Abstract
The rapid development in the last 10-15 years of microarray technologies, such as oligonucleotide array Comparative Genomic Hybridization (CGH) and Single Nucleotide Polymorphisms (SNP) genotyping array, has improved the identification of fine chromosomal structural variants, ranging in length from kilobases (kb) to megabases (Mb), as an important cause of genetic differences among healthy individuals and also as disease-susceptibility and/or disease-causing factors. Structural genomic variations due to unbalanced chromosomal rearrangements are known as Copy-Number Variants (CNVs) and these include variably sized deletions, duplications, triplications and translocations. CNVs can significantly contribute to human diseases and rearrangements in several dosage-sensitive genes have been identified as an important causative mechanism in the molecular aetiology of Charcot-Marie-Tooth (CMT) disease and of several CMT-related disorders, a group of inherited neuropathies with a broad range of clinical phenotypes, inheritance patterns and causative genes. Duplications or deletions of the dosage-sensitive gene PMP22 mapped to chromosome 17p12 represent the most frequent causes of CMT type 1A and Hereditary Neuropathy with liability to Pressure Palsies (HNPP), respectively. Additionally, CNVs have been identified in patients with other CMT types (e.g., CMT1X, CMT1B, CMT4D) and different hereditary poly- (e.g., giant axonal neuropathy) and focal- (e.g., hereditary neuralgic amyotrophy) neuropathies, supporting the notion of hereditary peripheral nerve diseases as possible genomic disorders and making crucial the identification of fine chromosomal rearrangements in the molecular assessment of such patients. Notably, the application of advanced computational tools in the analysis of Next-Generation Sequencing (NGS) data has emerged in recent years as a powerful technique for identifying a genome-wide scale complex structural variants (e.g., as the ones resulted from balanced rearrangements) and also smaller pathogenic (intragenic) CNVs that often remain beyond the detection limit of most conventional genomic microarray analyses; in the context of inherited neuropathies where more than 70 disease-causing genes have been identified to date, NGS and particularly Whole-Genome Sequencing (WGS) hold the potential to reduce the number of genomic assays required per patient to reach a diagnosis, analyzing with a single test all the Single Nucleotide Variants (SNVs) and CNVs in the genes possibly implicated in this heterogeneous group of disorders.
Keywords: Copy number variants, Structural variations, Inherited peripheral neuropathies, Charcot marie tooth, Array analysis, Whole genome sequencing
1. INTRODUCTION
Genomic duplication was first observed in the 1930s by Calvin Bridges who showed that a phenotype consisting of a reduction in eye size in Drosophila melanogaster was due to a duplication of the Bar locus at cytological band X16A [1]. However, at that time technological limitations (and a limited knowledge of the genomic rearrangements mechanisms) made the understanding of this phenomenon difficult. In more recent times, the BarH1 has been identified as the dosage-sensitive gene associated with the Bar phenotype observed in Drosophila [2].
In the last 10-15 years, the rapid development of microarray technologies, including oligonucleotide array Comparative Genomic Hybridization (CGH) and Single Nucleotide Polymorphisms (SNP) genotyping arrays, has led to the possibility of investigating structural variants of limited size [3].
Chromosomal structural variants with gain or loss of genetic sequences of at least 50 bp in size compared to a reference genome are commonly defined as Copy Number Variants (CNVs) contrasting with Single-Nucleotide Variants (SNVs), which affect only one single nucleotide base [4].
CNVs can be rare (< 1% population) or common (> 5% population), large (> 1 Mb) or small (< 500 bp) and include deletions, when there is a loss of genetic material with respect to the reference, and duplications with gain of a specific sequence that can be inserted contiguously or elsewhere in the genome [4, 5]. Other genomic variations that can be regarded as structural variants (possibly associated also with variations in copy number) include isochromosomes, segmental uniparental disomies and ring chromosomes [5]. More complex rearrangements include inversions or translocations flanked by CNVs and chromothripsis, a process characterized by the simultaneous occurrence of various sub-microscopic chromosomal rearrangements, possibly causing in the same individual clustered deletion, translocation and inversion events [5, 6].
Normal DNA Sanger sequencing or conventional cytogenetic analysis cannot detect most CNVs, therefore specific techniques are needed to investigate these genomic rearrangements.
Even though several methods have been described to detect CNVs in the human genome, there is still not a single test for accurately genotyping all the CNV types [6-8].
In the present review, we will describe the most utilized techniques for CNVs detection and will summarize both the clinical and the genetic implications of sub-microscopic structural variants in inherited neuropathies with examples of these defects.
2. GENERATION OF STRUCTURAL VARIANTS AND METHODS FOR CNVS DETECTION
Several mutational mechanisms can lead to the generation of CNVs, including (a) recombination errors such As Non-Allelic Homologous Recombination (NAHR); (b) errors generated in DNA break repair such as Non-Homologous End Joining (NHEJ); (c) errors occurring during DNA replication, such as Serial Replication Slippage (SRS) [6, 7]. NAHR is generally mediated by low-copy repeats and can occur in different ways (e.g., on the same chromatid or between homologous chromosomes). NHEJ is a repair mechanism for double-stranded breaks and results in junctions with micro-homology of a few base pairs and/or insertion of other sequences. In SRS the replication machinery slips backwards along the chromosome, resulting in tandem duplication, or slips forward, creating a deletion. This slippage can also create complex rearrangements when repeated, or multiple slippage events occur, creating blocks of deleted or duplicated sequence with intervening single-copy sequence [7, 8]. Different techniques can be used to detect human structural variations, with each of these approaches having its own strength and weakness (Table 1).
Table 1.
Laboratory techniques for CNVs detection.
| Chromosome banding: Chromosomes are prepared from dividing cells, stained, and viewed with a microscope. Large deletions, duplications, and translocations are detected if the banding pattern or chromosome structure is altered. However, smaller microdeletions and microduplications are not observed |
| FISH: The technique consist in fluorescent-labeled DNA probes that hybridize to metaphase or interphase cells to visualize a locus on a chromosome and determine copy number. FISH can determine the location of chromosomal segments identified by microarray, NGS, and WGS |
| qPCR: This is considered a high throughput technique for identifying CNVs. The cycle number (Ct) is proportional to the amount of starting template so the Ct values of the target gene can be compared to unrelated reference sequences that do not differ in copy content, so generating data which are used for CNVs analysis |
| MLPA: This method is able to analyze in a single reaction up to 50 DNA sequences and to detect copy number variation of specific genes, including small intragenic rearrangements. It has been utilised in recent years for the detection of CNVs and/or the validation of array-CGH results |
| Microarrays: Array-CGH detects copy-number differences between abnormal and reference genomes, but lack in the identification of balanced chrosmosomal rearrangements. SNP-array detect changes in copy-number and allelic ratios. Microarray analysis is not the best method to determine CNVs location and SV organization |
| WGS: Sequencing the whole genome provides the most comprehensive SV analysis. Breakpoints of CNV and copy-neutral SV are detectable by paired-end reads that have discordant mappings to the reference genome. Complex genomic structures identified by WGS may require FISH or chromosome banding to place rearranged segments |
CGH = Comparative Genomic Hybridization; CNVs = Copy Number Variants; FISH = Fluorescence In Situ Hybridization; qPCR = Quantitative PCR; SNP = Single Nucleotide Polymorphisms; SV = Structural Variants; WGS = Whole Genome Sequencing.
