Abstract
Pancreatic Ductal AdenoCarcinoma (PDAC) is one of the most lethal malignancies of all solid cancers. Precancerous lesions for PDAC include PanIN, IPMNs and MCNs. PDAC has a poor prognosis with a 5-year survival of approximately 6%. Whereas Periampulary AdenoCarcinoma (PAC) having four anatomic subtypes, pancreatic, Common Bile Duct (CBD), ampullary and duodenum shows relative better prognosis. The highest incidence of PDAC has been reported with black with respect to white population. Similarly, incidence rate of PAC also differs with different ethnic populations. Several lifestyle, environmental and occupational exposures including long-term diabetes, obesity, and smoking, have been linked to PDAC, however, for PAC the causal risk factors were poorly described. It is now clear that PDAC and PAC are a multi-stage process resulting from the accumulation of genomic alterations in the somatic DNA of normal cells as well as inherited mutations. Approximately 10% of PDAC have a familial inheritance. Germline mutations in CDKN2A, BRCA2, STK11, PALB2, PRSS1, etc., as well as certain syndromes have been well associated with predisposition to PDAC. KRAS, CDKN2A, TP53 and SMAD4 are the 4 “mountains” (high-frequency driver genes) which have been known to earliest somatic alterations for PDAC while relatively less frequent in PAC. Our understanding of the molecular carcinogenesis has improved in the last few years due to extensive research on PDAC which was not well explored in case of PAC. The genetic alterations that have been identified in PDAC and different subgroups of PAC are important implications for the development of genetic screening test, early diagnosis, and prognostic genetic markers. The present review will provide a brief overview of the incidence and prevalence of PDAC and PAC, mainly, increased risk in India, the several kinds of risk factors associated with the diseases as well as required genetic alterations for disease initiation and progression.
Keywords: Pancreatic ductal adenocarcinoma, Periampullary adenocarcinoma, Familial pancreatic cancer, High frequency mutations, Low frequency mutations, Molecular carcinogenesis
1. INTRODUCTION
The pancreas is an endoderm-derived organ having exocrine as well as endocrine function. Regulating protein and carbohydrate digestion, as well as glucose homeostasis, is its primary function. Eighty percent tissue mass of the pancreas is exocrine in function. Digestive zymogens are produced and delivered by acinar and ductal cells branching network into gastrointestinal (GI) tract [1]. Pancreatic Cancers (PC) fall into two major categories: (1) cancers arising from the endocrine cells of pancreas; and (2) cancers arising from the exocrine cells of pancreas. Islet cell cancers are rare and grow slowly compared to exocrine PCs. Exocrine PCs develop from the cells lining the system of ducts delivering enzymes to the small intestine and are commonly referred to as pancreatic adenocarcinomas. Pancreatic Ductal AdenoCarcinoma (PDAC), nomenclature derives from its histological features of ductal cells is the most common pancreatic neoplasms and accounts for >85% of all PC [2]. It can arise anywhere in the pancreas and periampullary region. Whereas, Periampulary AdenoCarcinoma (PAC) is a heterogeneous group of neoplasms that originates from within 2 cm of the major duodenal papilla. Periampullary tumor includes tumor originating from the head of the pancreas (60%), the ampulla of Vater (20%), distal common bile duct (10%), and the duodenum (10%) [3]. The macroscopic presentation of PAC includes: a) intramural tumors, which occur inside the ampulla, without any protuberance inside the duodenum; b) extramural tumors are polypoid tumors that swell through the ampullary aperture into the duodenum, c) ulcerative cancers of the ampulla, which is related with the worst prevision [4]. Microscopically, there are two main histological types of PAC namely, the “intestinal type” which is similar to tubular carcinoma of the stomach or the colon and show columnar tumor cells with elongated cigar-shaped basally located nuclei and nuclear stratification [5, 6]; and the “pancreatobiliary type”, resembling pancreatic carcinoma or cholangiocarcinoma. Pancreatobiliary type features papillary projections with mere fibrous cores [5, 6]. The presence of pre-invasive adenomas or areas of dysplasia can help in the distinction of ampulla and the intestinal subtype of PAC [5]. The mode of presentation and treatment options for ampullary and periampullary tumors is similar, their prognosis is quite different with that for PDAC [7].
2. WORLDWIDE INCIDENCE OF PDAC AND PAC
At the onset of the 21st century, the estimated number of PC worldwide was 110,000, with an estimated global mortality rate of 98% [8]. Pancreatic ductal adenocarcinoma of pancreas is the fourth most common cause of cancer related mortality across the world [1, 9-12]. It was reported that 48,960 new cases and 40,560 deaths for PDAC occurred in 2015 [13]. The risk is almost 20 times higher for individuals who are above 50 years as compared to the younger population. According to GLOBOCAN 2012, the Age Standardized Rate (ASR) of PDAC incidence data is 4.9 per 1,00,000 in men and 3.6 per 1,00,000 women. Worldwide ASR (ASR-W) for incidence and mortality of PDAC is 4.2% and 4.0% respectively [14]. According to the Cancer fact sheet 2016, it is estimated that in 2016, the above figures will raise up to 53,070 new cases and 41,780 deaths with respect to PDAC, which is 3.1% of all cancer. Based on data from SEER (Surveillance, Epidemiology, and End Result Programme) 18 2006-2012, the 5 year survivability for PDAC is 7.7%. According to SEER 2009-13, the median age of diagnosis of PDAC is 70 and the median age of death is 72. Using statistical models for analysis, rates for new pancreatic cancer cases have been rising on average 0.6% each year over the last 10 years. By 2030, PDAC will become the second dominant cause of cancer-related death in the US after lung cancer, outpacing colorectal, breast, and prostate cancers [15]. In the Indian subcontinent, there were no detailed and concrete reports of incidence and prevalence of PDAC and PAC diseases on the basis of recent databases. The incidence of PDAC is low (0.5-2.4 per 100,000 men and 0.2-1.8 per 100,000 women) in most parts of India. Higher rates are seen in the male urban populations of western and northern India. The incidence of PC in India is 0.5-2.4 per 100,000 men and 0.2-1.8 per 100,000 women [16]. The incidence of PAC is approximately 0.5-2% of all gastrointestinal malignancies and 20% of all tumors of the extrahepatic biliary tree [17-19]. Geographic and ethnic variations in the incidence of the 4 morphological subtypes of PAC also exist, which provide potential insights into their etiologies. He and co-workers reported a 3-decade study of 2564 resected PACs, in which the proportion of pancreatic, ampullary, biliary and duodenal carcinomas was 66%, 16%, 12% and 6%, respectively [20]. A Chinese series of 501 PACs showed a high proportion of ampullary cancers in their population with 34% accounting for pancreatic, 50% for ampullary, 10% for biliary and 5% accounting for duodenal cancers [21]. Australian series had a higher percentage of PDAC (56%), followed by 25% of ampullary cancer, then 15% of biliary and finally only 4% of duodenal cancers [22]. Although various genetic mutations have implicated in PAC, the primary molecular alterations leading to tumor initiation and progression remain unclear [23-25]. In a longitudinal study of over 450,000 participants, a healthy lifestyle reduces the risk of PC by nearly 60% [8]. The variable incidences of 4 morphological subtypes in the different countries may relate to geographic differences in the epidemiology of these cancers. Pancreatic cancer has a poorer survival rate compared to ampullary, duodenal, biliary cancers. Five years estimated survival in patients of PAC undergoing resection has been poor and ranges from approximately 20% in pancreatic adenocarcinoma to 50% in duodenal adenocracinoma. Among environmental risk factors diabetes, obesity, smoking, alcohol, increased oil fat consumption, coffee intake, decreased physical activity are associated with increased risk of PDAC. The proportion of PDAC that may be attributable to occupational exposures have been estimated to be 12% [26]. Among occupational risk factors, chlorinated hydrocarbons, organochlorines, ionizing radiation, airborne particles, nitrosamines have been associated with increased risk of PDAC. Other risk factors for PC include Chronic Pancreatitis (CP), allergies, periodontal diseases, several infections like HBV, H. pylori, Cholecystochemy and Cholelithiasis, Pernicious Anemia, NonSteroidal Anti-Inflammatory Drugs (NSAIDs). Smoking and diabetes mellitus, and smoking with family history of PC had the highest correlation to PC incidences. The joint effect of smoking and either risk factor seems to be additive, making a person who has both of these risk factors at a much higher risk to develop some form of PDAC [27]. Patients with a history of partial gastrectomy for ulcer have a high risk of PDAC [28, 29]. Distal common bile duct cancers are associated with several known host factors in addition to advanced age, like inflammatory bowel disease, sclerosing cholangitis, choledochal cysts, and intrahepatic or common bile duct stones. In addition, a geographic association has been noted for bile duct cancers, with clusters observed in some parts of the United States. Duodenal and ampullary cancers occur with heightened frequency in patients with hereditary polyposis syndromes including Hereditary Non Polyposis Colorectal Carcinoma (HNPCC), Peutz-Jeghers Syndrome (PJS), Familial Adenomatous Polyposis (FAP), and Gardner’s Syndrome (GS) [30].
2.1. Precursor Lesions of PDAC
The precursor lesion for PDAC is known as Pancreatic Intraepithelial Neoplasms (PanIN). It is categorized as either low grade (PanIN-1a or 1B), intermediate (PanIN-2), or high grade (PanIN-3) lesions [31]. Although PanINs are the most characterized precursors, other non-invasive lesions are also thought to precede PDAC formation. These are Intraductal Papillary Mucinous Neoplasms (IPMNs) and Mucinous Cystic Neoplasms (MCNs) named as they produce mucins [32]. PanINs are columnar, mucinous epithelium and with increasing architectural disorganization. IPMNs are characterized by larger size and involvement of the main ductal pancreatic duct or larger ductal branches. MCNs do not arise in the main pancreatic ducts and represent by their associated ovarian type stroma with a variable degree of epithelial dysplasia and focal regions of invasion. MCN is not connected with the ductal system and commonly occurs in female [33].
The aim of this review is to summarize all the genetic alterations that occur during PDAC initiation, progression, and maintenance along with inherited mutations describing standard approaches as well as genetic alterations of different anatomical subtypes of PAC. In addition, this review also focuses on ethnic variation, lifestyle, environmental, occupational, and genetic factors for the risk of PDAC and PAC. We described somatic high frequency and low-frequency gene mutations in PDAC and PAC patients. This review report also emphasizes on the incidence and prevalence of PDAC and PAC worldwide and in the Indian subcontinent.
2.2. Genetic Susceptibility
Development of PDAC may be associated with inherited mutations in specific genes. These mutations are thus the cause of a number of familial cancer syndromes, accounting for 5-10% of PDAC cases [34]. Of the remaining sporadic PDACs, some could be attributed to polymorphic mutations in genes encoding tobacco and food metabolizing enzymes, as well as to DNA repair genes. One recent study detected certain detoxifying genes in 455 patients with PDAC [35, 36]. Gene that is reported to have increased the risk of PDAC is GSTM1, while genes such as CYP1B1-4390-GG and uridine 5’-diphospho glucoronosyl transferase have been reported to have reduced the risk of PDAC [36].
2.3. Germline Mutation in PDAC
Approximately 5-10% of PDAC has a familial basis of inheritance [34, 37]. However, few studies have suggested lower penetrance of inheritance for PDAC (1.9-2.7%) [38, 39]. Inherited predisposition of PC can be described in two classes; Hereditary Syndromes associated with PDAC and Familial Pancreatic Cancer (FPC) [40]. FPC families can be further divided into 2 groups: “pure” PDAC families (40%) and another 1 associated with other tumor types (60%) [41]. Hereditary PDAC is defined as a genetic syndrome with an identifiable gene mutation associated with an increased risk of PDAC, whereas FPC is defined as family with at least one pair of first-degree relatives (parent-child or sibling pair) with PC without an identifiable syndrome, or two or more first degree relatives with PDAC that do not fulfill the criteria of any other inherited tumor syndrome [34]. The National Familial Pancreas Tumor Registry (NFPTR) at Johns Hopkins reported a 6.8 fold (95% CI =4.54-9.75) increased risk of PDAC in the first degree relatives of FPC patient compared to general United States Population [42].
The PanIN, IPMN, and MCN, have been found to be well defined precursor lesions of PDAC. PanIN is the most frequent precursor lesion of sporadic and FPC. Studies of resected pancreatic lesions from patients with FPC showed the presence of multifocal precursor lesions in most specimens. PanIN3 is the most common one in between several forms of PanIN [43, 44]. IPMNs are more frequent in familial cases than sporadic PC. The branch duct of IPMN is a potential indicator of PDAC in an individual with FPC [45].
2.4. Hereditary Syndromes
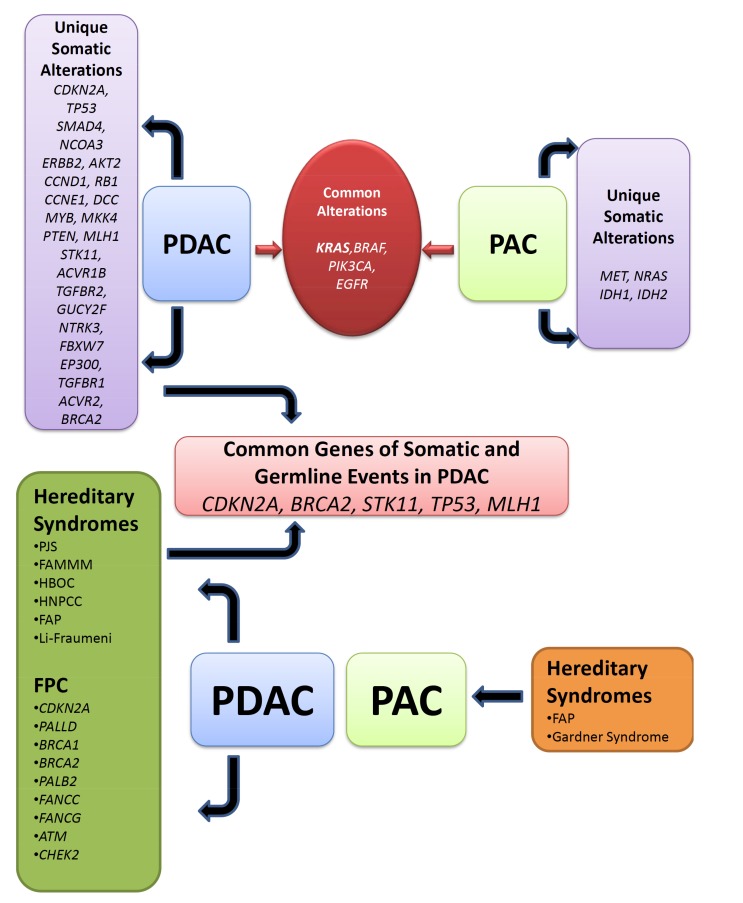
Hereditary Syndromes associated with PDAC are those where an inherited genetic syndrome with an identifiable gene mutation is associated with increased risk of PDAC. Common syndromes that are associated with PDAC are (i) PJS, (ii) Familial Atypical Multiple Mole Melanoma (FAMMM), (iii) Hereditary Breast-Ovarian Cancer (HBOC), (iv) HNPCC alternatively called Lynch Syndrome (LS), (v) FAP, (vi) Li-Fraumeni Syndrome (LFS) [45-47] (Table 1) (Fig. 1).
Table 1.
Syndromes and genes associated with Pancreatic ductal adenocarcinoma.
| Syndrome | Genes | PDAC Risk |
|---|---|---|
| Hereditary Syndromes | - | - |
| Peutz-Jeghers Syndrome | LKB1 | 35% lifetime |
| FAMMM Syndrome | CDKN2A, CDK4 | 13-22X |
| HBOC | BRCA1, BRCA2 | 2-3.5X |
| LI-Fraumeni Syndrome | TP53 | 7.3X |
| HNPCC FAP |
MLH1, MSH2 APC |
3.7% 4.5X |
| Syndromes with Chronic Inflammation | - | - |
| Hereditary Pancreatitis | PRSS1, SPINK1 | 30-40% |
| Cystic Fibrosis | CFTR | <5% |
| FPC Syndrome | PALB2, BRCA1, BRCA2,ATM, FANCC, FANCG, CDKN2A, PALLD, CHECK2 | 2 FDR: 8-12% >3 FDR: 16-38% |
Fig. (1).
Unique and common germline and somatic genomic alteration network in pancreatic ductal and periampullary adenocarcinoma. Unique somatic alterations in several genes are listed in PDAC and PAC separately. Common somatic alterations are listed together. Different hereditary syndromes were unique for PDAC and PAC and are listed individually. FPC genes are mutated and segregated in families risk for PDAC are only observed and grouped together. Common germline and somatic genomic alterations are assigned for PDAC risk.
2.4.1. Peutz-Jeghers Syndrome (STK11/LKB1) (PJS)
PJS is an autosomal dominant genetic disorder characterized by the development of benign hamartoma polyps in the gastrointestinal tract and hyper pigmented macules on the lips and oral mucosa [48, 49]. Almost half of the patients with PJS harbor germline mutations in tumor suppressor gene STK11/LKB1. There is a 132-fold increased risk of developing PDAC in patients with PJS. Affected individuals have 36% cumulative lifetime risk of developing PC [50]. One comprehensive Dutch study of PJS patients revealed a 26% increased risk of PDAC by the age of 70 and Relative Risk (RR) 76 [48].
