Abstract
Bipolar disorder (BD) and schizophrenia (SCZ) are highly heritable diseases that affect more than 3% of individuals worldwide. Genome-wide association studies have strongly and repeatedly linked risk for both of these neuropsychiatric diseases to a 100 kb interval in the third intron of the human calcium channel gene CACNA1C. However, the causative mutation is not yet known. We have identified a human-specific tandem repeat in this region that is composed of 30 bp units, often repeated hundreds of times. This large tandem repeat is unstable using standard polymerase chain reaction and bacterial cloning techniques, which may have resulted in its incorrect size in the human reference genome. The large 30-mer repeat region is polymorphic in both size and sequence in human populations. Particular sequence variants of the 30-mer are associated with risk status at several flanking single-nucleotide polymorphisms in the third intron of CACNA1C that have previously been linked to BD and SCZ. The tandem repeat arrays function as enhancers that increase reporter gene expression in a human neural progenitor cell line. Different human arrays vary in the magnitude of enhancer activity, and the 30-mer arrays associated with increased psychiatric disease risk status have decreased enhancer activity. Changes in the structure and sequence of these arrays likely contribute to changes in CACNA1C function during human evolution and may modulate neuropsychiatric disease risk in modern human populations.
Keywords: bipolar disorder, schizophrenia, CACNA1C, tandem repeat, minisatellite, copy-number variation, GWAS, human evolution, chimpanzee, psychiatric disease
Main Text
More than 3% of the global population has bipolar disorder (BD) or schizophrenia (SCZ), and both diseases are among the top 25 causes of disability worldwide.1, 2, 3 Along with the disability cost, both disorders are associated with an increased risk of suicide.4, 5 There are limited treatment options for BD and SCZ, and the burden of these diseases may be increasing.6 Improved diagnosis and treatments may come from a better understanding of the molecular pathways that contribute to disease risk.
Both BD and SCZ are highly heritable. While they are classified as different diseases based on their clinical symptoms, they share a similar set of genomic risk variants.7 Genome-wide association studies (GWASs) for BD and SCZ have consistently implicated risk variants in or near genes involved in calcium signaling.8, 9, 10, 11, 12, 13, 14 Calcium signaling-related genes are also enriched for rare variants in families multiply affected by BD15 and in individuals with SCZ,16, 17 suggesting that calcium signaling plays an important role in both BD and SCZ etiology.
Some of the strongest and best-replicated associations for BD and SCZ map within CACNA1C, which encodes the pore-forming subunit of the CaV1.2 calcium channel.18 Disease-associated single-nucleotide polymorphisms (SNPs) are in strong linkage disequilibrium with each other and contained within a 100 kb region of the gene’s third intron.8, 9, 10, 11, 12, 13, 14, 19 Underscoring the importance of this genomic region for psychiatric disease in humans, these disease-associated SNPs have also been associated with anxiety, depression-related symptoms, obsessive-compulsive symptoms, decreased performance in memory-related tasks, major depression, and autism.13, 20, 21, 22, 23, 24, 25, 26, 27, 28
The causative variants at loci identified by GWASs could be the assayed SNPs themselves29 or other variants tightly linked to the SNP markers.30 Previous studies have investigated the functional consequences of the genotyped SNPs in CACNA1C and other closely linked SNPs.31, 32 However, the mutation responsible for the association between SNPs within CACNA1C and human neuropsychiatric diseases is still unknown.
Given the difficulty in identifying causal mutations at CACNA1C over the past 10 years,8 we considered whether there might be additional structural variants at the locus that are not easily detected using current genotyping and sequencing methods. For example, copy-number variants and expansions and contractions of micro- and mini-satellite sequences can be difficult to identify with short-read or Sanger sequencing technologies. Nevertheless, these types of mutations have been implicated in a wide range of neurological diseases, including Huntington disease, spinocerebellar ataxia, fragile X syndrome, depression, and aggression and impulsivity behaviors,33, 34, 35, 36, 37, 38, 39, 40 and likely contribute to undetected variation and missing heritability for additional human traits.41, 42
To search for unrecognized copy-number variants at the CACNA1C locus, we examined regions of the genome where no mutations were identified by large-scale sequencing projects such as the 1000 Genomes Project,43 yet DNA sequencing reads consistently differed from the reference human assembly. We identified one such region (hg38; chr12:2255791–2256090) within the 100 kb interval associated with BD and SCZ. In the most recent human reference genome (hg38), this region is assembled as a tandem repeat composed of ten 30 bp units. Chimpanzees and other non-human primates have a single instance of a homologous 30 bp sequence at this location (Figure 1A), suggesting that an ancestral 30-mer sequence has expanded in the CACNA1C intron during human evolution. Strikingly, the number of reads from individuals in the 1000 Genomes Project that map to this 300 bp segment is 3–379× greater than expected based on the reference assembly, and these reads contain multiple base substitutions. Read depth coverage and composition in Neanderthal and Denisovan genomes44, 45 fall within the range observed among modern human populations (Figure S1). Further investigation identified a longer (3.3 kb) repeat array at this location in the Venter assembly (HuRef)46 and an approximately 6 kb repeat array in the genome of a hydatidiform mole sequenced to 40× coverage with long-read technology.47 Collectively, these data suggest that hominins have a large and variable tandem repeat in the neuropsychiatric risk-associated region of CACNA1C. The size of the tandem array is likely under-represented in the human reference genome by one or two orders of magnitude based on empirical estimates from read depth coverage (see Supplemental Methods).
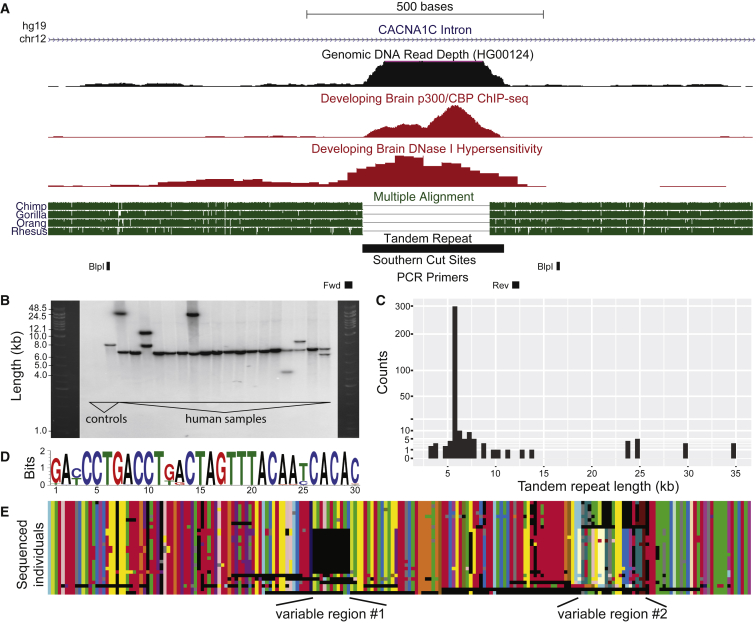
Figure 1.
