Abstract
Plastid genomes are not normally celebrated for being large. But researchers are steadily uncovering algal lineages with big and, in rare cases, enormous plastid DNAs (ptDNAs), such as volvocine green algae. Plastome sequencing of five different volvocine species has revealed some of the largest, most repeat-dense plastomes on record, including that of Volvox carteri (∼525 kb). Volvocine algae have also been used as models for testing leading hypotheses on organelle genome evolution (e.g., the mutational hazard hypothesis), and it has been suggested that ptDNA inflation within this group might be a consequence of low mutation rates and/or the transition from a unicellular to multicellular existence. Here, we further our understanding of plastome size variation in the volvocine line by examining the ptDNA sequences of the colonial species Yamagishiella unicocca and Eudorina sp. NIES-3984 and the multicellular Volvox africanus, which are phylogenetically situated between species with known ptDNA sizes. Although V. africanus is closely related and similar in multicellular organization to V. carteri, its ptDNA was much less inflated than that of V. carteri. Synonymous- and noncoding-site nucleotide substitution rate analyses of these two Volvox ptDNAs suggest that there are drastically different plastid mutation rates operating in the coding versus intergenic regions, supporting the idea that error-prone DNA repair in repeat-rich intergenic spacers is contributing to genome expansion. Our results reinforce the idea that the volvocine line harbors extremes in plastome size but ultimately shed doubt on some of the previously proposed hypotheses for ptDNA inflation within the lineage.
Keywords: Eudorina, genome expansion, mutational hazard hypothesis, Volvox, Yamagishiella
Introduction
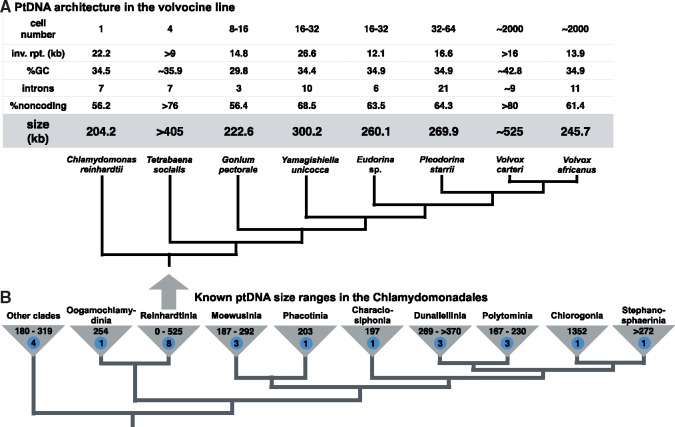
When thinking of genome expansion, plastid DNAs (ptDNAs) may not immediately come to mind. Indeed, of the more than 2,800 complete plastid genome sequences available in GenBank, 98% are under 200 kilobases (kb) and harbor modest amounts (<50%) of intergenic and intronic DNA. But there are a few known algal lineages whose members can have surprisingly large ptDNAs with an abundance of noncoding nucleotides (Brouard et al. 2010; de Vries et al. 2013; Del Cortona et al. 2017; Muñoz-Gómez et al. 2017; Bauman et al. 2018). One such lineage is the Chlamydomonadales (fig. 1B), an order of primarily freshwater flagellates within the chlorophycean class of the Chlorophyta (Nakada et al. 2008).
Fig. 1.
—Variation in plastid genome size across the volvocine line (A) and chlamydomonadalean algae (B). Organismal and plastid genome features are shown in the table above the tree in (A) (inverted repeat region; inv. rpt.). Branching orders based on published phylogenetic analyses, including Nozaki et al. (2006, 2015), Nakada et al. (2008), Herron et al. (2009), and Lemieux et al. (2015). Genome sizes based on published and/or GenBank data. Sample size shown in blue circle.