We hereby summarize the most utilized methods for CNVs detection.
2.1. Micro-array Analysis
A robust method of CNVs detection is array-CGH, a molecular cytogenetic method for analyzing CNVs relative to ploidy level in the DNA of a test sample compared to a reference genome using intensity hybridization ratios of two differentially dyed DNAs against the same target oligonucleotides. Different algorithms (e.g., circular binary segmentation, genome alteration detection algorithm) allow to obtain CNVs information from the data generated by the array [8-10].
Array-CGH generally uses slides arrayed with small segments of DNA; probes can vary in size from large-insert clones (40-200 kb in size), small insert clones (1.5-4.5 kb), cDNA clones (0.5-2 kb), genomic PCR products (100 bp-1.5 kb) and oligonucleotides (25-80 bp) [9].
The level of resolution is determined by considering both probe size and the genomic distance between DNA probes; as a consequence, a microarray with probes selected from regions across the genome that are 5 Mb apart will be unable to detect CNVs of the intervening sequence [10]. However, the ability to detect copy-number changes depends on the signal-to-noise ratio and the probe response characteristics [9]. The main advantage of CGH is the capacity to simultaneously detect sub-microscopic CNVs (e.g., deletions, duplications) of any locus on a genome-wide scale; however, cytogeneticists often prefer to use targeted arrays instead of the whole-genome scan in order to avoid the analysis of the regions of uncertain clinical significance [10]. One of the limitations of array-CGH array is that this technique is able only to detect imbalances between two individual genomes and thus an absolute copy number cannot be provided; additionally, array CGH cannot detect the balanced chromosomal rearrangements (i.e., with no gain nor loss of genetic material) such as reciprocal inversions, translocations and ring chromosomes as these do not affect copy number [9, 10].
Several algorithms have been developed in the last decade with the aim of extracting CNVs information also from SNP genotyping data [11]. These algorithms (e.g., PennCNV, Birdsuite) utilize SNP information (i.e., the sum of intensities of the two alleles, the intensity ratio of one allele relative to the other) to assess possible allele copy number variations. Despite resolution and accuracy of array-CGH in calling and detecting CNVs which are better compared to the SNP-array, the latter allow in some cases the detection of balanced structural variants (e.g., inversions, translocations) which affect the linkage disequilibrium pattern and leave a detectable signature [10, 11].
2.2. Multiplex Ligation-dependent Probe Amplification
The Multiplex Ligation-dependent Probe Amplification (MLPA) assay is a useful technique able to detect intragenic CNVs also of very limited size [3, 6]. This multiplex PCR assay utilizes up to 40 probes specific for a different DNA sequence (mainly exons of a specific gene of interest) in order to evaluate the presence of CNVs in each DNA sequence. Additionally, MLPA allows identification of mutations present at a low-level mosaicism state, as well as the presence of abnormalities in DNA methylation [12]. Because of the high reliability for identifying and quantitating CNVs in human disease-causing genes MLPA has been increasingly utilized in recent years as a robust and cost-effective technique in genetic laboratories both for diagnostic purposes and for research [13].
A limitation of MLPA is the difficulty to detect balanced genomic rearrangements [12, 13]. MLPA assay has been indicated as an effective method in the detection of CNVs in diagnosis of the PMP22-related inherited peripheral neuropathies with a cost-effective approach for the genetic diagnosis of some of these diseases [14].
2.3. Real-time Quantitative Polymerase-chain-reaction
Quantitative PCR (qPCR) is considered a high throughput technique for identifying CNVs [3, 6, 15]. The cycle number (Ct) is proportional to the amount of starting template so the Ct values of the target gene can be compared to unrelated reference sequences that do not differ in copy content, so generating ΔCt values which are used for CNV calculation [15]. Data analysis is conducted using the comparative Ct method (ΔΔCt) where the Ct values of the gene of interest and the reference gene are compared between a control sample and an unknown, with the equation presented as 2- ΔΔCt. This strategy has been used before to measure large-scale CNVs for conditions including Crohn’s disease, celiac disease and psoriasis [16]. However, the CNVs resulting from this method have been questioned in some cases as it has been shown that a small change in the amplification efficiency could result in a high error in the ΔCt calculations [15-17].
2.4. Southern Blotting and Pulse-field Gel Electrophoresis
Southern blotting uses a probe to detect a definite sequence in the genomic DNA that has been digested with a restriction enzyme, separated by gel electrophoresis and then transferred to a membrane [18]. Changes in the intensity of the bands show a gain or loss of the related genomic locus.
The efficacy of SB is greatly improved if used in combination with Pulse-Field Gel Electrophoresis (PFGE), a technique that measures large DNA molecules by applying to a gel matrix an electric field that periodically changes direction [18, 19]. The use of SB and PFGE can be extremely useful in analysing complex regions, difficult to be investigated by using other approaches [19].
2.5. Next Generation Sequencing
Current Next Generation Sequencing (NGS) platforms generate billions of bases of accurate nucleotide sequences in short reads using reversible sequencing chemistries, thus rapidly expanding our ability to understand both single nucleotide variations and structural variation in the human genome. The most utilized microarrays (i.e., CGH and SNP arrays) contain hundreds-of-thousands to millions of probes are able to detect CNVs ranging in size from 1 to 10 Kb up to several megabases (Mb) [6, 8]. Therefore, many smaller pathogenic (intragenic) variations are known to occur but often remain beyond the detection limit of the clinical genomic microarray. In this context, Next Generation Sequencing (NGS), including Whole Exome Sequencing (WES) and Whole Genome Sequencing (WGS) has led to a dramatic improvement in the identification of structural variations comprising of very small deletions and duplications of <50 bp (micro InDels) which were often missed by traditional Sanger sequencing as well as by microarray analyses [4, 8].
The comparison between whole genome assembly and a high-quality reference genome is currently the most precise strategy for discovering CNVs using NGS data and the detection of these variations mostly relies on the read lengths utilized in the sequencing platform as well as on the accuracy of base calls. The direct, reference-free, assembly of a complete genome of an individual from raw sequence read WGS data is theoretically the best method for CNVs identification [7, 8].
