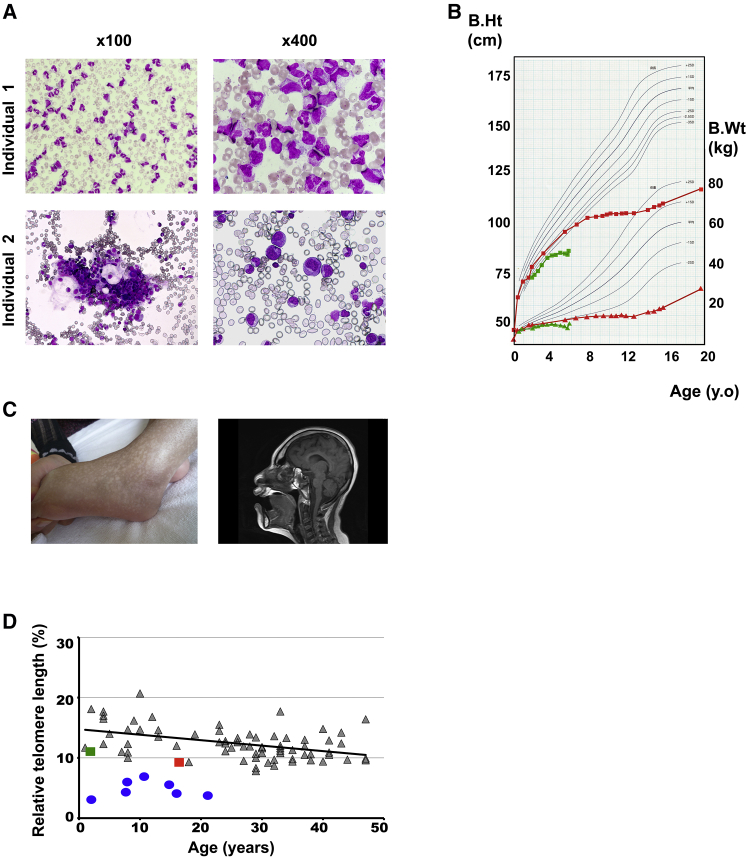
Figure 1.
Clinical Features of the Individuals
(A) Bone-marrow smears from individual 1 (top) and individual 2 (bottom) show selective erythroid hypoplasia.
(B) Growth curves of individual 1 (red) and individual 2 (green) exhibit marked growth retardation.
(C) Reticular skin pigmentation and severe microcephaly as demonstrated by magnetic resonance imaging noted in individual 1 at 16 years of age.
(D) Telomere lengths in peripheral blood lymphocytes from individual 1 (red) and individual 2 (green) compared to normal individuals (gray) (median age, 29 years; range, 1–47 years) and an individual with DC (blue); telomere lengths were estimated by flow-cytometry fluorescent in situ hybridization (flow-FISH) and expressed as the FISH signals relative to the signal from a normal control (1301 cell line).
Table 1.
Clinical Characteristics of ΔCTD Mutant Mice and the Individuals Carrying a TP53-ΔCTD Allele
|
Individuals |
Mice (No. Analyzed) |
Reported Frequencies in |
||||
|---|---|---|---|---|---|---|
| 1 | 2 | p53Δ31/Δ31 22 | p53Δ24/Δ24 23 | DBA1, 4, 11 | DC1, 8, 9, 11 | |
| Age at diagnosis | 2 m | 15 d | NA | NA | NA | NA |
| Gender | M | M | NA | NA | NA | NA |
| Bone-Marrow Failure | ||||||
| PRCA | + | + | − | − | ∼100% | +d |
| Pancytopenia | − | − | + | + | − | >95% |
| Aplasia in BM | +a | +a | + (8/8) | + | +a | + |
| Spontanous remission | + | − | − | − | 20% | ND |
| Physical Anomalies | ||||||
| Microcephaly | + | + | + | + | reported | 6% |
| Growth retardation | + | + | + (16/25) | + | 30% | 20% |
| Mental retardation | + | + | ND | ND | reported | 25% |
| Hypogonadism | + | ND | + (10/11) | ND | reported | 6% |
| Physical Anomalies Associated with DC | ||||||
| Pulmonary fibrosis | − | − | + (7/8) | ND | ND | 20% |
| Nail dystrophy | − | − | + (2/8) | ND | ND | 88% |
| Oral leukoplakia | − | − | + (8/8) | ND | ND | 78% |
| Skin hyperpigmentation | + | − | + (25/25) | + | ND | 89% |
| Teeth abnormal | + | − | ND | ND | ND | 17% |
| Abnormal Laboratory Findings | ||||||
| Hypogammaglobulinemia | + | + | ND | ND | rare | common |
| Telomere shortening | +b | +b | + | ND | +b | 100%c |
Abbreviations are as follows; d: day, m: month, y: year, M; male, NA: not applicable, PRCA: pure red cell aplasia, ND: not described.
