Abstract
Shigella sonnei is responsible for the majority of shigellosis infections in the US with over 500,000 cases reported annually. Here, we present the complete genome of the clinical multidrug resistant (MDR) strain 866, which is highly susceptible to bacteriophage infections. The strain has a circular chromosome of 4.85 Mb and carries a 113 kb MDR plasmid. This IncB/O/K/Z-type plasmid, termed p866, confers resistance to five different classes of antibiotics including ß-lactamase, sulfonamide, tetracycline, aminoglycoside, and trimethoprim. Comparative analysis of the plasmid architecture and gene inventory revealed that p866 shares its plasmid backbone with previously described IncB/O/K/Z-type Shigella spp. and Escherichia coli plasmids, but is differentiated by the insertion of antibiotic resistance cassettes, which we found associated with mobile genetic elements such as Tn3, Tn7, and Tn10. A whole genome-derived phylogenetic reconstruction showed the evolutionary relationships of S. sonnei strain 866 and the four established Shigella species, highlighting the clonal nature of S. sonnei.
Keywords: Shigella sonnei strain 866, multidrug resistance (MDR), whole genome sequencing, comparative phylogenomics
Introduction
Shigella sonnei, together with Shigella flexneri, is responsible for more than 90% of shigellosis cases in the US (Scallan et al. 2011), causing an estimated 500,000 infections, 6,000 hospitalizations and 70 deaths annually (Mead et al. 1999; Gupta et al. 2004). Historically, S. sonnei has been responsible for bacillary dysentery in developed countries but its recent emergence and spread into developing countries over the last decades has raised major public health concerns (Vinh et al. 2009). Unlike other Shigella spp., S. sonnei is genetically homogenous (Karaolis et al. 1994) and descended from a clonal, rapid-evolving multidrug resistant (MDR) ancestor that diversified into distinct lineages (Holt et al. 2012). MDR strains that confer resistance to different classes of antibiotics have been described (Jain et al. 2005; Kozyreva et al. 2016).
An important virulence factor in Shigella spp. is the Shiga toxin, first discovered in the 19th century in a Shigella dysenteriae clinical isolate by Kiyoshi Shiga (Trofa et al. 1999), which is also a hallmark of Shiga toxin-producing Escherichia coli (STEC) (Eppinger et al. 2011; Sadiq et al. 2014). Although not all Shigella spp. or S. sonnei isolates are shigatoxigenic, Stx can be laterally acquired through Stx phage infection, either from (STEC) or Stx-producing Shigella strains (Strauch et al. 2001; Nyholm et al. 2015). Other key virulence determinants of Shigella are long polar fimbriae (lpfA), P-related fimbria regulatory gene (prfB), Shigella IgA-like protease homolog (sigA), plasmid-encoded enterotoxin (senB), glutamate decarboxylase (gad), invasion protein S. flexneri (ipaD), or VirF transcriptional activator (virF), among others as reviewed in Mattock and Blocker (2017).
Strain 866 has been found to be highly susceptible to infection by a range of phages, including Cytolethal Distending Toxin and Stx-converting prophages found in STEC, as previously demonstrated (Muniesa 2004; Allue-Guardia et al. 2011; Imamovic and Muniesa 2011). This observation highlights the potential epidemiological role of this particular strain in the acquisition and spread of phage-borne pathogenicity factors into larger host-pathogen populations by means of bacteriophage transduction. The availability of the closed high-quality 866 genome presented in this study is foundational to further elucidate genome characteristics correlated with its virulence, resistance, and bacteriophage susceptibility.
Materials and Methods
Shigella sonnei 866
The clinical S. sonnei strain 866 was isolated from human feces in September of 2000 in Hospital de la Santa Creu i Sant Pau in Barcelona, Spain. Species identification was confirmed by an array of biochemical tests (API 20E, BioMérieux, Marcy-l'Étoile, France) (supplementary table 1), following the manufacturer’s instructions. As demonstrated previously, strain 886 is highly susceptible to lysogenic phage infection and prophage genome incorporation (Muniesa 2004; Allue-Guardia et al. 2011; Imamovic and Muniesa 2011).