2.4.2. Familial Atypical Multiple Mole Melanoma (CDKN2A)
Familial Atypical Multiple Mole Melanoma syndrome is an autosomal dominant genodermatosis characterized by multiple melanocytic nevi, usually more than 50, and a family history of melanoma. It is associated with mutations in the tumor suppressor gene CDKN2A and shows reduced penetrance and variable expressivity. CDKN2A is a potential risk factor for inherited PDAC [51-53]. It is a cell cycle regulator gene coding for the p16 protein product, and has functional effects in melanoma and PDAC cell lines. An important isoform of CDKN2A is p16INK4a, located on Chromosome 9p21.3 Approximately 10% of melanomas have a familial basis; mutations in CDKN2A reported approximately 40% in these families [54]. In some FAMMM kindreds with CDNK2A mutations, the prevalence of PDAC has been observed in a very high frequency, thus can be separated from hereditary tumor syndrome and referred to as PC melanoma syndrome or FAMMM-PC [55]. Individuals with this syndrome have a RR of 20-34% with a 17% life time risk of developing PDAC [56]. Sixty to 90% of melanoma risk by the age of 80 and 20% of PDAC risk was observed by the age of 75, being associated with CDKN2A mutations [47]. Studies from different ethnic backgrounds showed the different spectrum of data on mutation frequencies with RR of PDAC and Melanoma. A specific genetic background of FAMMM kindreds showed 25% PC risk whereas a cohort study observed 13-20 fold increase of PDAC [57].
2.4.3. Hereditary Breast-Ovarian Cancer (HBOC) (BRCA1 and BRCA2)
The incidence of HBOC in the general population is approximately 1 in 500 individuals. The majority of the cases of HBOC are due to mutations in the BRCA1 and BRCA2 genes [58, 59]. The HBOC syndrome is an autosomal dominant disorder with increased risks for breast cancer (47-55% by age 70), ovarian cancer (17-39%), and other cancers including prostate, male breast, melanoma and pancreas, associated with germline mutations in BRCA1 and BRCA2 [60-62]. Carrier frequency is increased among patients with Ashkenazi Jewish ethnicity (1 out of 40). Specifically, there are 3 founder mutations in this population: 185delAG and 5382insC in BRCA1 and 6174delT in BRCA2 [63].
Mutations in BRCA1 are primarily associated with early onset of breast and ovarian cancer risks, though other cancer risks do occur at higher rates than expected in the general population including PDAC. A study reported an RR of 2.8 compared to the general population risk of 1.3% for PDAC in BRCA1 mutation carriers [64]. However, in a study of British (n=268) no significant association of increased risk of PDAC has been found with BRCA1-associated HBOC families [65].
In a study of Ashkenazi Jewish population of North America (n=858), 1% carries the germline BRCA2 6174delT mutation, which is associated with 10 fold risk of PDAC [66, 67]. Overall BRCA2 mutation carriers have an estimated 5% life risk of developing PDAC [61]. Analysis of 222 BRCA2 families identified a statistically significant increased risk for PDAC (RR 4.1, 95% CI 1.9-7.8) [65]. In a study of 173 families with germline BRCA2 mutations and breast or ovarian cancers, representing a cohort of 3728 individuals, the RR of PC was 3.51 [68]. The Breast Cancer Linkage Consortium indicated that BRCA2 mutation carriers have a 3.5 RR compared to non-mutation carriers (5-7% lifetime risk) for developing PDAC. Animal studies confirm a major role for BRCA2 in a mouse model of FPC [69]. Mice with the BRCA2 null background showed genomic instability throughout the exocrine pancreatic cells, with the development of pancreatic intraepithelial neoplasia (PanIN lesions) in most mice and invasive PDAC in about 15% of mice. BRCA2 defective cells exhibited impaired Homologous Recombination (HR) repair, growth and proliferation arrest, abnormal cell cycle, radio resistant, DNA synthesis, genomic instability, and hypersensitivity to DNA damaging agents. Combining BRCA2 null and the Trp53 (R172H) variants promoted tumor formation. Absence of BRCA2 function predisposes the exocrine pancreas to profound DNA damage. The frequency of invasive neoplasia is accentuated by the concomitant deregulation of p53. Taken together, these data suggested that patients with BRCA2 mutations are at increased risk for PDAC, but this pathway is not essential or common in sporadic PDACs [69].
2.4.4. Herditary Nonpolyposis Colorectal Carcinoma /Lynch Syndrome (HNPCC/LS) (DNA Mismatch Repair Genes)
Lynch Syndrome or HNPCC is an autosomal dominant hereditary disease alternatively called HNPCC characterized by early onset of colon cancer due to germline mutations in one of the DNA mismatch repair genes (hMLH1, hPMS1, hPMS2, hMSH2, or hMSH6/GTBP) [49]. This syndrome is caused by a mutation in the MisMatch Repair (MMR) genes MSH2, MLH1, MSH6, and PMS2. In a study of 147 families with germline MMR gene mutations, the cumulative risk of PDAC was 1.31%, up to age 50 and 3.68% up to age 70yr, which is 8.6-fold (95% CI =4.7-15.7) higher as compared to the general population [70].
2.4.5. Familial Adenomatous Polyposis (FAP) (APC)
Familial Adenomatous Polyposis is an inherited disorder characterized by cancer of the large intestine (colon) and rectum. People with the classic type of FAP may begin to develop multiple noncancerous (benign) growths (polyps) in the colon as early as their teenage years. Inherited mutations in the tumor suppressor gene, APC, account for the majority of cases. FAP has an increased risk of PDAC, with calculated increased RR of 4.6% (95% CI 1.2-11.4) in affected individuals. Small bowel cancers occur in 50-90% of patients with FAP and are usually periampullary [71, 72].
Gardner’s Syndrome is a variant of FAP, and individuals with GS have a 100 fold increase risk of developing PAC when compared with general population [72] (Fig. 1).
2.4.6. Li-Fraumeni Syndrome (TP53)
Though very rare but there are reports suggesting that LFS carries a mutation in tumor suppressor gene TP53, thus increasing the risk of PDAC compared to general population. In addition, very rarely patients affected by AT develop PDAC [73].
2.5. Syndrome with Chronic Inflammation of the Pancreas
2.5.1. Hereditary Pancreatitis
Among the entire inherited cancer predisposition syndrome, Hereditary Pancreatitis (HP) is only one where PC is the sole cancer risk factor. HP is a rare form of CP, which is an autosomal dominant inherited disorder and highly penetrant (>80%) and typically occur prior to the age of 30, although dependent on ancestral origin in the family [74]. Germline mutations in PRSS1 cause an autosomal dominant form of the disease, whereas germline mutations in SPINK1 lead to an autosomal recessive pattern of inheritance [75, 76].
The PRSS1 gene, provides instructions for making an enzyme called cationic trypsinogen. Cationic trypsinogen is produced in the pancreas and helps with the digestion of food. Premature activation of trypsinogen leads to acute pancreatitis [75]. R117H germline mutation frequently observed germline mutation in PRSS1 [47]. As a result of this mutation, the enzyme is not able to be broken down, even when it is no longer bound to calcium. However, this mutation eliminates the trypsin hydrolysis site. Few rare mutations were also observed in cationic trypsinogen. The reported cationic trypsinogen mutations are listed here: N29I, A16V, D22G, K23R, A121T, and R122C. R116C is a rare mutation that may result in misfolding of the protein and present an alternative cause of activation [77]. Patients with HP have recurrent acute pancreatitis, which can often evolve into CP. Their lifetime risk for PDAC is 35-fold (or more) by ages 70-75 [78].
Modeling of HP has revealed that pancreata from Elastase-R122H (mPRSS1) transgenic mice display early-onset acinar cell injury and inflammatory cell infiltration [79].
2.5.2. Chronic Pancreatitis
Chymotrypsin C (CTRC) gene mutations can be found in chronic pancreatitis. CTRC mutations appear to boost the effects of the mutant forms of cationic trypsinogen [79]. The two most frequent variants, where disease association reached statistical significance, were c.760C>T (p.R254W) and c.738_761del24 (p.K247_R254del), both located in exon 7 of CTRC [79]. The effect sizes of these mutations, as measured by the Odds Ratio (OR), were 3.3 and 11.5, respectively. Functional analysis of the CTRC variants showed impaired activity and/or reduced secretion, which resulted in “removal” of the normal negative degradative influence of CTRC on trypsin levels [47, 79].
Pathogenic variants in SPINK1 gene, encoding serine protease inhibitor, Kazel-type 1, can lead to autosomal recessive pancreatitis. In the United States, Europe, and India a high-risk haplotype containing SPINK1 p.Asn34Ser is common, with a minor allele frequency as high as 3%. In China, Japan, and Korea the SPINK1 splice variant (c.194+2T>C, also known as IVS3+2T>C) is common. It is not known if biallelic SPINK1 pathogenic variants alone are sufficient to cause recurrent acute or CP, but multiple families with biallelic SPINK1 pathogenic variants in affected family members have been documented [80].
Heterozygous mutations in the Cystic Fibrosis Transmembrane Receptor (CFTR) gene, may be found in a subset of these patients. CFTR is an ion channel involved in the transport of chloride and thiocyanate. While CFTR gene mutations are traditionally associated with cystic fibrosis, they may also contribute to chronic pancreatitis [81]. Dysfunction of CFTR results in acute pancreatitis. The risk ratio of PDAC in patients with cystic fibrosis in the USA and Europe during 1985-2005 was 5.3 (95% CI 2.4-10.1) [82].
2.6. Familial Pancreatic Cancer
Familial Pancreatic Cancer is defined as a family with at least one pair of first-degree relatives (parent-child or sibling pair) with PC without an identifiable syndrome in the family [83]. Germline mutations of different DNA repair genes have been identified in familial PC. The reason behind the mechanism is that accumulation of damaged DNA can trigger different states-uncontrolled cell division, apoptosis, and senescence that may transform to cancerous cell. Genes that are associated with FPC are; PALLD, BRCA1, BRCA2, PALB2, FANCC, FANCG ATM, CDKN2A, and CHEK2 [45, 73] (Table 1) (Fig. 1).
Palladin is a protein that in humans is encoded by the PALLD gene. Palladin is a component of actin-containing microfilaments that control cell shape, adhesion, and contraction. In a study of linkage analysis of large FPC pedigree characterized by autosomal dominant inheritance of PC with early onset and high penetrance showed significant linkage to chromosome 4q32-34 [84]. After few years an oncogenic germline mutation (Pro239Ser) was identified in the affected members of the family [85]. Thus it was suggested PALLD gene is a major susceptible for PDAC.
Germline mutations in BRCA1 reported in small number of patients with FPC reported are very rare. BRCA1 mutations are mainly related with breast and ovarian cancers in female. A 2.8 RR was observed with respect to general population, 1.3% of PDAC was detected in BRCA1 mutation carriers. However, another study did not notice any increased risk of PDAC among BRCA1 mutation carriers of 268 BRCA1 families. A large scale study conducted by Breast Cancer linkage Consortium, found a 2.26 fold (95% CI=1.26-4.06) increased risk of PC in BRCA1 mutation carriers. Whereas, in a study of 66 FPC patients from NFPTR kindred with 3 or more relatives with PDAC did not identify any deleterious BRCA1 germline mutation in these patients [86]. In another study of 145 Ashkenazi Jewish PDAC patients, no increase in frequency of BRCA1 was found [87]. Most of the studies did not find any increased risk of PDAC in BRCA1 mutated patients; however, Thompson and co-workers reported 2 to 2.5 fold increased risk of PDAC in BRCA1 mutated patients [88].
BRCA2 is the commonest altered genes identified in FPC even in the absence of breast and/or ovarian cancer. But mutations in BRCA2 have been long reported and it is the most frequently identified alterations in FPC even in the absence of breast cancer. Approximately 10% of FPC carries BRCA2 mutations, although age of onset may not be particularly early in life. The majority of BRCA2 germline mutations are nonsense or frameshift mutations, such as the 6174delT and other exon 11 mutations, these are almost 80% in the total mutations [89]. Approximately 4% to 10% of Ashkhenazi Jews with PDAC carry a germline BRCA2 mutation [66, 87]. It was estimated that 1% of the Ashkenazi Jewish population in North America harbors germline BRCA2 6174delT founder mutation, which has been associated with a 10-fold increased risk of developing pancreatic, breast, prostate, and ovarian cancers [66, 67]. Studies from German and British FPC families and North American FPC families suggested 15% and 17% rate of BRCA2 mutation respectively [90]. However, in a study of moderate risk and high risk FPC families only 6% of deleterious BRCA2 mutations were detected [91]. The overall prevalence of BRCA2 mutations in moderate risk and high risk PDAC families is approximately 6% with frequency ranging from 3% to 15% for families depending on the number of affected family members. Partner and localizer of BRCA2, also known as PALB2 or FANCN, located on chromosome 16p12.2, is a protein which in humans is encoded by the PALB2 gene. Germline mutations in PALB2 interacting domain of BRCA2 could not bind strongly with PALB2 thereby disrupting the BRCA2 functions in DNA damage sites. PALB2 and BRCA2 deficient cells showed identical phenotypes. In HBOC syndrome and Fanconi Anemia families, mutation of PALB2 is identified. In a study of Whole Genome Sequencing (WGS) of North American FPC families, PALB2 reported as a new PDAC susceptible gene, as a truncating mutation occurred in 3.1% of the people [92]. In a study of German and British FPC families, PALB2 mutation was identified in 3.7% [93]. However, a Dutch study could not detect any mutation in PALB2 in a study of 28 FPC families without BRCA1/BRCA2 mutation [94]. The observation noted that PALB2 mutation in FPC might affect a small subset of European descent families with an addition of breast cancer. A Whole Exome Sequencing (WES) study from Jones and coworkers described that PALB2 identified as a susceptible gene in FPC. Studies suggested that PALB2 mutation carriers in FPC families have 10 to 32-fold increased risk for the development of PDAC depending on the number of affected family members [92, 93]. Indeed PALB2 is increasingly considered a good candidate for clinical testing in BRCA1 and BRCA2-negative HBOC families. FANCC and FANCG are the genes in the BRCA2 pathway. Germline mutations in these genes have been linked to early-age of onset PDAC, whereas segregating germline mutation is not reported in FPC [95].
Whole genome analysis of nearly 170 families with FPC revealed heterozygous germline mutation of the ATM gene, in two kindreds with FPC and four families had deleterious homozygous ATM mutations [96]. Mutation segregation with the disease in the both kindreds and analyzed tumors revealed loss of heterozygosity of the wild type allele. These can be defined by classic two-hit model in tumor suppressor genes. The risk of breast and PDACs observed carriers of monoallelic ATM mutations. Possible importance of ATM in PDAC was highlighted in BxPC-3 cells (with wild type K-ras) which were treated with curcumin [97]. Curcumin resulted in phosphorylation of ATM at Ser-1981, G2/M cell cycle arrest and apoptosis of the tumor cells [97]. Additional analysis of 166 patients with FPC identified another four deleterious ATM germline mutations [96]. However, no mutations were detected in 190 healthy spouses. The prevalence of ATM mutations in the whole FPC cohort was 2.4%, whereas prevalence of ATM mutation was 4.6% when families with 3 or more affected member were considered [96].
Several studies suggest that CDKN2A (p16INK4a) mutation occurs in FPC without metachronous or synchronous occurrence of melanoma in the family. Study from Italian and Dutch population suggests that CDKN2A may be FPC susceptible gene and CDKN2A testing may be appropriate in FPC when melanoma does not occur in the family [98]. In a study of 1537 North American unselected patients with PC found 0.6% patients carried CDKN2A mutations. Among the 120 FPC cases in that study, 3.3% were CDKN2A positive. It was concluded these mutations are penetrate among smokers [99]. CDKN2A mutations are mostly missense and located in exon1 and exon2. In a study germline testing was performed for CDKN2A in a series of unselected PDAC patients and found 4% of the patients are CDKN2A positive [100]. In that extended study of 225 PDAC patients and controls, the CDKN2A mutation rate was 5.7%, ranging from 2.6% in patients without a family history of PDAC or melanoma, to 17% when two cancers occurred in the index patients or FDR’s, and to 45% when three or more cancers occurred. Sixteen probands of FPC families were identified, defined for having at least two FDR’s affected by PDAC, and no other manifestation of hereditary cancer syndromes. Deleterious CDKN2A mutations were found in five of the probands (31%) [98]. The mutation frequency ranged from 20% in FPC families with two affected members to 50% in families with 3, and was comparable to the mutation rate in melanoma families [101]. In a study performed by Harnick and colleagues of CDKN2A mutation in 28 FPC families, CDKN2A mutations were identified in 3 of these families who are melanoma positive. The selection criteria included presence of melanoma and indeed melanoma also occurred in four of these families (14%). The prevalence of CDKN2A mutation in their FPC families with no occurrence of melanoma was 12% (n=3) [91, 98, 102, 103]. However, the likelihood of identifying a CDKN2A mutation may also be high in families with 2 or more instances of PDAC or with one instance of PDAC and 1 of melanoma among FDRs, because it was found that 17% of such kindreds were positive for CDKN2A mutations [98]. CHEK2 is a multi-organ cancer susceptibility gene associated with a predisposition to breast, prostate and colon cancer. Analysis of 35 index patients of German FPC families identified 1 CHEK2 mutation (1170delC) (3%) [104].
2.7. Genetic Progression of PDAC
The evolution of PDAC from normal duct is well understood. The molecular changes can be categorized in different precursor lesions of PDAC. Point mutation in KRAS is the first hit mutation for development of PDAC from normal ductal cells [105, 106]. Inactivation of several other tumor suppressor genes, such as those encoding p16INK4A, p53, and SMAD4, also contributes to the evolution of histologically defined precursor lesions into infiltrating cancer [107]. The progression has been model experimentally supported using transgenic mice expressing mutant Kras in the pancreas, alone [108] or in combination with inactivation of the tumor suppressor genes [109, 110].