Human-Specific Tandem Repeat Region Is Composed of 30-mer Sequence Units Repeated Head-to-Tail in Multi-kilobase Arrays
(A) The tandem repeat is located in the third intron of CACNA1C. The human reference assembly predicts ten copies of the 30 bp segment while chimpanzees and other simians have a single copy of the 30 bp segment. More distantly related placental mammals, out to Afrotheria, have a region that aligns to the 30 bp segment, but with insertions or deletions. There is an abnormally large number of genomic DNA sequencing reads mapping to the tandem repeat region, consistent with this repeat being further expanded in human individuals. The repeat region also shows enrichment for p300/CBP binding and DNase I hypersensitivity in the developing human brain.
(B) We performed Southern blot analysis on 18 human individuals by probing for the 30 bp repeat after digesting with BlpI. We also included two controls: mouse DNA (no orthologous sequence) and the 8 kb vector from which the probe was transcribed. The human reference genome predicts a BlpI fragment of approximately 900 bp. In contrast, all humans tested show much larger BlpI fragment sizes (4,000 to 35,000 bp), and many individuals show dual bands indicating distinct alleles at the locus.
(C) Frequency distribution of 362 repeat allele lengths detected by Southern blot analysis.
(D) The 30-mer sequence logo calculated from the 30-mer variants present in human repeat arrays that were sequenced with long-read (PacBio) technology. Some positions are nearly invariant, whereas others vary from 30-mer to 30-mer.
(E) Structure and composition of tandem repeat arrays sequenced by PacBio long-read technology. Each row represents a different sequenced array, and each color represents a distinct 30-mer variant. Black regions indicate gaps that we have introduced to maximize repeat alignments between arrays. Many regions are organized similarly in all arrays, but common variable regions distinguish array subtypes.
To further characterize the size of the tandem arrays using independent methods, we examined DNA from humans and our closest living relative, the chimpanzee. Polymerase chain reaction (PCR) amplification and sequencing from six chimpanzees confirmed a single instance of a 30-mer sequence, which exactly matches the chimpanzee reference genome. In contrast, when we performed Southern blots on human DNA (see Supplemental Methods), we found restriction fragment sizes consistent with repeat arrays of 3,000 to 30,000+ bp, with the majority of human repeat arrays showing sizes of approximately 6,000 bp (Figures 1B and 1C). We never observed a band size consistent with the human reference genome (hg38). The smallest band size seen in the 181 human samples we assayed (362 alleles) was 10 times larger than the repeat size annotated in the reference assembly (300 bp), while the largest was more than 100 times larger.
To understand why the human genome assembly appears to have a version of the tandem repeat that is not representative of the human population, we examined four bacterial artificial chromosome (BAC) clones derived from a single individual that were used in the sequencing and assembly of the human genome.48 One BAC clone matched the length and sequence present in the assembly (300 bp). A BAC library made from a single individual should have at most two alleles; however, the four BACs all gave different tandem repeat lengths. Compounding this anomalous result, colonies picked from a single BAC clone are expected to be identical, but two of the four BACs produced subclones with varying tandem repeat lengths (Figure S2). In our experience, multi-kilobase tandem repeats, whose size was determined by Southern blot, reduced in length after amplification by PCR under routine conditions or when propagated using standard circular vectors in bacteria. We propose that the human reference assembly is based on a BAC clone that was correctly sequenced and assembled but that the sequence present in the BAC is an artifact of the instability of this tandem repeat when cloned and propagated using standard methods. Given the large size disparity between the version represented in the current human genome assembly and the alleles detected by Southern blot, as well as the instability of this region, we believe that the allele present in the current human genome assembly is not present in humans.
In addition to variation in the length of this repeat region in the human population, the 30-mer units that comprise each array also show sequence changes. For example, the array in the reference assembly is composed of four identical 30-mer units and six unique units that each contain a small number of SNPs. This variability in 30-mers is also seen in the large number of reads from the 1000 Genomes Project that map to this area. To better understand this variation in tandem repeat arrays, even for arrays of the same length, we performed long-read (PacBio) sequencing of repeat arrays amplified from 20 individuals using optimized PCR conditions (see Supplemental Methods). The size of the resulting PCR fragments using our optimized conditions matched the corresponding repeat lengths determined from Southern blots. The sequenced arrays were entirely composed of 30-mer units repeated head-to-tail. Some positions in the 30-mer unit appear to be largely invariant (e.g., position 2 is almost always an “A”), whereas other positions are more variable (Figures 1D and S3). For instance, the most common 30-mer unit (31%) is 5′-GACCCTGACCTGACTAGTTTACAATCACAC-3′ and the second most common (17%) is 5′-GATCCTGACCTGACTAGTTTACAATCACAC-3′ (difference underlined). When aligning tandem repeat array variants, the structural organization of the 30-mer units within each array emerged (Figure 1E). Across all PacBio-sequenced repeat arrays, certain regions, such as the beginning and the end of each repeat array, contain the same 30-mer units organized almost identically. However, other regions (marked in Figure 1E) are more variable and contain specific patterns of 30-mer units that are consistently found in only a subset of the sequenced arrays.