Genome Sequencing, Assembly, and Annotation
The strain was cultured in Luria-Bertani broth (Fisher Scientific, Thermo Fisher Scientific, Asheville, NC, USA) overnight at 37 °C in a shaker (180 rpm). Total genomic DNA was extracted from the overnight culture using the QIAamp DNA Mini Kit (Qiagen, Inc., Valencia, CA, USA) according to the manufacturer's protocol. To close the genome, we pursued a hybrid approach using long-read PacBio RS II and short-read Illumina MiSeq sequencing. Briefly, for PacBio sequencing, genomic DNA was sheared into 20-kb fragments using g-TUBE (Covaris, Inc., Woburn, MA, USA). The library was prepared based on the 20-kb PacBio sample preparation protocol and sequenced using P6/C4 chemistry on four single-molecule real-time (SMRT) cells with a 240-min collection time. The continuous long-read data were de novo assembled using the PacBio hierarchical genome assembly process (HGAP version 3.0) (Chin et al. 2013) with default parameters in SMRT Analysis v2.3.0, including consensus polishing with Quiver (Chin et al. 2013). For Illumina sequencing, a paired-end library was prepared using the NxSeq AmpFREE Low DNA Library Kit (Lucigen) and sequenced with 250-bp read length using the MiSeq Reagent kit v2 500-cycle (Illumina) following the manufacturer’s guidelines. Illumina reads were utilized for PacBio sequence error correction using Pilon (Walker et al. 2014). The chromosomal and plasmid origin of replication, oriC (http://tubic.tju.edu.cn/Ori-Finder/; last accessed August 06, 2018) (Gao and Zhang 2008) and repA, respectively, were determined and designated as the zero point of the closed molecules prior to annotation using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) (Tatusova et al. 2016).
Comparative Phylogenomics
After assembly and annotation, virulence and antibiotic resistance (AR) genes in the 866 chromosome and plasmid were detected in silico by VirulenceFinder (https://cge.cbs.dtu.dk/services/VirulenceFinder/; last accessed August 06, 2018) (Joensen et al. 2014) and ResFinder (https://cge.cbs.dtu.dk/services/ResFinder/; last accessed August 06, 2018) (Zankari et al. 2012), respectively. Prophages and plasmid incompatibility groups were identified using PHASTER (Zhou et al. 2011; Arndt 2016) and PlasmidFinder (https://cge.cbs.dtu.dk/services/PlasmidFinder/; last accessed August 06, 2018) (Carattoli et al. 2014), respectively. BLASTn (Altschul et al. 1990) of the complete p866 sequence against the NCBI non-redundant database identified a number of Shigella and E. coli IncB/O/K/Z-type plasmids as closest phylogenetic relatives. Respective plasmid gene inventories were compared with BLAST atlas in the GView server (https://server.gview.ca; last accessed August 06, 2018) (Petkau et al. 2010). To further establish the phylogenetic relatedness of strain 866 within Shigella, we selected a total of 26 representative S. sonnei, S. flexneri, S. dysenteriae, and S. boydii genomes from NCBI (supplementary table 2) to capture the plasticity within the four established Shigella species (Sahl et al. 2015). The phylogeny was inferred from the whole genome alignment using Mugsy (Angiuoli and Salzberg 2011) and RAxML (Stamatakis 2014) with 100 bootstrap replicates. The tree was visualized in Geneious (Kearse et al. 2012) and decorated with strain-associated metadata in Evolview (Zhang et al. 2012; He et al. 2016).
Results and Discussion
Pathogenome Architecture and Inventory
A hybrid strategy using long-read PacBio and short-read Illumina technologies allowed us to sequence the S. sonnei strain 866 genome to closure with high chromosomal and plasmid coverage of 107x and 134x. The genome is comprised of a 4,849,628 bp chromosome and a 113,079 bp MDR plasmid, termed p866, with an average GC content of 51% for both molecules. Strain 866 lacks the large virulence plasmid pSS, which encodes the wbg Phase I O antigen cluster, and thus features a rough colony morphology (Kopecko et al. 1980; Caboni et al. 2015).
Annotation with PGAP (Tatusova et al. 2016) predicted a total of 4,482 coding sequences (CDS), 96 tRNAs, and 22 rRNAs. The annotated chromosome and plasmid p866 have been deposited in GenBank under accession numbers CP022672 and CP022673, respectively.