The genetic changes of PanINs have been studied in details. It can be explained as a bona fide precursor of PDAC. Invasive PDAC and PanIN can share almost similar kinds of mutation pattern [111]. KRAS and p16/CDKN2A mutations have been observed in the early stage of PanIN with low and intermediate grade dysplasia, whereas TP53 and SMAD4 mutations exhibit in late stage, occurring in PanINs with high grade dysplasia and in invasive cancer [111, 112]. KRAS mutations observed in most of the cases of PanIN could have been used as markers for the presence of PanIN for early and are present in most cases of PanINs [113]. These can be explained as founder mutations as they appear in every sample from a given patient. However, the presence of TP53, or SMAD4 gene mutations would suggest that a high grade precursor or an invasive carcinoma [113]. Moreover, analysis of PanINs reveal a stepwise accumulation of mutations as the lesions progress toward invasive adenocarcinoma. Molecular analysis of PanINs has shown that it harbours similar genetic abnormalities as infiltrating PDAC [111]. Activating point mutations in codon 12 of the KRAS gene typically occur in early low grade PanIN lesions (PanIN-1), and overexpression of Her2/neu are initial events whereas inactivating mutations in the p16INK4A/CDKN2A gene occur in intermediate lesions (PanIN-2), and inactivating mutations in SMAD4, TP53, and BRCA2 occur in late lesions (PanIN-3) [111, 114]. Telomere shortening is also an early event, that occurring in PanIN-1 lesions, contribute accumulation of chromosomal abnormalities in PanINs. Notably, telomere shortening was detectable at the beginning of PDAC formation and was observed in low grade PanINs [115], IPMNs [116], and PanIN-associated acinar-to-ductal metaplasias [117]. In PDAC, telomere length was short compared to those of normal chromosomes, that leads to severe telomere dysfunctions and chromosomal abnormalities including the inactivation of several tumor suppressor genes [118].
IPMNs also contain the KRAS point mutations and inactivating mutations in genes coding TP53 [119] and p16INK4A [120]. The SMAD4 gene is inactivated in a small number of IPMNs (3%) [121], whereas loss of the STK11/LKB1 gene product is significantly common [122].
The genetic alterations of MCNs are not extensively studied but it appears that they can have many of the same abnormalities found in infiltrating ductal adenocarcinoma of the pancreas, but at a lower frequency [111]. Like PanINs and IPMNs, MCNs also contain mutations in KRAS, TP53 and CDKN2A [107]. Some advanced MCNs may lose SMAD4 activity [123]. It is evident that some non-invasive IPMNs and some MCNs of the pancreas progress to invasive adenocarcinoma over time. Patients with these non-invasive lesions are on average 2-5 years younger than patients with the corresponding invasive lesions [124].
2.8. Somatic Alterations in PDAC
PDAC develops via Acinar-Ductal Metaplasia (ADM) and neoplastic precursor lesions; PanINs, IPMNs and MCNs. During this progression, PDAC genome harbours several genomic alterations that drive the tumor to metastasis. The most important paradigms to emerge from more than two decades of study in PDAC have been understood. PDAC is a disease of a combination of inherited and somatic mutations. According to the rate of occurrence, these somatic alterations can be subdivided into high-frequency alterations and low-frequency alterations. However, a comparison of the characteristics founder versus progressor gene alterations revealed that majority of the deleterious genetic alterations present in the PDAC including activating mutations of KRAS and inactivating mutations in CDKN2A, TP53, and SMAD4 are consistent with the progression model of pancreatic carcinogenesis (Fig. 1). Genomic alterations of CDKN2A, BRCA2, MLH1, TP53 and STK11 genes were mutated somatically in PDAC in addition to that FPC germline mutations were also shown in these genes, by Knudson’s two-hit hypothesis model these genes are probably the most significant effects in PDAC (Fig. 1). Although oncogenic mutations in KRAS occur frequently in PDAC, there are no explanations where PDAC showed with the wildtype KRAS allele. A minority of the patients showed mutated BRAF where KRAS is wildtype. It can be explained by mutual exclusive property of cancer mutation theory.
2.9. High Frequency Alterations
2.9.1. KRAS
KRAS is a proto-oncogene which encodes a ~21 kDa small GTPase that mediate a range of cellular functions, including proliferation, cell survival, cytoskeletal remodelling, etc. The switch to the active state of Ras protein is promoted by Guanine nucleotide Exchange Factors (GEFs), which aids exchange of GDP for GTP. KRAS inactivation occurs through guanosine-triphosphatase-activating proteins, which promotes GTP hydrolysis to the diphosphate GDP and attenuate signaling [125].
KRAS Activating mutations are found in more than 90% human PDAC. This leads to impair intrinsic GTPase activity of KRAS protein and can block the interaction between KRAS and GAPs. This leads to constitutive activation of KRAS and persistent stimulation of downstream signaling pathways that drive many of the hallmarks of cancer, sustained proliferation, metabolic reprogramming, anti-apoptosis, remodeling of tumor microenvironment, evasion of the immune response, cell migration and metastasis [126]. The spectrum of KRAS2 gene mutations in PDAC includes alteration in codon 12, 13, and 61 [125]. The frequency and specific substitutions show cancer type differences, with 98% of KRAS mutations in PDAC occurring at position G12. Of the 8 different substitutions found at this position, the predominant substitution is G12D. Mutations in codon 12 comprise 98% of all mutations in KRAS whereas frequencies of codon 13, and codon 61 mutations were 1% for both [127, 128]. Out of the G12 mutations 51% are G12D, (30%) G12V, (12%) G12R, (2%) G12S, (2%) G12C, (2%) G12A and <1% were G12L/F. Out of the G13 mutations 76% were G13D, 10% for both G13S and G13C, and 4% were G13P. Regarding Q61 mutations, 82% are Q61H, 11% Q61R, and 7% were Q61K [128].
Targeting of mutant KrasG12D or KrasG12V specifically to the murine pancreas is sufficient to initiate development of ADM, PanINs, IPMNs, and AFLs, which progress with long latency to invasive metastatic PDAC, thus recapitulating the disease [108, 126, 129, 130]. In transgenic mice, inducible KRAS G12D mutant was not shown only to initiate neoplastic lesions but was also involved in tumor maintenance [131]. In a study, it was found that KRAS mutation was associated with reduced patient survival in both malignant exocrine and PDAC [132]. Patients with PDAC carrying KRAS mutations showed a median survival of 17 months compared to 30 months for those who do not carry KRAS mutation (log-rank P=0.07) with a multivariate Hazard Ratio (HR) of 2.19 (95%CI 1.09-4.42) [132]. Recent studies in mouse models where Kras can be switched off and on demonstrated that continuous oncogenic Kras signaling is essential for both progression and maintenance of PDAC [131, 133]. Also, it was seen that sustained oncogenic Kras signaling is essential for growth and maintenance of metastasis in PDAC [131].
Activated KRAS engages multiple effecter pathways, notably the Raf/Mek/Erk, PI3K/Pdk1/Akt, and Ral guanine nucleotide exchange factor pathways [134-136]. Recent findings indicate Kras-PI3K-Pdk1 signaling drive PDAC initiation, progression and maintenance. Genetic proof of the importance of PI3K-Pdk1 signaling was shown in classical KrasG12D PDAC model [136]. Studies in PC cell lines have revealed crosstalk between oncogenic KRAS and the Hedgehog signaling pathway, suggesting that oncogenic KRAS plays an important role in activating Hedgehog signaling through the RAF/MEK/MAPK pathway in the absence of hedgehog ligands during pancreatic tumorigenesis [137]. Activating mutations of KRAS leads to increase such as proliferation, cell division, survival, and gene expression through the traditional targets of KRAS signaling, such as the phosphoinositide 3-kinase pathway and the RAF-mitogen activated protein kinase (MAPK) pathway. Oncogene KRAS also activate the proliferation of the desmoplastic stroma. The stroma has an important role in cell proliferation and invasion of PDAC development [131].
2.9.2. P16/CDKN2A/INK4A
The second most important frequently mutated gene in PDAC is CDKN2A/p16INK4A. CDKN2A regulates the cell cycle through the p16/Rb pathway and controls progression through the G1/S transition. CDKN2A is inactivated in PC through germline mutations as well as by somatic alterations, whereas inactivation of this tumor suppressor gene occurs in 80-95% sporadic cases. Disruption or inactivation of CDKN2A occurs through multiple mechanisms including nearly by homozygous deletion (40%), intragenic mutations with LOH (40%), and the remainder (15-20%) by epigenetic promoter silencing [137-139]. CDKN2A loss is generally seen in moderately advanced lesions that show features of dysplasia. CDKN2A encodes two tumor suppressors - INK4A and ARF - via distinct first exons and alternative reading frames in shared downstream errors [140]. Many PC sustain a loss of both the INK4A and ARF transcripts, thereby disrupting both the retinoblastoma (RB) and p53 tumor suppressor pathways. The cause of CDKN2A loss is due to the deletion of chromosome 9q21 in PDAC. This position encodes two tumor suppressor loci INK4A and ARF. This gene product stabilizes the p53 tumor suppressor through the neutralization of MDM2, it induces the ubiquitination and subsequent degradation of p53. INK4A seems to be the more important PDAC suppressor at this locus, as germline and sporadic mutations have been identified the target INK4A, but spare ARF [139, 141]. Though several different KRAS mutations detected in individual PanIN lesions, loss of INK4A usually occurs in later stages of pancreatic neoplasia. Immunohistochemical analysis confirmed the loss of p16 expression in affected tumor, specifies that p16 genetic events have functional importance. Homozygous deletion resulting inactivation of the p16INK4A/CDKN2A gene also inactivate an adjacent gene on chromosome 9p, MTAP (methylthioadenosine phosphorylase), which is located 100 kb telomeric and plays important role in the synthesis of adenosine. MTAP function is completely lost in 30% of PDAC [112, 137, 142].
2.9.3. TP53
The p53 protein encoded by TP53 gene is responsible for modulating cellular responses to cytotoxic stress by maintaining genome stability. TP53 is responsible for regulation of the G1/S cell cycle check points, maintenance of G2/M arrest, and induction of apoptosis, and protection against genomic rearrangement and accumulation of mutations. Almost 50-70% of the PDAC contains genomic alterations in DNA binding domain of TP53 [139]. Both oncogenic activation and loss of tumor suppressor pathways associated with TP53. The mutations, mainly the missense mutations, in coding sequences of DNA binding domain of p53, are often accompanied by loss of the wild-type allele [143]. Loss of heterozygosity of TP53 is detected in PanIN-3 lesions; resulting impaired p53 functions occur in late in the progression of PDAC. TP53 inactivation is the most common somatic alterations in most cancers. Mostly, the inactivation of TP53 detected by point mutations and homozygous deletions. The loss of p53 function leads to increase in cell growth, proliferation and cell division. Alterations of p53 are associated with K-ras mutations suggesting an effect in tumorigenesis [144, 145]. TP53 dependent fail-safe program of PDAC was explained by the upregulation of p21Cip1 in PanIN1 lesions, where P53 functions in senescence pathway. p21Cip1 acts as a cyclin-dependent kinase inhibitor. Loss of p21 functions through lack of transactivation has been detected in approximately 30-60% of the PDAC cases [146-148]. Loss of p53 functions results in aneuploidy, and genomic instability, a common observation in PDAC development [149, 150].
2.9.4. DPC4/SMAD4
Another frequently mutated tumor suppressor gene in PDAC is SMAD4/DPC4. The Smad4 protein plays a critical role in signaling through the transforming growth factor-beta (TGF-β) pathway. The TGF-β pathway is activated when TGF-β protein binds to specific cell surface receptors. This triggers an intracellular cascade that results in phosphorylation and nuclear localization of Smad transcription factors Smad2/3, complex with SMAD4. Inactivation of SMAD4 occurs in 50-60% of the PDAC patients [151]. Mainly SMAD4 gene is mutated or deleted and these events occur in the late stage of the tumor progression. In 30-35% cases, the gene is activated by homozygous deletion and in 20-30% cases loss of heterozygosity is observed [151]. Smad4 proteins can transduce the TGF-β signal from the cell surface to the nucleus [152]. During the early stages of pancreatic neoplastic progression TGFβ receptor or activin was mutated and abnormal expression also noticed in the neoplastic clone, which offers a selective advantage [153]. Whereas, in the late stage of PanIN, decreased expression of SMAD4, multiple genetic defects in cell cycle checkpoints and other regulatory systems were detected [154]. Smad4 loss interferes with the intracellular signaling cascade downstream from TGF-β, resulting in decreased growth inhibition via loss of pro-apoptotic signaling or via inappropriate G1/S transition [155]. Loss of SMAD4 nuclear labelling by IHC is observed late in pancreatic carcinogenesis, such as in PanIN-3 precursor lesions and infiltrating adenocarcinoma [121, 123, 154]. Loss of SMAD4 is also associated with poor prognosis of PDAC and with the development of widespread metastases in PDAC [156]. Many cancers, including PC, harbour defects in TGF-β and are resistant to TGF-β mediated growth suppression. The TGF-β type I receptor is also mutated in PDAC. Roles of activins and BMPs are reported in cancers. A recent study reported that activin type IB receptor gene (ACVR1B) is also mutated in PDAC. The TGF-β pathway is highly mutated in PDAC, with an overall mutation rate of >80% [157].
2.9.5. Mucin
The two proposed classifications of mucins based on cellular expression pattern, are membrane-bound mucins and secretory mucins. The secretory mucins, lack a transmembrane domain and are secreted directly into the extracellular spaces, include MUC2, MUC5AC, MUC5B, MUC6, MUC7, MUC8 and MUC19. The membrane-bound class of mucins are type I membrane proteins with single transmembrane domains and different lengths of cytoplasmic tail at the C-terminus. The membrane-bound class includes MUC1, MUC3A, MUC3B, MUC4, MUC12, MUC13, MUC15, MUC16, MUC17 and MUC20 [158]. Membrane-bound mucins can be released from cells through proteolytic cleavage, and many are produced in secreted forms that result from alternative mRNA splice forms in which the transmembrane domains are eliminated. All known mucins indicate the presence of positively charged lysine-arginine-rich motifs in the region juxtaposed to the plasma membrane. This motif is present in MUC1 (sequence RRK), MUC3 (RRGR), MUC12 (RKRHR), MUC15 (KRK), MUC16 (RRRKK) and MUC17 (RSKR). Another conserved motif in the cytoplasmic tails of many transmembrane mucins is the tyrosine-based sorting signal for clathrin-mediated endocytosis. This motif [YXX(L/M/V/I/F)] has been shown to be crucial for clathrin-mediated endocytosis of MUC1 (YHPM). Similar motifs are found in MUC3 (YVAL), MUC12 (YNNF) and MUC17 (YSNF). MUC1 intracellular signaling regulates other signaling pathways including MAPK, NF-kB, JAK-STAT, HIF, Wnt, TP53, Era and c-Src. In different cancer, MUC1 also regulates the processes such as growth, differentiation, apoptosis, cell fate, oxidative stress death protection, immunosurveillance, adhesion, polarity, inflammation, colonization and metabolism. Mucins are somatically mutated and dysregulated in PDAC. MUC1 T112P mutation was observed in 31.25% of all stages of PDAC. T4373 deletion mutation of MUC5B also identified in stage II PDAC. MUC6 and MUC16 have a relatively high mutation in PDAC. Forty-three percent of MUC16 mutations were nonsynonymous mutations, with 14.9% of these mutations resulting in frameshifts or deletions in PDAC [159]. MUC1 is a polymorphic, highly glycosylated, type I transmembrane protein, which engages in signal transduction through extracellular domain-mediated ligand binding or by interacting with receptors for growth and differentiation factors. Differential glycosylation patterns on the Tandem repeat may affect the adhesion properties that results in an increased ability to tumor cell metastasize [160].
2.10. Low Frequency Alterations
2.10.1. NCOA3/AIB1
The nuclear receptor coactivator 3 also known as NCOA3 is a protein that, in humans, is encoded by the NCOA3 gene. NCOA3 assists nuclear receptors in the up regulation of gene expression. NCOA3 is a transcriptional coactivator protein that contains several nuclear receptor interacting domains and an intrinsic histone acetyltransferase activity. NCOA3 is recruited to DNA promotion sites by ligand-activated nuclear receptors. NCOA3, in turn, acylates histones, which makes downstream DNA more accessible to transcription. High level of amplification for AIB I reported in ~10% of breast cancer tissues and four of nine PDAC cell lines [161, 162]. In archvial PDAC tissues copy number gains of AIB I gene observed in >37% [163].
2.10.2. ERBB2/Her2
The ErbB family consists of 4 plasma membrane-bound receptor tyrosine kinases. One of which is erbB-2, and the other members being epidermal growth factor receptor, erbB-3 (neuregulin-binding; lacks kinase domain), and erbB-4. HER2 can heterodimerise with any of the other three receptors and is considered to be the preferred dimerisation partner of the other ErbB receptors. Dimerisation results in the autophosphorylation of tyrosine residues within the cytoplasmic domain of the receptors and initiates a variety of signaling pathways like MAPK, PI3K/AKT, etc. In PDAC, ERBB2 amplification observed with variable incidence in 10-60% of cases with increased expression [164, 165].
2.10.3. AKT2
AKT2 is 1 of 3 closely related serine/threonine-protein kinases (AKT1, AKT2 and AKT3) called the AKT kinase, and which regulate many processes including metabolism, proliferation, cell survival, growth and angiogenesis. AKT2 encodes a serine and/or threonine kinase that act as a downstream effector of the PI3-AKT pathway. This gene is amplified and overexpressed in approximately 11% (3/28) to 20% (7/35) of PDAC [166, 167]. Some studies suggested 10-60% amplification in PDAC [1, 137]. AKT signaling also linked to enhance IGF-IR expression in PDAC by promoting invasive potential of cells [1].
2.10.4. BRAF
B-Raf is a member of the Raf kinase family of growth signal transduction protein kinases. This protein plays a role in regulating the MAP kinase/ERKs signaling pathway, which affects cell division, differentiation, and secretion. It is mutated in 33% of PC that has wild-type KRAS [168]. The mutational occurrence of KRAS and BRAF is suggested to be mutually exclusive. Some studies suggest that 1 mutation in 1 of these 2 genes results in retention of wild-type copies of the other. These observations suggest oncogenic regulation of KRAS or BRAF is a very important step in the development of PDAC [112, 137].