The presence of a large and variable repeat region in the third intron of human CACNA1C raises the possibility that variation in the tandem array contributes to functional changes at the locus. To test whether the length or sequence of the tandem repeat region shows any association with genomic risk markers for BD and SCZ, we examined whole-genome sequence reads from individuals in the 1000 Genomes Project. We limited our analysis to individuals of European or East Asian descent, the two groups in which BD and SCZ risk status has been previously associated with four SNPs clustered in the third intron of the gene (rs2007044, rs1006737, rs4765905, and rs4765913; Figure 2A).8, 9, 10, 11, 12, 13, 14, 19 We first identified all sequencing reads from this repeat region. To infer the length of the repeat array (average of the individual’s two alleles), we used the fraction of all sequencing reads for the individual that are from the repeat region (see Supplemental Methods). To estimate the sequence composition of the repeat array (averaged over the two alleles), we calculated the fraction of all 30-mers in the sequence reads identical to each observed sequence variant of the 30-mer unit (see Supplemental Methods). Since our length and composition statistics represent a mixture of the two alleles present in each person, we limited our analysis to individuals that are homozygous risk or protective at each of the four SNPs commonly associated with BD and SCZ (Figure 2A). These SNPs are all tightly linked and define risk and protective haplotypes, making it possible to study repeat structures associated with risk or protective genotypes at CACNA1C.
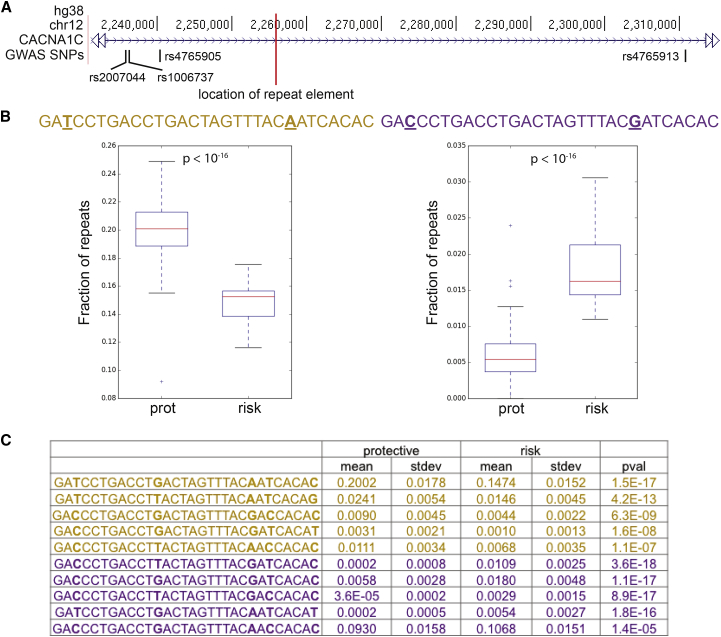
Figure 2.
30-mer Repeat Variants Are Associated with Protective or Risk Status at GWAS SNPs Linked to Neuropsychiatric Disease
(A) Genome browser view of the third intron of CACNA1C. A red line marks the location of the repeat region. The human-specific 30-mer repeats are embedded in a region defined by four SNPs that are repeatedly associated with BD and SCZ.
(B) We identified individuals from the 1000 Genomes Project that have the protective genotype at all four GWAS SNPs (protective haplotype) and individuals with the risk genotype at all four GWAS SNPs (risk haplotype). We used only European and East Asian individuals because GWASs have only been done with these populations. For each possible 30-mer repeat unit, we determined what fraction of 30-mers in the reads that map to this locus in each individual exactly match that particular variant. The 30-mer sequence on the left is significantly associated with the protective haplotype (“prot”), whereas the 30-mer variant on the right is significantly associated with the risk haplotype (“risk”). Base pair differences between the two 30-mer variants presented here are underlined. Shown are standard box-and-whisker plots where the box represents the lower quartile, median, and upper quartile, and the whiskers represent the range of the measurements. Outliers (“+”) are data points that are outside the nearest quartile + 1.5× the interquartile range.
(C) The table lists the mean and standard deviation of the fraction of reads that exactly match a given 30-mer for individuals with the protective or risk haplotype. Repeats enriched in the protective haplotype group are shown in yellow, and repeats enriched in the risk haplotype group are shown in purple. The p values were calculated using the Wilcoxon rank-sum test with Bonferroni correction (see Supplemental Methods).
We first tested whether repeat length is consistently associated with genotype status at the four GWAS SNPs. None of the four SNPs show a significant association with repeat length and the direction of effect is not consistent (Figure S4). It does not appear that repeat array length is associated with the protective or risk genotypes at the GWAS SNPs, at least not in a simple manner.
We then tested whether specific sequence variants of the 30-mer unit are associated with the risk or protective alleles at the four GWAS SNPs. For each sequence variant of the 30-mer unit, we tested whether its propensity to appear in reads from this repeat region differs between individuals that are homozygous for risk or protective genotypes at the four SNPs (Figure 2B). We identified a number of 30-mer units that are consistently associated with a genotypic class across all four SNPs (Figure S5). When considering only individuals that are homozygous protective at all four GWAS SNPs (“protective haplotype”) or homozygous risk at all four GWAS SNPs (“risk haplotype”), five 30-mer variants are associated with the protective haplotype, and five 30-mer variants are associated with the risk haplotype (Figure 2C). These particular 30-mer units tend to be located in the variable regions observed when aligning the PacBio-sequenced repeat arrays (marked in Figure 1E). Thus, while there is no straightforward association between the overall length of repeat arrays and the risk or protective haplotype, the abundance of particular 30-mer units is significantly and consistently associated with markers for psychiatric disease risk at CACNA1C.
The repeat region gives a significant signal for p300 enrichment in ChIP-seq experiments performed on tissue from the developing human brain49 and also shows an open chromatin signal during human brain development (Figures 1A and S6).50 Both of these results are consistent with the repeat region acting as a distal enhancer element during brain development. To experimentally test whether the 30-mer repeat arrays show enhancer activity in developing neural cells, we cloned the single 30-mer sequence found in chimpanzees (30 bp), as well as 21 different human repeat arrays (3.2–6 kb), upstream of a basal promoter and a luciferase reporter gene (Figure 3A). We used a linear cloning vector that greatly improved repeat stability, and we confirmed clone stability via comparison to the expected size and sequence as determined from Southern blot analysis and PacBio sequencing, respectively (see Supplemental Methods). We then transfected each construct into a human neural progenitor cell line (ReNcell Cx) and measured luciferase activity (see Supplemental Methods). The chimpanzee construct, containing a single 30 bp unit, weakly enhanced luciferase activity relative to the empty vector (p = 0.01), while the much larger repeat arrays found in humans significantly enhanced luciferase activity compared to both the empty vector (p < 10−8) and the chimpanzee construct (p < 10−8, Figure 3B). We additionally tested three individual 30-mer sequence variants that are commonly observed in humans. Like the single 30 bp unit found in chimpanzees, these 30-mer variants acted as weak enhancers in the luciferase assay (Figure S7). These results suggest that the expansion of a single 30 bp unit to hundreds of tandem repeats at the CACNA1C locus during human evolution has strengthened an existing enhancer element.