Whole Genome Phylogenetic Analysis
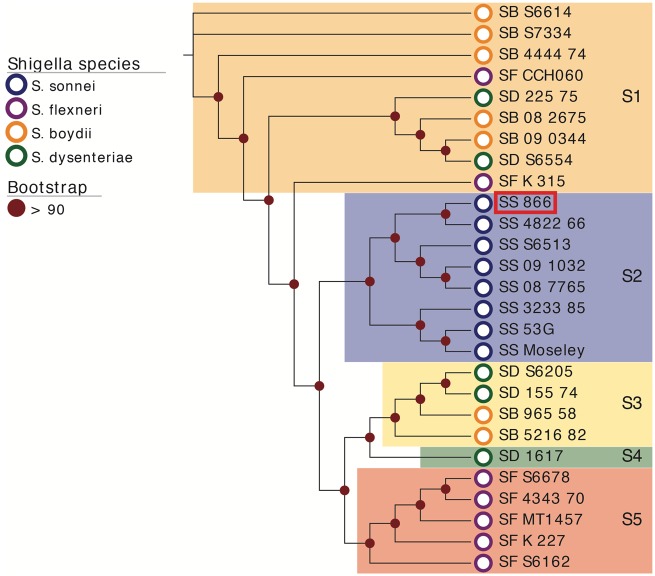
To determine the evolutionary relationship of S. sonnei strain 866 within Shigella, we established a phylogenetic framework by including representative S. sonnei, S. flexneri, S. boydii, and S. dysenteriae genomes (supplementary table 2) to capture diversity among the four species (fig. 1). The resulting whole genome-based phylogeny positions strain 866 into clade S2, a group exclusively comprised of S. sonnei isolates, indicative of the clonal nature of S. sonnei (Karaolis et al. 1994). The genetic homogeneity of this cluster was further confirmed by whole genome comparison, as evidenced by the high degree of the overall nucleotide sequence similarity in the genomes (supplementary fig. 1). The tree topology partitions the isolates into five distinct phylogenetic clusters (S1–S5), as previously established (Sahl et al. 2015). With the exception of the monophyletic S. sonnei clade S2 (fig. 1), we note here that the Shigella species designation, historically based on clinical outcome, often does not corroborate with the phylogenetic clustering, as different species make-up clades S1 and S3–S5. Further, pathovar boundaries in Shigella and E. coli are clearly obstructed by mobile virulence and resistance factors that can be acquired or lost through independent evolutionary events (Ke et al. 2011; Holt et al. 2012; Sadiq et al. 2014; Parajuli et al. 2017).
Fig. 1.—
Whole genome phylogeny of representative Shigella spp. strains. Genomes of a total of 27 Shigella strains (S. sonnei SS, blue; S. boydii SB, orange; S. flexneri SF, purple; and S. dysenteriae SD, green) including S. sonnei 866, highlighted by a red box, were aligned with Mugsy (Angiuoli and Salzberg 2011). The phylogenetic tree was inferred using RAxML (Stamatakis 2014) with a 100 bootstrap replicates and decorated with the corresponding metadata in Evolview (He et al. 2016). Only bootstrap values above 90 are shown in the tree. The tree topology partitions the isolates into five distinct clusters (S1–S5). Unlike the monophyletic S. sonnei clade S2, the phylogenetic clustering does not corroborate with the Shigella species designation.
Virulence and Resistance Profile of S. sonnei 866
The Shigella genome is highly dynamic, and mobile elements are major drivers of the S. sonnei pathogenome evolution (Jiang et al. 2005; Chang et al. 2011; Juhas 2015). Phage profiling by PHASTER (Arndt 2016) showed that strain 866 carries a highly diverse inventory of 17 prophages, including seven complete and several predicted prophage remnants. Details of respective prophage chromosomal position, length, GC content, and number of predicted CDS are provided in supplementary table 3. Identified prophage regions show partial homologies to several enterobacteria phages, such as, for example, phiP27, P1, Gifsy-2, lambda, or SEN34 (Parajuli et al. 2017). These phage-borne loci encode important virulence determinants, such as the invasion plasmid antigen IpaH9.8, an E3 ubiquitin ligase that interferes with the host’s ubiquitination pathways and facilitates colonization (Okuda et al. 2005). We also noted that the majority of the 866 prophages carry transposons and IS elements that are known drivers of bacteriophage diversification (Eppinger et al. 2011; Yin et al. 2015). Bacteria often carry an array of prophages in their genomes. The benefits conferred by polylysogeny are poorly understood, but previous reports show that in mixed infections with phage-free bacteria, phage carriage, and especially multiple phage carriage, is highly beneficial and provides a fitness advantage during mixed infections by mediating bacteria–bacteria competition (Burns et al. 2015). Moreover, many bacteria use their prophages as weapons against competitors. Because the different temperate phages are likely to have different host ranges, this expands the range of bacterial competitors that the polylysogen can kill (James et al. 2012).