A number of chlamydomonadalean species have been shown to have ptDNAs in excess of 300 kb (fig. 1B), including Haematococcus lacustris (Bauman et al. 2018), Carteria cerasiformis (Lemieux et al. 2015), and Dunaliella salina CONC-001 (Del Vasto et al. 2015). Plastid genome expansion is particularly prevalent within the volvocine line of the Chlamydomonadales, which spans the gamut of cellular complexity from unicellular species (e.g., Chlamydomonas reinhardtii) to colonial forms (e.g., Gonium pectorale) to complex multicellular taxa with cell differentiation and specialization (e.g., Volvox carteri) (fig. 1A) (Herron et al. 2009; Coleman 2012). Plastome sequencing of five different volvocine algae has uncovered a more than a 2-fold variation in ptDNA size and some of the largest, most repeat-dense plastomes on record (fig. 1A) (Smith 2018). For example, the ptDNA of the four-celled Tetrabaena socialis is >405 kb (Featherston et al. 2016) and that of V. carteri is ∼525 kb (Smith and Lee 2010).
Volvocine algae have also been used as models for testing leading hypotheses on organelle genome evolution, such as the mutational hazard hypothesis (MHH) (Lynch et al. 2006; Smith 2016). It has been suggested that ptDNA inflation within this group might be a consequence of low mutation rates (Smith and Lee 2010) and/or the transition from a unicellular to multicellular existence (resulting in a decrease in effective population size) (Smith et al. 2013), both of which are consistent with the MHH. More recently, analyses of large mitochondrial DNAs (mtDNAs) in land plants have indicated that organelle genome expansion can result from error-prone DNA repair mechanisms within intergenic regions, such as break-induced replication (BIR), which are exacerbated by high numbers of repeats (Christensen 2013, 2017). But such a model is yet to be applied to volvocine ptDNAs.
Here, we extend our understanding of plastid genome size in the volvocine line by examining the ptDNAs of three previously unexplored species, namely the colonial (16–32 cells) Yamagishiella unicocca and Eudorina sp. NIES-3984 and the multicellular (∼2000 cells) Volvox africanus, which are phylogenetically positioned between species with known ptDNA sizes (fig. 1A). We then use a subset of these data to investigate if error-prone DNA repair is contributing to genome expansion within this group. Our results reinforce the idea that the volvocine line harbors extremes in plastome size but shed doubt on some of the previously proposed hypotheses for ptDNA inflation within the lineage.
Materials and Methods
Yamagishiella unicocca strain NIES-3982 (mating type plus), Eudorina sp. strain NIES-3984 (female), and V. africanus strain 2013-0703-VO4 (NIES-3780) were grown in 300 mL SVM medium (Kirk and Kirk 1983) at 25°C on a 14:10 h light–dark cycle (150–180 mmol photons/m2/sec). Total DNA from each species was isolated following the protocol of Miller et al. (1993) and sequenced using PacBio (RS II system; SMRTbell 10- and 20-kb library preparations) and Illumina (HiSeq 2000 and 2500 systems; paired-end TruSeq library prep kit) technologies. The Eudorina sp. PacBio subreads were mapped to the C. reinhardtii and G. pectorale plastid genomes using BLASR (Pacific Biosciences) and then assembled de novo with HGAP3 (Pacific Biosciences). The Illumina data were then mapped against the PacBio assembly using BWA-MEM Release 0.7.7 (Li and Durbin 2010), including error correction with the samtools/bcftools/vcfutils.pl program v0.1.19 (http://samtools.sourceforge.net/), ultimately giving a complete Eudorina sp. plastid genome sequence (GenBank accession MH267732). Similarly, the Y. unicocca and V. africanus PacBio subreads were mapped against the Eudorina sp. ptDNA using BLAST v2.2.26, and then assembled and corrected as described earlier, yielding complete Y. unicocca and V. africanus plastome sequences (GenBank accession MH285950 and MH285951, respectively). Any unmapped regions were PCR-amplified from genomic DNA and sequenced using an ABI 3730xl DNA Analyzer (Applied Biosystems).