The accurate identification of CNVs requires information on copy number, their content and positional information regarding the chromosomal breakpoint where the structural variants arise.
There are four approaches to detect CNVs using NGS data: (i) Depth Of Coverage (DOC) that is more suitable for detecting larger structural variants and repetitive regions but does not provide accurate information on the breakpoint site; (ii) split-read methods that provide breakpoint information but are able to identify only small CNVs; (iii) read pair methods can be used to detect inversions and translocations by using information on the orientation in which the read pairs are mapped to the reference; and (iv) assembly-based methods that can be used with efficacy in identifying the breakpoint as well as novel inserted genomic sequences [7, 8, 16, 20]. All four approaches can be utilized for CNV detection in WGS data, whereas the DOC method is the most suitable approach in the case where CNVs have to be extracted from WES data [8, 20].
Long tandem repeats are still difficult to be visualized on WGS data using the above methods and the accuracy of identifying such repetitive regions is mostly influenced by the length of the sequence reads; sequencing with a read length (>5-10 kb) makes possible the identification of repetitive regions (and of complex inversions occurring at SV breakpoints) but such technology currently remains unavailable in most genetic laboratories [8].
3. CNVS IN INHERITED PERIPHERAL NEUROPATHIES
With the evolution of both microarray and NGS technologies in the last decade, CNVs have been discovered to be responsible for many different human abnormal phenotypes, including neurodevelopmental disorders (e.g., autism, schizophrenia), congenital anomalies (often associated with complex genetic deletion-duplication syndromes) and various neurological disorders affecting both the central and the peripheral nervous system [3-6].
One of the first genomic disorders discovered to be associated with a variation of copy number in a gene was Charcot-Marie-Tooth (CMT) type 1A, where a heterozygous duplication of the PMP22 gene has been identified as the cause of the disease in approximately 70% of CMT1 patients [21].
Subsequently, GJB1 was the second CMT gene, in addition to PMP22, for which gene deletions were reported, stressing the importance of CMT as a genomic disorder [22].
Since these discoveries, the presence of sub-microscopic chromosomal rearrangements in different disease-causing genes has been ascertained in a high proportion of individuals affected by inherited neuropathies.
In the present review, we summarize the current knowledge on the role of CNVs in causing inherited peripheral neuropathies.
3.1. Charcot Marie Tooth (CMT) Disease Type 1A
CMT disease occurs with an estimated prevalence of 1/2,500 and is variably characterized by progressive degeneration of peripheral nerves resulting in muscle weakness and wasting (predominantly in distal limbs, feet and hands) and sensory loss [22]. Onset varies from childhood to adulthood and clinical severity ranges from mild to severe between affected individuals. The classification of various CMT phenotypes was initially based on peculiar neuropathological and electrophysiological features, with CMT type 1 and CMT type 2 representing demyelinating and axonal neuropathies, respectively [22, 23]. Subsequently, the great advances in the understanding of the molecular aetiology underlying different CMT-related inherited peripheral neuropathies led to the discovery of more than 70 causative genes in the last 20 years, making CMT one of the most genetically heterogeneous neurological phenotype, with all the possible monogenic inheritance patterns (e.g., autosomal dominant, X-linked, and autosomal recessive) observed [21-23].
The CMT type 1A (CMT1A; OMIM #118220) can be regarded as the most frequent hereditary motor-sensory polyneuropathy. This Autosomal Dominant (AD) disease usually start in late childhood or adolescence manifesting with peroneal muscular atrophy, mild to moderate distal loss of sensation, pes cavus and marked slowing of nerve conduction velocities; the pathological process is characterized by a hypertrophic neuropathy with segmental de-remyelination, loss of large diameter fibres and complex onion bulbs (i.e., Schwann cell cytoplasmic processes surrounding residual fibres) [22, 23].
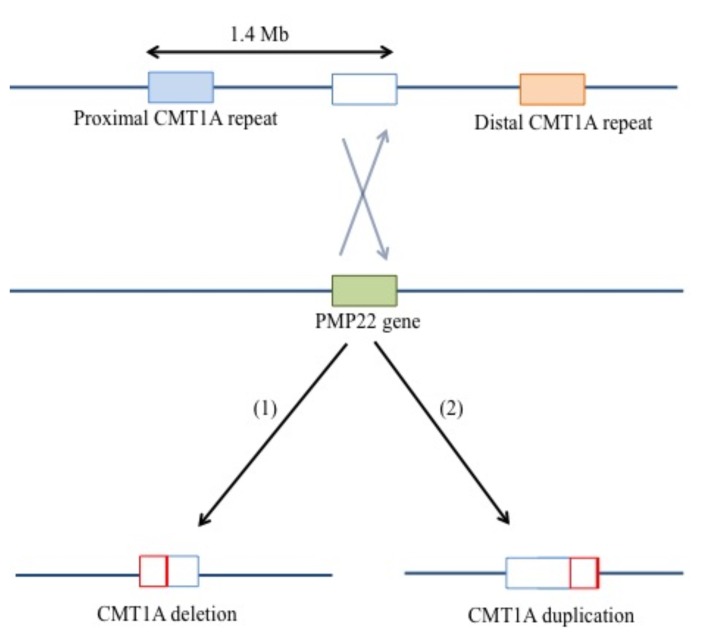
The CMT1A locus on chromosome 17p11.2-p12 was mapped in 1990 and a tandem-duplication of 1.4 megabases (Mb) on chromosome band 17p12 was identified in 1991 as the most recurrent genetic cause of CMT1A, which is known to represent around 70% of demyelinating CMT1 neuropathy cases [24-26]. A deletion of the same chromosomal region in 17p12 resulted in hereditary neuropathy with liability to pressure palsy (HNPP; OMIM #162500), a distinct inherited disease of the peripheral nerves (3.3) [27]. The CMT1A duplication and HNPP deletion locus is a 1.4 Mb chromosomal region that was found to be flanked by two 24 kb homologous Low Copy Repeats (LCRs) called the proximal and distal CMT1A-REPs [28, 29].
The proximal and distal copies of the CMT1A-REP repeats have 99% sequence identity potentially leading to misalignment of the chromosome 17 homologs during meiosis [27, 28].
