Abstract
Background:
Artificial Neural Networks (ANNs) can be used to classify tumor of Hepatocellular carcinoma based on their gene expression signatures. The neural network is trained with gene expression profiles of genes that were predictive of recurrence in liver cancer, the ANNs became capable of correctly classifying all samples and distinguishing the genes most suitable for the organization. The ability of the trained ANN models in recognizing the Cancer Genes was tested as we analyzed additional samples that were not used beforehand for the training procedure, and got the correctly classified result in the validation set. Bootstrapping of training and analysis of dataset was made as external justification for more substantial result.
Result:
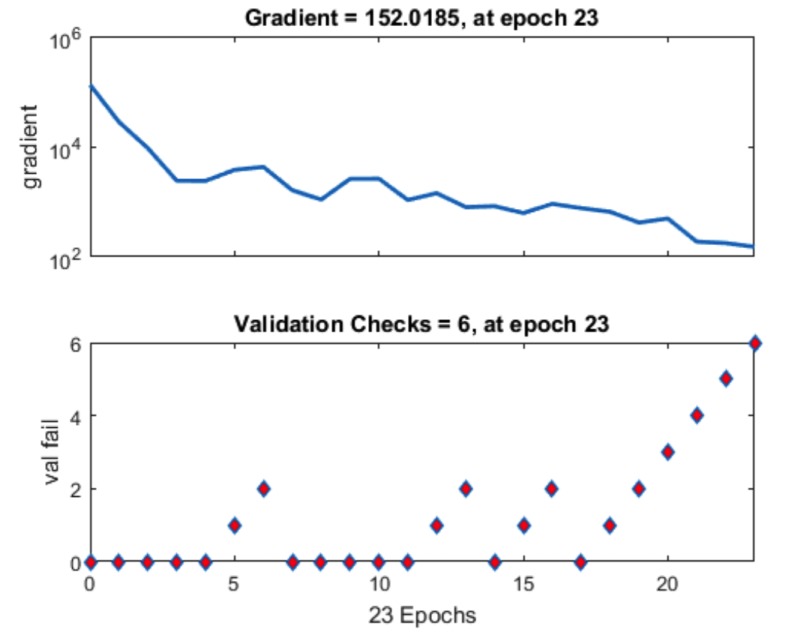
The best result achieved when the number of hidden layers was 10. The R2 value with training is 0.99136, R2 value obtained with testing is 0.80515, R2 value obtained after validation is 0.76678 and finally, with the total number of sets the R2 value is 0.93417. Performance was reported on the basis of graph plotted between Mean Squared Error (MSE) and 23 epoch. The value of gradient of the curve was 152 after 6 validation checks and 23 iterations.
Conclusion:
A successful attempt at developing a method for diagnostic classification of tumors from their gene-expression autographs that efficiently classify tumors and helps in decision making for providing appropriate treatment to the patients suffering from Hepatocellular carcinoma has been carried out.
Keywords: Gene database, Artificial neural network, Gene signatures, Classification, Hepatocellular carcinoma, Liver cancer
1. INTRODUCTION
Liver cancer is the 6th most common form of cancer and 782,000 were diagnosed with it in 2012. It is basically a form of chronic cancer which develops in liver [1-3]. Liver cancer can be divided into 2 phases: benign and malignant [4]. Liver cancer can also be divided into many forms, of which Hepatocelluar Carcinoma (HCC) is the most common followed by Intra-Hepatic Cholangiocarcinoma (IHCC) [5-9]. The onset of liver cancer includes symptoms like upper abdominal pain, unintended weight forfeiture, appetite loss, feebleness, jaundice etc. Some people with metastasis do not have symptoms [10-17]. Their metastasis is found by X- rays or other tests. Enlargement of the liver or jaundice can indicate that cancer has spread to the liver. When the cancer originates in liver and subsequently spread into other organs in the body then it is called metastasis liver cancer condition. It is a rare condition. There are many treatments available for liver cancer, most of which depends upon how much the cancer has spread and the overall health of the liver and the patient.
Recent advances in the field of molecular biology have led to an explosion of data generation which has compelled the researchers around the globe to find a way to organize, manage and to analyze this data. This led to the emergence of the interdisciplinary field of bioinformatics. Bioinformatics is the amalgamation of biology, statistical mathematics and computer science to organize, analyze and to understand large molecular biology databases. Hand, Heikk Mannila and Smyth defined Data mining as “the analysis of observational data (often large) sets to find unsuspected relations and to abridge the data in novel ways that are both understandable and useful to the information owner” [18].