2.10.5. CCND1
The protein encoded by this gene belongs to the highly conserved cyclin family, whose members are characterized by a dramatic periodicity in protein abundance throughout the cell cycle. CCND1 forms a complex with and functions as a regulatory subunit of CDK4 or CDK6, whose activity is required for cell cycle G1/S transition. Southern analysis showed CCND1 amplification present in 25% PDAC which is correlated with increased expression [169].
2.10.6. RB1
Function of pRb is to prevent excessive cell growth by inhibiting cell cycle progression until a cell is ready to divide. When the cell is ready to divide, pRb is phosphorylated, becomes inactive and allows cell cycle progression. It is also a recruiter of several chromatin remodeling enzymes such as methylases and acetylases. In many cancers, RB gene is frequently mutated but in PDAC rare mutations (<6%) are reported [170].
2.10.7. CCNE1
Cyclins function as regulators of CDK kinases which contribute to the temporal coordination of each mitotic event. CCNE1 forms a complex with and functions as a regulatory subunit of CDK2, whose activity is required for cell cycle G1/S transition. Amplification and overexpression of CCNE1 reported in various cancers include colon adenocarcinoma, metastatic ductal carcinoma of the breast, serous carcinoma of the ovary, and adenocarcinoma of the stomach. Immunohistochemistry (IHC) studies of CCNE1 suggest overexpression of CCNE1 in ~6% of PDAC with gene amplification and mutational inactivation of FBXW7 [171].
2.10.8. DCC
Deleted in Colorectal Carcinoma (DCC) is a tumor suppressor gene named due to its rare homozygous deletion in colorectal carcinomas. In a study, 6% of PDAC is reported to have this mutation in DCC gene, suggesting a role in PDAC [172, 173].
2.10.9. MYB
MYB proto-oncogene acts as a transcriptional activator in humans. This gene encodes a protein with 3 HTH DNA-binding domains that functions as a transcription regulator. This protein plays an essential role in the regulation of haematopoiesis. MYB amplification is reported in 4-6% of PDAC cell lines and tissues [171, 174, 175]. Studies indicate that the c-myb oncogene was overexpressed not only in the amplified samples but also in the majority of the examined PC tissues and cell lines, suggesting that amplification is one of the mechanisms leading to overexpression. Genetic alterations of c-myb were mainly found in advanced tumors, indicating a possible correlation between tumor progression and aggressive tumor phenotypes.
2.10.10. MAP2K4/MKK4
MAP2K4 encodes a member of the Mitogen-Activated Protein Kinase (MAPK) family involved in a wide variety of cellular processes such as proliferation, differentiation, transcription regulation, and development. Studies showed deletion and mutation in MKK4 in human PC cell lines may have an additional role of MKK4 as tumor suppressor genes. Homozygous deletions in MKK4 were detected in 2-4% of PDAC [176, 177]. In addition, in a panel of 45 PDAC prescreened for loss of heterozygosity, one somatic missense mutation of MKK4 was observed. The finding of a somatic missense mutation in the absence of any other nucleotide polymorphisms or silent nucleotide changes continues to favor MKK4 as a mutational targeted tumor suppressor gene [176].
2.10.11. EGFR/ERBB1/Her1
The protein encoded by Epidermal Growth Factor Receptor (EGFR) is a transmembrane glycoprotein that is member of the protein kinase superfamily. EGFR is a cell surface protein that binds to epidermal growth factor. Binding of the protein to a ligand induces receptor dimerization and tyrosine autophosphorylation and leads to cell proliferation. In a study of 55 PDAC patients, 2 patients (3.6%) showed mutation delE746-A750 in tyrosine kinase domain, though few reports suggest no occurrence of EGFR mutations in PDAC [178].
2.10.12. PTEN
The PTEN is a tumor suppressor gene that provides instructions for making an enzyme that is found in almost all tissues in the body. The enzyme acts as a tumor suppressor, which means that it helps regulate cell division by keeping cells from growing and dividing too rapidly or in an uncontrolled way. Disease progression of PDAC has been associated with frequent loss of tumor suppressors like PI3K/PTEN pathway. Aberrant expression and deletion of the PTEN gene has been frequently reported in PDAC [179].
2.10.13. MLH1
It is a MMR gene homolog of the E. coli DNA, mutL. MLH1 mediates protein-protein interactions during mismatch recognition, strand discrimination, and strand removal. Defects in MLH1 are associated with the MicroSatellite Instability (MSI) observed in HNPCC. Genetic alterations in MLH1 have been reported in 3-15% of PDAC [180, 181].
2.10.14. PIK3CA
PI3K-AKT pathway is a key effector of Ras dependent transformation of many cell types and play role in cell survival, cell proliferation, and other growth related processes. The PIK3CA gene provides instructions for making the p110 alpha (p110α) protein, which is one piece (subunit) of an enzyme called phosphatidylinositol 3-kinase (PI3K). The p110α protein is called the catalytic subunit because it performs the action of PI3K, while the other subunit (produced by a different gene) regulates the enzyme's activity. Activating mutations of PIK3CA, gene encoding PI3K, have been reported in subset of PDAC. It was found 11% of IPMNs carries PIK3CA mutations [182].
2.10.15. STK11/LKB1
The STK11/LKB1 gene, which encodes a member of the serine/threonine kinase family, regulates cell polarity and functions as a tumor suppressor. Though STK11/LKB1 gene mutations are associated with PJS that increase risk of PDAC, sporadic mutations observed in 5% cases of PDAC particularly those arise in association of IPMN. LOH has been observed in 25% of patients with IPMN who lack PJS features [183].
2.10.16. ACVR1B
This gene encodes an activin A type IB receptor. Activins are dimeric growth and differentiation factors which belong to the transforming growth factor-beta (TGF-beta) superfamily of structurally related signaling proteins. Xenograft model studies showed homozygous deletion of exon 8 (657bp) and 6 bp deletion in exon 7. Sequencing analysis of the MADH4 gene revealed a nonsense mutation in the exon 5 (codon 245) of the MADH4 gene in that same xenograft model suggests coexistence of mutations of ACVR1B and MADH4 in PDAC [153].
2.10.17. TGFBR2
The TGFBR2 gene provides instructions for making a protein called transforming growth factor-beta (TGF-β) receptor type 2. This receptor transmits signals from the cell surface into the cell through a process called signal transduction. Through this type of signaling, the environment outside the cell affects activities inside the cell such as stimulation of cell growth and division. Somatic mutations in TGFBR2 were identified in 4.1% which includes homozygous deletion in PDAC [157].
2.10.18. GUCY2F
Retinal guanylyl cyclase 2 also known as guanylate cyclase F (GUCY2F) is a protein that in humans is encoded by the GUCY2F gene. The protein encoded by this gene is a guanylyl cyclase found predominantly in photoreceptors in the retina. The encoded protein is thought to be involved in resynthesis of cGMP after light activation of the visual signal transduction cascade, allowing a return to the dark state. In a study of 60 PDAC samples, GUCY2F mutation was found in 2 in percentage samples (p.Lys672Thr, p.Lys1063 Gln) [184].
2.10.19. NTRK3
This gene encodes a member of the Neurotrophic Tyrosine Receptor Kinase (NTRK) family. This kinase is a membrane-bound receptor that, upon neurotrophin binding, phosphorylates itself and members of the MAPK pathway. Signaling through this kinase leads to cell differentiation and may play a role in the development of proprioceptive neurons that sense body position. A report showed a mutation in NTRK3 gene (p.His599Ty) that was detected in 1.6% of PDAC cases [184].
2.10.20. FBXW7/CDC4
F-box/WD repeat-containing protein 7 protein in humans is encoded by the FBXW7 gene. This gene encodes a member of the F-box protein family which is characterized by an approximately 40 amino acid motif, the F-box. The F-box proteins constitute 1 of the 4 subunits of ubiquitin protein ligase complex called SCFs (SKP1-cullin-F-box), which function in phosphorylation-dependent ubiquitination. It was reported that mutation in FBXW7 found in 1% of PDAC [171].
2.10.21. EP300
This gene encodes the adenovirus E1A-associated cellular p300 transcriptional co-activator protein. The protein functions as histone acetyltransferase that regulates transcription via chromatin remodeling, and is important in the processes of cell proliferation and differentiation. It mediates cAMP-gene regulation by binding specifically to phosphorylated CREB protein. In a study of epithelial tumors, 2 pancreatic cell lines found to have a missence mutations and in frame insertion [185].
2.10.22. ALK5/TGFBR1
The protein encoded by this gene forms a heteromeric complex with type II TGF-β receptors when bound to TGF-β, transducing the TGF-β signal from the cell surface to the cytoplasm. The encoded protein is a serine/threonine protein kinase. A homozygous deletion in ALK5 gene was identified in 1% of PDAC patient [157].
2.10.23. ACVR2
The Activin type 2 receptors modulate signals for ligands belonging to the transforming growth factor beta superfamily of ligands. It functions in TGFβ signaling pathway. ACTVR2 mutations were identified in pancreatic cell lines, pancreatic tumors with both MSI positive and negative cases. All of the ACVR2 mutations are truncating mutations and defective ACVR2 result in the reduction of activin mediated cell signaling. These are involved in a host of physiological processes including, growth, cell differentiation, homeostasis, osteogenesis, apoptosis and many other functions. In a study, ACVR2A was found to be mutated in all MSI pancreatic tumors [186].
2.10.24. BRCA2/FANCD1
Though BRCA2 mutations are generally inherited germline mutations, but only one case of sporadic pancreatic cancer due to the somatic inactivation of both BRCA2 alleles in PDAC is reported [187]. In a few cases, BRCA2 is mutated in late tumor progression. In that scenario, the DNA repair pathway is altered (Table 2).
Table 2.
High frequency and low frequency somatic alterations in pancreatic ductal adenocarcinoma.
| Gene Name | Type of Alterations | Frequency | Function |
|---|---|---|---|
| KRAS | Point Mutation | 80-90% | GTPase |
| CDKN2A | Deletion, LOH | 80% | Regulate cell cycle |
| TP53 | Point Mutation, LOH | 80% | Regulate DNA repair, Cell cycle, apoptosis |
| SAMD4/DPC4 | Deletion, LOH | 50-60% | Signal transducer |
| MUC1 | Point Mutation | 10-31% | Growth, differentiation, anti apoptotic |
| AIB1/NCOA3 | Amplification | 10% | Transcriptional coactivator |
| ERBB2/HER2/EGFR2 | Amplification | 10-60% | Signal Transduction |
| AKT2 | Amplification | 10-60% | Serine/threonine-protein kinase |
| BRAF | Point Mutation | 0-33% | Growth signal transduction protein kinase |
| CCND1 | Amplification | 25% | Regulate Cell Cycle |
| RB1 | Mutation | <6% | Regulate Cell Cycle |
| CCNE1 | Amplification | 6% | Regulate Cell Cycle |
| DCC | Mutation | 6% | Transmembrane receptor protein |
| MYB | Amplification | 4-6% | Transcription factor |
| MKK4/MAP2K4 | Deletion | 2-4% | Ser/Thr protein kinase |
| EGFR/HER1/ERBB1 | Deletion | 3.6% | Cell surface receptors |
| MLH1/hMLH1 | Mutation | 3-15% | DNA mismatch repair |
| PIK3CA/PI3K | Mutation | 11% | Phosphorylate phosphatidylinositols |
| STK11/LKB1 | Mutation | 5% | Serine/threonine kinase |
| TGFBR2 | Deletion | 4.1% | Serine/threonine protein kinase, and TGF receptor subfamily |
| GUCY2F | Mutation | 1.2% | Resynthesis of cGMP after light activation of the visual signal transduction cascade |
| NTRK3 | Mutation | 0.6% | Receptor tyrosine kinase |
| FBXW7 | Mutation | 1% | Targets cyclin E for ubiquitin-mediated degradation |
| TGFBR1/ALK5 | Mutation | 1% | Serine/threonine protein kinase |
2.11. Somatic Alterations in Periampullary Adenocarcinoma
Genomic and molecular studies have not been extensively studied worldwide for PACs. Though few studies reported somatic key alterations in PACs, which is less common with PC. The rate of KRAS mutation in ampullary subtype is 40-50% [188]. In a study of 52 ampullary cancers, where 25 were intestinal subtype and 24 were pancreatobiliary subtype, KRAS mutation observed in 52% intestinal subtype, whereas, 42% of the pancreatobiliary subtype had KRAS mutation [188]. Schultz et al. found that 80% of resected PDACs and 67% of ampullary cancers in chemotherapy-naïve pancreatic and ampullary cancers had KRAS mutation [189]. KRAS mutation in the biliary subtype is much lower than ampullary subtype. A study reported 33% KRAS mutation in biliary subtype [190]. Another study of oncogenic mutations in KRAS, PIK3CA, MET, BRAF, EGFR, and NRAS of 94 resected cholangiocarcinoma reported 25 mutations predominantly in KRAS, PIK3CA, MET, BRAF, EGFR, and NRAS [191]. Borger et al. reported that mutations in IDH1 and IDH2 were only found in only intrahepatic, but not in extrahepatic cholangiocarcinomas. The incidence of KRAS mutation was 5% in intrahepatic and 23% in extrahepatic cholangiocarcinomas [192]. In a study, the incidence of KRAS mutation was found to be 35% in duodenal cancer subtype [193]. Seventy-four percent of the mutations were G>A transitions. The same mutation was associated with late-stage and poor differentiation [193]. In another study, KRAS mutation was found in 32% of the patients but no BRAF mutation was observed [194]. Interestingly, in a study by Schonleben et al., BRAF mutations (V600E and G469A) were observed in 66% (2/3) of PAC of duodenal subtype [24]. They also identified 28.6% KRAS mutation in all PAC. No mutation was detected in HRAS, NRAS, and PIK3CA [24]. Oliveira-Cunha et al. reported 41% of KRAS mutation incidence in 68 cases of PDAC and PAC [195] (Table 3) (Fig. 1).
Table 3.
Somatic alterations in periampullary adenocarcinoma.
| Gene | Type of Alterations | Frequency | Function |
|---|---|---|---|
| KRAS | Mutation | 30-60% | GTPase |
| BRAF | Mutation | 0-66% | Growth signal transduction protein kinase |
| PIK3CA | Mutation | 5.3% | Signal transduction |
| MET | Mutation | 4.25% | Receptor tyrosine kinase |
| EGFR/HER1/ERBB1 | Mutation | 1% | Cell surface receptors |
| NRAS | Mutation | 1% | GTPase |
2.12. Common Germline and Somatic Alterations of PDAC and PAC
In families with PC where the gene mutation is known, genetic counseling and testing could predict the PDAC risk of individuals. It has clinical implications for the affected individuals in addition to at-risk individuals in the family. Germline mutations of CDKN2A, STK11, TP53, BRCA2 and MLH1 are established as causative genes in hereditary syndrome associated with PDAC and FPC. CDKN2A and TP53 genes also somatically altered in PDAC in higher alteration frequencies; however STK11, BRCA2 and MLH1 genes showed lower alteration frequencies in PDAC. This observation notifies that the roles of these genes are very important in PDAC development pathways, and are
commonly altered in PDAC. These genes might be candidate genes for PDAC progression and development. No single genetic change has been identified as the catalyst for tumorigenesis in PDAC. Pancreatic and periampullary are common tumors, the origin of these tumors are in very close proximity. Somatic mutations of KRAS, BRAF, PIK3CA and EGFR genes are common in between PDAC and PAC (Fig. 1). There are certain genes altered are unique for PAC. Pancreatic ductal adenocarcinoma also arises from pancreatic head and body. Periampullary adenocarcinomas located in the pancreatic head may arise from the ampulla, the periampullary duodenum, the distal bile duct, or the pancreatic tissue itself. Pancreatobiliary subtype of PAC might share similar kind of mutation pattern though the mutation spectra of PAC are not well studied. Due to morphological origin variations, the mutation pattern of several PAC subtypes might differ between each other. These reports indicate that different mechanisms might play the key role in disease pathogenesis. KRAS mutations are more common in pancreatobiliary malignancies and PDAC indicate that they might follow similar mechanism of action. This would suggest that interactions among the identified common genes may confer selective growth advantages for neoplastic transformation. In PAC, this type of phenomena could not be observed as the study reports are not present in details. More research needs to be conducted on the genomic characteristics of PAC. Identification and characterization of somatic and/or germline genetic alterations would provide the mechanistic foundation of these genetically complex diseases.
CONCLUSION
Pancreatic ductal adenocarcinoma and PAC continue to be a devastating disease. These cancers are fatal and difficult to treat. In India, the incidence rate of these cancers is increasing very fast in the last two decades. We do not have recent reports on PDAC and PAC from different parts of Indian subcontinent. Major understanding of the earliest histologic and molecular changes has been well understood. Critical risk factors of PDAC like, lifestyle, environmental and occupational risk factors are extremely defined for western population, whereas in Indian patient population data is very limited and the cause of the disease is poorly understood. The exact molecular architecture of different stages of PDAC is well established in Western countries however, in Indian scenario, the genomic alteration in PDAC and different subtypes of PAC has not been explored yet. Screening of high-frequency FPC genes among families with PC can identify high-risk individuals among families. Though early drivers of PDAC and their molecular mechanism are completely understood, targeting them for therapeutic intervals is yet not possible. Advancement of sequencing technology has already revealed novel genetic alterations and probable pathways. Novel therapeutics target new drivers or combination of drivers is in clinical trials. More research needs to be done on genomic alterations of PAC to identify the key or early drivers for different histopathological subtypes. As PDAC and PAC are fast growing and detected in advance stages, the current goal of research should be focused on identification of early biomarkers as well as novel targeted therapeutics. Novel strategies based on genomic information seem likely to revolutionize PDAC therapy over the next few years, and may ultimately lead to fully personalized or precision medicine.