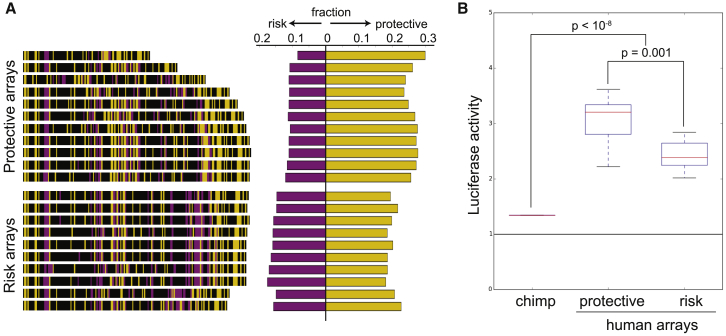
Figure 3.
Human-Specific Repeat Arrays Act as Enhancers in Neural Cells
(A) The single 30-mer found in chimpanzees (30 bp) and 21 different human repeat arrays (3.5–6 kb) were cloned upstream of a minimal promoter driving expression of the luciferase reporter gene. 30-mer variants significantly associated with the protective haplotype are colored yellow, 30-mer variants significantly associated with the risk haplotype are purple, and non-significant variants are black. The fraction of total 30-mer variants associated with either the risk or the protective haplotype varies between the protective-associated and risk-associated repeat arrays as expected, although the differences are subtle.
(B) Constructs were assayed for luciferase activity in a human neural progenitor cell line (ReNcell Cx), as described in Supplemental Methods. Human repeat alleles drove significantly higher luciferase activity compared to the single 30-mer found in chimpanzees (p < 10−8). Protective arrays drove significantly higher luciferase activity than risk arrays (p = 0.001). The p values were calculated using the Wilcoxon rank-sum test.
Although all of the tested human repeat arrays consistently acted as enhancers, there was substantial quantitative variation in enhancer strength among human repeat arrays (Figure S8). To test whether enhancer strength varied for arrays linked to protective or risk GWAS SNPs for neuropsychiatric disease, we determined the genotypes of the individuals from which these human repeat arrays were cloned. Repeat arrays derived from individuals with the protective haplotype were classified as “protective,” and repeat arrays derived from individuals with the risk haplotype were classified as “risk.” For repeat arrays derived from individuals who are heterozygous at the GWAS SNPs, we determined the proportion of protective- and risk-associated 30-mer variants from PacBio sequencing. We then asked whether these proportions most closely resembled individuals with the protective haplotype or individuals with the risk haplotype in the 1000 Genomes Project and designated the ambiguous repeat arrays accordingly (Figure S9, see Supplemental Methods). The repeat arrays characteristic of the protective haplotype drove significantly higher luciferase activity than repeat arrays characteristic of the risk haplotype (p = 0.001, Figure 3). In contrast, we did not observe an association between repeat length in the human repeat arrays we tested (3.2–6 kb) and luciferase activity (Figure S10). These data show that compositional differences between human repeat arrays lead to functional differences in enhancer activity and suggest that differences in the repeat arrays may be causative genomic changes underlying the association between linked CACNA1C markers and susceptibility to neuropsychiatric disease.
While the transcriptional enhancer is located within the CACNA1C locus, the enhancer might also affect the expression of other linked genes.51 In human brain samples, the enhancer is present in a topologically associating domain (TAD) that contains both CACNA1C and seven downstream genes.52, 53 In human dorsolateral prefrontal cortex, more than 90% of Hi-C associations found within 5 kb of the enhancer map to other locations within CACNA1C, including locations near CACNA1C transcription start sites.52 These results are consistent with the enhancer regulating the expression of CACNA1C.
Previous studies have tested whether risk and protective genotypes at the CACNA1C locus lead to higher or lower CACNA1C expression in the brain. Studies in the dorsolateral prefrontal cortex and cerebellum reported decreased CACNA1C expression in individuals with risk variants at human GWAS SNPs.20, 54 In contrast, studies in the superior temporal gyrus and fibroblast-derived induced human neurons reported increased CACNA1C expression in individuals with risk variants at human GWAS SNPs.32, 55 Our data show that risk-associated repeat arrays have reduced enhancer activity in the particular human neural progenitor cell line we tested. We note that differences in human repeat arrays could also underlie more complex expression differences at other tissues or developmental time points. The base pair changes seen in particular 30-mer motifs that are associated with risk or protective genotypes alter the predicted binding sites for a number of potential trans-regulatory factors (Table S1). These factors themselves vary in expression and abundance in different brain regions,56, 57, 58, 59, 60 which could in turn lead to differential effects of repeat variants at different times or places in vivo.
Previous studies of coding region mutations suggest that both loss-of-function and gain-of-function alterations in CACNA1C can lead to behavioral changes in mice and humans with similarities to BD and SCZ. For example, in mouse models where CACNA1C expression levels are either globally reduced or ablated only in specific brain regions, mice display increased anxiety and depression in behavioral tests such as the elevated plus maze, light-dark box, and learned helplessness test.18, 61, 62, 63 Conversely, gain-of-function mutations in CACNA1C lead to Timothy syndrome (TS) in humans, an autosomal-dominant disease where afflicted individuals display autism-like symptoms in addition to a host of non-neurological pathologies.64, 65 Although TS is normally lethal in young children, a rare individual with TS who survived into his late teens developed BD.54 These studies suggest that modulating CACNA1C expression levels, such as through human variation at the repeat arrays we report in this study, could result in behavioral changes associated with BD and SCZ.