Even though most of the world still utilizes the conventional method to identify, classify and diagnose cancer, these conventional techniques have largely been inaccurate and ambiguous. Molecular level diagnostics such as microarray gene expression profiles are viable alternative for cancer identification and classification which are not only efficient but also accurate. Hong-Hee Won has proposed the collection of network classifiers learned from the undesirably correlated physiognomies of cancer genes to accurately classify the cancer disease and systematically estimate the acts of the planned technique on the datasets. Recent studies have indicated that the negatively correlated classifiers have the most excellent recognition rate on the datasets [19]. Machine learning algorithms can be classified into a classification based on the preferred outcome of the algorithm or the category of input available during training the machine. Hu proposed and analyzed in 1994 the classification performances of cancer cell by using unsupervised and supervised learning methods [20]. Supervised learning produces a function that can map the inputs to preferred outputs (also called labels). In unsupervised learning, the input given to the learner is not labelled. For supervised learning, a single hidden layer fuzzy neural network with back propagation training is developed and implemented [1]. So considering all the above discussed reasons, we need to ponder and have to come up with an alternate, more efficient technique which is reliable and accurate. We propose a computer based method for early detection and prediction of Hepatocellular carcinoma.
DNA microarrays is the sole option to rapidly assess the global expression picture of thousands of genes in any given time opinion and associate the detailed combinatory analysis results of global expression profiles for normal and malignant cells at various useful stages or distinct experimental conditions. Acquisition of such “genetic portraits” allows searching for orderliness and difference in countenance patterns of certain genes, understanding their function and pathological prominence, and eventually developing the “molecular nosology” of cancer.
Neural network technique has evolved as an efficient classifier and also as a program writing practice for optimization of systems [21, 22]. For the evaluation of gene-expression information, neural networks have been exactly tested for their capability to precisely distinguish among cancers belonging to several diagnostic categories. The high mortality rate is attributed to lack of conspicuous symptoms and the internal location inside the body; it is difficult to be diagnosed at an early stage. In order to get rid of this problem, new practices for early detection of Hepatocellular carcinoma are needed to determine the condition of the liver and to give patients at the early stage treatment. Surely, the decisions made by the medical experts are the most important factors in cancer diagnosis but human beings are always prone to commit errors because of their boundaries. Thus, most of the researchers nowadays have proposed Artificial Intelligence (AI) classification procedures for cancer diagnosis. These methods have been proved in supporting the experts to facilitate their decision making process. Artificial neural network is an artificial analogous model of human nervous system and is inspired from the architecture of brain. The human brain is a complex network of neurons, which is highly organized and has an incredible decision making capability. Like a human brain, the ANN learns by already worked out examples. Once they are sufficiently experienced they can work the examples out by themselves. The neural network which is used in this paper is based on Leven marquardt algorithm.
2. METHODS
2.1. Data Collection
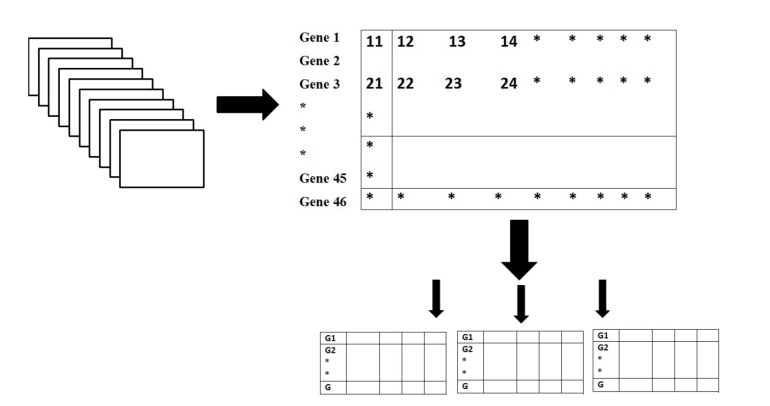
We first tried to obtain the nucleotide sequence of the genes which showed alterations of copy number in Hepatocellular carcinoma (HCC). Bacterial Artificial Chromosome (BAC) clone DNA or oligonucleotide probe arrays (microarray-based relative genomic hybridization) have been used in a number of studies to search for copy number changes in liver cancer. The gene name of the above mentioned genes are obtained from Tatsuhiro Shibata and Hiroyuki Aburatani [23]. Total 46 gene names were obtained and the coding sequences of these genes were obtained from National Centre for Biotechnology Information (NCBI) database Genbank. 30 of these genes were recurrently deleted genes and 16 of them were recurrently amplified genes in HCC. The genes which were analyzed in our work are listed below in Table 1. The gene Database we prepared is shown schematically in Fig. (1).
Table 1.