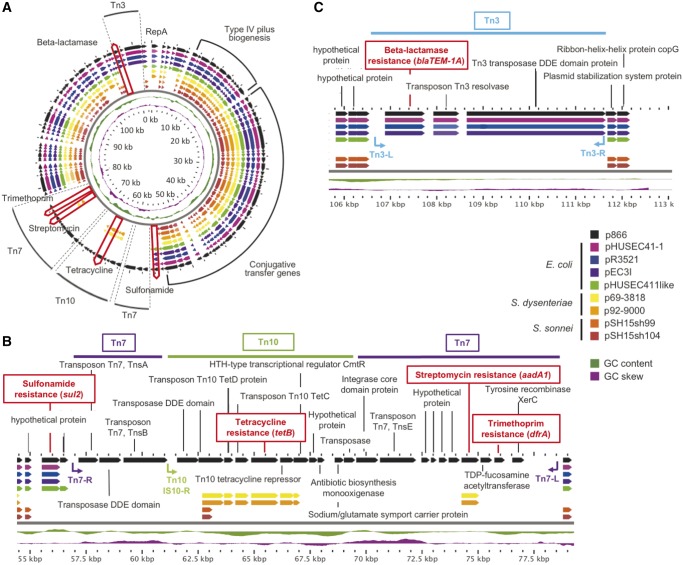
This strain features virulence hallmarks of S. sonnei such as lpfA, ipaH9.8, gad, senB, and sigA (Okuda et al. 2005; Al-Hasani et al. 2009; Torres et al. 2009; Joensen et al. 2014; Mattock and Blocker 2017). We further identified an arsenal of five AR loci on the MDR-plasmid p866 (fig. 2A), which renders this strain ß-lactamase (blaTEM-1A), sulfonamide (sul2), tetracycline (tetB), aminoglycoside (aadA1), and trimethoprim (dfrA) resistant (Zankari et al. 2012; Miranda et al. 2016).
Fig. 2.—
Comparison of Shigella sonnei p866 plasmid architecture and inventory with phylogenetically related Escherichia coli and Shigella plasmids. BLAST atlas (Petkau et al. 2010) comparison of related E. coli and Shigella plasmids, as determined by BLASTn inferred nucleotide sequence similarities (Altschul et al. 1990). (A) Respective gene inventories are referenced to the 113 kb MDR-plasmid p866 (black, outer circle). CDS are presented as arrows and color-coded according to plasmid origin as depicted in the legend. GC-content and -skew of p866 are depicted in the two innermost circles, in green and purple, respectively. Plasmid locations of the type IV pilus biogenesis, conjugal transfer, and five antimicrobial resistance loci, which are part of transposable elements Tn3, Tn7, and Tn10 are highlighted. The other two figures correspond to a linear representation of the five coded antimicrobial resistance loci in p866. Four of the five resistances were introduced by insertion of three transposable elements, with the notable exception of sulfonamide locus, which is an integral part of the plasmid backbone. Tn3-, Tn7-, and Tn10-associated resistance cassettes are inserted at two different locations. (B) Tn7 and Tn10 elements form a composite transposon, in which Tn10 disrupts Tn7, and confers tetracycline, streptomycin, and trimethoprim resistance. (C) Transposon Tn3 confers ß-lactamase resistance. Transposon boundaries and inverted repeats, if detected, are shown as arrows. CDS are presented as arrows and color-coded according to plasmid origin as depicted in the legend. GC-content and -skew of p866 are depicted, in green and purple, respectively.
Phylogenetic Relatedness and Gene Inventory of MDR-Plasmid p866
To elucidate phylogenetic relatedness of the MDR plasmid, we performed a nucleotide BLASTn (Altschul et al. 1990) search of the complete p866 plasmid sequence against the non-redundant database at NCBI. We, hereby, identified several plasmids of diverse E. coli and Shigella pathovars that share high similarity at the nucleotide level (>99%) with a coverage of the plasmid backbones ranging from 72% to 79%. Namely, E. coli plasmids pHUSEC41-1, from an EAEC O104: H4 hybrid strain from the 2011 outbreak in Germany (Kunne et al. 2012), pEC3I (KU932021.1), pR3521 (Papagiannitsis et al. 2011), and pHUSEC411, from ExPEC O18: K1 strain PMV-1 (Peris-Bondia et al. 2013). Further, S. dysenteriae 1 plasmids p69-3818 (strain 69-3818, Guatemala 1969) and p92-9000 (strain 92-9000, Panama 1991) (Njamkepo et al. 2016). Here, we note that the phylogenetic closest S. sonnei plasmids in the NCBI repository show only ∼68% sequence coverage, as seen for plasmids pSH15sh99 (KY471628.1) or pSH15sh104 (KY471629.1).