We note that the 30-mer repeat arrays might have additional functional effects beyond the enhancer activities we characterize here. For example, the most common 30-mer sequences have open reading frames in both directions (Figure S11A). Previous studies have shown that some tandem repeats are transcribed and translated even in the absence of conventional ATG start codons.66, 67, 68 The tandem repeat also contains canonical splice site consensus sequences, including a donor site, an acceptor site, branch sites, and a polypyrimidine tract (Figure S11B). Intriguingly, the single 30-mer found in chimpanzees has an “A” at the 17th position, whereas the vast majority of human 30-mers (99.94%) have a “G” at that position. This single base pair difference means that chimpanzees do not have canonical splice donor or acceptor sites at this locus. Finally, when organized in head-to-tail fashion, the 30-mers also form a CpG site located between the “C” that ends most 30-mers and the “G” that begins the next 30-mer (Figure S11B). The tandem repeat arrays may affect translation, splicing, or methylation, in addition to forming a functional enhancer sequence within CACNA1C.
Tandem repeats have previously been proposed as a possible causal basis for the evolution of both species-specific traits and individual-to-individual variation in complex phenotypes such as neurological functions in humans.69 Our studies have identified a dramatic expansion of a 30-mer sequence that generates human-specific tandem arrays in a key gene related to calcium signaling, gene expression, and behavior. The human-specific repeat arrays show enhancer activity in human neural progenitor cells, and risk-associated versions of the tandem repeat have less enhancer activity than protective-associated versions. We hypothesize that generation of these repeat arrays has modified CaV1.2 function during human evolution and that structural and compositional differences of the 30-mer repeats among humans represent causal genomic changes that modify risk of neuropsychiatric disease in modern populations.
Many diseases that are particularly common in human populations occur at body locations that have also undergone dramatic and relatively recent evolution in the human lineage. For example, humans have a high incidence of lower back, knee, and foot problems, likely due to the recent evolutionary transition to upright bipedal walking.70 More than 70% of young adults develop impacted third molars (wisdom teeth), likely due to evolutionary reduction of jaw size in the human lineage and modern changes in diet.71, 72 Similarly, the high prevalence of neurological diseases in modern humans may be, in part, due to recent evolutionary changes in genes controlling brain size, connectivity, and function in humans compared to other primates.73, 74 Tandem repeat expansions provide a particularly interesting class of genomic variants in evolution and disease studies because the generation of new tandem repeats can not only alter gene functions between species, but also make the same genes prone to variation and diversity among individuals of the same species.69
Producing new cellular and animal models that carry either chimpanzee or various human 30-mer repeat arrays at the CACNA1C locus should make it possible to further characterize both the evolutionary and disease effects of this repeat region. In addition, the sequence differences in the 30-mer repeats can now be used as a feature to group affected individuals into distinct genetic subtypes. Further stratification of individuals based on CACNA1C repeat genotypes may prove useful for refined disease association studies or for identifying affected individuals who are likely to show favorable responses to drugs targeting calcium channel activity. These drugs have long been available but have produced mixed results as treatments for psychiatric diseases.75, 76
Finally, our research illustrates how characterizing hidden variation in the human genome can uncover variants associated with both human evolution and disease. SNPs are still the most commonly studied type of variant in most genotyping and trait association studies. However, structural variants and repeat sequences make up a substantial fraction of the human genome, show abundant variation both within and between species, and may contribute to key phenotypic traits and disease susceptibilities in humans and other organisms.77
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We wish to thank members of the Kingsley Lab for useful discussions and comments on the manuscript. Research reported in this publication was supported in part by the NIDCR of the National Institutes of Health (K25DE025316 to C.B.L.), and by a National Science Foundation Graduate Research Fellowship and a Stanford Graduate Fellowship (J.H.T.S.). D.M.K. is an Investigator of the Howard Hughes Medical Institute. DNA or tissue samples were obtained through the NIH Neurobiobank from the Human Brain and Spinal Fluid Resource Center (VA West Los Angeles Healthcare Center), the University of Maryland Brain and Tissue Bank, the Harvard Brain Tissue Resource Center, the University of Miami Brain Endowment Bank, the Mt. Sinai Brain Bank, and the Brain Tissue Donation Program at the University of Pittsburgh. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Published: August 9, 2018
Footnotes
Supplemental Data include Supplemental Methods, 11 figures, and 1 table and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.07.011.
Accession Numbers
The accession numbers for the repeat array sequences reported in this paper are GenBank: MH645925–MH645951.
Supplemental Data
References
- 1.Saha S., Chant D., Welham J., McGrath J. A systematic review of the prevalence of schizophrenia. PLoS Med. 2005;2:e141. doi: 10.1371/journal.pmed.0020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merikangas K.R., Jin R., He J.-P., Kessler R.C., Lee S., Sampson N.A., Viana M.C., Andrade L.H., Hu C., Karam E.G. Prevalence and correlates of bipolar spectrum disorder in the world mental health survey initiative. Arch. Gen. Psychiatry. 2011;68:241–251. doi: 10.1001/archgenpsychiatry.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.GBD 2015 Disease and Injury Incidence and Prevalence Collaborators Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1545–1602. doi: 10.1016/S0140-6736(16)31678-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krishnan K.R. Psychiatric and medical comorbidities of bipolar disorder. Psychosom. Med. 2005;67:1–8. doi: 10.1097/01.psy.0000151489.36347.18. [DOI] [PubMed] [Google Scholar]