Plastid genes were aligned with MUSCLE (Edgar 2004), implemented through Geneious v10.2.4 (Biomatters Ltd., Auckland, NZ) and using the “translational align” setting (default parameters). Start and stop codons as well as any gapped sites were removed from each alignment, ensuring not to alter the reading frame. Synonymous and nonsynonymous substitutions were measured with the CODEML program of PAML v4.3 (Yang 2007), employing the maximum likelihood method and the F3x4 codon model. We initially attempted to pairwise align the entire intergenic regions end-to-end with MUSCLE (default parameters) and then with the Geneious aligner, first selecting global alignment (Needleman–Wunsch) and then trying again using a global alignment with free end gaps (cost-matrix = 93% similarity). Local alignments of the intergenic regions were performed with the Geneious aligner using the Smith–Waterman approach and a 93% similarity cost-matrix. All of the gaps were removed from the local alignments prior to measuring the intergenic ptDNA substitution rates, which were estimated with BASEML of PAML using the HKY85 model.
Results and Discussion
Multiple Instances of ptDNA Expansion/Contraction across the Volvocine Line
The complete ptDNA sequences of Y. unicocca, Eudorina sp., and V. africanus show that volvocine ptDNA length is even more dynamic and diverse than previously thought, varying significantly across the entire lineage and among closely related species (fig. 1A). With sizes of 300, 260, and 246 kb, respectively, and with >60% noncoding DNA, all three of these newly sequenced plastid genomes are large and bloated, but it is the length of the V. africanus ptDNA that is, perhaps, the most surprising.
Volvox africanus is closely affiliated with V. carteri (fig. 1A), which has the largest plastid genome observed from the volvocine line (∼525 kb; >80% noncoding) (Smith and Lee 2010). Thus, one might have expected V. africanus also to have a very large ptDNA, but, in fact, its plastome size is half that of V. carteri, despite these two species having the same number of genes. Or, put differently, the V. carteri plastome contains ∼279 kb more noncoding DNA than that of V. africanus. The comparatively small size of the V. africanus ptDNA casts doubt on the idea that volvocine plastome size scales positively with organismal complexity (Smith et al. 2013).
As the first volvocine ptDNAs (and mtDNAs) were sequenced an interesting trend was observed: the larger the cell number, the bigger and more repeat-rich the ptDNA (Smith et al. 2013). It was argued that this trend was at least partly the result of increased random genetic drift, due to smaller effective population sizes in species with greater cell numbers, supporting the hypothesis of Lynch and Conery (2003). Now, with the availability of more data from diverse volvocine species with varying levels of cell number and complexity, one can easily see that this trend breaks down (fig. 1A). Species with similar cell numbers and levels of complexity (e.g., V. carteri and V. africanus) can have very different plastome sizes and repeat contents (fig. 2A and C), and in some cases taxa with small numbers of cells (e.g., T. socialis) have much larger ptDNAs than those with many cells (e.g., Pleodorina starrii) (fig. 1A). The same is also true when looking at volvocine mtDNAs (Hamaji et al. 2017). Moreover, researchers are beginning to find an equally large variation in ptDNA size in chlamydomonadalean clades outside the volvocine line (fig. 1B), which are made up entirely of unicellular taxa.
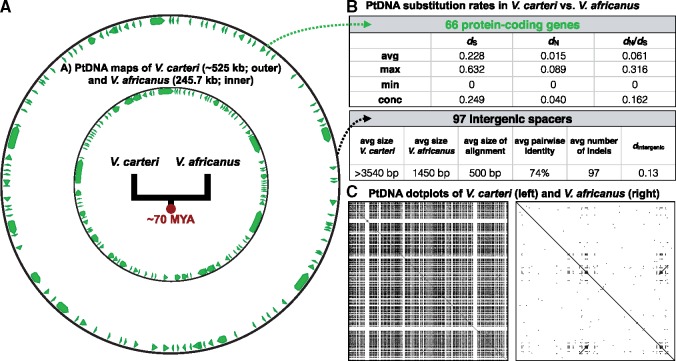
Fig. 2.