We compared both the p866 coding capacity and plasmid architecture among these phylogenetically related plasmids, which are all of the IncB/O/K/Z incompatibility group (Kozyreva et al. 2016). Nucleotide-level analysis using BLAST atlas (Petkau et al. 2010) shows a largely syntenic shared plasmid backbone of ∼86 kb that is highly conserved (fig. 2A). It encodes operons for type IV pilus biogenesis, conjugal transfer, and plasmid maintenance, which altogether account for more than 50% of the shared plasmid backbone. Our analysis clearly shows that microevolution in this MDR plasmid is driven by transposon (Tn) insertions into the plasmid backbone. As shown in figure 2A, four of the five AR loci were introduced by insertion of three transposable elements that disrupt plasmid synteny: Tn3 (blaTEM-1A), Tn7 (aadA1 and dfrA), and Tn10 (tetB) (Bailey et al. 2011; Nogrady et al. 2013; Kozyreva et al. 2016), although the sul2 gene is part of the E. coli and S. sonnei p866 plasmid backbones. The prototypical Tn7 is plastic in its carried resistance gene cassettes and encodes seven genes (tnsABCDE) required for transposition (Peters and Craig 2001). In case of p866 Tn7, we found that Tn7tnsCD loci are missing, likely due to a secondary insertion of Tn10 in-between the Tn7tnsBE (fig. 2B). Transposon Tn3 appears intact while Tn10 lacks the left IS10 repeat, which might impair its mobility (Chalmers et al. 2000; Lawley et al. 2000; Haniford and Ellis 2015). Details of the respective Tn sequence compositions including inverted repeats can be found in figure 2B and C.
Comparison of AR profiles among these plasmids shows that trimethoprim (dfrA) resistance is uniquely found in p866 as an integral component of Tn7 (fig. 2A). Both p866 and the two S. dysenteriae plasmids confer aadA1 and tetB resistance, though the respective streptomycin loci are found in different transposon contexts; Tn7aadA1 in p866 and Tn21aadA1 (Liebert et al. 1999), in case of S. dysenteriae (fig. 2B). Strain 866 carries another transposon-encoded resistance on Tn3blaTEM-1A (fig. 2C), a feature shared by all compared E. coli plasmids with the exception of pHUSEC411. In contrast, sulfonamideR (fig. 2B) was identified as an integral part of the E. coli plasmid backbone. The observed prevalence and mosaic structure of the transposon resistance cassettes stresses their role as major drivers of IncB/O/K/Z-type plasmid evolution. Plasmid spread into broader host populations crossing E. coli and Shigella pathovar boundaries is likely facilitated by the conjugal transfer system found on its backbone (fig. 2A).
The emergence of MDR bacteria has been identified by the World Health Organization as a pressing global public health problem. An emerging threat is posed by the spread of antibiotic resistant S. sonnei isolates (Puzari et al. 2017), the major culprit of human shigellosis within last decades (Sivapalasingam et al. 2006; Kahsay and Muthupandian 2016; Taneja and Mewara 2016), which has an exceptional ability to laterally acquire and transfer resistance genes (Ke et al. 2011; Holt et al. 2012; Thompson et al. 2015; Sváb et al. 2017; Rajpara et al. 2018). Using whole genome sequencing and typing strategies, we have elucidated the genomic make-up and phylogenetic relationship of the clinical S. sonnei strain 866 and identified the genomic basis of AR against five classes of antibiotics on the carried MDR-plasmid p866. The microevolution of this IncB/O/K/Z-type plasmid is driven by different Tn insertions that render strain 866 MDR. The carried conjugal transfer backbone of p866 makes it an ideal vehicle for the lateral spread of resistance loci into diverse E. coli and Shigella pathovars (fig. 2A) and potentially other enterics of clinical importance. Monitoring emerging trends in the S. sonnei pathogenome evolution is thus critical to translate sequence-based information into actionable countermeasures to detect and prevent the spread of MDR resistant Shigella clones. Identified genome signatures of such emerging highly virulent and AR resistant clones are readily applicable for improved biosurveillance, risk assessment, and informed and improved therapeutic strategies (Oany et al. 2017; Pinaud et al. 2017).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the South Texas Center of Emerging Infectious Diseases (STCEID) and the Department of Biology at the University of Texas at San Antonio; the National Institute of Allergy and Infectious Diseases of the National Institutes of Health [SC2AI120941]; and the US Department of Homeland Security [2014-ST-062-000058] to M.E. The contributions of J.L.B. were funded by the Agricultural Research Service of the U.S. Department of Agriculture (ARS-USDA). We would like to acknowledge Ferran Navarro (Hospital de la Santa Creu i Sant Pau, Microbiology service, Barcelona, Spain) for providing strain 866. We would like to thank Heidi Gildersleeve (UTSA), Sandy Fryda-Bradley (USDA), and Kristin Kuhn (USDA) for providing technical assistance. The mention of a trade name, proprietary product, or specific equipment does not constitute a guarantee or warranty by the USDA and does not imply approval to the exclusion of other products that might be suitable. The United States Meat Animal Research Center is an equal opportunity employer and provider.