Aplasia is limited to erythroid lineage in BM.
Telomere length is between the 1st and 10th percentile.
Most patients with DC have very short telomeres, less than 1st percentile (< 1%).
In most cases with DC, the initial hematologic abnormalities are thrombocytopenia or macrocytic anemia followed by pancytopenia.
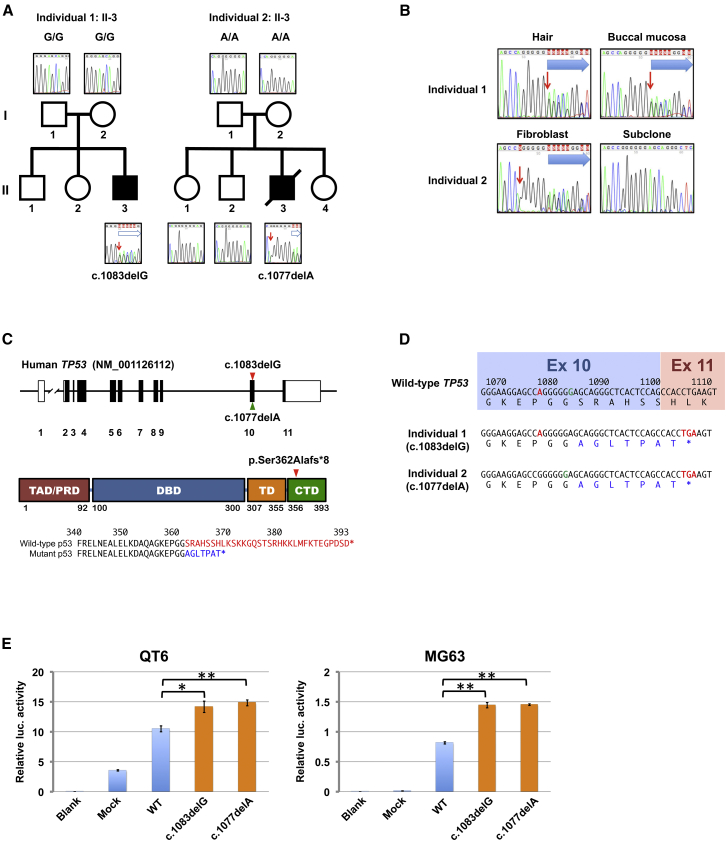
Figure 2.
TP53 Gene Alteration Observed in Individuals with Inherited Bone-Marrow-Failure Syndrome
(A) Pedigrees for individuals 1 (left) and 2 (right). Each individual had a de novo germline mutation. The red arrows indicate the positions of the single-nucleotide deletions. Blue arrows indicate the frameshift signals.
(B) The TP53 mutation in different tissues from individuals 1 and 2. The right lower panel shows the DNA sequences of the subcloned molecule derived from individual 2, demonstrating the deletion of c.1077A. Red arrows indicate positions of the single-nucleotide deletion. Blue arrows indicate the frameshift signals.
(C) Structure of the TP53 locus in which the locations of the mutations in individual 1 (red) and individual 2 (green) are indicated by arrowheads (top). Structure of human p53 consisting of domains for transactivation (TAD), specific DNA binding (DBD), and tetramerization (TD), as well as proline-rich (PRD) and C-terminal (CTD) domains (middle). The two heterozygous frameshift variants in TP53 in individuals 1 and 2 resulted in identical C-terminal truncations (arrowhead). A comparison of the amino acid sequences of the C termini of the wild-type and mutant p53s is shown (bottom). Deleted or altered sequences in p53 variants in both individuals are shown in red and blue, respectively.