—Comparisons between the Volvox carteri and V. africanus plastid genomes, including genomic maps (A), genetic divergence (B), and repeat content (C). Coding regions shown in green. Substitution rates statistics of intergenic regions based on local alignments. Dotplot was generated using dottup (EMBOSS 6.5.7) using a word size of 20.
When ignoring differences in size, the available volvocine ptDNAs have highly conserved architectures, characterized by circular mapping structures, low GC contents (29.8–42.8%), a pair of large inverted repeats, near identical gene repertoires and gene orders (when ignoring nonstandard genes), and modest numbers of introns (3–21) (fig. 1A). In fact, of the three newly sequenced volvocine ptDNAs presented here, it is hard to find any noteworthy differences among them and previously published volvocine plastomes, save for a moderately inflated inverted repeat element in Y. unicocca, which, at 26.6 kb, is 4.4–14.5 kb larger than those of its close relatives (fig. 1A). The increased size of this repeat is a consequence of it having more noncoding nucleotides rather than additional genes.
Given all of these similarities, why have some volvocine ptDNAs been pushed to such extremes in size and why are their lengths (i.e., the amounts of noncoding ptDNA) so variable across the lineage? The answer to these two questions, as described below, might lie in the nature of the noncoding ptDNA itself.
Substitution Rates Are Low at Synonymous Sites but Potentially High within Intergenic Regions
To better understand the underlying plastid mutation rates within the volvocine lineage, we measured the number of nucleotide substitutions between the V. carteri and V. africanus ptDNAs (fig. 2B andsupplementary table S1, Supplementary Material online). These two genomes, which represent the most closely related pair among the currently available volvocine plastomes (fig. 1A), have an identical gene order and content, allowing us to explore substitutions within coding and intergenic regions (fig. 2A and B). Alignments of the 66 protein-coding genes (totalling 78,288 ungapped bases) revealed an average synonymous-site substitution rate (dS) of 0.228, ranging from 0 (psbI and psbN) to 0.632 (petD) (fig. 2B). The nonsynonymous substitution rate (dN) was much lower, averaging 0.015 and maxing out at 0.089 (ftsH). The dN/dS ratio, which can give insights into the directionality and intensity of natural selection, averaged 0.061 (maximum = 0.36 rpoC2), suggesting that most of the genes in the V. carteri and V. africanus plastid are under strong purifying selection. Substitution rate analyses of an alignment of the concatenated protein-coding genes gave similar values (fig. 2B). [Note: plastid synonymous substitution rates between the other available volvocine ptDNAs was at or close to saturation (data not shown).]
If synonymous sites are assumed to be neutral, dS can provide an entrée into mutation rate (Kimura 1983). When considering that the lineages giving rise to V. carteri and V. africanus are estimated to have diverged from one another ∼70 Ma (Herron et al. 2009), an average synonymous site divergence of ∼0.2 is arguably far from high. For comparison, the average level of dS for seed plant ptDNA was estimated to be ∼0.6 in gymnosperms and ∼0.4 in angiosperms, and ∼0.2 for seed plant mtDNAs, which are renowned for having extremely low genic mutation rates (Drouin et al. 2008; but see Christensen 2013). The estimated divergence times for most major seed plant groups is between 50 and 150 Myr (Lu et al. 2014; Zeng et al. 2014). In other green algal genera, dS has been found to range from 0.09 (Dunaliella) to 0.30 (Chlamydomonas) to 0.76 (Ostreococcus) (Smith 2015); unfortunately, associated divergence times are lacking for these data, but mutation accumulation experiments in C. reinhardtii are indicative of a low ptDNA mutation rate (Ness et al. 2016). Finally, in animal mtDNA, the number of substitutions per synonymous site per million years has been shown to range from 0.01 to 0.18 (Nabholz et al. 2008). All of this points to a low/moderate level of dS in Volvox as compared with other organelle systems, which could be indicative of a low to moderate mutation rate. However, it should be stressed that dS can vary greatly within lineages (Bromham et al. 2015; Park et al. 2017) and even within individual organelle genomes (Zhu et al. 2014), not to mention the problems associated with assuming neutrality of synonymous sites and the many challenges of calculating accurate divergence times.