This unique genomic architecture enhances the risk of recurrent rearrangements (e.g., NAHR) with one recombination pathway leading to the CMT1A tandem duplication and the reciprocal recombination pathway causing the HNPP deletion (Fig. 1). Thus, both CMT1A and HNPP are due to a gene-dosage mechanism: the CMT1A duplication chromosome has gained a 1.4-Mb segment and harbors an additional copy of the PMP22 gene; conversely, the HNPP-deleted chromosome has lost a 1.4-Mb segment from chromosome 17, including a copy of the PMP22 gene [30, 31]. Notably, triplications of the 17p12 locus have been also described in some CMT1A patients with a more severe phenotype and other atypically sized (longer/shorter than 1.4-Mb) CNVs have been rarely reported in patients with CMT1A/HNPP [32, 33]. In this regard, analyzing the atypical genomic rearrangements using high-density oligonucleotide-based CHG array, researchers showed that non-NAHR recombination mechanisms (e.g., non-homologous end joining, microhomology-mediated break-induced replication) may also occur as additional potential causative mechanisms of the disease in some patients [34]. In addition, also point mutations in PMP22 can cause CMT1A [20].
Fig. (1).
Mechanisms of deletion/duplication at chromosome 17p12. The proximal and distal copies of the CMT1A-REP repeats have 99% sequence identity potentially leading to misalignment of the chromosome 17 homologs during meiosis [24, 25]. This genomic architecture at chromosome 17p12 (with proximal and distal copies of the CMT1A-REP repeats having 99% sequence increasing risk of misalignment of the chromosome during meiosis) can lead to the 1.4-Mb HNPP deletion (1) or the CMT1A tandem duplication (2).
3.2. CNVs in Other CMT-related Inherited Neuropathies
CNVs in non-CMT1A inherited neuropathies have been more rarely reported in the literature.
The X-linked form of CMT disease (CMT1X; OMIM #302800) represents the second most common form of this genetically heterogeneous group of peripheral nerves diseases, after CMT1A.
The disorder is caused by mutations in the GJB1 gene, which encode for connexin 32 (CX32) [35, 36]. Affected males usually have onset of symptoms in childhood/adolescence with a variable combination of difficulties in walking and running, sensory loss, muscular atrophy and weakness (predominantly distal). Atrophy of the muscles (especially of the intrinsic hand muscles) and loss of sensation can be regarded as more prominent features of CMT1X compared to CMT1A [22, 36].
The disorder is considered to be an X-linked dominant trait because it affects also female carriers who usually develop a (late-onset) milder version of the same phenotype. Most of the GJB1 mutations causative for CMT1X are missense variants, but nonsense mutations and frameshift deletions (that lead to truncated forms of the protein) have been reported.
Genomic rearrangements of GJB1 have been also described in association with CMT1X, including reports of patients with complete deletion of the GJB1 gene resulting in a neuropathy with features of concomitant severe demyelination and axonal degeneration [37, 38].
In a series of 5 CMTX patients, Gonzaga-Jauregui et al. reported three variably sized deletions (spanning from 12 to 48 kb) involving the GJB1 gene [39]. The gene deletions were initially ascertained by MLPA analysis of the gene and further characterised by array CGH, having the breakpoint junctions refined by PCR and sequencing. Analyses of these breakpoints showed that GJB1 deletions were produced by different mechanisms, including NAHR and NHEJ [39].
CMT type 1B (CMT1B; OMIM #118200) is another inherited peripheral neuropathy caused by mutations in the MPZ gene which encodes for the myelin protein zero. The MPZ protein is expressed by Schwann cells (comprising approximately 50% of all myelin proteins in the peripheral nerves) and is necessary for both normal myelin function and structure [22, 40]. Mutations in MPZ appear to either disrupt myelination during development (leading to severe early-onset neuropathies), or to disrupt axo-glial interactions leading to adult-onset peripheral neuropathies [40].
CNVs in MPZ have been reported as the molecular cause of CMT1B.
Hoyer et al. used MLPA and confirmatory breakpoint-PCR analyses to identify a MPZ pathogenic duplication segregating in multiple affected members of a Norwegian family with CMT1B phenotype [41]. Speevak et al. described a kindred with early-onset peripheral demyelinating neuropathy and a disruptive MPZ duplication that was also assessed functionally by quantitative DNA and cDNA sequencing studies [42]. These analyses revealed a consistent finding of five copies of MPZ in affected members, thought to have arisen due to repeated interallelic cross-overs [41, 42].
CMT type 4D (CMT4D; OMIM #601455) is an Autosomal Recessive (AR) peripheral neuropathy firstly described in 14 affected individuals from the Gypsy community of Lom in Bulgaria, in association with homozygous mutations in the NDRG1 gene [22]. The disorder was lately reported in families from Italy, Slovenia, Germany, Spain, France, and Romania, patients with CMT4D classically present muscle weakness and wasting, tendon areflexia, foot deformities and sensory loss; deafness can also appear, usually in the third decade [22, 43].
Okamoto et al. firstly reported CNVs in CMT4D describing a Turkish family where a ~6.25 kb region of increased copy number encompassing exons 6-8 of the NDRG1 gene was identified as the cause of the phenotype [44]. Molecular studies in the family showed segregation of the homozygous duplication in the affected individuals, whereas the heterozygous duplication was present in the unaffected carriers. Functional studies were performed in the family, which included cDNA sequencing of lymphoblastoid cell line RNA from both affected and unaffected individuals, and researchers showed that the duplication in the patients led to a non-sense mutation at codon 223 in the mRNA [44].
3.3. CNVs in Inherited Focal Episodic Neuropathies
Inherited, focal episodic neuropathies are a group of genetic disorders affecting the peripheral nerves characterized by paroxysmal attacks of motor/sensor neuropathy of segmental distribution and with the absence of a complete (or full) recovery between the episodes [45].
The two most important disorders belonging to this group are HNPP and hereditary neuralgic amyotrophy (HNA; OMIM #162100).
HNPP is an AD episodic focal motor/sensory neuropathy with attacks usually triggered by a minor compression and/or trauma of the affected peripheral nerve. The usual presentation is as an acute mononeuropathy and most sensitive sites include wrist, elbow, knee and shoulder, affecting the median, ulnar, and peroneal nerve and brachial plexus, respectively [22, 46]. More rare non-focal presentations (e.g., chronic sensorimotor polyneuropathy) have been also described [47, 48]. Neurophysiological investigations in HNPP show focal slowing of nerve conduction velocities at common entrapment sites and diffuse mild slowing of nerve conduction velocities with prolonged distal motor latencies.
A deletion of the chromosomal region in 17p12, the same locus of the CMT1A duplication, is the most frequent genetic cause of the disorder, occurring in more than 80% HNPP patients (3.1) [49].
The HNPP deletion locus is a 1.4 Mb chromosomal region flanked by two LCRs (the proximal and distal CMT1A-REPs) and this unique genomic architecture enhances the risk of NAHR which is responsible for the HNPP deletion (and the CMT1A duplication); analysis of the genomic region nearby the CMT1A-REPs yielded an evolutionary mechanism for the creation of novel genes by DNA rearrangement as causative of the formation of the CMT1A-REPs [29, 48].