Author Contributions
A.A.G., S.K.K., and M.E. performed comparative phylogenomics analyses. J.L.B. provided PacBio sequences and assembly. M.M. and P.Q. provided S. sonnei strain 866 and biochemical characterizations. A.A.G. and M.E. drafted the manuscript. All authors read and approved the final manuscript.
Literature Cited
- Al-Hasani K, Navarro-Garcia F, Huerta J, Sakellaris H, Adler B.. 2009. The immunogenic SigA enterotoxin of Shigella flexneri 2a binds to HEp-2 cells and induces fodrin redistribution in intoxicated epithelial cells. PLoS One 412:e8223.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allue-Guardia A, Garcia-Aljaro C, Muniesa M.. 2011. Bacteriophage-encoding cytolethal distending toxin type V gene induced from nonclinical Escherichia coli isolates. Infect Immun. 798:3262–3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 2153:403–410. [DOI] [PubMed] [Google Scholar]
- Angiuoli SV, Salzberg SL.. 2011. Mugsy: fast multiple alignment of closely related whole genomes. Bioinformatics 273:334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt D. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44(W1):W16–W21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JK, Pinyon JL, Anantham S, Hall RM.. 2011. Distribution of the blaTEM gene and blaTEM-containing transposons in commensal Escherichia coli. J Antimicrob Chemother. 664:745–751. [DOI] [PubMed] [Google Scholar]
- Burns N, James CE, Harrison E.. 2015. Polylysogeny magnifies competitiveness of a bacterial pathogen in vivo. Evol Appl. 84:346–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caboni M, et al. 2015. An O antigen capsule modulates bacterial pathogenesis in Shigella sonnei. PLoS Pathog. 113:e1004749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A, et al. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 587:3895–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers R, Sewitz S, Lipkow K, Crellin P.. 2000. Complete nucleotide sequence of Tn10. J Bacteriol. 18210:2970–2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CY, et al. 2011. Integron types, gene cassettes, antimicrobial resistance genes and plasmids of Shigella sonnei isolates from outbreaks and sporadic cases in Taiwan. J Med Microbiol. 602:197–204. [DOI] [PubMed] [Google Scholar]
- Chin CS, et al. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 106:563–569. [DOI] [PubMed] [Google Scholar]
- Eppinger M, Mammel MK, Leclerc JE, Ravel J, Cebula TA.. 2011. Genomic anatomy of Escherichia coli O157:H7 outbreaks. Proc Natl Acad Sci U S A. 10850:20142–20147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Zhang CT.. 2008. Ori-Finder: a web-based system for finding oriCs in unannotated bacterial genomes. BMC Bioinformatics 91:79.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Polyak CS, Bishop RD, Sobel J, Mintz ED.. 2004. Laboratory-confirmed shigellosis in the United States, 1989-2002: epidemiologic trends and patterns. Clin Infect Dis. 3810:1372–1377. [DOI] [PubMed] [Google Scholar]
- Haniford DB, Ellis MJ.. 2015. Transposons Tn10 and Tn5. Microbiol Spectr. 31:MDNA3-0002-2014. [DOI] [PubMed] [Google Scholar]
- He Z, et al. 2016. Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 44(W1):W236–W241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt KE, et al. 2012. Shigella sonnei genome sequencing and phylogenetic analysis indicate recent global dissemination from Europe. Nat Genet. 449:1056–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamovic L, Muniesa M.. 2011. Quantification and evaluation of infectivity of shiga toxin-encoding bacteriophages in beef and salad. Appl Environ Microbiol. 7710:3536–3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain SK, Gupta A, Glanz B, Dick J, Siberry GK.. 2005. Antimicrobial-resistant Shigella sonnei: limited antimicrobial treatment options for children and challenges of interpreting in vitro azithromycin susceptibility. Pediatr Infect Dis J. 246:494–497. [DOI] [PubMed] [Google Scholar]
- James CE, et al. 2012. Differential infection properties of three inducible prophages from an epidemic strain of Pseudomonas aeruginosa. BMC Microbiol. 121:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, et al. 2005. The complete sequence and analysis of the large virulence plasmid pSS of Shigella sonnei. Plasmid 542:149–159. [DOI] [PubMed] [Google Scholar]