Further indication of a potentially low genic mutation rate in Volvox has come from previous experiments showing very low levels of within-species synonymous-site genetic diversity for V. carteri (Smith and Lee 2010). These data are in line with the mutational hazard hypothesis, which argues that a low mutation rate is more conducive for organelle genome expansion than a high one because the burden of harboring excess noncoding DNA is reduced (Lynch et al. 2006; Smith 2016). If true, one would also expect to find a low mutation rate—that is, a low substitution rate—in the intergenic DNA of expanded organelle genomes. However, our substitution rate analyses of the intergenic regions from V. carteri and V. africanus found the opposite.
When ignoring duplicates, there are 97 orthologous intergenic regions in the V. carteri and V. africanus ptDNAs (fig. 2A). But the average intergenic length of these two genomes differs dramatically (>3540 bp for V. carteri vs. 1450 bp in V. africanus) (fig. 2B), as does the number of repeats, with the V. carteri intergenic ptDNA harboring many more than that of V. africanus (fig. 2C). Not surprisingly given these differences in size and repeat content, our attempts to do pairwise global (i.e., end-to-end) alignments of the noncoding ptDNA failed for all but 30 of the 97 intergenic spacers. And the regions that did successfully align had an average pairwise identity of only 60% as well as dozens to hundreds of insertions and deletions (indels).
We then carried out pairwise local alignments of the 97 intergenic spacers to look for shorter regions of similarity between the corresponding noncoding regions. These, too, gave poor results (fig. 2B andsupplementary table S2, Supplementary Material online): the average alignment length was just 500 bp (∼400 bp with gaps removed), ranging from 29 bp (trnI-trnA spacer) to 3100 bp (psaA-psbJ spacer); the average pairwise identity was 74%; and the average number of indels was 97. Nevertheless, we trimmed and polished each local alignment and measured the per-base substitution rate (dintergenic), which averaged 0.13, with a max and min of 0.18 and 0.01 (fig. 2B). In other words, when looking only at the short, conserved segments of the intergenic ptDNA, and when ignoring indels, the substitution rate of noncoding sites was about half that of synonymous sites (0.13 vs. 0.23). This is a gross underestimate of the true substitution rate of the intergenic ptDNA, which in reality is likely ≫0.23, and altogether these data suggest that the intergenic regions from the V. carteri and V. africanus plastomes have much higher rates of mutation than the synonymous sites of coding regions.
These findings parallel previous work on angiosperm mtDNAs, which found low and high level of nucleotide substitution at synonymous and intergenic sites, respectively (Christensen 2013, 2017). It is not known why the observed substitution rates differed so significantly between these two types of silent sites, but Christensen (2013, 2017) believed it might be a reflection of different types of double-strand break DNA repair occurring in land plant mitochondria. He proposed that coding regions are repaired using long homology-based repair mechanisms, such as gene conversion or homologous recombination, which can be very accurate and do not normally result in genomic expansion. But he argued that intergenic regions were repaired using short-, micro-, or nonhomology-based repair pathways, such as nonhomologous end-joining or microhomology-mediated break-induced replication, which are error prone and can often lead to genomic expansion and rearrangements and are exacerbated by repeats, thus explaining why land plant mtDNA is so prone to inflation.
These hypotheses about land plant mtDNA repair mechanisms and their impact on genome size fit nicely with the available data on volvocine ptDNAs. They not only explain the differences in plastid substitution rates (when factoring in indels and alignment length) between the synonymous and intergenic sites of V. carteri and V. africanus but can also account for why volvocine plastome size is so large and variable (fig. 1A). Moreover, the cooccurrence of repeats with particularly large ptDNA lengths (fig. 2C) (Smith et al. 2013) further supports the view that error-prone repair of noncoding regions, often resulting in genome expansion, is intensified by the presence of repetitive DNA. Plastome nucleotide divergence data from more closely related volvocine species will help to further address this hypothesis, particularly from species that are closely enough related to provide more accurate alignments of the noncoding regions. On this front, it is worth noting that the previously described plastid nucleotide diversity data from V. carteri did not uncover a single polymorphism at synonymous sites but observed multiple polymorphisms and, in some cases, massive indels (>360 bp) in the intergenic regions (Smith and Lee 2010), indicating that the repeat-rich noncoding DNA is truly a hotspot for mutations in volvocine algae.