ACKNOWLEDGEMENTS
B. Roy from Human Genetics Unit, Indian Statistical Institute gave critical comments for this review. N. Sikdar is funded by Ramaligaswami Re-entry Fellowship (BT/RLF/ Re-Entry/05/2012) from Department of Biotechnology, Government of India. Gourab Saha is supported by DST Inspire fellowship from Department of Science and Technology, Government of India. S. Ghosh is supported by Medical College and Hospital, Kolkata, India, S.V. Shrikhande is supported by Tata Memorial Centre, Mumbai, India and S. Banerjee is supported by Tata Medical Hospital, Newtown, Rajarhat, Kolkata, India.
List of Abbreviations
- ASR
Age Standardized Rate
- ATM
Ataxia Teleangiectasia
- CFTR
Cystic Fibrosis Transmembrane Receptor
- CP
Chronic Pancreatitits
- FAMMM
Familial Atypical Multiple Mole Melanoma
- FAP
Famililal Adenomatous Polyposis
- FDR
First-Degree Relative
- FPC
Fammilial Pancreatic Cancer
- HBOC
Hereditary Breast-Ovarian Cancer
- HNPCC
Herditary Nonpolyposis Colorectal Carcinoma
- HP
Hereditary Pancreatitis
- IHC
Immunohistochemistry
- IPMN
Intraductal Papillary Mucinous Neoplasms
- LFS
Li-Fraumeni Syndrome
- LOH
Loss Of Heterozygosity
- LS
Lynch Syndrome
- MCN
Mucinous Cystic Neoplasms
- MSI
Microsattelite Instability
- PAC
Periampullary Adenocarcinoma
- PanIN
Pancreatic Intraepithelial Neoplasms
- PC
Pancreatic Cancer
- PDAC
Pancreatic Ductal Adenocarcinoma
- PJS
Peutz-Jeghers Syndrome
- RR
Relative Risk
- TGF-β
Transforming Growth Factor-beta
- TSG
Tumor Supressor Gene
Consent for Publication
Not applicable.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
AUTHORS’ CONTRIBUTIONS
Sikdar N. designed the outline, performed the majority of the writing and supervised the review. Saha G., Dutta A., Ghosh S., Shrikhande V.S., and Banerjee S. performed writing and editing of the review.
REFERENCES
- 1.Hezel A.F., Kimmelman A.C., Stanger B.Z., Bardeesy N., Depinho R.A. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20(10):1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 2.Warshaw A.L., Fernandez-del Castillo C. Pancreatic carcinoma. N. Engl. J. Med. 1992;326(7):455–465. doi: 10.1056/NEJM199202133260706. [DOI] [PubMed] [Google Scholar]
- 3.Kimura W., Futakawa N., Zhao B. Neoplastic diseases of the papilla of Vater. J. Hepatobiliary Pancreat. Surg. 2004;11(4):223–231. doi: 10.1007/s00534-004-0894-7. [DOI] [PubMed] [Google Scholar]
- 4.Carter J.T., Grenert J.P., Rubenstein L., Stewart L., Way L.W. Tumors of the ampulla of vater: Histopathologic classification and predictors of survival. J. Am. Coll. Surg. 2008;207(2):210–218. doi: 10.1016/j.jamcollsurg.2008.01.028. [DOI] [PubMed] [Google Scholar]
- 5.Kumari N., Prabha K., Singh R.K., Baitha D.K., Krishnani N. Intestinal and pancreatobiliary differentiation in periampullary carcinoma: The role of immunohistochemistry. Hum. Pathol. 2013;44(10):2213–2219. doi: 10.1016/j.humpath.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 6.de Paiva Haddad L.B., Patzina R.A., Penteado S., Montagnini A.L., da Cunha J.E., Machado M.C., Jukemura J. Lymph node involvement and not the histophatologic subtype is correlated with outcome after resection of adenocarcinoma of the ampulla of vater. J. Gastrointest. Surg. 2010;14(4):719–728. doi: 10.1007/s11605-010-1156-4. [DOI] [PubMed] [Google Scholar]
- 7.Ramfidis V.S., Syrigos K.N., Saif M.W. Ampullary and periampullary adenocarcinoma: New challenges in management of recurrence. JOP. 2013;14(2):158–160. doi: 10.6092/1590-8577/1471. [DOI] [PubMed] [Google Scholar]
- 8.Raimondi S., Maisonneuve P., Lowenfels A.B. Epidemiology of pancreatic cancer: An overview. Nat. Rev. Gastroenterol. Hepatol. 2009;6(12):699–708. doi: 10.1038/nrgastro.2009.177. [DOI] [PubMed] [Google Scholar]
- 9.Ying H., Dey P., Yao W., Kimmelman A.C., Draetta G.F., Maitra A., DePinho R.A. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2016;30(4):355–385. doi: 10.1101/gad.275776.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verma M. Pancreatic cancer biomarkers and their implication in cancer diagnosis and epidemiology. Cancers (Basel) 2010;2(4):1830–1837. doi: 10.3390/cancers2041830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider G., Siveke J.T., Eckel F., Schmid R.M. Pancreatic cancer: basic and clinical aspects. Gastroenterology. 2005;128(6):1606–1625. doi: 10.1053/j.gastro.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Lewis Z.K., Frost C.J., Venne V.L. Pancreatic cancer surveillance among high-risk populations: Knowledge and intent. J. Genet. Couns. 2009;18(3):229–238. doi: 10.1007/s10897-008-9205-9. [DOI] [PubMed] [Google Scholar]
- 13.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2015. CA Cancer J. Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 14.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 15.Rahib L., Smith B.D., Aizenberg R., Rosenzweig A.B., Fleshman J.M., Matrisian L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 16.Dhir V., Mohandas K.M. Epidemiology of digestive tract cancers in India IV. Gall bladder and pancreas. Indian J. Gastroenterol. 1999;18(1):24–28. [PubMed] [Google Scholar]
- 17.Benhamiche A.M., Jouve J.L., Manfredi S., Prost P., Isambert N., Faivre J. Cancer of the ampulla of Vater: Results of a 20-year population-based study. Eur. J. Gastroenterol. Hepatol. 2000;12(1):75–79. doi: 10.1097/00042737-200012010-00014. [DOI] [PubMed] [Google Scholar]
- 18.Qiao Q.L., Zhao Y.G., Ye M.L., Yang Y.M., Zhao J.X., Huang Y.T., Wan Y.L. Carcinoma of the ampulla of Vater: Factors influencing long-term survival of 127 patients with resection. World J. Surg. 2007;31(1):137–143. doi: 10.1007/s00268-006-0213-3. [DOI] [PubMed] [Google Scholar]
- 19.Berberat P.O., Kunzli B.M., Gulbinas A., Ramanauskas T., Kleeff J., Muller M.W., Wagner M., Friess H., Buchler M.W. An audit of outcomes of a series of periampullary carcinomas. Eur. J. Surg. Oncol. 2009;35(2):187–191. doi: 10.1016/j.ejso.2008.01.030. [DOI] [PubMed] [Google Scholar]
- 20.He J., Ahuja N., Makary M.A., Cameron J.L., Eckhauser F.E., Choti M.A., Hruban R.H., Pawlik T.M., Wolfgang C.L. 2564 resected periampullary adenocarcinomas at a single institution: Trends over three decades. HPB. 2014;16(1):83–90. doi: 10.1111/hpb.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen S.C., Shyr Y.M., Wang S.E. Longterm survival after pancreaticoduodenectomy for periampullary adenocarcinomas. HPB. 2013;15(12):951–957. doi: 10.1111/hpb.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandrasegaram M.D., Chiam S.C., Chen J.W., Khalid A., Mittinty M.L., Neo E.L., Tan C.P., Dolan P.M., Brooke-Smith M.E., Kanhere H., Worthley C.S. Distribution and pathological features of pancreatic, ampullary, biliary and duodenal cancers resected with pancreaticoduodenectomy. World J. Surg. Oncol. 2015;13:85. doi: 10.1186/s12957-015-0498-5. https://wjso.biomedcentral.com/ articles/10.1186/s12957-015-0498-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallinger S., Vivona A.A., Odze R.D., Mitri A., O’Beirne C.P., Berk T.C., Bapat B.V. Somatic APC and K-ras codon 12 mutations in periampullary adenomas and carcinomas from familial adenomatous polyposis patients. Oncogene. 1995;10(9):1875–1878. [PubMed] [Google Scholar]
- 24.Schonleben F., Qiu W., Allendorf J.D., Chabot J.A., Remotti H.E., Su G.H. Molecular analysis of PIK3CA, BRAF, and RAS oncogenes in periampullary and ampullary adenomas and carcinomas. J. Gastrointest. Surg. 2009;13(8):1510–1516. doi: 10.1007/s11605-009-0917-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rashid A., Ueki T., Gao Y.T., Houlihan P.S., Wallace C., Wang B.S., Shen M.C., Deng J., Hsing A.W. K-ras mutation, p53 overexpression, and microsatellite instability in biliary tract cancers: A population-based study in China. Clin. Cancer Res. 2002;8(10):3156–3163. [PubMed] [Google Scholar]
- 26.Ojajarvi I.A., Partanen T.J., Ahlbom A., Boffetta P., Hakulinen T., Jourenkova N., Kauppinen T.P., Kogevinas M., Porta M., Vainio H.U., Weiderpass E., Wesseling C.H. Occupational exposures and pancreatic cancer: A meta-analysis. Occup. Environ. Med. 2000;57(5):316–324. doi: 10.1136/oem.57.5.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hassan M.M., Bondy M.L., Wolff R.A., Abbruzzese J.L., Vauthey J.N., Pisters P.W., Evans D.B., Khan R., Chou T.H., Lenzi R., Jiao L., Li D. Risk factors for pancreatic cancer: Case-control study. Am. J. Gastroenterol. 2007;102(12):2696–2707. doi: 10.1111/j.1572-0241.2007.01510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ross A.H., Smith M.A., Anderson J.R., Small W.P. Late mortality after surgery for peptic ulcer. N. Engl. J. Med. 1982;307(9):519–522. doi: 10.1056/NEJM198208263070902. [DOI] [PubMed] [Google Scholar]
- 29.Mack T.M., Yu M.C., Hanisch R., Henderson B.E. Pancreas cancer and smoking, beverage consumption, and past medical history. J. Natl. Cancer Inst. 1986;76(1):49–60. [PubMed] [Google Scholar]
- 30.Virnig B.A., Shamliyan T., Tuttle T.M., Kane R.L., Wilt T.J. Diagnosis and management of ductal carcinoma in situ (DCIS). Evid. Rep. Technol. Assess. 2009;(185):1–549. [PMC free article] [PubMed] [Google Scholar]
- 31.Hruban R.H., Adsay N.V., Albores-Saavedra J., Compton C., Garrett E.S., Goodman S.N., Kern S.E., Klimstra D.S., Kloppel G., Longnecker D.S., Luttges J., Offerhaus G.J. Pancreatic intraepithelial neoplasia: A new nomenclature and classification system for pancreatic duct lesions. Am. J. Surg. Pathol. 2001;25(5):579–586. doi: 10.1097/00000478-200105000-00003. [DOI] [PubMed] [Google Scholar]
- 32.Brugge W.R., Lauwers G.Y., Sahani D., Fernandez-del Castillo C., Warshaw A.L. Cystic neoplasms of the pancreas. N. Engl. J. Med. 2004;351(12):1218–1226. doi: 10.1056/NEJMra031623. [DOI] [PubMed] [Google Scholar]
- 33.Thompson L.D., Becker R.C., Przygodzki R.M., Adair C.F., Heffess C.S. Mucinous cystic neoplasm (mucinous cystadenocarcinoma of low-grade malignant potential) of the pancreas: A clinicopathologic study of 130 cases. Am. J. Surg. Pathol. 1999;23(1):1–16. doi: 10.1097/00000478-199901000-00001. [DOI] [PubMed] [Google Scholar]
- 34.Brand R.E., Lerch M.M., Rubinstein W.S., Neoptolemos J.P., Whitcomb D.C., Hruban R.H., Brentnall T.A., Lynch H.T., Canto M.I. Advances in counselling and surveillance of patients at risk for pancreatic cancer. Gut. 2007;56(10):1460–1469. doi: 10.1136/gut.2006.108456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jang J.H., Cotterchio M., Borgida A., Gallinger S., Cleary S.P. Genetic variants in carcinogen-metabolizing enzymes, cigarette smoking and pancreatic cancer risk. Carcinogenesis. 2012;33(4):818–827. doi: 10.1093/carcin/bgs028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pandol S.J., Apte M.V., Wilson J.S., Gukovskaya A.S., Edderkaoui M. The burning question: Why is smoking a risk factor for pancreatic cancer? Pancreatology. 2012;12(4):344–349. doi: 10.1016/j.pan.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hruban R.H., Canto M.I., Goggins M., Schulick R., Klein A.P. Update on familial pancreatic cancer. Adv. Surg. 2010;44(1):293–311. doi: 10.1016/j.yasu.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bartsch D.K., Kress R., Sina-Frey M., Grutzmann R., Gerdes B., Pilarsky C., Heise J.W., Schulte K.M., Colombo-Benkmann M., Schleicher C., Witzigmann H., Pridohl O., Ghadimi M.B., Horstmann O., von Bernstorff W., Jochimsen L., Schmidt J., Eisold S., Estevez-Schwarz L., Hahn S.A., Schulmann K., Bock W., Gress T.M., Zugel N., Breitschaft K., Prenzel K., Messmann H., Endlicher E., Schneider M., Ziegler A., Schmiegel W., Schafer H., Rothmund M., Rieder H. Prevalence of familial pancreatic cancer in Germany. Int. J. Cancer. 2004;110(6):902–906. doi: 10.1002/ijc.20210. [DOI] [PubMed] [Google Scholar]
- 39.Hemminki K., Li X. Familial and second primary pancreatic cancers: A nationwide epidemiologic study from Sweden. Int. J. Cancer. 2003;103(4):525–530. doi: 10.1002/ijc.10863. [DOI] [PubMed] [Google Scholar]
- 40.Solomon S., Das S., Brand R., Whitcomb D.C. Inherited pancreatic cancer syndromes. Cancer J. 2012;18(6):485–491. doi: 10.1097/PPO.0b013e318278c4a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider R., Slater E.P., Sina M., Habbe N., Fendrich V., Matthai E., Langer P., Bartsch D.K. German national case collection for familial pancreatic cancer (FaPaCa): Ten years experience. Fam. Cancer. 2011;10(2):323–330. doi: 10.1007/s10689-010-9414-x. [DOI] [PubMed] [Google Scholar]
- 42.Brune K.A., Lau B., Palmisano E., Canto M., Goggins M.G., Hruban R.H., Klein A.P. Importance of age of onset in pancreatic cancer kindreds. J. Natl. Cancer Inst. 2010;102(2):119–126. doi: 10.1093/jnci/djp466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brune K., Abe T., Canto M., O’Malley L., Klein A.P., Maitra A., Volkan Adsay N., Fishman E.K., Cameron J.L., Yeo C.J., Kern S.E., Goggins M., Hruban R.H. Multifocal neoplastic precursor lesions associated with lobular atrophy of the pancreas in patients having a strong family history of pancreatic cancer. Am. J. Surg. Pathol. 2006;30(9):1067–1076. [PMC free article] [PubMed] [Google Scholar]
- 44.Shi C., Klein A.P., Goggins M., Maitra A., Canto M., Ali S., Schulick R., Palmisano E., Hruban R.H. Increased prevalence of precursor lesions in familial pancreatic cancer patients. Clin. Cancer Res. 2009;15(24):7737–7743. doi: 10.1158/1078-0432.CCR-09-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartsch D.K., Gress T.M., Langer P. Familial pancreatic cancer-current knowledge. Nat. Rev. Gastroenterol. Hepatol. 2012;9(8):445–453. doi: 10.1038/nrgastro.2012.111. [DOI] [PubMed] [Google Scholar]
- 46.Reznik R., Hendifar A.E., Tuli R. Genetic determinants and potential therapeutic targets for pancreatic adenocarcinoma. Front. Physiol. 2014;5:87. doi: 10.3389/fphys.2014.00087. https:// www.frontiersin.org/articles/10.3389/fphys.2014.00087/full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rustgi A.K. Familial pancreatic cancer: Genetic advances. Genes Dev. 2014;28(1):1–7. doi: 10.1101/gad.228452.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korsse S.E., Harinck F., van Lier M.G., Biermann K., Offerhaus G.J., Krak N., Looman C.W., van Veelen W., Kuipers E.J., Wagner A., Dekker E., Mathus-Vliegen E.M., Fockens P., van Leerdam M.E., Bruno M.J. Pancreatic cancer risk in Peutz-Jeghers syndrome patients: A large cohort study and implications for surveillance. J. Med. Genet. 2013;50(1):59–64. doi: 10.1136/jmedgenet-2012-101277. [DOI] [PubMed] [Google Scholar]
- 49.Rustgi A.K. The genetics of hereditary colon cancer. Genes Dev. 2007;21(20):2525–2538. doi: 10.1101/gad.1593107. [DOI] [PubMed] [Google Scholar]
- 50.Giardiello F.M., Welsh S.B., Hamilton S.R., Offerhaus G.J., Gittelsohn A.M., Booker S.V., Krush A.J., Yardley J.H., Luk G.D. Increased risk of cancer in the Peutz-Jeghers syndrome. N. Engl. J. Med. 1987;316(24):1511–1514. doi: 10.1056/NEJM198706113162404. [DOI] [PubMed] [Google Scholar]
- 51.Whelan A.J., Bartsch D., Goodfellow P.J. Brief report: A familial syndrome of pancreatic cancer and melanoma with a mutation in the CDKN2 tumor-suppressor gene. N. Engl. J. Med. 1995;333(15):975–977. doi: 10.1056/NEJM199510123331505. [DOI] [PubMed] [Google Scholar]