- Joensen KG, et al. 2014. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J Clin Microbiol. 525:1501–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M. 2015. Horizontal gene transfer in human pathogens. Crit Rev Microbiol. 411:101–108. [DOI] [PubMed] [Google Scholar]
- Kahsay AG, Muthupandian S.. 2016. A review on Sero diversity and antimicrobial resistance patterns of Shigella species in Africa, Asia and South America, 2001-2014. BMC Res Notes 91:422.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaolis DK, Lan R, Reeves PR.. 1994. Sequence variation in Shigella sonnei (Sonnei), a pathogenic clone of Escherichia coli, over four continents and 41 years. J Clin Microbiol. 323:796–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke X, Gu B, Pan S, Tong M.. 2011. Epidemiology and molecular mechanism of integron-mediated antibiotic resistance in Shigella. Arch Microbiol. 19311:767–774. [DOI] [PubMed] [Google Scholar]
- Kearse M, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2812:1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecko DJ, Washington O, Formal SB.. 1980. Genetic and physical evidence for plasmid control of Shigella sonnei form I cell surface antigen. Infect Immun. 29:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozyreva VK, et al. 2016. Recent outbreaks of Shigellosis in California caused by two distinct populations of Shigella sonnei with either increased virulence or fluoroquinolone resistance. mSphere 16: doi:10.1128/mSphere.00344-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunne C, et al. 2012. Complete sequences of plasmids from the hemolytic-uremic syndrome-associated Escherichia coli strain HUSEC41. J Bacteriol. 1942:532–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Burland V, Taylor DE.. 2000. Analysis of the complete nucleotide sequence of the tetracycline-resistance transposon Tn10. Plasmid 433:235–239. [DOI] [PubMed] [Google Scholar]
- Liebert CA, Hall RM, Summers AO.. 1999. Transposon Tn21, flagship of the floating genome. Microbiol Mol Biol Rev. 633:507–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattock E, Blocker AJ.. 2017. How do the virulence factors of Shigella work together to cause disease? Front Cell Infect Microbiol. 7:64.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead PS, et al. 1999. Food-related illness and death in the United States. Emerg Infect Dis. 55:607–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda A, et al. 2016. Emergence of plasmid-borne dfrA14 trimethoprim resistance gene in Shigella sonnei. Front Cell Infect Microbiol. 6:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniesa M. 2004. Diversity of stx2 converting bacteriophages induced from Shiga-toxin-producing Escherichia coli strains isolated from cattle. Microbiology 1509:2959–2971. [DOI] [PubMed] [Google Scholar]
- Njamkepo E, et al. 2016. Global phylogeography and evolutionary history of Shigella dysenteriae type 1. Nat Microbiol. 14:16027. [DOI] [PubMed] [Google Scholar]
- Nogrady N, et al. 2013. Antimicrobial resistance and genetic characteristics of integron-carrier shigellae isolated in Hungary (1998-2008). J Med Microbiol. 6210:1545–1551. [DOI] [PubMed] [Google Scholar]
- Nyholm O, et al. 2015. Characterization of Shigella sonnei isolate carrying Shiga Toxin 2-producing gene. Emerg Infect Dis. 215:891–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oany AR, et al. 2017. Vaccinomics approach for designing potential peptide vaccine by targeting Shigella spp. serine protease autotransporter subfamily protein SigA. J Immunol Res. 2017:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda J, et al. 2005. Shigella effector IpaH9.8 binds to a splicing factor U2AF(35) to modulate host immune responses. Biochem Biophys Res Commun. 3332:531–539. [DOI] [PubMed] [Google Scholar]
- Papagiannitsis CC, Tzouvelekis LS, Kotsakis SD, Tzelepi E, Miriagou V.. 2011. Sequence of pR3521, an IncB plasmid from Escherichia coli encoding ACC-4, SCO-1, and TEM-1 beta-lactamases. Antimicrob Agents Chemother. 551:376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parajuli P, Adamski M, Verma NK.. 2017. Bacteriophages are the major drivers of Shigella flexneri serotype 1c genome plasticity: a complete genome analysis. BMC Genomics 181:722.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris-Bondia F, Muraille E, Van Melderen L.. 2013. Complete genome sequence of the Escherichia coli PMV-1 strain, a model extraintestinal pathogenic E. coli strain used for Host-Pathogen Interaction Studies. Genome Announc. 15: doi: 10.1128/genomeA.00913-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JE, Craig NL.. 2001. Tn7: smarter than we thought. Nat Rev Mol Cell Biol. 211:806–814. [DOI] [PubMed] [Google Scholar]