HNA is an AD disorder characterized by paroxysmal episodes of severe pain, weakness and sensory loss, predominantly affecting peripheral nerves within the brachial plexus distribution [50]. The disease usually begins in the second decade of life and is characterized by attacks of variable duration (lasting from days to weeks or sometimes months); patients may complain between these attacks some residual weakness and sensory disturbances in the affected limbs. Different events (e.g., infections, immunisations, trauma, psychological stress) can trigger the HNA-related symptoms, also suggesting a potential role of immuno-mediated processes in eliciting the disease [50, 51].
In 2005, a locus for HNA was mapped to chromosome 17q25 and mutations in the SEPT9 gene were then identified as the molecular cause of the disease in 6 HNA families [52].
Subsequently, non-recurrent genomic rearrangements in the SEPT9 gene have been reported to cause HNA also [53, 54].
Landsverk et al. identified using array CGH a 38 kb intragenic microduplication within the SEPT9 gene in several HNA kindreds that were known to harbor a common founder haplotype [53]. Collie et al. reported six (variably sized) intragenic duplications of the SEPT9 gene in different kindreds with HNA [54]. Five of these novel duplications were intragenic and resulted in larger transcript and protein products, as the researchers demonstrated by Western blotting studies.
One duplication spanned the entire SEPT9 gene and did not generate aberrant transcripts and proteins. The breakpoints of all the duplications were unique, containing regions of microhomology sized from 2 to 9 bp, but all the duplicated regions in the six families encompassed a proline rich region of the gene. The majority of the SEPT9 proline-rich region is encoded by a 645 bp exon in which the two most recurrent missense HNA-linked mutations are located, suggesting a crucial role of this region in the molecular pathogenesis of HNA as previously proposed [53-55].
3.4. Segmental Uniparental Disomy and Giant Axonal Neuropathy
Giant Axonal Neuropathy (GAN; OMIM #256850) is a rare AR genetic disease affecting both the central and the peripheral nervous system and characterised by the presence of enlarged axons with an accumulation of neurofilaments and progressive axonal loss. Clinically the disorder begins in childhood in most of the cases and presenting symptoms include the variable combination of distal limb weakness, areflexia and gait disturbances [56].
The GAN locus was identified by homozygosity mapping in several consanguineous families at chromosome 16q24.1 and since then more than 40 different compound heterozygous or homozygous mutations (e.g., missense, nonsense, frameshift) have been identified in the GAN gene [56, 57].
Additionally, CNVs of the gene have been associated with the disease, described in association with segmental uniparental disomies [6-8].
Uniparental disomy (UPD) is defined as the inheritance of a pair of chromosomes or segments from only one parent, while uniparental isodisomy (UPiD) is the duplication of a single parental allele [58]. UPiD harboring a homozygous mutation can be a genetic cause of autosomal recessive disorders in the case when only one of the two parents carry a pathogenic monoallelic mutation. In recent years, segmental UPiD has been described in patients with GAN [59].
CONCLUSION
CNVs significantly contribute to the molecular aetiology of several inherited neuropathies. Therefore, the integration of techniques such MLPA, microarray analysis and NGS sequencing studies (either panel of genes or WES) is nowadays crucial in the correct molecular assessment of patients with inherited disorders of the peripheral nerves. In the near future, WGS could potentially reduce the number of genomic assays required per patient to reach a diagnosis, analysing with a single test all the Single Nucleotide Variants (SNVs) and the CNVs in the many different genes known to be implicated in the molecular pathogenesis of inherited peripheral neuropathies.
CONSENT FOR PUBLICATION
Not applicable.
ACKNOWLEDGEMENTS
This study review was supported by the Medical Research Council (MRC UK), The Wellcome Trust (equipment and the Synaptopathies strategic award (104033)), The UK HSP Society, The Muscular Dystrophy Association (MDA) and the EU FP7/2007-2013 under grant agreement number 2012-305121 (NEUROMICS). We are also supported by the National Institute for Health Research (NIHR) University College London Hospitals (UCLH) Biomedical Research Centre (BRC).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Bridges C.B. The bar “gene” a duplication. Science. 1936;83(2148):210–211. doi: 10.1126/science.83.2148.210. science.sciencemag.org /content/83/2148/210 [DOI] [PubMed] [Google Scholar]
- 2.Kojima T., Ishimaru S., Higashijima S., Takayama E., Akimaru H., Sone M., Emori Y., Saigo K. Identification of a different-type homeobox gene, BarH1, possibly causing Bar (B) and Om(1D) mutations in Drosophila. Proc. Natl. Acad. Sci. USA. 1991;88(10):4343–4347. doi: 10.1073/pnas.88.10.4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carter N.P. Methods and strategies for analyzing copy number variation using DNA microarrays. Nat. Genet. 2007;39(7) Suppl.:S16–S21. doi: 10.1038/ng2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alkan C., Coe B.P., Eichler E.E. Genome structural variation discovery and genotyping. Nat. Rev. Genet. 2011;12(5):363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weckselblatt B., Rudd M.K. Human structural variation: Mechanisms of chromosome rearrangements. Trends Genet. 2015;31(10):587–599. doi: 10.1016/j.tig.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Escaramís G., Docampo E., Rabionet R. A decade of structural variants: Description, history and methods to detect structural variation. Brief. Funct. Genomics. 2015;14(5):305–314. doi: 10.1093/bfgp/elv014. [DOI] [PubMed] [Google Scholar]