Gene sequences with their accession numbers and outputs.
| Sequence Name | Accession Number | Function | Output | ||
|---|---|---|---|---|---|
| Mdm4 | JN794077 | Tumor protein p53 pathway | 1 | ||
| BCL9 | KP345733 | Embryonic development | 2 | ||
| ARNT | M69238 | Xenobiotic metabolism | 3 | ||
| ABL2 | DQ084361 | Proliferation | 4 | ||
| MET | NM_001127500 | Embryonic development, organogenesis and wound healing | 5 | ||
| COPS5 | NM_006837 | Regulator in multiple signaling pathways | 6 | ||
| MTDH | NM_178812 | Angiogenesis | 7 | ||
| COX6C | AH006984 | Mitochondrial respiratory chain | 8 | ||
| MYC | NM_002467 | Cell cycle progression, apoptosis and cellular transformation | 9 | ||
| CCND1 | NM_053056 | Proliferation | 10 | ||
| FGF19 | AY358302 | FGF family | 11 | ||
| EEF1A2 | NM_001958 | Enzymatic delivery of aminoacltRNAs to the ribosome | 12 | ||
| TNFRSF14 | NM_003820 | TNF-receptor superfamily | 13 | ||
| CDKN2C | NM_001262 | Cell growth regulator | 14 | ||
| ARID1A | NM_006015 | Regulate transcription | 15 | ||
| TNFAIP3 | NM_001270508 | NF-κB pathway | 16 | ||
| CSMD1 | AY358174 | Tumor suppressor | 17 | ||
| DLC1 | NM_182643 | Human hepatocellular carcinoma | 18 | ||
| SORBS3 | NM_005775 | Migration | 19 | ||
| WRN | AF091214 | Cell division and repairing DNA | 20 | ||
| SH2D4A | NM_022071 | Proliferation | 21 | ||
| PROSC | NM_007198 | Unknown | 22 | ||
| CDKN2A | NM_001195132 | Tumor suppressor by regulating the cell cycle | 23 | ||
| CDKN2B | NM_004936 | Tumor suppressor by regulating the cell cycle | 24 | ||
| PTEN | AH007803 | Tumor suppressor by inhibition of the AKT signaling pathway | 25 | ||
| SPRY2 | NM_005842 | Proliferation | 26 | ||
| BRCA2 | U43746 | Repair damaged DNA | 27 | ||
| RB1 | NM_000321 | Tumor suppressor | 28 | ||
| XPO4 | NM_022459 | Nuclear export | 29 | ||
| SMAD4 | NM_005359 | Proliferation or differentiation | 30 | ||
| HNF1A | HM449088.1 | Encoded protein involved in the regulation of the expression of several liver-specific genes | 31 | ||
| APC | M74088.1 | APC protein is a negative regulator that controls beta-catenin concentrations and interacts with E-cadherin, which are involved in cell adhesion | 32 | ||
| PIK3CA | KF921496.1 | integral part of the PI3K pathway | 33 | ||
| TP53 | NC_000017.11 | Encodes P53 which can activate DNA repair proteins when DNA has sustained damage | 34 | ||
| COL1A1 | NM_000088.3 | Encodes a Janus Kinase protein which has been implicated in signaling by members of the type II cytokine receptor family | 35 | ||
| Sequence Name | Accession Number | Function | Output | ||
| TCS2 | NM_000548.4 | Encodes tumor suppressor and is able to stimulate specific GTPases | 36 | ||
| CTNNB1 | NM_001904.3 | Encodes beta-catenin | 37 | ||
| SMARCA4 | NM_001128849.1 | Encodes protein is part of the large ATP-dependent chromatin remodeling complex, which is required for transcriptional activation of genes normally repressed by chromatin | 38 | ||
| AXIN1 | NM_003502.3 | Encodes a cytoplasmic protein which contains a regulation of G-protein signaling domain | 39 | ||
| CHD1L | NM_004284.5 | Encodes a Protein involved in DNA repair | 40 | ||
| JAK2 | NM_004972.3 | Encodes a Janus Kinase protein which has been implicated in signaling by members of the type II cytokine receptor family | 41 | ||
| BRAF | NM_004333.5 | Encoded protein is involved in sending signals inside cells which are involved in directing cell growth | 42 | ||
| TCS2 | NM_000548.4 | The gene product is believed to be a tumor suppressor and is able to stimulate specific GTPases | 43 | ||
| NFE2L2 | NM_006164.4 | Encodes protein that regulates the expression of antioxidant proteins that protect against oxidative damage triggered by injury and inflammation | 44 | ||
| ARID2 | NM_152641.3 | Encodes PBAF chromatin-remodeling complex which facilitates ligand-dependent transcriptional activation by nuclear receptors | 45 | ||
| ATM | U82828.1 | Encodes a kinase that phosphorylates several key proteins that initiate activation of the DNA damage checkpoint | 46 | ||
Fig. (1).
Gene Database.