- Petkau A, Stuart-Edwards M, Stothard P, Van Domselaar G.. 2010. Interactive microbial genome visualization with GView. Bioinformatics 2624:3125–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinaud L, et al. 2017. Identification of novel substrates of Shigella T3SA through analysis of its virulence plasmid-encoded secretome. PLoS One 1210:e0186920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puzari M, Sharma M, Chetia P.. 2017. Emergence of antibiotic resistant Shigella species: a matter of concern. J Infect Public Health doi: 10.1016/j.jiph.2017.09.025. [DOI] [PubMed] [Google Scholar]
- Rajpara N, et al. 2018. Molecular analysis of multidrug resistance in clinical isolates of Shigella spp. from 2001-2010 in Kolkata, India: role of integrons, plasmids, and topoisomerase mutations. Infect Drug Resist. 11:87–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadiq SM, Hazen TH, Rasko DA, Eppinger M.. 2014. EHEC genomics: past, present, and future. Microbiol Spectr. 24:EHEC-0020-2013. [DOI] [PubMed] [Google Scholar]
- Sahl JW, et al. 2015. Defining the phylogenomics of Shigella species: a pathway to diagnostics. J Clin Microbiol. 533:951–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan E, et al. 2011. Foodborne illness acquired in the United States–major pathogens. Emerg Infect Dis. 171:7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivapalasingam S, et al. 2006. High prevalence of antimicrobial resistance among Shigella isolates in the United States tested by the National Antimicrobial Resistance Monitoring System from 1999 to 2002. Antimicrob Agents Chemother. 501:49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 309:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch E, Lurz R, Beutin L.. 2001. Characterization of a Shiga toxin-encoding temperate bacteriophage of Shigella sonnei. Infect Immun. 6912:7588–7595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sváb D, Bálint B, Vásárhelyi B, Maróti G, Tóth I.. 2017. Comparative genomic and phylogenetic analysis of a Shiga Toxin Producing Shigella sonnei (STSS) strain. Front Cell Infect Microbiol. 7:229.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja N, Mewara A.. 2016. Shigellosis: epidemiology in India. Indian J Med Res. 1435:565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova T, et al. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 4414:6614–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CN, Duy PT, Baker S.. 2015. The rising dominance of Shigella sonnei: an Intercontinental Shift in the Etiology of Bacillary Dysentery. PLoS Negl Trop Dis. 96:e0003708.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres AG, et al. 2009. Genes related to long polar fimbriae of pathogenic Escherichia coli strains as reliable markers to identify virulent isolates. J Clin Microbiol. 478:2442–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trofa AF, Ueno-Olsen H, Oiwa R, Yoshikawa M.. 1999. Dr. Kiyoshi Shiga: discoverer of the dysentery bacillus. Clin Infect Dis. 295:1303–1306. [DOI] [PubMed] [Google Scholar]
- Vinh H, et al. 2009. A changing picture of shigellosis in southern Vietnam: shifting species dominance, antimicrobial susceptibility and clinical presentation. BMC Infect Dis. 91:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 911:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S, et al. 2015. Escherichia coli O157:H7 strains harbor at least three distinct sequence types of Shiga toxin 2a-converting phages. BMC Genomics 161:733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E, et al. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 6711:2640–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gao S, Lercher MJ, Hu S, Chen WH.. 2012. EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 40(W1):W569–W572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS.. 2011. PHAST: a fast phage search tool. Nucleic Acids Res. 39(Suppl):W347–W352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.