(D) The position of TP53 c.1077A is shown in red and c.1083G in green. c.1083G was deleted in individual 1, c.1077A was deleted in individual 2, and both translated products had identical amino acid sequences. Altered amino acid residues are indicated in blue. Both variants in exon 10 resulted in the introduction of premature terminal codons in the last coding exon (exon 11).
(E) Luciferase activities from a CDKN1A promoter co-transduced with indicated constructs in QT6 (left) and MG63 (right) cell lines. Blank: no DNA. MOCK: mock vector. WT: wild-type TP53. c.1083delG: c.1083delG TP53 variant. c.1077delA: c.1077delA TP53 variant. ∗p < 0.01; ∗∗p < 0.001, t test. Error bars indicate standard deviation.
TP53 is a tumor-suppressor gene most frequently mutated in human cancers; occasional germline variants also occur in Li-Fraumeni cancer-predisposition syndrome (LFS [MIM: 151623]).14, 15 Most of these mutations affect the core DNA-binding domain, leading to compromised transcriptional activities.16 In contrast, the variants found in two individuals showed the same protein truncations, resulting in the loss of 32 residues from the C-terminal domains (CTDs) (Figure 2C). Significantly, no mutations involving the CTD have been reported in human cancers or LFS according to the International Agency for Research on Cancer TP53 database v.R18, which compiles >29,000 somatic mutations in cancer and >880 germline mutations.16 Furthermore, we could not find IBFMS cases with the CTD-truncating mutations of TP53 in the published literature. To test whether the loss of the CTD affected the transcriptional activities of p53, we first assessed the transcriptional activity of the p53 mutant seen in two individuals by luciferase assay involving the promoter of CDKN1A as described in the Supplemental Material and Methods. The transcriptional activation of the mutant was significantly higher than that of wild-type p53 (Figure 2E), suggesting that the variants found in both individuals were activating mutations.
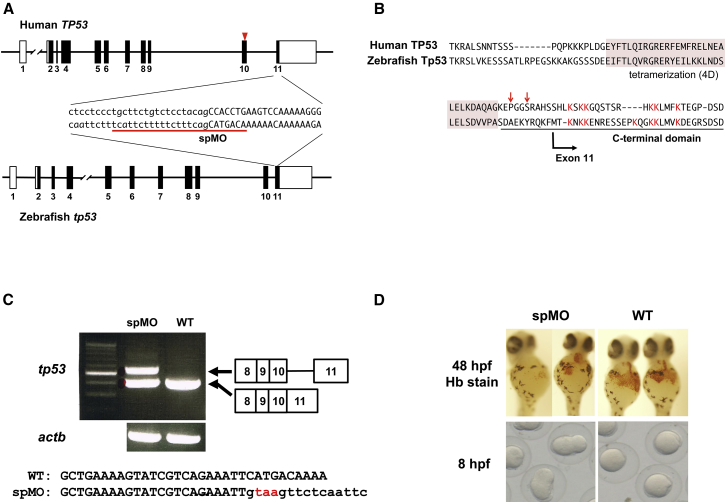
To investigate the biological effects of the TP53 mutations in vivo, we developed zebrafish expressing a CTD-truncated p53. Thus, using one-cell-stage embryos, we injected a Morpholino antisense oligo (MO) that targeted the 3′ splice site of intron 10 (spMO) (Figure 3A and Supplemental Material and Methods). The MO-injected embryos were grown at 28.5°C. Total RNA was isolated from wild-type and MO-injected embryos at 8 hr post-fertilization (hpf). Reverse transcription (RT)-PCR was used for distinguishing normal and intron-containing sizes of the tp53 transcripts. Injection of spMO into the one-cell-stage embryos perturbed the splicing of intron 10 and introduced a premature stop codon just after exon 10, leading to the elimination of the lysine-rich CTD in Tp53 (Figures 3B and 3C). The spMO-injected embryos presented developmental defects with severe morphological abnormalities and died by 96 hpf. Hemoglobin staining at 48 hpf reflected reduced erythrocyte production in the cardial vein of the embryos (Figure 3D). These results suggest that the CTD-truncated p53 perturbed the early development and erythrocyte production of zebrafish.