2.2. Data Preparation
Data encoding plays an essential function in improving the network performance. Neural networks are capable of processing many diverse forms of data presented to the network in a suitable format. There are different classes of inputs which are properly distinguished by the neural network, and hence data encoding methodology is used to represent data. DNA sequences of Homo sapiens are made up of four bases, A, T, G and C. These four bases should be represented in numerical vector form for training and use a neural network for order cataloging. The numerical values for encoding the input data sequences were 0, 0.5, 1 and 1.5 for the bases A, T, G and C, respectively and to represent the output value chosen so as to satisfy the training, prediction and classification needs of the neural network. A C-program was made for the conversion of the gene nucleotide sequence into an excel file with extension .xls. The program directed successive nucleotide code into successive rows with 1 common column.The entire data of 30 genes were compiled into a matrix of 10000 * 30.
2.3. Experimental Protocol
First we collected all the required sequences, retrieved from gene bank and made a data collection. Then we arranged all the sequences according to our requirement, we completed the sequence up to 10000 places. The gene sequences having more than 10000 nucleotides were trimmed short up to 10000 nucleotides, and for the sequences having less than 10000 nucleotides, we copied the first 1-9 nucleotide of same sequence and pasted it up to 10000. We made two files one for input data (ipp) and another for output (opp). Further, we opened matlab and ran the program for maximum output.
2.4. Neural Network Methodology
There are many methodologies for analysis of Hepatocellular Carcinoma, some of them are - Preliminaries of ANN, Multi-layered Feed forward Artificial Neural Network (MLFANN), Gradient Descent Algorithm (GDA), Conjugate Gradient Descent Algorithm, Leven marquardt (LM)
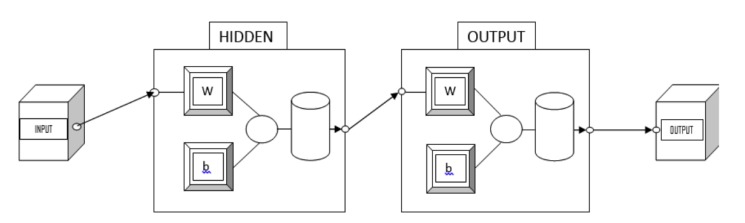
algorithm etc. We used LM algorithm for the work performed in this article. A particular type of “Artificial Neural Network (ANN)”, viz. Multifaceted Feed-forward Artificial Neural Network (MLFANN) has been labeled. To train such a system, we have contemplated two types of learning algorithms, namely Gradient Descent Algorithm (GDA) and Conjugate Gradient Descent Algorithm (CGDA). The LM method is a normal technique used to solve nonlinear least squares problems. Nonlinear least quadrangles glitches arise when the function is not linear in the parameters. Nonlinear least squares method includes an iterative progress to parameter values in order to decrease the sum of the quadrangles of the errors between the purpose and the sedate data points. The Levenberg-Marquardt curve-fitting method is actually a combination of two minimization methods: the gradelineage method and the Gauss-Newton technique. In the incline descent method, the sum of the squared errors is reduced by updating the parameters in the steepest-descent direction. In the Gauss-Newton method, the sum of the squared errors is abridged by assuming the minimum squares function which is locally quadratic, and finding the minimum of the quadratic. The Levenberg-Marquardt technique acts more similar a gradient-descent method when the parameters are far from their optimal value, and acts more like the Gauss-Newton method when the strictures are close to their optimal value. The working of Levenberg-Marquardt method is explained diagrammatically below, here first we have to take the input (ipp) which we have to train, an output (opp) for after training we get. Hidden layer plays an important role in the training process. Actually, the number of hidden nodes is one of the significant parameters responsible for the final output. Here, the process of training, testing and validation occurs. Two networks run parallel, network 1 and network 2. The neural network structure is shown in Fig. (2).
Fig. (2).
Neural network architecture.
3. RESULTS
Artificial neural networks are used due to their ability to tackle enormous amount of data with a good convergence rate and the ease with which they can be trained resulting in prudent modeling of large datasets. This in turn helps in support and prediction of clinical diagnosis of a disease. The principle aim of this work is to present a neural network methodology for accurate classification of candidate genes causing Hepatocellular Carcinoma (HCC) and to deduce a relationship between them.