- 52.Gruis N.A., van der Velden P.A., Sandkuijl L.A., Prins D.E., Weaver-Feldhaus J., Kamb A., Bergman W., Frants R.R. Homozygotes for CDKN2 (p16) germline mutation in Dutch familial melanoma kindreds. Nat. Genet. 1995;10(3):351–353. doi: 10.1038/ng0795-351. [DOI] [PubMed] [Google Scholar]
- 53.Lynch H.T., Brand R.E., Hogg D., Deters C.A., Fusaro R.M., Lynch J.F., Liu L., Knezetic J., Lassam N.J., Goggins M., Kern S. Phenotypic variation in eight extended CDKN2A germline mutation familial atypical multiple mole melanoma-pancreatic carcinoma-prone families: The familial atypical mole melanoma-pancreatic carcinoma syndrome. Cancer. 2002;94(1):84–96. doi: 10.1002/cncr.10159. [DOI] [PubMed] [Google Scholar]
- 54.Goldstein A.M., Chan M., Harland M., Gillanders E.M., Hayward N.K., Avril M.F., Azizi E., Bianchi-Scarra G., Bishop D.T., Bressac-de Paillerets B., Bruno W., Calista D., Cannon Albright L.A., Demenais F., Elder D.E., Ghiorzo P., Gruis N.A., Hansson J., Hogg D., Holland E.A., Kanetsky P.A., Kefford R.F., Landi M.T., Lang J., Leachman S.A., Mackie R.M., Magnusson V., Mann G.J., Niendorf K., Newton Bishop J., Palmer J.M., Puig S., Puig-Butille J.A., de Snoo F.A., Stark M., Tsao H., Tucker M.A., Whitaker L., Yakobson E. High-risk melanoma susceptibility genes and pancreatic cancer, neural system tumors, and uveal melanoma across GenoMEL. Cancer Res. 2006;66(20):9818–9828. doi: 10.1158/0008-5472.CAN-06-0494. [DOI] [PubMed] [Google Scholar]
- 55.Bartsch D.K., Langer P., Habbe N., Matthai E., Chaloupka B., Sina M., Hahn S.A., Slater E.P. Clinical and genetic analysis of 18 pancreatic carcinoma/melanoma-prone families. Clin. Genet. 2010;77(4):333–341. doi: 10.1111/j.1399-0004.2009.01352.x. [DOI] [PubMed] [Google Scholar]
- 56.Vasen H.F., Gruis N.A., Frants R.R., van Der Velden P.A., Hille E.T., Bergman W. Risk of developing pancreatic cancer in families with familial atypical multiple mole melanoma associated with a specific 19 deletion of p16 (p16-Leiden). Int. J. Cancer. 2000;87(6):809–811. [PubMed] [Google Scholar]
- 57.Lynch H.T., Fusaro R.M., Lynch J.F., Brand R. Pancreatic cancer and the FAMMM syndrome. Fam. Cancer. 2008;7(1):103–112. doi: 10.1007/s10689-007-9166-4. [DOI] [PubMed] [Google Scholar]
- 58.Claus E.B., Schildkraut J.M., Thompson W.D., Risch N.J. The genetic attributable risk of breast and ovarian cancer. Cancer. 1996;77(11):2318–2324. doi: 10.1002/(SICI)1097-0142(19960601)77:11<2318::AID-CNCR21>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 59.Pal T., Permuth-Wey J., Betts J.A., Krischer J.P., Fiorica J., Arango H., LaPolla J., Hoffman M., Martino M.A., Wakeley K., Wilbanks G., Nicosia S., Cantor A., Sutphen R. BRCA1 and BRCA2 mutations account for a large proportion of ovarian carcinoma cases. Cancer. 2005;104(12):2807–2816. doi: 10.1002/cncr.21536. [DOI] [PubMed] [Google Scholar]
- 60.Iqbal J., Ragone A., Lubinski J., Lynch H.T., Moller P., Ghadirian P., Foulkes W.D., Armel S., Eisen A., Neuhausen S.L., Senter L., Singer C.F., Ainsworth P., Kim-Sing C., Tung N., Friedman E., Llacuachaqui M., Ping S., Narod S.A. The incidence of pancreatic cancer in BRCA1 and BRCA2 mutation carriers. Br. J. Cancer. 2012;107(12):2005–2009. doi: 10.1038/bjc.2012.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Asperen C.J., Brohet R.M., Meijers-Heijboer E.J., Hoogerbrugge N., Verhoef S., Vasen H.F., Ausems M.G., Menko F.H., Gomez Garcia E.B., Klijn J.G., Hogervorst F.B., van Houwelingen J.C., van’t Veer L.J., Rookus M.A., van Leeuwen F.E. Cancer risks in BRCA2 families: estimates for sites other than breast and ovary. J. Med. Genet. 2005;42(9):711–719. doi: 10.1136/jmg.2004.028829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cancer risks in BRCA2 mutation carriers. J. Natl. Cancer Inst. 1999;91(15):1310–1316. doi: 10.1093/jnci/91.15.1310. [DOI] [PubMed] [Google Scholar]
- 63.Struewing J.P., Hartge P., Wacholder S., Baker S.M., Berlin M., McAdams M., Timmerman M.M., Brody L.C., Tucker M.A. The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among Ashkenazi Jews. N. Engl. J. Med. 1997;336(20):1401–1408. doi: 10.1056/NEJM199705153362001. [DOI] [PubMed] [Google Scholar]
- 64.Brose M.S., Rebbeck T.R., Calzone K.A., Stopfer J.E., Nathanson K.L., Weber B.L. Cancer risk estimates for BRCA1 mutation carriers identified in a risk evaluation program. J. Natl. Cancer Inst. 2002;94(18):1365–1372. doi: 10.1093/jnci/94.18.1365. [DOI] [PubMed] [Google Scholar]
- 65.Moran A., O’Hara C., Khan S., Shack L., Woodward E., Maher E.R., Lalloo F., Evans D.G. Risk of cancer other than breast or ovarian in individuals with BRCA1 and BRCA2 mutations. Fam. Cancer. 2012;11(2):235–242. doi: 10.1007/s10689-011-9506-2. [DOI] [PubMed] [Google Scholar]
- 66.Ozcelik H., Schmocker B., Di Nicola N., Shi X.H., Langer B., Moore M., Taylor B.R., Narod S.A., Darlington G., Andrulis I.L., Gallinger S., Redston M. Germline BRCA2 6174delT mutations in Ashkenazi Jewish pancreatic cancer patients. Nat. Genet. 1997;16(1):17–18. doi: 10.1038/ng0597-17. [DOI] [PubMed] [Google Scholar]
- 67.Oddoux C., Struewing J.P., Clayton C.M., Neuhausen S., Brody L.C., Kaback M., Haas B., Norton L., Borgen P., Jhanwar S., Goldgar D., Ostrer H., Offit K. The carrier frequency of the BRCA2 6174delT mutation among Ashkenazi Jewish individuals is approximately 1%. Nat. Genet. 1996;14(2):188–190. doi: 10.1038/ng1096-188. [DOI] [PubMed] [Google Scholar]
- 68.Goldgar D.E. Analysis of familial breast cancer in genetic analysis workshop 9: Summary of findings. Genet. Epidemiol. 1995;12(6):833–836. doi: 10.1002/gepi.1370120650. [DOI] [PubMed] [Google Scholar]
- 69.Feldmann G., Karikari C., dal Molin M., Duringer S., Volkmann P., Bartsch D.K., Bisht S., Koorstra J.B., Brossart P., Maitra A., Fendrich V. Inactivation of Brca2 cooperates with Trp53(R172H) to induce invasive pancreatic ductal adenocarcinomas in mice: A mouse model of familial pancreatic cancer. Cancer Biol. Ther. 2011;11(11):959–968. doi: 10.4161/cbt.11.11.15534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kastrinos F., Mukherjee B., Tayob N., Wang F., Sparr J., Raymond V.M., Bandipalliam P., Stoffel E.M., Gruber S.B., Syngal S. Risk of pancreatic cancer in families with Lynch syndrome. JAMA. 2009;302(16):1790–1795. doi: 10.1001/jama.2009.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kadmon M., Tandara A., Herfarth C. Duodenal adenomatosis in familial adenomatous polyposis coli. A review of the literature and results from the Heidelberg Polyposis Register. Int. J. Colorectal Dis. 2001;16(2):63–75. doi: 10.1007/s003840100290. [DOI] [PubMed] [Google Scholar]
- 72.Giardiello F.M., Offerhaus G.J., Lee D.H., Krush A.J., Tersmette A.C., Booker S.V., Kelley N.C., Hamilton S.R. Increased risk of thyroid and pancreatic carcinoma in familial adenomatous polyposis. Gut. 1993;34(10):1394–1396. doi: 10.1136/gut.34.10.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ghiorzo P. Genetic predisposition to pancreatic cancer. World J. Gastroenterol. 2014;20(31):10778–10789. doi: 10.3748/wjg.v20.i31.10778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Whitcomb D.C. Genetic risk factors for pancreatic disorders. Gastroenterology. 2013;144(6):1292–1302. doi: 10.1053/j.gastro.2013.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Whitcomb D.C., Gorry M.C., Preston R.A., Furey W., Sossenheimer M.J., Ulrich C.D., Martin S.P., Gates L.K., Jr, Amann S.T., Toskes P.P., Liddle R., McGrath K., Uomo G., Post J.C., Ehrlich G.D. Hereditary pancreatitis is caused by a mutation in the cationic trypsinogen gene. Nat. Genet. 1996;14(2):141–145. doi: 10.1038/ng1096-141. [DOI] [PubMed] [Google Scholar]
- 76.Witt H., Luck W., Hennies H.C., Classen M., Kage A., Lass U., Landt O., Becker M. Mutations in the gene encoding the serine protease inhibitor, Kazal type 1 are associated with chronic pancreatitis. Nat. Genet. 2000;25(2):213–216. doi: 10.1038/76088. [DOI] [PubMed] [Google Scholar]
- 77.Kereszturi E., Szmola R., Kukor Z., Simon P., Weiss F.U., Lerch M.M., Sahin-Toth M. Hereditary pancreatitis caused by mutation-induced misfolding of human cationic trypsinogen: A novel disease mechanism. Hum. Mutat. 2009;30(4):575–582. doi: 10.1002/humu.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lowenfels A.B., Maisonneuve P., DiMagno E.P., Elitsur Y., Gates L.K., Jr, Perrault J., Whitcomb D.C. Hereditary pancreatitis and the risk of pancreatic cancer. International Hereditary Pancreatitis Study Group. J. Natl. Cancer Inst. 1997;89(6):442–446. doi: 10.1093/jnci/89.6.442. [DOI] [PubMed] [Google Scholar]
- 79.Rosendahl J., Witt H., Szmola R., Bhatia E., Ozsvari B., Landt O., Schulz H.U., Gress T.M., Pfutzer R., Lohr M., Kovacs P., Bluher M., Stumvoll M., Choudhuri G., Hegyi P., te Morsche R.H., Drenth J.P., Truninger K., Macek M., Jr, Puhl G., Witt U., Schmidt H., Buning C., Ockenga J., Kage A., Groneberg D.A., Nickel R., Berg T., Wiedenmann B., Bodeker H., Keim V., Mossner J., Teich N., Sahin-Toth M., Chymotrypsin C. CTRC) variants that diminish activity or secretion are associated with chronic pancreatitis. Nat. Genet. 2008;40(1):78–82. doi: 10.1038/ng.2007.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.LaRusch J., Solomon S., Whitcomb D.C. Pancreatitis Overview. In: Adam M.P., Ardinger H.H., Pagon R.A., editors. GeneReviews. Seattle, WA: University of Washington; 1993. [Google Scholar]
- 81.Zoller H., Egg M., Graziadei I., Creus M., Janecke A.R., Loffler-Ragg J., Vogel W. CFTR gene mutations in pancreatitis: Frequency and clinical manifestations in an Austrian patient cohort. Wien. Klin. Wochenschr. 2007;119(17-18):527–533. doi: 10.1007/s00508-007-0849-5. [DOI] [PubMed] [Google Scholar]
- 82.Maisonneuve P., Marshall B.C., Lowenfels A.B. Risk of pancreatic cancer in patients with cystic fibrosis. Gut. 2007;56(9):1327–1328. doi: 10.1136/gut.2007.125278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hruban R.H., Petersen G.M., Goggins M., Tersmette A.C., Offerhaus G.J., Falatko F., Yeo C.J., Kern S.E. Familial pancreatic cancer. Ann. Oncol. 1999;10(Suppl. 4):69–73. [PubMed] [Google Scholar]
- 84.Eberle M.A., Pfutzer R., Pogue-Geile K.L., Bronner M.P., Crispin D., Kimmey M.B., Duerr R.H., Kruglyak L., Whitcomb D.C., Brentnall T.A. A new susceptibility locus for autosomal dominant pancreatic cancer maps to chromosome 4q32-34. Am. J. Hum. Genet. 2002;70(4):1044–1048. doi: 10.1086/339692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pogue-Geile K.L., Chen R., Bronner M.P., Crnogorac-Jurcevic T., Moyes K.W., Dowen S., Otey C.A., Crispin D.A., George R.D., Whitcomb D.C., Brentnall T.A. Palladin mutation causes familial pancreatic cancer and suggests a new cancer mechanism. PLoS Med. 2006;3(12):e516. doi: 10.1371/journal.pmed.0030516. http:// journals.plos.org/plosmedicine/article?id=10.1371/journal.pmed.0030516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Axilbund J.E., Argani P., Kamiyama M., Palmisano E., Raben M., Borges M., Brune K.A., Goggins M., Hruban R.H., Klein A.P. Absence of germline BRCA1 mutations in familial pancreatic cancer patients. Cancer Biol. Ther. 2009;8(2):131–135. doi: 10.4161/cbt.8.2.7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ferrone C.R., Levine D.A., Tang L.H., Allen P.J., Jarnagin W., Brennan M.F., Offit K., Robson M.E. BRCA germline mutations in Jewish patients with pancreatic adenocarcinoma. J. Clin. Oncol. 2009;27(3):433–438. doi: 10.1200/JCO.2008.18.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thompson D., Easton D.F. Cancer incidence in BRCA1 mutation carriers. J. Natl. Cancer Inst. 2002;94(18):1358–1365. doi: 10.1093/jnci/94.18.1358. [DOI] [PubMed] [Google Scholar]
- 89.Hahn S.A., Greenhalf B., Ellis I., Sina-Frey M., Rieder H., Korte B., Gerdes B., Kress R., Ziegler A., Raeburn J.A., Campra D., Grutzmann R., Rehder H., Rothmund M., Schmiegel W., Neoptolemos J.P., Bartsch D.K. BRCA2 germline mutations in familial pancreatic carcinoma. J. Natl. Cancer Inst. 2003;95(3):214–221. doi: 10.1093/jnci/95.3.214. [DOI] [PubMed] [Google Scholar]
- 90.Murphy K.M., Brune K.A., Griffin C., Sollenberger J.E., Petersen G.M., Bansal R., Hruban R.H., Kern S.E. Evaluation of candidate genes MAP2K4, MADH4, ACVR1B, and BRCA2 in familial pancreatic cancer: Deleterious BRCA2 mutations in 17%. Cancer Res. 2002;62(13):3789–3793. [PubMed] [Google Scholar]
- 91.Slater E.P., Langer P., Fendrich V., Habbe N., Chaloupka B., Matthai E., Sina M., Hahn S.A., Bartsch D.K. Prevalence of BRCA2 and CDKN2a mutations in German familial pancreatic cancer families. Fam. Cancer. 2010;9(3):335–343. doi: 10.1007/s10689-010-9329-6. [DOI] [PubMed] [Google Scholar]
- 92.Jones S., Hruban R.H., Kamiyama M., Borges M., Zhang X., Parsons D.W., Lin J.C., Palmisano E., Brune K., Jaffee E.M., Iacobuzio-Donahue C.A., Maitra A., Parmigiani G., Kern S.E., Velculescu V.E., Kinzler K.W., Vogelstein B., Eshleman J.R., Goggins M., Klein A.P. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324(5924):217. doi: 10.1126/science.1171202. http://science.sciencemag.org/content/ 324/5924/217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Slater E.P., Langer P., Niemczyk E., Strauch K., Butler J., Habbe N., Neoptolemos J.P., Greenhalf W., Bartsch D.K. PALB2 mutations in European familial pancreatic cancer families. Clin. Genet. 2010;78(5):490–494. doi: 10.1111/j.1399-0004.2010.01425.x. [DOI] [PubMed] [Google Scholar]
- 94.Harinck F., Kluijt I., van Mil S.E., Waisfisz Q., van Os T.A., Aalfs C.M., Wagner A., Olderode-Berends M., Sijmons R.H., Kuipers E.J., Poley J.W., Fockens P., Bruno M.J. Routine testing for PALB2 mutations in familial pancreatic cancer families and breast cancer families with pancreatic cancer is not indicated. Eur. J. Hum. Genet. 2012;20(5):577–579. doi: 10.1038/ejhg.2011.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Couch F.J., Johnson M.R., Rabe K., Boardman L., McWilliams R., de Andrade M., Petersen G. Germ line Fanconi anemia complementation group C mutations and pancreatic cancer. Cancer Res. 2005;65(2):383–386. [PubMed] [Google Scholar]
- 96.Roberts N.J., Jiao Y., Yu J., Kopelovich L., Petersen G.M., Bondy M.L., Gallinger S., Schwartz A.G., Syngal S., Cote M.L., Axilbund J., Schulick R., Ali S.Z., Eshleman J.R., Velculescu V.E., Goggins M., Vogelstein B., Papadopoulos N., Hruban R.H., Kinzler K.W., Klein A.P. ATM mutations in patients with hereditary pancreatic cancer. Cancer Discov. 2012;2(1):41–46. doi: 10.1158/2159-8290.CD-11-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sahu R.P., Batra S., Srivastava S.K. Activation of ATM/Chk1 by curcumin causes cell cycle arrest and apoptosis in human pancreatic cancer cells. Br. J. Cancer. 2009;100(9):1425–1433. doi: 10.1038/sj.bjc.6605039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ghiorzo P., Fornarini G., Sciallero S., Battistuzzi L., Belli F., Bernard L., Bonelli L., Borgonovo G., Bruno W., De Cian F., Decensi A., Filauro M., Faravelli F., Gozza A., Gargiulo S., Mariette F., Nasti S., Pastorino L., Queirolo P., Savarino V., Varesco L., Scarra G.B. CDKN2A is the main susceptibility gene in Italian pancreatic cancer families. J. Med. Genet. 2012;49(3):164–170. doi: 10.1136/jmedgenet-2011-100281. [DOI] [PubMed] [Google Scholar]