Figure 3.
Functional Analyses Based on a Zebrafish Model
(A) The structures of human TP53 and zebrafish tp53. The sequences at the intron 10/exon 11 boundary regions are indicated. Uppercase and lowercase letters show the exon and intron sequences, respectively. The MO target site is underlined. The arrowhead indicates the position of the mutated nucleotides in the affected individuals.
(B) The amino acid sequences of the C-terminal regions of human TP53 and zebrafish Tp53. The conserved lysine residues in the CTD for human and zebrafish are shown in red. The arrows indicate the positions of the mutant nucleotides in the individuals. The tetramerization domain is shaded in pink.
(C) RT-PCR-based detection of tp53 and actb in MO-injected and wild-type embryos. A larger transcript retaining intron 10 is detected in the MO-injected embryos. Injection of spMO perturbed the splicing of intron 10 and introduced a premature stop codon just after exon 10, leading to the elimination of the lysine-rich CTD in Tp53.
(D) Representative hemoglobin staining of cardial veins at 48 hpf and cell divisions at 8 hpf found in MO-injected and wild-type embryos. Approximately 40% of the embryos injected with spMO at 2.5 μg/μL show a severe to moderate reduction of erythrocytes. The injection of MO at 5 μg/μL led to developmental arrest at 8–9 hpf.
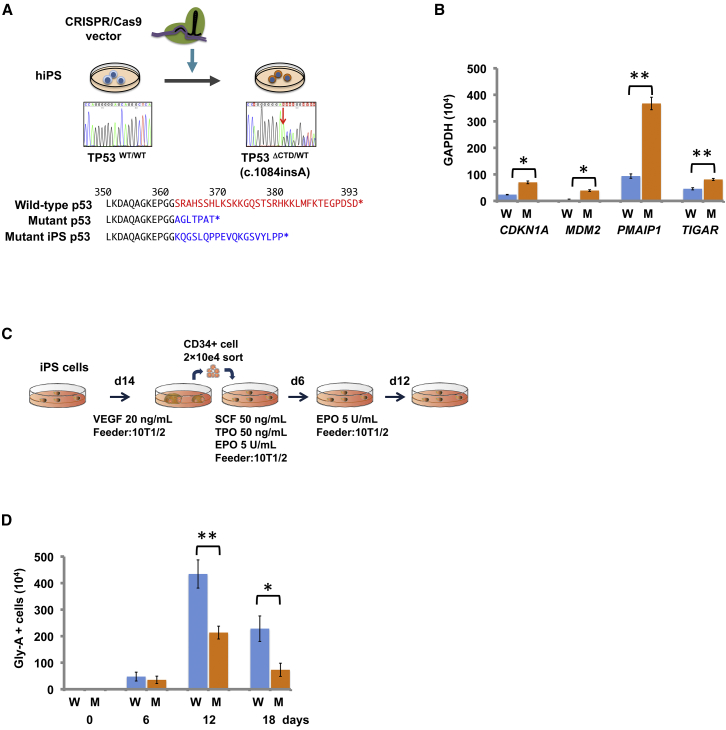
To further examine the effects of the mutated TP53 in human cells, we used CRISPR/Cas9-mediated gene editing (Supplemental Material and Methods) to establish a human-induced pluripotent stem cell (hiPSC) line carrying a heterozygous TP53 mutant allele (Figure 4A). We prepared the plasmids expressing CRISPR guide RNA and Cas9 by ligating oligos into the BbsI site of pX330 (Addgene no. 42230). We designed the candidate sequences for CRISPR guide RNA by using UCSC Genome Browser to introduce frameshift mutations in the CTD of TP53. The targeting constructs for genome editing and neomycin resistance gene expression plasmids were transfected into hiPSCs #817 and selected with neomycin (G418). We confirmed significantly elevated expression of the downstream targets of p53 in the mutant hiPSCs compared to isogeneic wild-type iPSCs, for all the targets tested, including CDKN1A (OMIN: 116899), MDM2 (OMIN: 164785), PMAIP1 (OMIM: 604959), and TIGAR (OMIN: 610775) (Figure 4B). We then assessed the ability of the mutant hiPSCs to produce erythroid lineage cells upon induced erythroid differentiation as previously reported (Figure 4C).17 Twelve and 18 days after induction, the number of CD71- and glycophorin-A-positive cells was significantly lower in mutant hiPSCs than in the isogenic control (Figure 4D and Table S3). These results suggest that hiPSCs carrying the heterozygous TP53 mutations possess enhanced p53 activity and impaired erythroid differentiation.