3.1. Neural Network Training Results
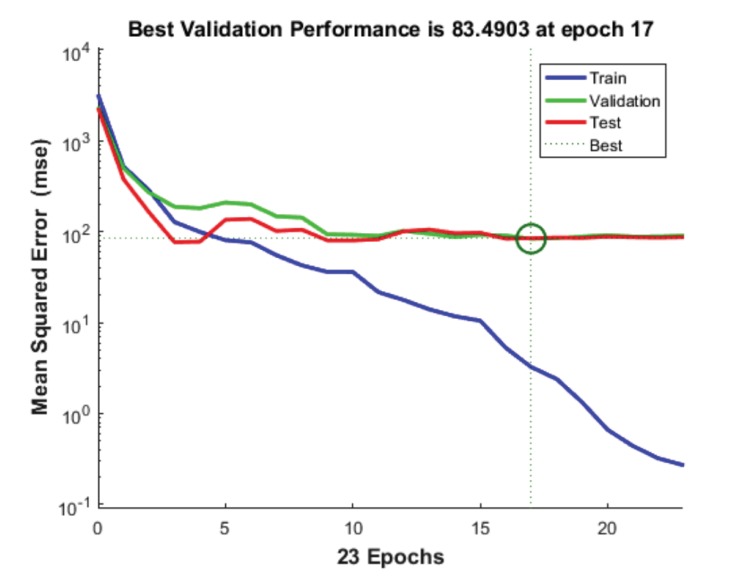
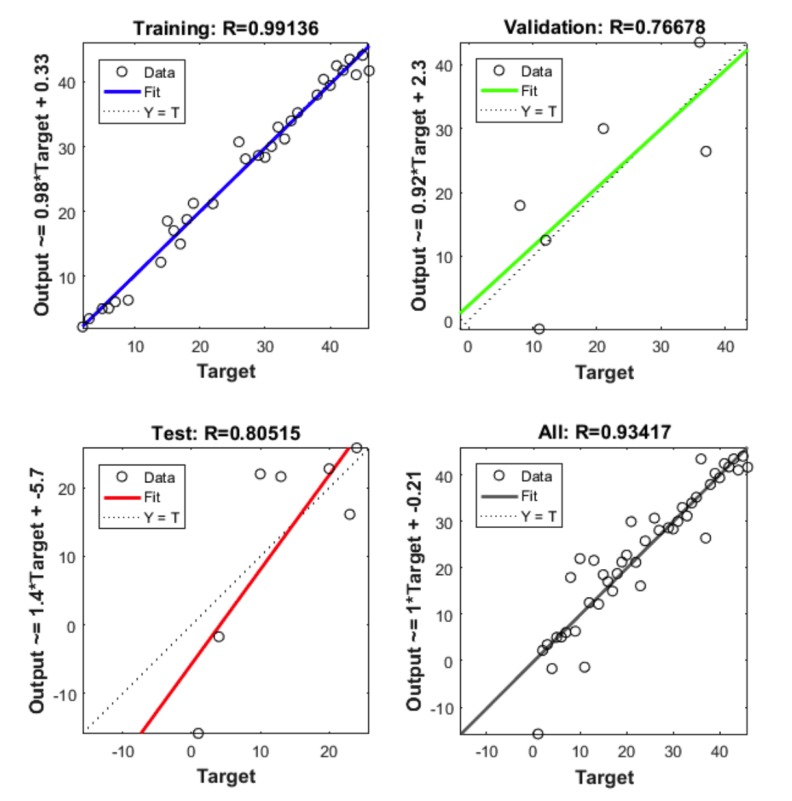
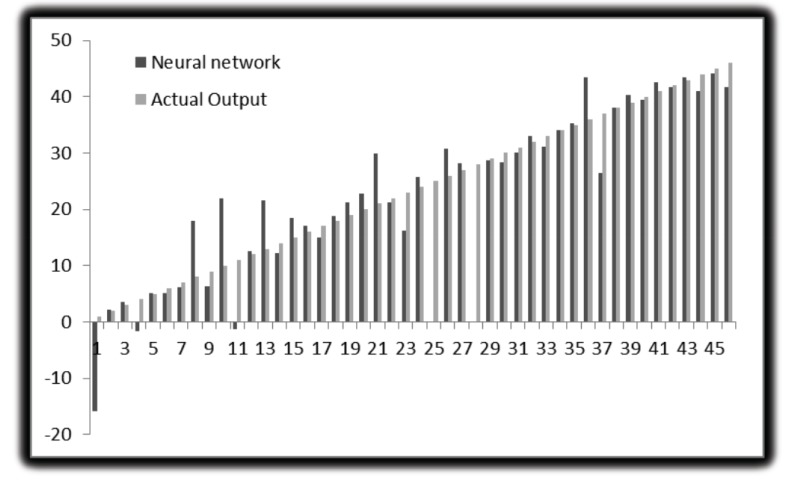
The following parameters are set in MATLAB 2012 for neural network. For molecular marker of liver cancer 46 experimental data used to develop ANN models. Out of them, 32 are used to train the network, 7 for validation and 7 for testing. The network performance measures such as MSE and R2 are shown in the figures below. This is performed with the 70%, 15% and 15% of the total 46 genes being used for training, validation and testing, respectively. Like this, the training, testing and validation are performed with all the possible combinations. Here we used different numbers of hidden layers and feasible mixture to generate net architecture which provides the least error. The network trained with many types of hidden nodes and validation, the testing sets were changed every time. After rigorous training, the best network output is shown in the figures given below. From these figures, we can depict that 10 hidden nodes with testing set 15% and validation set 15% is giving optimum performance. The total sets were 46, out of that, 20 were used for training, 5 were used for testing and 5 were used for validation. The R2 value with training is 0.99136, R2 value obtained with testing is 0.80515, R2 value obtained after validation is 0.76678 and with the total number of sets the R2 value is 0.93417. The results show that experimental results are closer to the neural network predicted results. Fig. (5) shows the best validation performance. The figure is drawn against MSE vs. epochs. 23 epochs were taken for this. From the Figure, 5th Error histogram can be seen. The dashed line in each plot signifies the perfect result - outputs = targets and the solid line denotes the best fit linear regression line between outputs and targets. The R2 value indicates the relationship between the outputs and targets. Greater the value of R2 (close to 1) greater the accuracy in the linear relationship between outputs and targets. The value of R2 and MSE are for training, validation, test and overall data is given in Fig. (3). In the Fig. (4), we can see training state, how the error is decreasing with the iterations. Fig. (6) shows the Actual and Predicted output of the neural network training. The results of ANN modeling are comparable to that of RSM modeling. But in our study, we are representing the advanced methodology of neural network methodology as a molecular marker for liver cancer. The Epoch with 1, 2, 3, 4…………….9 hidden node and different iterations were trained, we couldn’t get good output, however, in 10 hidden layers 23 epochs showed good output and optimum result. The performance is as follows, the algorithm used was random data division (dividerand), Training was scaled conjugated Gradient(tracing) and Performance was reported on the basis of graph plotted between Mean Squared error (MSE) and 20 epoch. Derivative used was Default (defaultderiv). Progress was as follows, Epoch with 9 iteration (maximum sated- 1000), Time: 0.00.06 second. Performance: Gradient: 152 and 6 Validation Checks. R2 Values in the regression curve quantize the correlation between outputs and targets. An R2 value of 1 means a close relationship, 0 means a random relationship. Mean Squared Error is the regular squared difference between productions and targets. The network is adjusted according to its error. These parameters are applied to measure network generalization, and to stop training when generalization stops improving. These have no effect on training and thus provide an independent measure of network performance during and after training for 30 sample output. The sample output was 70%, validation of 5 samples gave 15% output and testing of 