- 7.Stankiewicz P., Lupski J.R. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. https://www.annualreviews.org/doi/pdf/ 10.1146/annurev-med-100708-204735 [DOI] [PubMed] [Google Scholar]
- 8.Hehir-Kwa J.Y., Pfundt R., Veltman J.A. Exome sequencing and whole genome sequencing for the detection of copy number variation. Expert Rev. Mol. Diagn. 2015;15(8):1023–1032. doi: 10.1586/14737159.2015.1053467. [DOI] [PubMed] [Google Scholar]
- 9.Bejjani B.A., Theisen A.P., Ballif B.C., Shaffer L.G. Array-based comparative genomic hybridization in clinical diagnosis. Expert Rev. Mol. Diagn. 2005;5(3):421–429. doi: 10.1586/14737159.5.3.421. [DOI] [PubMed] [Google Scholar]
- 10.Lucito R., Healy J., Alexander J., Reiner A., Esposito D., Chi M., Rodgers L., Brady A., Sebat J., Troge J., West J.A., Rostan S., Nguyen K.C., Powers S., Ye K.Q., Olshen A., Venkatraman E., Norton L., Wigler M. Representational oligonucleotide microarray analysis: A high-resolution method to detect genome copy number variation. Genome Res. 2003;13(10):2291–2305. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruno D.L., Stark Z., Amor D.J., Burgess T., Butler K., Corrie S., Francis D., Ganesamoorthy D., Hills L., James P.A., O’Rielly D., Oertel R., Savarirayan R., Prabhakara K., Salce N., Slater H.R. Extending the scope of diagnostic chromosome analysis: Detection of single gene defects using high-resolution SNP microarrays. Hum. Mutat. 2011;32(12):1500–1506. doi: 10.1002/humu.21581. [DOI] [PubMed] [Google Scholar]
- 12.Schouten J.P., McElgunn C.J., Waaijer R., Zwijnenburg D., Diepvens F., Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30(12):e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kozlowski P., Jasinska A.J., Kwiatkowski D.J. New applications and developments in the use of multiplex ligation-dependent probe amplification. Electrophoresis. 2008;29(23):4627–4636. doi: 10.1002/elps.200800126. [DOI] [PubMed] [Google Scholar]
- 14.Slater H., Bruno D., Ren H., La P., Burgess T., Hills L., Nouri S., Schouten J., Choo K.H. Improved testing for CMT1A and HNPP using multiplex ligation-dependent probe amplification (MLPA) with rapid DNA preparations: Comparison with the interphase FISH method. Hum. Mutat. 2004;24(2):164–171. doi: 10.1002/humu.20072. [DOI] [PubMed] [Google Scholar]
- 15.Fernandez-Jimenez N., Castellanos-Rubio A., Plaza-Izurieta L., Gutierrez G., Irastorza I., Castaño L., Vitoria J.C., Bilbao J.R. Accuracy in copy number calling by qPCR and PRT: A matter of DNA. PLoS One. 2011;6(12):e28910. doi: 10.1371/journal.pone.0028910. journals.plos.org/plosone/article?id=10.1371/journal.pone.0028910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mills R.E., Walter K., Stewart C., Handsaker R.E., Chen K., Alkan C., Abyzov A., Yoon S.C., Ye K., Cheetham R.K., Chinwalla A., Conrad D.F., Fu Y., Grubert F., Hajirasouliha I., Hormozdiari F., Iakoucheva L.M., Iqbal Z., Kang S., Kidd J.M., Konkel M.K., Korn J., Khurana E., Kural D., Lam H.Y., Leng J., Li R., Li Y., Lin C.Y., Luo R., Mu X.J., Nemesh J., Peckham H.E., Rausch T., Scally A., Shi X., Stromberg M.P., Stütz A.M., Urban A.E., Walker J.A., Wu J., Zhang Y., Zhang Z.D., Batzer M.A., Ding L., Marth G.T., McVean G., Sebat J., Snyder M., Wang J., Ye K., Eichler E.E., Gerstein M.B., Hurles M.E., Lee C., McCarroll S.A., Korbel J.O. 1000 Genomes Project. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470(7332):59–65. doi: 10.1038/nature09708. https://www.nature.com/articles/nature09708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patitucci A., Muglia M., Magariello A., Gabriele A.L., Peluso G., Sprovieri T., Conforti F.L., Mazzei R., Ungaro C., Condino F., Valentino P., Bono F., Rodolico C., Mazzeo A., Toscano A., Vita G., Quattrone A. Comparison of different techniques for detecting 17p12 duplication in CMT1A. Neuromuscul. Disord. 2005;15(7):488–492. doi: 10.1016/j.nmd.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 18.Notini A.J., Craig J.M., White S.J. Copy number variation and mosaicism. Cytogenet. Genome Res. 2008;123(1-4):270–277. doi: 10.1159/000184717. [DOI] [PubMed] [Google Scholar]
- 19.Bustamante C., Gurrieri S., Smith S.B. Towards a molecular description of pulsed-field gel electrophoresis. Trends Biotechnol. 1993;11(1):23–30. doi: 10.1016/0167-7799(93)90071-G. [DOI] [PubMed] [Google Scholar]
- 20.Narzisi G., O’Rawe J.A., Iossifov I., Fang H., Lee Y.H., Wang Z., Wu Y., Lyon G.J., Wigler M., Schatz M.C. Accurate de novo and transmitted indel detection in exome-capture data using microassembly. Nat. Methods. 2014;11(10):1033–1036. doi: 10.1038/nmeth.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rossor A.M., Polke J.M., Houlden H., Reilly M.M. Clinical implications of genetic advances in Charcot-Marie-Tooth disease. Nat. Rev. Neurol. 2013;9(10):562–571. doi: 10.1038/nrneurol.2013.179. [DOI] [PubMed] [Google Scholar]
- 22.Bird T.D. Charcot-Marie-Tooth Hereditary Neuropathy Overview. [DOI] [PubMed]
- 23.Timmerman V., Strickland A.V., Zuchner S. Genetics of Charcot-Marie-Tooth (CMT) disease within the frame of the human genome project success. Genes (Basel) 2014;5(1):13–32. doi: 10.3390/genes5010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Timmerman V., Raeymaekers P., de Jonghe P., de Winter G., Swerts L., Jacobs K., Gheuens J., Martin J.J., Vandenberghe A., van Broeckhoven C. Assignment of the Charcot-Marie-Tooth neuropathy type 1 (CMT 1a) gene to 17p11.2-p12. Am. J. Hum. Genet. 1990;47(4):680–685. [PMC free article] [PubMed] [Google Scholar]