Figure 4.
Functional Analyses Based on a Human iPSC Model
(A) An hiPSC line carrying a heterozygous TP53 mutant allele was generated by CRISPR/Cas9-mediated gene editing (upper). Comparison of the amino acid sequences of the C termini of wild-type and mutant p53s in the affected individuals and mutant hiPSC (bottom). Deleted or altered sequences in p53 variants in both individuals and the mutant hiPSC are shown in red and blue, respectively.
(B) Comparison of the expression of four downstream targets of p53 in the mutant hiPSC (M) and isogenic control (W) by real-time quantitative RT-PCR. Error bars indicate standard deviation.
(C) Scheme for differentiation of erythroid lineage from hiPSCs. To differentiate hiPSCs into erythroid cells, we used our previously established protocol.16
(D) Effects of the truncating p53 mutant on erythropoiesis as measured by the numbers of glycophorin-A (Gly-A)-positive cells after in vitro erythroid induction. The combined results of three independent experiments are shown. ∗p < 0.01; ∗∗p < 0.001, t test. Error bars indicate standard deviation.
In response to a variety of stresses, p53 undergoes extensive post-translational modification, including phosphorylation and acetylation. The CTD is subjected to multiple and diverse post-translational modifications that are thought to be essential for the regulation of p53 stability and activity.18 Although the precise roles and functions of the p53 CTD are not fully understood,19 the p53 CTD has been shown to act as a docking site for negative regulators such as Smyd2 and SET.20, 21 Thus, the deletion of the CTD might result in compromised binding of these p53 variants to the negative regulators and impairment of their transcriptional repression.
Recent studies using mouse models demonstrated that the CTD plays key roles in postnatal homeostasis of the hematopoietic compartment and development of the brain.22, 23 Simeonova et al. described a genetically engineered mouse carrying a mutant Tp53 knock-in allele that lacked the C-terminal 31 amino acids and was very similar to the mutations found in both individuals.22 Mice homozygous for the mutant allele showed bone-marrow aplasia, short telomere lengths, abbreviated life expectancy, microcephaly, a small body size, and pulmonary fibrosis, a collection of characteristics that are essentially a phenocopy of DC. Hamard et al. reported that another mouse line expressing a truncated p53 lacking the C-terminal 24 amino acids presented similar phenotypes.23 Both affected individuals described here shared several phenotypes, including bone marrow failure, microcephaly, and severe growth retardation (Table 1), with these mutant mice. However, there exist some differences between our cases and the mouse models. For example, telomere shortening was not prominent in either individual compared to typical DC-affected individuals with mutated TERT-related genes (Figure 1D).11 Notably, although the heterozygous mice appeared to show minimal phenotypes, both individuals developed severe phenotypes even with a heterozygous TP53 mutation. An hiPSC line carrying a heterozygous TP53 mutation also showed impairment of erythrocyte production. It is unclear how these truncations (and augmented p53 functions) lead to the observed phenotype and exactly which of many p53 functions are affected by these mutations. Further studies are warranted to clarify these issues.