- 5.Baldessarini R.J., Pompili M., Tondo L. Suicide in bipolar disorder: Risks and management. CNS Spectr. 2006;11:465–471. doi: 10.1017/s1092852900014681. [DOI] [PubMed] [Google Scholar]
- 6.Saha S., Chant D., McGrath J. A systematic review of mortality in schizophrenia: is the differential mortality gap worsening over time? Arch. Gen. Psychiatry. 2007;64:1123–1131. doi: 10.1001/archpsyc.64.10.1123. [DOI] [PubMed] [Google Scholar]
- 7.Forstner A.J., Hecker J., Hofmann A., Maaser A., Reinbold C.S., Mühleisen T.W., Leber M., Strohmaier J., Degenhardt F., Treutlein J. Identification of shared risk loci and pathways for bipolar disorder and schizophrenia. PLoS ONE. 2017;12:e0171595. doi: 10.1371/journal.pone.0171595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferreira M.A., O’Donovan M.C., Meng Y.A., Jones I.R., Ruderfer D.M., Jones L., Fan J., Kirov G., Perlis R.H., Green E.K., Wellcome Trust Case Control Consortium Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat. Genet. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ripke S., Sanders A.R., Kendler K.S., Levinson D.F., Sklar P., Holmans P.A., Lin D.Y., Duan J., Ophoff R.A., Andreassen O.A., Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ripke S., O’Dushlaine C., Chambert K., Moran J.L., Kähler A.K., Akterin S., Bergen S.E., Collins A.L., Crowley J.J., Fromer M., Multicenter Genetic Studies of Schizophrenia Consortium. Psychosis Endophenotypes International Consortium. Wellcome Trust Case Control Consortium 2 Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ripke S., Neale B.M., Corvin A., Walters J.T., Farh K.H., Holmans P.A., Lee P., Bulik-Sullivan B., Collier D.A., Huang H., Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sklar P., Ripke S., Scott L.J., Andreassen O.A., Cichon S., Craddock N., Edenberg H.J., Nurnberger J.I., Rietschel M., Blackwood D., Psychiatric GWAS Consortium Bipolar Disorder Working Group Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat. Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smoller J.W., Ripke S., Lee P.H., Neale B., Nurnberger J.I., Santangelo S., Sullivan P.F., Perlis R.H., Purcell S.M., Fanous A., Cross-Disorder Group of the Psychiatric Genomics Consortium Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruderfer D.M., Fanous A.H., Ripke S., McQuillin A., Amdur R.L., Gejman P.V., O’Donovan M.C., Andreassen O.A., Djurovic S., Hultman C.M., Schizophrenia Working Group of the Psychiatric Genomics Consortium. Bipolar Disorder Working Group of the Psychiatric Genomics Consortium. Cross-Disorder Working Group of the Psychiatric Genomics Consortium Polygenic dissection of diagnosis and clinical dimensions of bipolar disorder and schizophrenia. Mol. Psychiatry. 2014;19:1017–1024. doi: 10.1038/mp.2013.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ament S.A., Szelinger S., Glusman G., Ashworth J., Hou L., Akula N., Shekhtman T., Badner J.A., Brunkow M.E., Mauldin D.E., Bipolar Genome Study Rare variants in neuronal excitability genes influence risk for bipolar disorder. Proc. Natl. Acad. Sci. USA. 2015;112:3576–3581. doi: 10.1073/pnas.1424958112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Purcell S.M., Moran J.L., Fromer M., Ruderfer D., Solovieff N., Roussos P., O’Dushlaine C., Chambert K., Bergen S.E., Kähler A. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrade A., Hope J., Allen A., Yorgan V., Lipscombe D., Pan J.Q. A rare schizophrenia risk variant of CACNA1I disrupts CaV3.3 channel activity. Sci. Rep. 2016;6:34233. doi: 10.1038/srep34233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dedic N., Pöhlmann M.L., Richter J.S., Mehta D., Czamara D., Metzger M.W., Dine J., Bedenk B.T., Hartmann J., Wagner K.V. Cross-disorder risk gene CACNA1C differentially modulates susceptibility to psychiatric disorders during development and adulthood. Mol. Psychiatry. 2018;23:533–543. doi: 10.1038/mp.2017.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nie F., Wang X., Zhao P., Yang H., Zhu W., Zhao Y., Chen B., Valenzuela R.K., Zhang R., Gallitano A.L., Ma J. Genetic analysis of SNPs in CACNA1C and ANK3 gene with schizophrenia: A comprehensive meta-analysis. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2015;168:637–648. doi: 10.1002/ajmg.b.32348. [DOI] [PubMed] [Google Scholar]
- 20.Bigos K.L., Mattay V.S., Callicott J.H., Straub R.E., Vakkalanka R., Kolachana B., Hyde T.M., Lipska B.K., Kleinman J.E., Weinberger D.R. Genetic variation in CACNA1C affects brain circuitries related to mental illness. Arch. Gen. Psychiatry. 2010;67:939–945. doi: 10.1001/archgenpsychiatry.2010.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casamassima F., Hay A.C., Benedetti A., Lattanzi L., Cassano G.B., Perlis R.H. L-type calcium channels and psychiatric disorders: A brief review. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2010;153B:1373–1390. doi: 10.1002/ajmg.b.31122. [DOI] [PubMed] [Google Scholar]
- 22.Erk S., Meyer-Lindenberg A., Schnell K., Opitz von Boberfeld C., Esslinger C., Kirsch P., Grimm O., Arnold C., Haddad L., Witt S.H. Brain function in carriers of a genome-wide supported bipolar disorder variant. Arch. Gen. Psychiatry. 2010;67:803–811. doi: 10.1001/archgenpsychiatry.2010.94. [DOI] [PubMed] [Google Scholar]
- 23.Green E.K., Grozeva D., Jones I., Jones L., Kirov G., Caesar S., Gordon-Smith K., Fraser C., Forty L., Russell E., Wellcome Trust Case Control Consortium The bipolar disorder risk allele at CACNA1C also confers risk of recurrent major depression and of schizophrenia. Mol. Psychiatry. 2010;15:1016–1022. doi: 10.1038/mp.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y., Blackwood D.H., Caesar S., de Geus E.J., Farmer A., Ferreira M.A., Ferrier I.N., Fraser C., Gordon-Smith K., Green E.K., Wellcome Trust Case-Control Consortium Meta-analysis of genome-wide association data of bipolar disorder and major depressive disorder. Mol. Psychiatry. 2011;16:2–4. doi: 10.1038/mp.2009.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hori H., Yamamoto N., Fujii T., Teraishi T., Sasayama D., Matsuo J., Kawamoto Y., Kinoshita Y., Ota M., Hattori K. Effects of the CACNA1C risk allele on neurocognition in patients with schizophrenia and healthy individuals. Sci. Rep. 2012;2:634. doi: 10.1038/srep00634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Q., Shen Q., Xu Z., Chen M., Cheng L., Zhai J., Gu H., Bao X., Chen X., Wang K. The effects of CACNA1C gene polymorphism on spatial working memory in both healthy controls and patients with schizophrenia or bipolar disorder. Neuropsychopharmacology. 2012;37:677–684. doi: 10.1038/npp.2011.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He K., An Z., Wang Q., Li T., Li Z., Chen J., Li W., Wang T., Ji J., Feng G. CACNA1C, schizophrenia and major depressive disorder in the Han Chinese population. Br. J. Psychiatry. 2014;204:36–39. doi: 10.1192/bjp.bp.113.126979. [DOI] [PubMed] [Google Scholar]