5 samples gave 15% output. The correlation between outputs and targets were measured by the mean squared error and Regression R2 Values of the performance measure. If the R2 value is 0 then it means a random relationship and if it is 1 it means a close relationship. Mean Squared Error is defined as the average squared difference between targets and outputs. If we train multiple times, they will generate different results due to their different initial sampling and conditions. If the generalization stops improving further, the training automatically stops as it indicates an increase in mean square error of the validation samples. 10 hidden nodes are used in network 1 and network 2 and these two networks consist of varying number of hidden nodes. 10 hidden nodes gave optimal results. During training, the networks present the training nodes and the adjustment of network is done according to its error. Network generalization are measured by the testing nodes and training stops when generalization stops improving. The training is not affected by the Validation nodes and it will provide an independent measure of network performance during and after training.
Fig. (5).
Performance.
Fig. (3).
Regression, training validation.
Fig. (4).
Training state.
Fig. (6).
Actual and Predicted output (exact prediction of data set was observed except of 1, 8, 10, 21, 36 may be due to large amount of data).
4. DISCUSSION
Cancer is currently identified by histology and immune histochemistry, based on their morphology and protein countenance, respectively. However, poorly distinguished cancers can be difficult to identify by monotonous histopathology. In addition, the histological arrival of a growth cannot reveal the underlying genetic deviations or biological procedures that contribute to the process. Here, we developed a method of analytical organization of cancers from their gene-expression autographs and recognized the genes that contributed to this classification. Monitoring global gene-expression levels by cDNA microarrays delivers a supplementary tool for elucidating cancer biology as well as for molecular diagnostic classification of cancer [24, 25]. Currently, classification and gathering tools by means of gene-expression data have not been thoroughly tested for analytical classification of more than two groups. Other methods that share the parametric nature of ANNs and have been used to categorize gene-expression profiles include Support Vector Machines [23].
Even though ANN analysis leads to documentation of genes exact for a cancer with insinuations for biology and diagnosis, strength of this technique is that it does not need genes to be exclusively related with a single cancer type [26]. This allows for organization based on multifaceted gene-expression designs. As the main drive of this study was to optimize the organization of these cancers, we used a stringent quality filter to include only the genetic factor for which there were good capacities for all samples. This may remove certain genes that are highly pronounced in some growths, but not expressed in other cancers, or may appear unexpressed because of an artefact in a specific cDNA spot. Nonetheless, we expect that this list can be long-drawn-out by the use of more comprehensive arrays and larger sample sets for training. Here we developed a method of analytical organization of cancers from their gene expression signatures using future applications of these approaches will comprise studies to classify cancers according to phase and biological behavior in order to predict forecast and straight therapy. We believe this offers another influential method for the detection of gene-expression autographs, and the discovery of novel genes that describe a problem-solving subgroup which may also identify new targets for therapy.