In conclusion, we identified germline TP53 mutations in two cases of IBMFS. These mutations have not been previously described in humans. The p53 varaint was shown to have augmented p53 functions, rather than loss of function. The close similarities of genotypes and phenotypes among the individuals and the zebrafish, hiPSC and mouse models strongly suggest that the clinical phenotypes in our individuals could be explained by activated p53 functions caused by truncation of the C terminus. These findings will provide important insights into the previously postulated connection between p53 and IBMFSs.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We would like to thank individuals 1 and 2 and their family members for making this work possible. This research used computational resources of the K computer provided by the RIKEN Advanced Institute for Computational Science through the HPCI System Research project (S.O.; hp150232). This work was supported by Practical Research Project for Rare/Intractable Diseases (JP17ek0109133), grants-in-aid (JP17ek0109099, JP16ck0106073, and JP17ek0109286s0101) from the Japan Agency for Medical Research and Development (AMED), and a research grant from the Ministry of Health, Labour, and Welfare of Japan (Research on Measures for Intractable Diseases) (201711031A) and Japan Society for the Promotion of Science KAKENHI grant number JP17K10093. S.O. was supported by the JSPS Core-to-Core Program, A. Advanced Research Networks.
Published: August 23, 2018
Footnotes
Supplemental Data include a Supplemental Case Report, Supplemental Material and Methods, one figure, and three tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.07.020.
Contributor Information
Seishi Ogawa, Email: sogawa-tky@umin.ac.jp.
Etsuro Ito, Email: eturou@hirosaki-u.ac.jp.
Accession Numbers
The accession numbers for the sequences reported in this paper are ClinVar: SUB4310928, SUB4307372, SCV000786665.
Web Resources
European Genome-Phenome Archive, https://www.ebi.ac.uk/ega/home
Human Genetic Variation Browser, http://www.genome.med.kyoto-u.ac.jp/SnpDB/
NHLBI Exome Sequencing Project, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
UCSC Genome Browser, https://genome.ucsc.edu/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
SIFT, http://sift.jcvi.org/
Supplemental Data
References
- 1.Bessler, M., Masson, P.J., Link, D.C., and Wilson, D.B. Inherited bone marrow failure syndrome. In Nathan and Oski’s Hematology of Infancy and Childhood, 7th edition. S.H. Orkin, D.G. Nathan, D. Ginsburg, A.T. Look, D.E. Fisher, and S.E. Lux, eds. (W.B. Saunders), pp. 307–395.
- 2.Dutt S., Narla A., Lin K., Mullally A., Abayasekara N., Megerdichian C., Wilson F.H., Currie T., Khanna-Gupta A., Berliner N. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood. 2011;117:2567–2576. doi: 10.1182/blood-2010-07-295238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Townsley D.M., Dumitriu B., Young N.S. Bone marrow failure and the telomeropathies. Blood. 2014;124:2775–2783. doi: 10.1182/blood-2014-05-526285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vlachos A., Ball S., Dahl N., Alter B.P., Sheth S., Ramenghi U., Meerpohl J., Karlsson S., Liu J.M., Leblanc T., Participants of Sixth Annual Daniella Maria Arturi International Consensus Conference Diagnosing and treating Diamond Blackfan anaemia: Results of an international clinical consensus conference. Br. J. Haematol. 2008;142:859–876. doi: 10.1111/j.1365-2141.2008.07269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sankaran V.G., Ghazvinian R., Do R., Thiru P., Vergilio J.A., Beggs A.H., Sieff C.A., Orkin S.H., Nathan D.G., Lander E.S., Gazda H.T. Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia. J. Clin. Invest. 2012;122:2439–2443. doi: 10.1172/JCI63597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Narla A., Ebert B.L. Ribosomopathies: Human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calado R.T., Young N.S. Telomere diseases. N. Engl. J. Med. 2009;361:2353–2365. doi: 10.1056/NEJMra0903373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jyonouchi S., Forbes L., Ruchelli E., Sullivan K.E. Dyskeratosis congenita: A combined immunodeficiency with broad clinical spectrum--a single-center pediatric experience. Pediatr. Allergy Immunol. 2011;22:313–319. doi: 10.1111/j.1399-3038.2010.01136.x. [DOI] [PubMed] [Google Scholar]