- 25.Lupski J.R., de Oca-Luna R.M., Slaugenhaupt S., Pentao L., Guzzetta V., Trask B.J., Saucedo-Cardenas O., Barker D.F., Killian J.M., Garcia C.A., Chakravarti A., Patel P.I. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell. 1991;66(2):219–239. doi: 10.1016/0092-8674(91)90613-4. [DOI] [PubMed] [Google Scholar]
- 26.Nelis E., van Broeckhoven C., de Jonghe P., Löfgren A., Vandenberghe A., Latour P., Le Guern E., Brice A., Mostacciuolo M.L., Schiavon F., Palau F., Bort S., Upadhyaya M., Rocchi M., Archidiacono N., Mandich P., Bellone E., Silander K., Savontaus M.L., Navon R., Goldberg-Stern H., Estivill X., Volpini V., Friedl W., Gal A. Estimation of the mutation frequencies in Charcot-Marie-Tooth disease type 1 and hereditary neuropathy with liability to pressure palsies: A European collaborative study. Eur. J. Hum. Genet. 1996;4(1):25–33. doi: 10.1159/000472166. [DOI] [PubMed] [Google Scholar]
- 27.Chance P.F., Alderson M.K., Leppig K.A., Lensch M.W., Matsunami N., Smith B., Swanson P.D., Odelberg S.J., Disteche C.M., Bird T.D. DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell. 1993;72(1):143–151. doi: 10.1016/0092-8674(93)90058-x. [DOI] [PubMed] [Google Scholar]
- 28.Reiter L.T., Murakami T., Koeuth T., Pentao L., Muzny D.M., Gibbs R.A., Lupski J.R. A recombination hotspot responsible for two inherited peripheral neuropathies is located near a mariner transposon-like element. Nat. Genet. 1996;12(3):288–297. doi: 10.1038/ng0396-288. [DOI] [PubMed] [Google Scholar]
- 29.Kennerson M.L., Nassif N.T., Dawkins J.L., DeKroon R.M., Yang J.G., Nicholson G.A. The Charcot-Marie-Tooth binary repeat contains a gene transcribed from the opposite strand of a partially duplicated region of the COX10 gene. Genomics. 1997;46(1):61–69. doi: 10.1006/geno.1997.5012. [DOI] [PubMed] [Google Scholar]
- 30.Matsunami N., Smith B., Ballard L., Lensch M.W., Robertson M., Albertsen H., Hanemann C.O., Müller H.W., Bird T.D., White R., Chance P.F. Peripheral myelin protein-22 gene maps in the duplication in chromosome 17p11.2 associated with Charcot-Marie-Tooth 1A. Nat. Genet. 1992;1(3):176–179. doi: 10.1038/ng0692-176. [DOI] [PubMed] [Google Scholar]
- 31.Lupski J.R., Wise C.A., Kuwano A., Pentao L., Parke J.T., Glaze D.G., Ledbetter D.H., Greenberg F., Patel P.I. Gene dosage is a mechanism for Charcot-Marie-Tooth disease type 1A. Nat. Genet. 1992;1(1):29–33. doi: 10.1038/ng0492-29. [DOI] [PubMed] [Google Scholar]
- 32.Palau F., Löfgren A., De Jonghe P., Bort S., Nelis E., Sevilla T., Martin J.J., Vilchez J., Prieto F., Van Broeckhoven C. Origin of the de novo duplication in Charcot-Marie-Tooth disease type 1A: Unequal nonsister chromatid exchange during spermatogenesis. Hum. Mol. Genet. 1993;2(12):2031–2035. doi: 10.1093/hmg/2.12.2031. [DOI] [PubMed] [Google Scholar]
- 33.Liu P., Gelowani V., Zhang F., Drory V.E., Ben-Shachar S., Roney E., Medeiros A.C., Moore R.J., DiVincenzo C., Burnette W.B., Higgins J.J., Li J., Orr-Urtreger A., Lupski J.R. Mechanism, prevalence, and more severe neuropathy phenotype of the Charcot-Marie-Tooth type 1A triplication. Am. J. Hum. Genet. 2014;94(3):462–469. doi: 10.1016/j.ajhg.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang F., Seeman P., Liu P., Weterman M.A., Gonzaga-Jauregui C., Towne C.F., Batish S.D., De Vriendt E., De Jonghe P., Rautenstrauss B., Krause K.H., Khajavi M., Posadka J., Vandenberghe A., Palau F., Van Maldergem L., Baas F., Timmerman V., Lupski J.R. Mechanisms for nonrecurrent genomic rearrangements associated with CMT1A or HNPP: Rare CNVs as a cause for missing heritability. Am. J. Hum. Genet. 2010;86(6):892–903. doi: 10.1016/j.ajhg.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taioli F., Cabrini I., Cavallaro T., Acler M., Fabrizi G.M. Inherited demyelinating neuropathies with micromutations of peripheral myelin protein 22 gene. Brain. 2011;134(Pt 2):608–617. doi: 10.1093/brain/awq374. [DOI] [PubMed] [Google Scholar]
- 36.Scherer S.S., Kleopa K.A. X-linked Charcot-Marie-Tooth disease. J. Peripher. Nerv. Syst. 2012;17(3):9–13. doi: 10.1111/j.1529-8027.2012.00424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakagawa M., Takashima H., Umehara F., Arimura K., Miyashita F., Takenouchi N., Matsuyama W., Osame M. Clinical phenotype in X-linked Charcot-Marie-Tooth disease with an entire deletion of the connexin 32 coding sequence. J. Neurol. Sci. 2001;185(1):31–37. doi: 10.1016/s0022-510x(01)00454-3. [DOI] [PubMed] [Google Scholar]
- 38.Ainsworth P.J., Bolton C.F., Murphy B.C., Stuart J.A., Hahn A.F. Genotype/phenotype correlation in affected individuals of a family with a deletion of the entire coding sequence of the connexin 32 gene. Hum. Genet. 1998;103(2):242–244. doi: 10.1007/s004390050812. [DOI] [PubMed] [Google Scholar]
- 39.Gonzaga-Jauregui C., Zhang F., Towne C.F., Batish S.D., Lupski J.R. GJB1/Connexin 32 whole gene deletions in patients with X-linked Charcot-Marie-Tooth disease. Neurogenetics. 2010;11(4):465–470. doi: 10.1007/s10048-010-0247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shy M.E. Peripheral neuropathies caused by mutations in the myelin protein zero. J. Neurol. Sci. 2006;242(1-2):55–66. doi: 10.1016/j.jns.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 41.Høyer H., Braathen G.J., Eek A.K., Skjelbred C.F., Russell M.B. Charcot-Marie-Tooth caused by a copy number variation in myelin protein zero. Eur. J. Med. Genet. 2011;54(6):e580–e583. doi: 10.1016/j.ejmg.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 42.Speevak M.D., Farrell S.A. Charcot-Marie-Tooth 1B caused by expansion of a familial myelin protein zero (MPZ) gene duplication. Eur. J. Med. Genet. 2013;56(10):566–569. doi: 10.1016/j.ejmg.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 43.Maeda M.H., Mitsui J., Soong B.W., Takahashi Y., Ishiura H., Hayashi S., Shirota Y., Ichikawa Y., Matsumoto H., Arai M., Okamoto T., Miyama S., Shimizu J., Inazawa J., Goto J., Tsuji S. Increased gene dosage of myelin protein zero causes Charcot-Marie-Tooth disease. Ann. Neurol. 2012;71(1):84–92. doi: 10.1002/ana.22658. [DOI] [PubMed] [Google Scholar]