CONCLUSION
The aim of this work is to present an efficient methodology for Hepatocellular Carcinoma classification and gene signature identification in patients based on the gene expression profile. A method of diagnostic classification of tumors from their gene-expression autographs has been developed that efficiently classify tumors and helps in decision making for providing appropriate treatment to the patients suffering from Hepatocellular carcinoma. Artificial neural networks (ANN) allow us to categorize the patients and recognize the low risk patients and avoid the unnecessary systemic adjuvant chemotherapy. It was concluded that varying the number of neurons in the hidden layer increase the performance of prediction result with the same computational time even though increase in the complexity of the algorithm. Apparently, the performance of prediction enhances in correspondence with the number of iterations i.e. the better the training of the ANN, better will be its accuracy but only up to a certain limit.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No Animals/Humans were used for studies that are the basis of this research.
CONSENT FOR PUBLICATION
Not applicable.
ACKNOWLEDGEMENTS
We are thankful to National Institute of Technology Raipur, India for providing the necessary facilities to prepare the manuscript and permission to publish it.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. 2015 doi: 10.1002/ijc.29210. https://onlinelibrary.wiley.com/doi/full/10.1002/ [DOI] [PubMed]
- 2.De Angelis R., Sant M., Coleman M.P., Francisci S., Baili P., Pierannunzio D., Trama A., Visser O., Brenner H., Ardanaz E., Bielska-Lasota M., Engholm G., Nennecke A., Siesling S., Berrino F., Capocaccia R., EUROCARE-5 Working Group Cancer survival in Europe 1999-2007 by country and age: Results of EUROCARE-5-a population-based study. Lancet Oncol. 2014;15(1):23–34. doi: 10.1016/S1470-2045(13)70546-1. [DOI] [PubMed] [Google Scholar]
- 3.Jelic S., Sotiropoulos G.C. Hepatocellular carcinoma: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2010;21(Suppl. 5):v59–v64. doi: 10.1093/annonc/mdq166. [DOI] [PubMed] [Google Scholar]
- 4.Dietrich C.F., Mertens J.C., Braden B., Schuessler G., Ott M., Ignee A. Contrast-enhanced ultrasound of histologically proven liver hemangiomas. Hepatology. 2007;45(5):1139–1145. doi: 10.1002/hep.21615. [DOI] [PubMed] [Google Scholar]
- 5.El-Serag H.B., Engels E.A., Landgren O., Chiao E., Henderson L., Amaratunge H.C., Giordano T.P. Risk of hepatobiliary and pancreatic cancers after hepatitis C virus infection: A population-based study of U.S. veterans. Hepatology. 2009;49(1):116–123. doi: 10.1002/hep.22606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Endo I., Gonen M., Yopp A.C., Dalal K.M., Zhou Q., Klimstra D. DʼAngelica, M.; DeMatteo, R.P.; Fong, Y.; Schwartz, L.; Kemeny, N.; O’Reilly, E.; Abou-Alfa, G.K.; Shimada, H.; Blumgart, L.H.; Jarnagin, W.R. Intrahepatic Cholangiocarcinoma. Ann. Surg. 2008;248(1):84–96. doi: 10.1097/SLA.0b013e318176c4d3. [DOI] [PubMed] [Google Scholar]
- 7.Malhi H., Gores G.J. Cholangiocarcinoma: Modern advances in understanding a deadly old disease. J. Hepatol. 2006;45(6):856–867. doi: 10.1016/j.jhep.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El-Serag H.B. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142(6):1264–1273. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu J., Shen J., Sun T., Zhang X., Wong N. Obesity, insulin resistance, NASH and hepatocellular carcinoma. Semin. Cancer Biol. 2013;23(6 Pt B):483–491. doi: 10.1016/j.semcancer.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Shaib Y., El-Serag H. The epidemiology of cholangiocarcinoma. Semin. Liver Dis. 2004;24(2):115–125. doi: 10.1055/s-2004-828889. [DOI] [PubMed] [Google Scholar]