- 99.McWilliams R.R., Wieben E.D., Rabe K.G., Pedersen K.S., Wu Y., Sicotte H., Petersen G.M. Prevalence of CDKN2A mutations in pancreatic cancer patients: implications for genetic counseling. Eur. J. Hum. Genet. 2011;19(4):472–478. doi: 10.1038/ejhg.2010.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ghiorzo P., Pastorino L., Bonelli L., Cusano R., Nicora A., Zupo S., Queirolo P., Sertoli M., Pugliese V., Bianchi-Scarra G. INK4/ARF germline alterations in pancreatic cancer patients. Ann. Oncol. 2004;15(1):70–78. doi: 10.1093/annonc/mdg498. [DOI] [PubMed] [Google Scholar]
- 101.Bruno W., Ghiorzo P., Battistuzzi L., Ascierto P.A., Barile M., Gargiulo S., Gensini F., Gliori S., Guida M., Lombardo M., Manoukian S., Menin C., Nasti S., Origone P., Pasini B., Pastorino L., Peissel B., Pizzichetta M.A., Queirolo P., Rodolfo M., Romanini A., Scaini M.C., Testori A., Tibiletti M.G., Turchetti D., Leachman S.A., Bianchi Scarra G. Clinical genetic testing for familial melanoma in Italy: A cooperative study. J. Am. Acad. Dermatol. 2009;61(5):775–782. doi: 10.1016/j.jaad.2009.03.039. [DOI] [PubMed] [Google Scholar]
- 102.Moskaluk C.A., Hruban H., Lietman A., Smyrk T., Fusaro L., Fusaro R., Lynch J., Yeo C.J., Jackson C.E., Lynch H.T., Kern S.E. Novel germline p16(INK4) allele (Asp145Cys) in a family with multiple pancreatic carcinomas. Mutations in brief no. 148. Online. Hum. Mutat. 1998;12(1):70. doi: 10.1002/(SICI)1098-1004(1998)12:1<70::AID-HUMU13>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 103.Bartsch D.K., Sina-Frey M., Lang S., Wild A., Gerdes B., Barth P., Kress R., Grutzmann R., Colombo-Benkmann M., Ziegler A., Hahn S.A., Rothmund M., Rieder H. CDKN2A germline mutations in familial pancreatic cancer. Ann. Surg. 2002;236(6):730–737. doi: 10.1097/00000658-200212000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bartsch D.K., Krysewski K., Sina-Frey M., Fendrich V., Rieder H., Langer P., Kress R., Schneider M., Hahn S.A., Slater E.P. Low frequency of CHEK2 mutations in familial pancreatic cancer. Fam. Cancer. 2006;5(4):305–308. doi: 10.1007/s10689-006-7850-4. [DOI] [PubMed] [Google Scholar]
- 105.Lohr M., Kloppel G., Maisonneuve P., Lowenfels A.B., Luttges J. Frequency of K-ras mutations in pancreatic intraductal neoplasias associated with pancreatic ductal adenocarcinoma and chronic pancreatitis: A meta-analysis. Neoplasia. 2005;7(1):17–23. doi: 10.1593/neo.04445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Moskaluk C.A., Hruban R.H., Kern S.E. p16 and K-ras gene mutations in the intraductal precursors of human pancreatic adenocarcinoma. Cancer Res. 1997;57(11):2140–2143. [PubMed] [Google Scholar]
- 107.Cowan R.W., Maitra A. Genetic progression of pancreatic cancer. Cancer J. 2014;20(1):80–84. doi: 10.1097/PPO.0000000000000011. [DOI] [PubMed] [Google Scholar]
- 108.Hingorani S.R., Petricoin E.F., Maitra A., Rajapakse V., King C., Jacobetz M.A., Ross S., Conrads T.P., Veenstra T.D., Hitt B.A., Kawaguchi Y., Johann D., Liotta L.A., Crawford H.C., Putt M.E., Jacks T., Wright C.V., Hruban R.H., Lowy A.M., Tuveson D.A. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4(6):437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 109.Aguirre A.J., Bardeesy N., Sinha M., Lopez L., Tuveson D.A., Horner J., Redston M.S., DePinho R.A. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17(24):3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kojima K., Vickers S.M., Adsay N.V., Jhala N.C., Kim H.G., Schoeb T.R., Grizzle W.E., Klug C.A. Inactivation of Smad4 accelerates Kras(G12D)-mediated pancreatic neoplasia. Cancer Res. 2007;67(17):8121–8130. doi: 10.1158/0008-5472.CAN-06-4167. [DOI] [PubMed] [Google Scholar]
- 111.Maitra A., Fukushima N., Takaori K., Hruban R.H. Precursors to invasive pancreatic cancer. Adv. Anat. Pathol. 2005;12(2):81–91. doi: 10.1097/01.pap.0000155055.14238.25. [DOI] [PubMed] [Google Scholar]
- 112.Maitra A., Kern S.E., Hruban R.H. Molecular pathogenesis of pancreatic cancer. Best Pract. Res. Clin. Gastroenterol. 2006;20(2):211–226. doi: 10.1016/j.bpg.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 113.Wolfgang C.L., Herman J.M., Laheru D.A., Klein A.P., Erdek M.A., Fishman E.K., Hruban R.H. Recent progress in pancreatic cancer. CA Cancer J. Clin. 2013;63(5):318–348. doi: 10.3322/caac.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Scarlett C.J., Salisbury E.L., Biankin A.V., Kench J. Precursor lesions in pancreatic cancer: Morphological and molecular pathology. Pathology. 2011;43(3):183–200. doi: 10.1097/PAT.0b013e3283445e3a. [DOI] [PubMed] [Google Scholar]
- 115.van Heek N.T., Meeker A.K., Kern S.E., Yeo C.J., Lillemoe K.D., Cameron J.L., Offerhaus G.J., Hicks J.L., Wilentz R.E., Goggins M.G., De Marzo A.M., Hruban R.H., Maitra A. Telomere shortening is nearly universal in pancreatic intraepithelial neoplasia. Am. J. Pathol. 2002;161(5):1541–1547. doi: 10.1016/S0002-9440(10)64432-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hashimoto Y., Murakami Y., Uemura K., Hayashidani Y., Sudo T., Ohge H., Fukuda E., Shimamoto F., Sueda T., Hiyama E. Telomere shortening and telomerase expression during multistage carcinogenesis of intraductal papillary mucinous neoplasms of the pancreas. J. Gastrointest. Surg. 2008;12(1):17–28. doi: 10.1007/s11605-007-0383-9. [DOI] [PubMed] [Google Scholar]
- 117.Hong S.M., Heaphy C.M., Shi C., Eo S.H., Cho H., Meeker A.K., Eshleman J.R., Hruban R.H., Goggins M. Telomeres are shortened in acinar-to-ductal metaplasia lesions associated with pancreatic intraepithelial neoplasia but not in isolated acinar-to-ductal metaplasias. Mod. Pathol. 2011;24(2):256–266. doi: 10.1038/modpathol.2010.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gisselsson D., Jonson T., Petersen A., Strombeck B., Dal Cin P., Hoglund M., Mitelman F., Mertens F., Mandahl N. Telomere dysfunction triggers extensive DNA fragmentation and evolution of complex chromosome abnormalities in human malignant tumors. Proc. Natl. Acad. Sci. USA. 2001;98(22):12683–12688. doi: 10.1073/pnas.211357798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sessa F., Solcia E., Capella C., Bonato M., Scarpa A., Zamboni G., Pellegata N.S., Ranzani G.N., Rickaert F., Kloppel G. Intraductal papillary-mucinous tumours represent a distinct group of pancreatic neoplasms: An investigation of tumour cell differentiation and K-ras, p53 and c-erbB-2 abnormalities in 26 patients. Virchows Arch. 1994;425(4):357–367. doi: 10.1007/BF00189573. [DOI] [PubMed] [Google Scholar]
- 120.Abe T., Fukushima N., Brune K., Boehm C., Sato N., Matsubayashi H., Canto M., Petersen G.M., Hruban R.H., Goggins M. Genome-wide allelotypes of familial pancreatic adenocarcinomas and familial and sporadic intraductal papillary mucinous neoplasms. Clin. Cancer Res. 2007;13(20):6019–6025. doi: 10.1158/1078-0432.CCR-07-0471. [DOI] [PubMed] [Google Scholar]
- 121.Iacobuzio-Donahue C.A., Klimstra D.S., Adsay N.V., Wilentz R.E., Argani P., Sohn T.A., Yeo C.J., Cameron J.L., Kern S.E., Hruban R.H. Dpc-4 protein is expressed in virtually all human intraductal papillary mucinous neoplasms of the pancreas: Comparison with conventional ductal adenocarcinomas. Am. J. Pathol. 2000;157(3):755–761. doi: 10.1016/S0002-9440(10)64589-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sahin F., Maitra A., Argani P., Sato N., Maehara N., Montgomery E., Goggins M., Hruban R.H., Su G.H. Loss of Stk11/Lkb1 expression in pancreatic and biliary neoplasms. Mod. Pathol. 2003;16(7):686–691. doi: 10.1097/01.MP.0000075645.97329.86. [DOI] [PubMed] [Google Scholar]
- 123.Iacobuzio-Donahue C.A., Wilentz R.E., Argani P., Yeo C.J., Cameron J.L., Kern S.E., Hruban R.H. Dpc4 protein in mucinous cystic neoplasms of the pancreas: Frequent loss of expression in invasive carcinomas suggests a role in genetic progression. Am. J. Surg. Pathol. 2000;24(11):1544–1548. doi: 10.1097/00000478-200011000-00011. [DOI] [PubMed] [Google Scholar]
- 124.Maitra A., Hruban R.H. Pancreatic cancer. Annu. Rev. Pathol. 2008;3:157–188. doi: 10.1146/annurev.pathmechdis.3.121806.154305. https://www.annualreviews.org/ doi/full/10.1146/annurev.pathmechdis.3.121806.154305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Eser S., Schnieke A., Schneider G., Saur D. Oncogenic KRAS signalling in pancreatic cancer. Br. J. Cancer. 2014;111(5):817–822. doi: 10.1038/bjc.2014.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pylayeva-Gupta Y., Grabocka E., Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat. Rev. Cancer. 2011;11(11):761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Caldas C., Kern S.E. K-ras mutation and pancreatic adenocarcinoma. Int. J. Pancreatol. 1995;18(1):1–6. doi: 10.1007/BF02825415. [DOI] [PubMed] [Google Scholar]
- 128.Bryant K.L., Mancias J.D., Kimmelman A.C., Der C.J. KRAS: feeding pancreatic cancer proliferation. Trends Biochem. Sci. 2014;39(2):91–100. doi: 10.1016/j.tibs.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Guerra C., Schuhmacher A.J., Canamero M., Grippo P.J., Verdaguer L., Perez-Gallego L., Dubus P., Sandgren E.P., Barbacid M. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11(3):291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 130.Seidler B., Schmidt A., Mayr U., Nakhai H., Schmid R.M., Schneider G., Saur D. A Cre-loxP-based mouse model for conditional somatic gene expression and knockdown in vivo by using avian retroviral vectors. Proc. Natl. Acad. Sci. USA. 2008;105(29):10137–10142. doi: 10.1073/pnas.0800487105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Collins M.A., Bednar F., Zhang Y., Brisset J.C., Galban S., Galban C.J., Rakshit S., Flannagan K.S., Adsay N.V., Pasca di Magliano M. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J. Clin. Invest. 2012;122(2):639–653. doi: 10.1172/JCI59227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rachakonda P.S., Bauer A.S., Xie H., Campa D., Rizzato C., Canzian F., Beghelli S., Greenhalf W., Costello E., Schanne M., Heller A., Scarpa A., Neoptolemos J.P., Werner J., Buchler M., Hoheisel J.D., Hemminki K., Giese N., Kumar R. Somatic mutations in exocrine pancreatic tumors: association with patient survival. PLoS One. 2013;8(4):e60870. doi: 10.1371/journal.pone.0060870. http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0060870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ying H., Kimmelman A.C., Lyssiotis C.A., Hua S., Chu G.C., Fletcher-Sananikone E., Locasale J.W., Son J., Zhang H., Coloff J.L., Yan H., Wang W., Chen S., Viale A., Zheng H., Paik J.H., Lim C., Guimaraes A.R., Martin E.S., Chang J., Hezel A.F., Perry S.R., Hu J., Gan B., Xiao Y., Asara J.M., Weissleder R., Wang Y.A., Chin L., Cantley L.C., DePinho R.A. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149(3):656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lim K.H., Baines A.T., Fiordalisi J.J., Shipitsin M., Feig L.A., Cox A.D., Der C.J., Counter C.M. Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell. 2005;7(6):533–545. doi: 10.1016/j.ccr.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 135.Feldmann G., Mishra A., Hong S.M., Bisht S., Strock C.J., Ball D.W., Goggins M., Maitra A., Nelkin B.D. Inhibiting the cyclin-dependent kinase CDK5 blocks pancreatic cancer formation and progression through the suppression of Ras-Ral signaling. Cancer Res. 2010;70(11):4460–4469. doi: 10.1158/0008-5472.CAN-09-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Eser S., Reiff N., Messer M., Seidler B., Gottschalk K., Dobler M., Hieber M., Arbeiter A., Klein S., Kong B., Michalski C.W., Schlitter A.M., Esposito I., Kind A.J., Rad L., Schnieke A.E., Baccarini M., Alessi D.R., Rad R., Schmid R.M., Schneider G., Saur D. Selective requirement of PI3K/PDK1 signaling for Kras oncogene-driven pancreatic cell plasticity and cancer. Cancer Cell. 2013;23(3):406–420. doi: 10.1016/j.ccr.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 137.Koorstra J.B., Hustinx S.R., Offerhaus G.J., Maitra A. Pancreatic carcinogenesis. Pancreatology. 2008;8(2):110–125. doi: 10.1159/000123838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Caldas C., Hahn S.A., da Costa L.T., Redston M.S., Schutte M., Seymour A.B., Weinstein C.L., Hruban R.H., Yeo C.J., Kern S.E. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat. Genet. 1994;8(1):27–32. doi: 10.1038/ng0994-27. [DOI] [PubMed] [Google Scholar]
- 139.Rozenblum E., Schutte M., Goggins M., Hahn S.A., Panzer S., Zahurak M., Goodman S.N., Sohn T.A., Hruban R.H., Yeo C.J., Kern S.E. Tumor-suppressive pathways in pancreatic carcinoma. Cancer Res. 1997;57(9):1731–1714. [PubMed] [Google Scholar]
- 140.Sherr C.J. The INK4a/ARF network in tumour suppression. Nat. Rev. Mol. Cell Biol. 2001;2(10):731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- 141.Liu L., Dilworth D., Gao L., Monzon J., Summers A., Lassam N., Hogg D. Mutation of the CDKN2A 5′ UTR creates an aberrant initiation codon and predisposes to melanoma. Nat. Genet. 1999;21(1):128–132. doi: 10.1038/5082. [DOI] [PubMed] [Google Scholar]
- 142.Hustinx S.R., Hruban R.H., Leoni L.M., Iacobuzio-Donahue C., Cameron J.L., Yeo C.J., Brown P.N., Argani P., Ashfaq R., Fukushima N., Goggins M., Kern S.E., Maitra A. Homozygous deletion of the MTAP gene in invasive adenocarcinoma of the pancreas and in periampullary cancer: a potential new target for therapy. Cancer Biol. Ther. 2005;4(1):83–86. doi: 10.4161/cbt.4.1.1380. [DOI] [PubMed] [Google Scholar]
- 143.Redston M.S., Caldas C., Seymour A.B., Hruban R.H., da Costa L., Yeo C.J., Kern S.E. p53 mutations in pancreatic carcinoma and evidence of common involvement of homocopolymer tracts in DNA microdeletions. Cancer Res. 1994;54(11):3025–3033. [PubMed] [Google Scholar]
- 144.Kalthoff H., Schmiegel W., Roeder C., Kasche D., Schmidt A., Lauer G., Thiele H.G., Honold G., Pantel K., Riethmuller G. p53 and K-RAS alterations in pancreatic epithelial cell lesions. Oncogene. 1993;8(2):289–298. [PubMed] [Google Scholar]
- 145.Pellegata N.S., Sessa F., Renault B., Bonato M., Leone B.E., Solcia E., Ranzani G.N. K-ras and p53 gene mutations in pancreatic cancer: ductal and nonductal tumors progress through different genetic lesions. Cancer Res. 1994;54(6):1556–1560. [PubMed] [Google Scholar]
- 146.Ahrendt S.A., Brown H.M., Komorowski R.A., Zhu Y.R., Wilson S.D., Erickson B.A., Ritch P.S., Pitt H.A., Demeure M.J. p21WAF1 expression is associated with improved survival after adjuvant chemoradiation for pancreatic cancer. Surgery. 2000;128(4):520–530. doi: 10.1067/msy.2000.108052. [DOI] [PubMed] [Google Scholar]
- 147.Song M.M., Nio Y., Dong M., Tamura K., Furuse K., Tian Y.L., He S.G., Shen K. Comparison of K-ras point mutations at codon 12 and p21 expression in pancreatic cancer between Japanese and Chinese patients. J. Surg. Oncol. 2000;75(3):176–185. doi: 10.1002/1096-9098(200011)75:3<176::aid-jso5>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 148.Garcea G., Neal C.P., Pattenden C.J., Steward W.P., Berry D.P. Molecular prognostic markers in pancreatic cancer: A systematic review. Eur. J. Cancer. 2005;41(15):2213–2236. doi: 10.1016/j.ejca.2005.04.044. [DOI] [PubMed] [Google Scholar]