Other Chlamydomonadalean Clades with Extremely Large ptDNAs
One of the main goals of this work was to identify and characterize giant plastomes from the volvocine line. This objective made sense in that volvocine algae have been shown to contain some of the largest ptDNAs within the Eukaryota (Smith 2018). However, recent explorations of other algal lineages have uncovered even more examples of extremely large plastomes (Muñoz-Gómez et al. 2017), and the volvocine line is now just one among many groups known to harbor prodigious ptDNAs. In fact, there are other chlamydomonadalean clades that might be even more diverse with respect to ptDNA size than the volvocine line.
Indeed, as we were analyzing the data for this study, we were taken off guard by the recently sequenced plastome of the beta-carotene-rich chlamydomonadalean Haematococcus lacustris, which, at 1.35 Mb (fig. 1B), is not only the biggest known green algal ptDNA but the biggest of all eukaryotes. This gigantic organellar genome sequence, generated by a small team of scientists from the private company Synthetic Genomics, was deposited in GenBank in late February 2018 (accession number NC_037007) and described in an article for Genome Announcements (Bauman et al. 2018), a nonpeer-reviewed journal publishing short reports (∼500 words) of new microbial genome sequences. Both the sequence and publication have likely gone unnoticed by many in the organelle genome community, and with such an unprecedently large size it is, in our opinion, unfortunate that the genome was not presented on a wider platform. Indeed, its closest rival in size, the large, intron-dense plastome of the red alga Corynoplastis japonica (1.13 Mb) was published in Current Biology in June 2017 (Muñoz-Gómez et al. 2017) and was the focus of a highlight article (Moreira and López-García 2017).
Unfortunately, the presently available annotation of the H. lacustris plastome, which was carried out using GeneMarkS (Besemer et al. 2001) and the Synthetic Genomics proprietary Archetype annotation pipeline (Gallone et al. 2016), is incomplete and contains some inaccuracies. The authors identified 125 protein-coding regions and 12 tRNAs, but they did not find any rRNAs. Approximately half of the annotated protein-coding regions are hypothetical or intronic open-reading frames (ORFs) and the other half includes multiple proteins not normally encoded in plastids (e.g., nicotine oxidoreductase) as well as partial or discontinuous annotations. These errors are most likely, in part, a reflection of the annotation pipeline that was used, rather than an unconventional coding content. When the H. lacustris ptDNA sequence is run through an organelle genome annotation program, such as GeSeq (Tillich et al. 2017) or MFannot (Lang et al. 2007), an ordinary and complete stock of plastid protein-coding genes and functional RNAs is easily identified, including 31 tRNAs. Most of these genes were previously annotated (and deposited in GenBank) by Lemieux et al. (2015), who generated a partial plastid genome assembly of H. lacustris.
This is not meant to be a critique of the scientists at Synthetic Genomics, who are focused on applied research rather than plastid genome evolution, and it is commendable that they took the time to publish and upload this sequence to GenBank. We mention the H. lacustris ptDNA merely to stress that one of the most interesting ptDNA sequences in GenBank is poorly annotated and merits further attention. If anything, the H. lacustris plastome underscores the fact that we are only just beginning to appreciate the true limits of plastid genome size within the Chlamydomonadales and to fully understand the forces underlying organelle DNA expansion. It will be interesting to see if the trends described here for V. carteri and V. africanus regarding the rate of mutation in coding versus noncoding regions also hold true for H. lacustris and other gigantic plastomes, such as those from red algae. Whatever the case, we undoubtedly still have a lot to learn about the diversity and evolution of plastomes, especially those at the upper end of the size spectrum.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by a Discovery Grant from the Natural Sciences and Engineering Research Council (NSERC) of Canada (to D.R.S.); a Grants-in-Aid for Scientific Research on Innovative Areas [grant numbers JP221S0002 (to A.T. and A.F.) and JP16H06279 (to A.T.)] and Scientific Research (A) (grant number JP16H02518; to H.N.); JSPS Research Fellow (grant number JP18J11391; to K.Y.); and Research Activity Start-up (grant number JP16H06734 to T.H.) from MEXT/JSPS KAKENHI.