- 9.Vulliamy T.J., Marrone A., Knight S.W., Walne A., Mason P.J., Dokal I. Mutations in dyskeratosis congenita: their impact on telomere length and the diversity of clinical presentation. Blood. 2006;107:2680–2685. doi: 10.1182/blood-2005-07-2622. [DOI] [PubMed] [Google Scholar]
- 10.Khan S., Pereira J., Darbyshire P.J., Holding S., Doré P.C., Sewell W.A., Huissoon A. Do ribosomopathies explain some cases of common variable immunodeficiency? Clin. Exp. Immunol. 2011;163:96–103. doi: 10.1111/j.1365-2249.2010.04280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alter B.P., Baerlocher G.M., Savage S.A., Chanock S.J., Weksler B.B., Willner J.P., Peters J.A., Giri N., Lansdorp P.M. Very short telomere length by flow fluorescence in situ hybridization identifies patients with dyskeratosis congenita. Blood. 2007;110:1439–1447. doi: 10.1182/blood-2007-02-075598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kunishima S., Okuno Y., Yoshida K., Shiraishi Y., Sanada M., Muramatsu H., Chiba K., Tanaka H., Miyazaki K., Sakai M. ACTN1 mutations cause congenital macrothrombocytopenia. Am. J. Hum. Genet. 2013;92:431–438. doi: 10.1016/j.ajhg.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hug N., Longman D., Cáceres J.F. Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res. 2016;44:1483–1495. doi: 10.1093/nar/gkw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kandoth C., McLellan M.D., Vandin F., Ye K., Niu B., Lu C., Xie M., Zhang Q., McMichael J.F., Wyczalkowski M.A. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malkin D., Garber J.E., Strong L.C., Friend S.H. CANCER. The cancer predisposition revolution. Science. 2016;352:1052–1053. doi: 10.1126/science.aag0832. [DOI] [PubMed] [Google Scholar]
- 16.Bouaoun L., Sonkin D., Ardin M., Hollstein M., Byrnes G., Zavadil J., Olivier M. TP53 variations in human cancers: New lessons from the IARC TP53 database and genomics data. Hum. Mutat. 2016;37:865–876. doi: 10.1002/humu.23035. [DOI] [PubMed] [Google Scholar]
- 17.Ochi K., Takayama N., Hirose S., Nakahata T., Nakauchi H., Eto K. Multicolor staining of globin subtypes reveals impaired globin switching during erythropoiesis in human pluripotent stem cells. Stem Cells Transl. Med. 2014;3:792–800. doi: 10.5966/sctm.2013-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng L., Lin T., Uranishi H., Gu W., Xu Y. Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol. Cell. Biol. 2005;25:5389–5395. doi: 10.1128/MCB.25.13.5389-5395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laptenko O., Tong D.R., Manfredi J., Prives C. The tail that wags the dog: How the disordered C-terminal domain controls the transcriptional activities of the p53 tumor-suppressor protein. Trends Biochem. Sci. 2016;41:1022–1034. doi: 10.1016/j.tibs.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang J., Perez-Burgos L., Placek B.J., Sengupta R., Richter M., Dorsey J.A., Kubicek S., Opravil S., Jenuwein T., Berger S.L. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 21.Wang D., Kon N., Lasso G., Jiang L., Leng W., Zhu W.G., Qin J., Honig B., Gu W. Acetylation-regulated interaction between p53 and SET reveals a widespread regulatory mode. Nature. 2016;538:118–122. doi: 10.1038/nature19759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simeonova I., Jaber S., Draskovic I., Bardot B., Fang M., Bouarich-Bourimi R., Lejour V., Charbonnier L., Soudais C., Bourdon J.C. Mutant mice lacking the p53 C-terminal domain model telomere syndromes. Cell Rep. 2013;3:2046–2058. doi: 10.1016/j.celrep.2013.05.028. [DOI] [PubMed] [Google Scholar]
- 23.Hamard P.J., Barthelery N., Hogstad B., Mungamuri S.K., Tonnessen C.A., Carvajal L.A., Senturk E., Gillespie V., Aaronson S.A., Merad M., Manfredi J.J. The C terminus of p53 regulates gene expression by multiple mechanisms in a target- and tissue-specific manner in vivo. Genes Dev. 2013;27:1868–1885. doi: 10.1101/gad.224386.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.