- 149.Harada T., Okita K., Shiraishi K., Kusano N., Kondoh S., Sasaki K. Interglandular cytogenetic heterogeneity detected by comparative genomic hybridization in pancreatic cancer. Cancer Res. 2002;62(3):835–839. [PubMed] [Google Scholar]
- 150.Gorunova L., Hoglund M., Andren-Sandberg A., Dawiskiba S., Jin Y., Mitelman F., Johansson B. Cytogenetic analysis of pancreatic carcinomas: Intratumor heterogeneity and nonrandom pattern of chromosome aberrations. Genes Chromosomes Cancer. 1998;23(2):81–99. doi: 10.1002/(sici)1098-2264(199810)23:2<81::aid-gcc1>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 151.Hahn S.A., Schutte M., Hoque A.T., Moskaluk C.A., da Costa L.T., Rozenblum E., Weinstein C.L., Fischer A., Yeo C.J., Hruban R.H., Kern S.E. 1996 doi: 10.1126/science.271.5247.350. http://science.sciencemag.org/content/271/5247/ [DOI] [PubMed]
- 152.Blobe G.C., Schiemann W.P., Lodish H.F. Role of transforming growth factor beta in human disease. N. Engl. J. Med. 2000;342(18):1350–1358. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- 153.Su G.H., Bansal R., Murphy K.M., Montgomery E., Yeo C.J., Hruban R.H., Kern S.E. ACVR1B (ALK4, activin receptor type 1B) gene mutations in pancreatic carcinoma. Proc. Natl. Acad. Sci. USA. 2001;98(6):3254–3257. doi: 10.1073/pnas.051484398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wilentz R.E., Iacobuzio-Donahue C.A., Argani P., McCarthy D.M., Parsons J.L., Yeo C.J., Kern S.E., Hruban R.H. Loss of expression of Dpc4 in pancreatic intraepithelial neoplasia: Evidence that DPC4 inactivation occurs late in neoplastic progression. Cancer Res. 2000;60(7):2002–2006. [PubMed] [Google Scholar]
- 155.Massague J., Blain S.W., Lo R.S. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103(2):295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 156.Heinmoller E., Dietmaier W., Zirngibl H., Heinmoller P., Scaringe W., Jauch K.W., Hofstadter F., Ruschoff J. Molecular analysis of microdissected tumors and preneoplastic intraductal lesions in pancreatic carcinoma. Am. J. Pathol. 2000;157(1):83–92. doi: 10.1016/S0002-9440(10)64520-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Goggins M., Shekher M., Turnacioglu K., Yeo C.J., Hruban R.H., Kern S.E. Genetic alterations of the transforming growth factor beta receptor genes in pancreatic and biliary adenocarcinomas. Cancer Res. 1998;58(23):5329–5332. [PubMed] [Google Scholar]
- 158.Singh P.K., Hollingsworth M.A. Cell surface-associated mucins in signal transduction. Trends Cell Biol. 2006;16(9):467–476. doi: 10.1016/j.tcb.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 159.King R.J., Yu F., Singh P.K. Genomic alterations in mucins across cancers. Oncotarget. 2017;8(40):67152–67168. doi: 10.18632/oncotarget.17934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Mehla K., Singh P.K. MUC1: A novel metabolic master regulator. Biochim. Biophys. Acta. 2014;1845(2):126–135. doi: 10.1016/j.bbcan.2014.01.001. https://www.sciencedirect.com/science/article/pii/S0304419X1400002X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Anzick S.L., Kononen J., Walker R.L., Azorsa D.O., Tanner M.M., Guan X.Y., Sauter G., Kallioniemi O.P., Trent J.M., Meltzer P.S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. 1997 doi: 10.1126/science.277.5328.965. http://science.sciencemag.org/content/277/5328/ [DOI] [PubMed]
- 162.Ghadimi B.M., Schrock E., Walker R.L., Wangsa D., Jauho A., Meltzer P.S., Ried T. Specific chromosomal aberrations and amplification of the AIB1 nuclear receptor coactivator gene in pancreatic carcinomas. Am. J. Pathol. 1999;154(2):525–536. doi: 10.1016/S0002-9440(10)65298-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Henke R.T., Haddad B.R., Kim S.E., Rone J.D., Mani A., Jessup J.M., Wellstein A., Maitra A., Riegel A.T. Overexpression of the nuclear receptor coactivator AIB1 (SRC-3) during progression of pancreatic adenocarcinoma. Clin. Cancer Res. 2004;10(18 Pt 1):6134–6142. doi: 10.1158/1078-0432.CCR-04-0561. [DOI] [PubMed] [Google Scholar]
- 164.Yamanaka Y., Friess H., Kobrin M.S., Buchler M., Kunz J., Beger H.G., Korc M. Overexpression of HER2/neu oncogene in human pancreatic carcinoma. Hum. Pathol. 1993;24(10):1127–1134. doi: 10.1016/0046-8177(93)90194-l. [DOI] [PubMed] [Google Scholar]
- 165.Hermanova M., Lukas Z., Nenutil R., Brazdil J., Kroupova I., Kren L., Pazourkova M., Ruzicka M., Dite P. Amplification and overexpression of HER-2/neu in invasive ductal carcinomas of the pancreas and pancreatic intraepithelial neoplasms and the relationship to the expression of p21(WAF1/CIP1). Neoplasma. 2004;51(2):77–83. [PubMed] [Google Scholar]
- 166.Cheng J.Q., Ruggeri B., Klein W.M., Sonoda G., Altomare D.A., Watson D.K., Testa J.R. Amplification of AKT2 in human pancreatic cells and inhibition of AKT2 expression and tumorigenicity by antisense RNA. Proc. Natl. Acad. Sci. USA. 1996;93(8):3636–3641. doi: 10.1073/pnas.93.8.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ruggeri B.A., Huang L., Wood M., Cheng J.Q., Testa J.R. Amplification and overexpression of the AKT2 oncogene in a subset of human pancreatic ductal adenocarcinomas. Mol. Carcinog. 1998;21(2):81–86. [PubMed] [Google Scholar]
- 168.Calhoun E.S., Jones J.B., Ashfaq R., Adsay V., Baker S.J., Valentine V., Hempen P.M., Hilgers W., Yeo C.J., Hruban R.H., Kern S.E. BRAF and FBXW7 (CDC4, FBW7, AGO, SEL10) mutations in distinct subsets of pancreatic cancer: Potential therapeutic targets. Am. J. Pathol. 2003;163(4):1255–1260. doi: 10.1016/S0002-9440(10)63485-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Gansauge S., Gansauge F., Ramadani M., Stobbe H., Rau B., Harada N., Beger H.G. Overexpression of cyclin D1 in human pancreatic carcinoma is associated with poor prognosis. Cancer Res. 1997;57(9):1634–1637. [PubMed] [Google Scholar]
- 170.Huang L., Lang D., Geradts J., Obara T., Klein-Szanto A.J., Lynch H.T., Ruggeri B.A. Molecular and immunochemical analyses of RB1 and cyclin D1 in human ductal pancreatic carcinomas and cell lines. Mol. Carcinog. 1996;15(2):85–95. doi: 10.1002/(SICI)1098-2744(199602)15:2<85::AID-MC1>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 171.Calhoun E.S., Hucl T., Gallmeier E., West K.M., Arking D.E., Maitra A., Iacobuzio-Donahue C.A., Chakravarti A., Hruban R.H., Kern S.E. Identifying allelic loss and homozygous deletions in pancreatic cancer without matched normals using high-density single-nucleotide polymorphism arrays. Cancer Res. 2006;66(16):7920–7928. doi: 10.1158/0008-5472.CAN-06-0721. [DOI] [PubMed] [Google Scholar]
- 172.Thiagalingam S., Lengauer C., Leach F.S., Schutte M., Hahn S.A., Overhauser J., Willson J.K., Markowitz S., Hamilton S.R., Kern S.E., Kinzler K.W., Vogelstein B. Evaluation of candidate tumour suppressor genes on chromosome 18 in colorectal cancers. Nat. Genet. 1996;13(3):343–346. doi: 10.1038/ng0796-343. [DOI] [PubMed] [Google Scholar]
- 173.Hilgers W., Song J.J., Haye M., Hruban R.R., Kern S.E., Fearon E.R. Homozygous deletions inactivate DCC, but not MADH4/DPC4/SMAD4, in a subset of pancreatic and biliary cancers. Genes Chromosomes Cancer. 2000;27(4):353–357. [PubMed] [Google Scholar]
- 174.Kauraniemi P., Hedenfalk I., Persson K., Duggan D.J., Tanner M., Johannsson O., Olsson H., Trent J.M., Isola J., Borg A. MYB oncogene amplification in hereditary BRCA1 breast cancer. Cancer Res. 2000;60(19):5323–5328. [PubMed] [Google Scholar]
- 175.Wallrapp C., Muller-Pillasch F., Solinas-Toldo S., Lichter P., Friess H., Buchler M., Fink T., Adler G., Gress T.M. Characterization of a high copy number amplification at 6q24 in pancreatic cancer identifies c-myb as a candidate oncogene. Cancer Res. 1997;57(15):3135–3139. [PubMed] [Google Scholar]
- 176.Su G.H., Hilgers W., Shekher M.C., Tang D.J., Yeo C.J., Hruban R.H., Kern S.E. Alterations in pancreatic, biliary, and breast carcinomas support MKK4 as a genetically targeted tumor suppressor gene. Cancer Res. 1998;58(11):2339–2342. [PubMed] [Google Scholar]
- 177.Xin W., Yun K.J., Ricci F., Zahurak M., Qiu W., Su G.H., Yeo C.J., Hruban R.H., Kern S.E., Iacobuzio-Donahue C.A. MAP2K4/MKK4 expression in pancreatic cancer: Genetic validation of immunohistochemistry and relationship to disease course. Clin. Cancer Res. 2004;10(24):8516–8520. doi: 10.1158/1078-0432.CCR-04-0885. [DOI] [PubMed] [Google Scholar]
- 178.Kwak E.L., Jankowski J., Thayer S.P., Lauwers G.Y., Brannigan B.W., Harris P.L., Okimoto R.A., Haserlat S.M., Driscoll D.R., Ferry D., Muir B., Settleman J., Fuchs C.S., Kulke M.H., Ryan D.P., Clark J.W., Sgroi D.C., Haber D.A., Bell D.W. Epidermal growth factor receptor kinase domain mutations in esophageal and pancreatic adenocarcinomas. Clin. Cancer Res. 2006;12(14 Pt 1):4283–4287. doi: 10.1158/1078-0432.CCR-06-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Asano T., Yao Y., Zhu J., Li D., Abbruzzese J.L., Reddy S.A. The PI 3-kinase/Akt signaling pathway is activated due to aberrant Pten expression and targets transcription factors NF-kappaB and c-Myc in pancreatic cancer cells. Oncogene. 2004;23(53):8571–8580. doi: 10.1038/sj.onc.1207902. [DOI] [PubMed] [Google Scholar]
- 180.Goggins M., Offerhaus G.J., Hilgers W., Griffin C.A., Shekher M., Tang D., Sohn T.A., Yeo C.J., Kern S.E., Hruban R.H. Pancreatic adenocarcinomas with DNA replication errors (RER+) are associated with wild-type K-ras and characteristic histopathology. Poor differentiation, a syncytial growth pattern, and pushing borders suggest RER+. Am. J. Pathol. 1998;152(6):1501–1507. [PMC free article] [PubMed] [Google Scholar]
- 181.Wilentz R.E., Goggins M., Redston M., Marcus V.A., Adsay N.V., Sohn T.A., Kadkol S.S., Yeo C.J., Choti M., Zahurak M., Johnson K., Tascilar M., Offerhaus G.J., Hruban R.H., Kern S.E. Genetic, immunohistochemical, and clinical features of medullary carcinoma of the pancreas: A newly described and characterized entity. Am. J. Pathol. 2000;156(5):1641–1651. doi: 10.1016/S0002-9440(10)65035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Schonleben F., Qiu W., Ciau N.T., Ho D.J., Li X., Allendorf J.D., Remotti H.E., Su G.H. PIK3CA mutations in intraductal papillary mucinous neoplasm/carcinoma of the pancreas. Clin. Cancer Res. 2006;12(12):3851–3855. doi: 10.1158/1078-0432.CCR-06-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sato N., Rosty C., Jansen M., Fukushima N., Ueki T., Yeo C.J., Cameron J.L., Iacobuzio-Donahue C.A., Hruban R.H., Goggins M. STK11/LKB1 Peutz-Jeghers gene inactivation in intraductal papillary-mucinous neoplasms of the pancreas. Am. J. Pathol. 2001;159(6):2017–2022. doi: 10.1016/S0002-9440(10)63053-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wood L.D., Calhoun E.S., Silliman N., Ptak J., Szabo S., Powell S.M., Riggins G.J., Wang T.L., Yan H., Gazdar A., Kern S.E., Pennacchio L., Kinzler K.W., Vogelstein B., Velculescu V.E. Somatic mutations of GUCY2F, EPHA3, and NTRK3 in human cancers. Hum. Mutat. 2006;27(10):1060–1061. doi: 10.1002/humu.9452. [DOI] [PubMed] [Google Scholar]
- 185.Gayther S.A., Batley S.J., Linger L., Bannister A., Thorpe K., Chin S.F., Daigo Y., Russell P., Wilson A., Sowter H.M., Delhanty J.D., Ponder B.A., Kouzarides T., Caldas C. Mutations truncating the EP300 acetylase in human cancers. Nat. Genet. 2000;24(3):300–303. doi: 10.1038/73536. [DOI] [PubMed] [Google Scholar]
- 186.Hempen P.M., Zhang L., Bansal R.K., Iacobuzio-Donahue C.A., Murphy K.M., Maitra A., Vogelstein B., Whitehead R.H., Markowitz S.D., Willson J.K., Yeo C.J., Hruban R.H., Kern S.E. Evidence of selection for clones having genetic inactivation of the activin A type II receptor (ACVR2) gene in gastrointestinal cancers. Cancer Res. 2003;63(5):994–999. [PubMed] [Google Scholar]
- 187.Schutte M., da Costa L.T., Hahn S.A., Moskaluk C., Hoque A.T., Rozenblum E., Weinstein C.L., Bittner M., Meltzer P.S., Trent J.M. Identification by representational difference analysis of a homozygous deletion in pancreatic carcinoma that lies within the BRCA2 region. Proc. Natl. Acad. Sci. USA. 1995;92(13):5950–5954. doi: 10.1073/pnas.92.13.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Mikhitarian K., Pollen M., Zhao Z., Shyr Y., Merchant N.B., Parikh A., Revetta F., Washington M.K., Vnencak-Jones C., Shi C. Epidermal growth factor receptor signaling pathway is frequently altered in ampullary carcinoma at protein and genetic levels. Mod. Pathol. 2014;27(5):665–674. doi: 10.1038/modpathol.2013.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Schultz N.A., Roslind A., Christensen I.J., Horn T., Hogdall E., Pedersen L.N., Kruhoffer M., Burcharth F., Wojdemann M., Johansen J.S. Frequencies and prognostic role of KRAS and BRAF mutations in patients with localized pancreatic and ampullary adenocarcinomas. Pancreas. 2012;41(5):759–766. doi: 10.1097/MPA.0b013e31823cd9df. [DOI] [PubMed] [Google Scholar]
- 190.Hsu M., Sasaki M., Igarashi S., Sato Y., Nakanuma Y. KRAS and GNAS mutations and p53 overexpression in biliary intraepithelial neoplasia and intrahepatic cholangiocarcinomas. Cancer. 2013;119(9):1669–1674. doi: 10.1002/cncr.27955. [DOI] [PubMed] [Google Scholar]
- 191.Voss J.S., Holtegaard L.M., Kerr S.E., Fritcher E.G., Roberts L.R., Gores G.J., Zhang J., Highsmith W.E., Halling K.C., Kipp B.R. Molecular profiling of cholangiocarcinoma shows potential for targeted therapy treatment decisions. Hum. Pathol. 2013;44(7):1216–1222. doi: 10.1016/j.humpath.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 192.Borger D.R., Tanabe K.K., Fan K.C., Lopez H.U., Fantin V.R., Straley K.S., Schenkein D.P., Hezel A.F., Ancukiewicz M., Liebman H.M., Kwak E.L., Clark J.W., Ryan D.P., Deshpande V., Dias-Santagata D., Ellisen L.W., Zhu A.X., Iafrate A.J. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17(1):72–79. doi: 10.1634/theoncologist.2011-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Fu T., Guzzetta A.A., Jeschke J., Vatapalli R., Dave P., Hooker C.M., Morgan R., Iacobuzio-Donahue C.A., Liu B., Ahuja N. KRAS G>A mutation favors poor tumor differentiation but may not be associated with prognosis in patients with curatively resected duodenal adenocarcinoma. Int. J. Cancer. 2013;132(11):2502–2509. doi: 10.1002/ijc.27910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Fu T., Pappou E.P., Guzzetta A.A., Jeschke J., Kwak R., Dave P., Hooker C.M., Morgan R., Baylin S.B., Iacobuzio-Donahue C.A., Wolfgang C.L., Ahuja N. CpG island methylator phenotype-positive tumors in the absence of MLH1 methylation constitute a distinct subset of duodenal adenocarcinomas and are associated with poor prognosis. Clin. Cancer Res. 2012;18(17):4743–4752. doi: 10.1158/1078-0432.CCR-12-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Oliveira-Cunha M., Hadfield K.D., Siriwardena A.K., Newman W. EGFR and KRAS mutational analysis and their correlation to survival in pancreatic and periampullary cancer. Pancreas. 2012;41(3):428–434. doi: 10.1097/MPA.0b013e3182327a03. [DOI] [PubMed] [Google Scholar]