- 11.Palmer W.C., Patel T. Are common factors involved in the pathogenesis of primary liver cancers? A meta-analysis of risk factors for intrahepatic cholangiocarcinoma. J. Hepatol. 2012;57(1):69–76. doi: 10.1016/j.jhep.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaib Y.H., Davila J.A., McGlynn K., El-Serag H.B. Rising incidence of intrahepatic cholangiocarcinoma in the United States: A true increase? J. Hepatol. 2004;40(3):472–477. doi: 10.1016/j.jhep.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi M., Ikeda K., Saitoh S., Suzuki F., Tsubota A., Suzuki Y., Arase Y., Murashima N., Chayama K., Kumada H. Incidence of primary cholangiocellular carcinoma of the liver in Japanese patients with hepatitis C virus-related cirrhosis. Cancer. 2000;88(11):2471–2477. doi: 10.1002/1097-0142(20000601)88:11<2471::aid-cncr7>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 14.Shaib Y.H., El-Serag H.B., Davila J.A., Morgan R., McGlynn K.A. Risk factors of intrahepatic cholangiocarcinoma in the United States: A case-control study. Gastroenterology. 2005;128(3):620–626. doi: 10.1053/j.gastro.2004.12.048. [DOI] [PubMed] [Google Scholar]
- 15.Donato F., Gelatti U., Tagger A., Favret M., Ribero M.L., Callea F., Martelli C., Savio A., Trevisi P., Nardi G. Intrahepatic Cholangiocarcinoma and Hepatitis C and B virus infection, alcohol intake, and Hepatolithiasis: A case-control study in Italy. Cancer Causes Control. 2001;12(10):959–964. doi: 10.1023/a:1013747228572. [DOI] [PubMed] [Google Scholar]
- 16.Welzel T.M., Graubard B.I., El-Serag H.B., Shaib Y.H., Hsing A.W., Davila J.A., McGlynn K.A. Risk factors for intrahepatic and extrahepatic Cholangiocarcinoma in the United States: A population-based case-control study. Clin. Gastroenterol. Hepatol. 2007;5(10):1221–1228. doi: 10.1016/j.cgh.2007.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Srivatanakul P., Parkin D.M., Jiang Y-Z., Khlat M., Kao-Ian U.T., Sontipong S., Wild C. The role of infection by Opisthorchis Viverrini, Hepatitis B virus, and Aflatoxin exposure in the etiology of liver cancer in Thailand. A correlation study. Cancer. 1991;68(11):2411–2417. doi: 10.1002/1097-0142(19911201)68:11<2411::aid-cncr2820681114>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 18.Hand D., Mannila H., Smyth P. Principles of Data Mining. Cambridge, MA, USA: MIT Press; 2001. [Google Scholar]
- 19.Won H-H., Cho S-B. Proceedings of the International Joint Conference on Neural Networks; Portland, OR, USA. July 20-24, 2003; pp. 1708–1713. [Google Scholar]
- 20.Hu Y., Ashenayi K., Veltri R., O’Dowd G., Miller G., Hurst R., Bonner R. Proceedings of 1994 IEEE International Conference on Neural Networks (ICNN’94); Orlando, FL, USA. June 27-29, 1994; pp. 3461–3466. [Google Scholar]
- 21.Venkateswarlu C., Kiran K., Eswari J.S. A hierarchical artificial neural system for genera classification and species identification in mosquitoes. Appl. Artif. Intell. 2012;26(10):903–920. [Google Scholar]
- 22.Eswari J.S., Venkateswarlu C. Multiobjective simultaneous optimization of biosurfactant process medium by integrating differential evolution with artificial neural networks. Ind. J. Chem. Tech. 2016;23(5):335–344. [Google Scholar]
- 23.Totoki Y., Tatsuno K., Covington K.R., Ueda H., Creighton C.J., Kato M., Tsuji S., Donehower L.A., Slagle B.L., Nakamura H., Yamamoto S., Shinbrot E., Hama N., Lehmkuhl M., Hosoda F., Arai Y., Walker K., Dahdouli M., Gotoh K., Nagae G., Gingras M.C., Muzny D.M., Ojima H., Shimada K., Midorikawa Y., Goss J.A., Cotton R., Hayashi A., Shibahara J., Ishikawa S., Guiteau J., Tanaka M., Urushidate T., Ohashi S., Okada N., Doddapaneni H., Wang M., Zhu Y., Dinh H., Okusaka T., Kokudo N., Kosuge T., Takayama T., Fukayama M., Gibbs R.A., Wheeler D.A., Aburatani H., Shibata T. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat. Genet. 2014;46(12):1267–1273. doi: 10.1038/ng.3126. [DOI] [PubMed] [Google Scholar]
- 24.Khan J., Simon R., Bittner M., Chen Y., Leighton S.B., Pohida T., Smith P.D., Jiang Y., Gooden G.C., Trent J.M., Meltzer P.S. Gene expression profiling of alveolar rhabdomyosarcoma with cDNA microarrays. Cancer Res. 1998;58(22):5009–5013. [PubMed] [Google Scholar]
- 25.Brown M.P., Grundy W.N., Lin D., Cristianini N., Sugnet C.W., Furey T.S., Ares M., Haussler D. Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc. Natl. Acad. Sci. USA. 2000;97(1):262–267. doi: 10.1073/pnas.97.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eswari J.S., Chandrakar N. Artificial neural network analysis of DNA-microarray based lymph node negative breast cancer. Korean J. Chem. Eng. 2016;33(4):1318–1324. [Google Scholar]