- 44.Wang W., Wang C., Dawson D.B., Thorland E.C., Lundquist P.A., Eckloff B.W., Wu Y., Baheti S., Evans J.M., Scherer S.S., Dyck P.J., Klein C.J. Target-enrichment sequencing and copy number evaluation in inherited polyneuropathy. Neurology. 2016;86(19):1762–1771. doi: 10.1212/WNL.0000000000002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Høyer H., Braathen G.J., Eek A.K., Nordang G.B., Skjelbred C.F., Russell M.B. Copy number variations in a population-based study of Charcot-Marie-Tooth disease. BioMed Res. Int. 2015;2015:960404. doi: 10.1155/2015/960404. https://www.hindawi.com/journals/ bmri/2015/960404/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ricard E., Mathis S., Magdelaine C., Delisle M.B., Magy L., Funalot B., Vallat J.M. CMT4D (NDRG1 mutation): Genotype-phenotype correlations. J. Peripher. Nerv. Syst. 2013;18(3):261–265. doi: 10.1111/jns5.12039. [DOI] [PubMed] [Google Scholar]
- 47.Okamoto Y., Goksungur M.T., Pehlivan D., Beck C.R., Gonzaga-Jauregui C., Muzny D.M., Atik M.M., Carvalho C.M., Matur Z., Bayraktar S., Boone P.M., Akyuz K., Gibbs R.A., Battaloglu E., Parman Y., Lupski J.R. Exonic duplication CNV of NDRG1 associated with autosomal-recessive HMSN-Lom/CMT4D. Genet. Med. 2014;16(5):386–394. doi: 10.1038/gim.2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chance P.F. Inherited focal, episodic neuropathies: Hereditary neuropathy with liability to pressure palsies and hereditary neuralgic amyotrophy. Neuromol. Med. 2006;8(1-2):159–174. doi: 10.1385/NMM:8:1:159. [DOI] [PubMed] [Google Scholar]
- 49.Nelis E., Van Broeckhoven C., De Jonghe P., Löfgren A., Vandenberghe A., Latour P., Le Guern E., Brice A., Mostacciuolo M.L., Schiavon F., Palau F., Bort S., Upadhyaya M., Rocchi M., Archidiacono N., Mandich P., Bellone E., Silander K., Savontaus M.L., Navon R., Goldberg-Stern H., Estivill X., Volpini V., Friedl W., Gal A. Estimation of the mutation frequencies in Charcot-Marie-Tooth disease type 1 and hereditary neuropathy with liability to pressure palsies: A European collaborative study. Eur. J. Hum. Genet. 1996;4(1):25–33. doi: 10.1159/000472166. [DOI] [PubMed] [Google Scholar]
- 50.Spagnoli C., Iodice A., Salerno G.G., Frattini D., Bertani G., Fusco C. Hereditary neuropathy with liability to pressure palsies in childhood: Case series and literature update. Neuromuscul. Disord. 2016;26(6):394. doi: 10.1016/j.nmd.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 51.Van Eijk J.J., Groothuis J.T., Van Alfen N. Neuralgic amyotrophy: An update on diagnosis, pathophysiology, and treatment. Muscle Nerve. 2016;53(3):337–350. doi: 10.1002/mus.25008. [DOI] [PubMed] [Google Scholar]
- 52.Kuhlenbäumer G., Hannibal M.C., Nelis E., Schirmacher A., Verpoorten N., Meuleman J., Watts G.D., De Vriendt E., Young P., Stögbauer F., Halfter H., Irobi J., Goossens D., Del-Favero J., Betz B.G., Hor H., Kurlemann G., Bird T.D., Airaksinen E., Mononen T., Serradell A.P., Prats J.M., Van Broeckhoven C., De Jonghe P., Timmerman V., Ringelstein E.B., Chance P.F. Mutations in SEPT9 cause hereditary neuralgic amyotrophy. Nat. Genet. 2005;37(10):1044–1046. doi: 10.1038/ng1649. [DOI] [PubMed] [Google Scholar]
- 53.Landsverk M.L., Ruzzo E.K., Mefford H.C., Buysse K., Buchan J.G., Eichler E.E., Petty E.M., Peterson E.A., Knutzen D.M., Barnett K., Farlow M.R., Caress J., Parry G.J., Quan D., Gardner K.L., Hong M., Simmons Z., Bird T.D., Chance P.F., Hannibal M.C. Duplication within the SEPT9 gene associated with a founder effect in North American families with hereditary neuralgic amyotrophy. Hum. Mol. Genet. 2009;18(7):1200–1208. doi: 10.1093/hmg/ddp014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Collie A.M., Landsverk M.L., Ruzzo E., Mefford H.C., Buysse K., Adkins J.R., Knutzen D.M., Barnett K., Brown R.H., Jr, Parry G.J., Yum S.W., Simpson D.A., Olney R.K., Chinnery P.F., Eichler E.E., Chance P.F., Hannibal M.C. Non-recurrent SEPT9 duplications cause hereditary neuralgic amyotrophy. J. Med. Genet. 2010;47(9):601–607. doi: 10.1136/jmg.2009.072348. [DOI] [PubMed] [Google Scholar]
- 55.Klein C.J., Wu Y., Cunningham J.M., Windebank A.J., Dyck P.J., Friedenberg S.M., Klein D.M., Dyck P.J. SEPT9 mutations and a conserved 17q25 sequence in sporadic and hereditary brachial plexus neuropathy. Arch. Neurol. 2009;66(2):238–243. doi: 10.1001/archneurol.2008.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hentati F., Hentati E., Amouri R. Giant axonal neuropathy. Handb. Clin. Neurol. 2013;115:933–938. doi: 10.1016/B978-0-444-52902-2.00052-7. [DOI] [PubMed] [Google Scholar]
- 57.Bomont P., Cavalier L., Blondeau F., Ben Hamida C., Belal S., Tazir M., Demir E., Topaloglu H., Korinthenberg R., Tüysüz B., Landrieu P., Hentati F., Koenig M. The gene encoding gigaxonin, a new member of the cytoskeletal BTB/kelch repeat family, is mutated in giant axonal neuropathy. Nat. Genet. 2000;26(3):370–374. doi: 10.1038/81701. [DOI] [PubMed] [Google Scholar]
- 58.Engel E. A new genetic concept: uniparental disomy and its potential effect, isodisomy. Am. J. Med. Genet. 1980;6(2):137–143. doi: 10.1002/ajmg.1320060207. [DOI] [PubMed] [Google Scholar]
- 59.Buysse K., Vergult S., Mussche S., Ceuterick-de Groote C., Speleman F., Menten B., Lissens W., Van Coster R. Giant axonal neuropathy caused by compound heterozygosity for a maternally inherited microdeletion and a paternal mutation within the GAN gene. Am. J. Med. Genet. A. 2010;152A(11):2802–2804. doi: 10.1002/ajmg.a.33508. [DOI] [PubMed] [Google Scholar]