Literature Cited
- Bauman N, et al. 2018. Next-generation sequencing of Haematococcus lacustris reveals an extremely large 1.35-megabase chloroplast genome. Genome Announc. 612:e00181–e00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer J, Lomsadze A, Borodovsky M.. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2912:2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromham L, Hua X, Lanfear R, Cowman PF.. 2015. Exploring the relationships between mutation rates, life history, genome size, environment, and species richness in flowering plants. Am Nat. 1854:507–524. [DOI] [PubMed] [Google Scholar]
- Brouard JS, Otis C, Lemieux C, Turmel M.. 2010. The exceptionally large chloroplast genome of the green alga Floydiella terrestris illuminates the evolutionary history of the Chlorophyceae. Genome Biol Evol. 2:240–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AC. 2013. Plant mitochondrial genome evolution can be explained by DNA repair mechanisms. Genome Biol Evol. 56:1079–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AC. 2017. Mitochondrial DNA repair and genome evolution. Ann Plant Rev. 50:11–31. [Google Scholar]
- Coleman AW. 2012. A comparative analysis of Volvocaceae (Chlorophyta). J Phycol. 483:491–513. [DOI] [PubMed] [Google Scholar]
- de Vries J, et al. 2013. Is ftsH the key to plastid longevity in sacoglossan slugs? Genome Biol Evol. 512:2540–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Cortona A, et al. 2017. The plastid genome in Cladophorales green algae is encoded by hairpin chromosomes. Curr Biol. 2724:3771–3782. [DOI] [PubMed] [Google Scholar]
- Del Vasto M, et al. 2015. Massive and widespread organelle genomic expansion in the green algal genus Dunaliella. Genome Biol Evol. 73:656–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin G, Daoud H, Xia J.. 2008. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol Phylogenet Evol. 493:827–831. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 325:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Featherston J, Arakaki Y, Nozaki H, Durand PM, Smith DR.. 2016. Inflated organelle genomes and a circular-mapping mtDNA probably existed at the origin of coloniality in volvocine green algae. Eur J Phycol. 514:369–377. [Google Scholar]
- Gallone B, et al. 2016. Domestication and divergence of Saccharomyces cerevisiae beer yeasts. Cell 1666:1397–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaji T, et al. 2017. Multiple independent changes in mitochondrial genome conformation in chlamydomonadalean algae. Genome Biol Evol. 94:993–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron MD, Hackett JD, Aylward FO, Michod RE.. 2009. Triassic origin and early radiation of multicellular volvocine algae. Proc Natl Acad Sci U S A. 1069:3254–3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. 1983. The neutral theory of molecular evolution. Cambridge (England: ): Cambridge University Press. [Google Scholar]
- Kirk DL, Kirk MM.. 1983. Protein synthetic patterns during the asexual life cycle of Volvox carteri. Dev Biol. 962:493–506. [DOI] [PubMed] [Google Scholar]
- Lang BF, Laforest MJ, Burger G.. 2007. Mitochondrial introns: a critical view. Trends Genet. 233:119–125. [DOI] [PubMed] [Google Scholar]
- Lemieux C, Vincent AT, Labarre A, Otis C, Turmel M.. 2015. Chloroplast phylogenomic analysis of chlorophyte green algae identifies a novel lineage sister to the Sphaeropleales (Chlorophyceae). BMC Evol Biol. 15:264.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2010. Fast and accurate long-read alignment with BurrowsWheeler transform. Bioinformatics 265:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Ran JH, Guo DM, Yang ZY, Wang XQ.. 2014. Phylogeny and divergence times of gymnosperms inferred from single-copy nuclear genes. PLoS One 99:e107679.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS.. 2003. The origins of genome complexity. Science 3025649:1401–1404. [DOI] [PubMed] [Google Scholar]