- 28.Li J., Zhao L., You Y., Lu T., Jia M., Yu H., Ruan Y., Yue W., Liu J., Lu L. Schizophrenia related variants in CACNA1C also confer risk of autism. PLoS ONE. 2015;10:e0133247. doi: 10.1371/journal.pone.0133247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guenther C.A., Tasic B., Luo L., Bedell M.A., Kingsley D.M. A molecular basis for classic blond hair color in Europeans. Nat. Genet. 2014;46:748–752. doi: 10.1038/ng.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Claussnitzer M., Dankel S.N., Kim K.H., Quon G., Meuleman W., Haugen C., Glunk V., Sousa I.S., Beaudry J.L., Puviindran V. FTO obesity variant circuitry and adipocyte browning in humans. N. Engl. J. Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roussos P., Mitchell A.C., Voloudakis G., Fullard J.F., Pothula V.M., Tsang J., Stahl E.A., Georgakopoulos A., Ruderfer D.M., Charney A. A role for noncoding variation in schizophrenia. Cell Rep. 2014;9:1417–1429. doi: 10.1016/j.celrep.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eckart N., Song Q., Yang R., Wang R., Zhu H., McCallion A.S., Avramopoulos D. Functional characterization of schizophrenia-associated variation in CACNA1C. PLoS ONE. 2016;11:e0157086. doi: 10.1371/journal.pone.0157086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gardiner S.L., van Belzen M.J., Boogaard M.W., van Roon-Mom W.M.C., Rozing M.P., van Hemert A.M., Smit J.H., Beekman A.T.F., van Grootheest G., Schoevers R.A. Huntingtin gene repeat size variations affect risk of lifetime depression. Transl. Psychiatry. 2017;7:1277. doi: 10.1038/s41398-017-0042-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gardiner S.L., van Belzen M.J., Boogaard M.W., van Roon-Mom W.M.C., Rozing M.P., van Hemert A.M., Smit J.H., Beekman A.T.F., van Grootheest G., Schoevers R.A. Large normal-range TBP and ATXN7 CAG repeat lengths are associated with increased lifetime risk of depression. Transl. Psychiatry. 2017;7:e1143. doi: 10.1038/tp.2017.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landefeld C.C., Hodgkinson C.A., Spagnolo P.A., Marietta C.A., Shen P.-H., Sun H., Zhou Z., Lipska B.K., Goldman D. Effects on gene expression and behavior of untagged short tandem repeats: the case of arginine vasopressin receptor 1a (AVPR1a) and externalizing behaviors. Transl. Psychiatry. 2018;8:72. doi: 10.1038/s41398-018-0120-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindblad K., Savontaus M.L., Stevanin G., Holmberg M., Digre K., Zander C., Ehrsson H., David G., Benomar A., Nikoskelainen E. An expanded CAG repeat sequence in spinocerebellar ataxia type 7. Genome Res. 1996;6:965–971. doi: 10.1101/gr.6.10.965. [DOI] [PubMed] [Google Scholar]
- 38.Renton A.E., Majounie E., Waite A., Simón-Sánchez J., Rollinson S., Gibbs J.R., Schymick J.C., Laaksovirta H., van Swieten J.C., Myllykangas L., ITALSGEN Consortium A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.The Huntington’s Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 40.Verkerk A.J., Pieretti M., Sutcliffe J.S., Fu Y.H., Kuhl D.P., Pizzuti A., Reiner O., Richards S., Victoria M.F., Zhang F.P. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 41.Hannan A.J. Tandem repeat polymorphisms: modulators of disease susceptibility and candidates for ‘missing heritability’. Trends Genet. 2010;26:59–65. doi: 10.1016/j.tig.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 42.Hannan A.J. Tandem repeats mediating genetic plasticity in health and disease. Nat. Rev. Genet. 2018;19:286–298. doi: 10.1038/nrg.2017.115. [DOI] [PubMed] [Google Scholar]
- 43.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meyer M., Kircher M., Gansauge M.-T., Li H., Racimo F., Mallick S., Schraiber J.G., Jay F., Prüfer K., de Filippo C. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prüfer K., Racimo F., Patterson N., Jay F., Sankararaman S., Sawyer S., Heinze A., Renaud G., Sudmant P.H., de Filippo C. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levy S., Sutton G., Ng P.C., Feuk L., Halpern A.L., Walenz B.P., Axelrod N., Huang J., Kirkness E.F., Denisov G. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chaisson M.J., Huddleston J., Dennis M.Y., Sudmant P.H., Malig M., Hormozdiari F., Antonacci F., Surti U., Sandstrom R., Boitano M. Resolving the complexity of the human genome using single-molecule sequencing. Nature. 2015;517:608–611. doi: 10.1038/nature13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Osoegawa K., Mammoser A.G., Wu C., Frengen E., Zeng C., Catanese J.J., de Jong P.J. A bacterial artificial chromosome library for sequencing the complete human genome. Genome Res. 2001;11:483–496. doi: 10.1101/gr.169601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Visel A., Taher L., Girgis H., May D., Golonzhka O., Hoch R.V., McKinsey G.L., Pattabiraman K., Silberberg S.N., Blow M.J. A high-resolution enhancer atlas of the developing telencephalon. Cell. 2013;152:895–908. doi: 10.1016/j.cell.2012.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kleinjan D.A., van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmitt A.D., Hu M., Jung I., Xu Z., Qiu Y., Tan C.L., Li Y., Lin S., Lin Y., Barr C.L., Ren B. A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep. 2016;17:2042–2059. doi: 10.1016/j.celrep.2016.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Won H., de la Torre-Ubieta L., Stein J.L., Parikshak N.N., Huang J., Opland C.K., Gandal M.J., Sutton G.J., Hormozdiari F., Lu D. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature. 2016;538:523–527. doi: 10.1038/nature19847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gershon E.S., Grennan K., Busnello J., Badner J.A., Ovsiew F., Memon S., Alliey-Rodriguez N., Cooper J., Romanos B., Liu C. A rare mutation of CACNA1C in a patient with bipolar disorder, and decreased gene expression associated with a bipolar-associated common SNP of CACNA1C in brain. Mol. Psychiatry. 2014;19:890–894. doi: 10.1038/mp.2013.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshimizu T., Pan J.Q., Mungenast A.E., Madison J.M., Su S., Ketterman J., Ongur D., McPhie D., Cohen B., Perlis R., Tsai L.H. Functional implications of a psychiatric risk variant within CACNA1C in induced human neurons. Mol. Psychiatry. 2015;20:162–169. doi: 10.1038/mp.2014.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bulfone A., Martinez S., Marigo V., Campanella M., Basile A., Quaderi N., Gattuso C., Rubenstein J.L., Ballabio A. Expression pattern of the Tbr2 (Eomesodermin) gene during mouse and chick brain development. Mech. Dev. 1999;84:133–138. doi: 10.1016/s0925-4773(99)00053-2. [DOI] [PubMed] [Google Scholar]
- 57.Lein E.S., Hawrylycz M.J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A.F., Boguski M.S., Brockway K.S., Byrnes E.J. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 58.Liu X., Bates R., Yin D.M., Shen C., Wang F., Su N., Kirov S.A., Luo Y., Wang J.Z., Xiong W.C., Mei L. Specific regulation of NRG1 isoform expression by neuronal activity. J. Neurosci. 2011;31:8491–8501. doi: 10.1523/JNEUROSCI.5317-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hawrylycz M.J., Lein E.S., Guillozet-Bongaarts A.L., Shen E.H., Ng L., Miller J.A., van de Lagemaat L.N., Smith K.A., Ebbert A., Riley Z.L. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller J.A., Ding S.L., Sunkin S.M., Smith K.A., Ng L., Szafer A., Ebbert A., Riley Z.L., Royall J.J., Aiona K. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dao D.T., Mahon P.B., Cai X., Kovacsics C.E., Blackwell R.A., Arad M., Shi J., Zandi P.P., O’Donnell P., Knowles J.A., Bipolar Genome Study (BiGS) Consortium Mood disorder susceptibility gene CACNA1C modifies mood-related behaviors in mice and interacts with sex to influence behavior in mice and diagnosis in humans. Biol. Psychiatry. 2010;68:801–810. doi: 10.1016/j.biopsych.2010.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jeon D., Kim S., Chetana M., Jo D., Ruley H.E., Lin S.Y., Rabah D., Kinet J.P., Shin H.S. Observational fear learning involves affective pain system and Cav1.2 Ca2+ channels in ACC. Nat. Neurosci. 2010;13:482–488. doi: 10.1038/nn.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee A.S., Ra S., Rajadhyaksha A.M., Britt J.K., De Jesus-Cortes H., Gonzales K.L., Lee A., Moosmang S., Hofmann F., Pieper A.A., Rajadhyaksha A.M. Forebrain elimination of cacna1c mediates anxiety-like behavior in mice. Mol. Psychiatry. 2012;17:1054–1055. doi: 10.1038/mp.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Splawski I., Timothy K.W., Sharpe L.M., Decher N., Kumar P., Bloise R., Napolitano C., Schwartz P.J., Joseph R.M., Condouris K. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119:19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 65.Splawski I., Timothy K.W., Decher N., Kumar P., Sachse F.B., Beggs A.H., Sanguinetti M.C., Keating M.T. Severe arrhythmia disorder caused by cardiac L-type calcium channel mutations. Proc. Natl. Acad. Sci. USA. 2005;102:8089–8096. doi: 10.1073/pnas.0502506102. discussion 8086–8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zu T., Gibbens B., Doty N.S., Gomes-Pereira M., Huguet A., Stone M.D., Margolis J., Peterson M., Markowski T.W., Ingram M.A. Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. USA. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cleary J.D., Ranum L.P. Repeat-associated non-ATG (RAN) translation in neurological disease. Hum. Mol. Genet. 2013;22(R1):R45–R51. doi: 10.1093/hmg/ddt371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bañez-Coronel M., Ayhan F., Tarabochia A.D., Zu T., Perez B.A., Tusi S.K., Pletnikova O., Borchelt D.R., Ross C.A., Margolis R.L. RAN translation in Huntington disease. Neuron. 2015;88:667–677. doi: 10.1016/j.neuron.2015.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nithianantharajah J., Hannan A.J. Dynamic mutations as digital genetic modulators of brain development, function and dysfunction. BioEssays. 2007;29:525–535. doi: 10.1002/bies.20589. [DOI] [PubMed] [Google Scholar]
- 70.Pennisi E. Evolutionary biology. The burdens of being a biped. Science. 2012;336:974. doi: 10.1126/science.336.6084.974. [DOI] [PubMed] [Google Scholar]
- 71.Stedman H.H., Kozyak B.W., Nelson A., Thesier D.M., Su L.T., Low D.W., Bridges C.R., Shrager J.B., Minugh-Purvis N., Mitchell M.A. Myosin gene mutation correlates with anatomical changes in the human lineage. Nature. 2004;428:415–418. doi: 10.1038/nature02358. [DOI] [PubMed] [Google Scholar]
- 72.Lieberman D. Pantheon Books; New York: 2013. The Story of the Human Body: Evolution, Health, and Disease. [PubMed] [Google Scholar]
- 73.Oksenberg N., Stevison L., Wall J.D., Ahituv N. Function and regulation of AUTS2, a gene implicated in autism and human evolution. PLoS Genet. 2013;9:e1003221. doi: 10.1371/journal.pgen.1003221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Srinivasan S., Bettella F., Mattingsdal M., Wang Y., Witoelar A., Schork A.J., Thompson W.K., Zuber V., Winsvold B.S., Zwart J.A., Schizophrenia Working Group of the Psychiatric Genomics Consortium, The International Headache Genetics Consortium Genetic markers of human evolution are enriched in schizophrenia. Biol. Psychiatry. 2016;80:284–292. doi: 10.1016/j.biopsych.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hollister L.E., Trevino E.S. Calcium channel blockers in psychiatric disorders: a review of the literature. Can. J. Psychiatry. 1999;44:658–664. doi: 10.1177/070674379904400702. [DOI] [PubMed] [Google Scholar]
- 76.Zamponi G.W. Targeting voltage-gated calcium channels in neurological and psychiatric diseases. Nat. Rev. Drug Discov. 2016;15:19–34. doi: 10.1038/nrd.2015.5. [DOI] [PubMed] [Google Scholar]
- 77.Kronenberg Z.N., Fiddes I.T., Gordon D., Murali S., Cantsilieris S., Meyerson O.S., Underwood J.G., Nelson B.J., Chaisson M.J.P., Dougherty M.L. High-resolution comparative analysis of great ape genomes. Science. 2018;360 doi: 10.1126/science.aar6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.