- Lynch M, Koskella B, Schaack S.. 2006. Mutation pressure and the evolution of organelle genomic architecture. Science 3115768:1727–1730. [DOI] [PubMed] [Google Scholar]
- Miller SM, Schmitt R, Kirk DL.. 1993. Jordan, an active Volvox transposable element similar to higher plant transposons. Plant Cell 59:1125–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira D, López-García P.. 2017. Evolution: king-size plastid genomes in a new red algal clade. Curr Biol. 2713:R651–R653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Gómez SA, et al. 2017. The new red algal subphylum Proteorhodophytina comprises the largest and most divergent plastid genomes known. Curr Biol. 2711:1677–1684. [DOI] [PubMed] [Google Scholar]
- Nabholz B, Glémin S, Galtier N.. 2008. Strong variations of mitochondrial mutation rate across mammals—the longevity hypothesis. Mol Biol Evol. 251:120–130. [DOI] [PubMed] [Google Scholar]
- Nakada T, Misawa K, Nozaki H.. 2008. Molecular systematics of Volvocales (Chlorophyceae, Chlorophyta) based on exhaustive 18S rRNA phylogenetic analyses. Mol Phylogenet Evol. 481:281–291. [DOI] [PubMed] [Google Scholar]
- Ness RW, Kraemer SA, Colegrave N, Keightley PD.. 2016. Direct estimate of the spontaneous mutation rate uncovers the effects of drift and recombination in the Chlamydomonas reinhardtii plastid genome. Mol Biol Evol. 333:800–808. [DOI] [PubMed] [Google Scholar]
- Nozaki H, Matsuzaki R, Yamamoto K, Kawachi M, Takahashi F.. 2015. Delineating a new heterothallic species of Volvox (Volvocaceae, Chlorophyceae) using new strains of “Volvox africanus”. PLoS One 1011:e0142632.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki H, Ott FD, Coleman AW.. 2006. Morphology, molecular phylogeny and taxonomy of two new species of Pleodorina (Volvoceae, Chlorophyceae). J Phycol. 425:1072–1080. [Google Scholar]
- Park S, et al. 2017. Contrasting patterns of nucleotide substitution rates provide insight into dynamic evolution of plastid and mitochondrial genomes of geranium. Genome Biol Evol. 96:1766–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR. 2015. Mutation rates in plastid genomes: they are lower than you might think. Genome Biol Evol. 75:1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR. 2016. The mutational hazard hypothesis of organelle genome evolution: 10 years on. Mol Ecol. 2516:3769–3775. [DOI] [PubMed] [Google Scholar]
- Smith DR. 2018. Plastid genomes hit the big time. New Phytol. 2192:491–495. [DOI] [PubMed] [Google Scholar]
- Smith DR, et al. 2013. Organelle genome complexity scales positively with organism size in volvocine green algae. Mol Biol Evol. 304:793–797. [DOI] [PubMed] [Google Scholar]
- Smith DR, Lee RW.. 2010. Low nucleotide diversity for the expanded organelle and nuclear genomes of Volvox carteri supports the mutational-hazard hypothesis. Mol Biol Evol. 2710:2244–2256. [DOI] [PubMed] [Google Scholar]
- Tillich M, et al. 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 248:1586–1591. [DOI] [PubMed] [Google Scholar]
- Zeng L, et al. 2014. Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nat Commun. 5:4956.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A, Guo W, Jain K, Mower JP.. 2014. Unprecedented heterogeneity in the synonymous substitution rate within a plant genome. Mol Biol Evol. 315:1228–1236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.