Abstract
Patients diagnosed with glioblastoma (GBM) continue to face a bleak prognosis. It is critical that new effective therapeutic strategies are developed. GBM stem cells have molecular hallmarks of neural stem and progenitor cells and it is possible to propagate both non-transformed normal neural stem cells and GBM stem cells, in defined, feeder-free, adherent culture. These primary stem cell lines provide an experimental model that is ideally suited to cell-based drug discovery or genetic screens in order to identify tumour-specific vulnerabilities. For many solid tumours, including GBM, the genetic disruptions that drive tumour initiation and growth have now been catalogued. CRISPR/Cas-based genome editing technologies have recently emerged, transforming our ability to functionally annotate the human genome. Genome editing opens prospects for engineering precise genetic changes in normal and GBM-derived neural stem cells, which will provide more defined and reliable genetic models, with critical matched pairs of isogenic cell lines. Generation of more complex alleles such as knock in tags or fluorescent reporters is also now possible. These new cellular models can be deployed in cell-based phenotypic drug discovery (PDD). Here we discuss the convergence of these advanced technologies (iPS cells, neural stem cell culture, genome editing and high content phenotypic screening) and how they herald a new era in human cellular genetics that should have a major impact in accelerating glioblastoma drug discovery.
Keywords: Neural stem cell, Glioblastoma stem cell, CRISPR/Cas9, Genome editing, Phenotypic screening, HCS
1. Introduction
The prognosis for children and adults suffering from high grade glioma is dismal. An improved understanding of disease biology is urgently needed. Gliomas are a heterogeneous group of tumours, but the higher grade tumours – more commonly known as glioblastoma (GBM) – are invariably driven by cells that display features of neural stem and progenitor cells (Lathia et al., 2015). Many putative genetic and epigenetic drivers of glioma have now been uncovered through systematic genome-wide molecular annotation, opening up a wealth of new directions for fundamental discovery and improved molecular classifications (Brennan et al., 2013; Sturm et al., 2012). This fundamental knowledge will ultimately lead to new treatments and enhanced patient outcomes; however, in the shorter term there remains an urgent unmet need to repurpose existing drugs for use in GBM as well as identify key molecular targets and develop new lead compounds.
During the past five years there have been remarkable advances across several technologies that will enhance glioma discovery research, including: 1) improved cellular models and stem cell culture conditions (iPS cell, neural stem cell and glioma stem cells), 2) CRISPR/Cas genome editing, and 3) cell phenotypic screening platforms. The emergence of these technologies, paralleled by improved understanding of cancer genetic and epigenetic disruptions, should drive development of novel patient-derived cellular models that can be channelled into cell-based chemical and genetic screens in vitro and xenotransplantation models in vivo. Here we discuss each of these areas, particularly how they intersect and might be deployed in the coming years to improve the prognosis for people living with GBM – one of the most lethal human cancers. We focus on chemical screens using patient-derived cellular models, and the opportunities for gene editing to underpin novel cell-based phenotypic assays. Use of CRISPR/Cas for genetic screens has been discussed elsewhere (Agrotis and Ketteler, 2015).
2. Sources of neural stem and progenitor cells
Much effort has been expended over the past few decades by developmental neurobiologists seeking to define the diversity of neural stem and progenitor cell types responsible for construction of the mammalian central nervous system (CNS) (Gage and Temple, 2013). Knowledge of mammalian brain development has largely come from studies of mouse developmental biology and several distinct categories of neural progenitor cells have been identified. The most primitive and earliest-born neural progenitors are termed neuroepithelial cells, and these likely retain the potential to differentiate into a variety of neuronal or glial subtypes. Neuroepithelial cells transit at the onset of neurogenesis into what are now termed apical progenitors (formerly radial glia) that generate neurons, and at later foetal stages glial cells (Taverna et al., 2014). These apical progenitors generate the wave of newborn neuronal populations, but do so via stepwise transitions along a series of distinct intermediate progenitors (Rowitch and Kriegstein, 2010). These major temporal transitions in neural progenitor states/subtypes are superimposed by well-understood patterning events that establish distinct positional identity: e.g. forebrain versus spinal cord, or cortex versus striatum. In the adult mouse brain, two regions have been uncovered in which new neurons are generated throughout adulthood: the hippocampus, and the walls of the forebrain ventricles. A subpopulation of apical progenitors are the founders of adult neural stem cells and these emerge postnatally (Merkle et al., 2004). The reader is pointed to other reviews which cover these topics in more detail (Bond et al., 2015; Kriegstein and Alvarez-Buylla, 2009).
Despite this progress, we still lack a comprehensive understanding of the full diversity of distinct immature populations and their differentiation potential and plasticity. New classes of progenitor are still being uncovered in the mouse (Pilz et al., 2013). Also, inevitably our understanding of the diversity of human neural stem and progenitors has lagged behind that of the mouse, and important species differences are now being uncovered in the repertoire of progenitors and their molecular regulation (Florio et al., 2015; Lui et al., 2011, 2014).
Considerable attention has focussed on the developing cortex, due to its importance in human biology and evolution, and a population of progenitors termed outer radial glia have been described that are thought to drive the massive expansion of the human (but not mouse) cortical surface area (Hansen et al., 2010). Application of single cell transcriptome analysis and epigenetic profiling are now providing a more complete picture of the full range of distinct cell types (Johnson et al., 2015). Open access databases such as Allen Brain Atlas that integrate neuroanatomical and gene expression datasets also provide a wealth of information to understand the genetic and cellular basis of CNS development in mouse and human (Miller et al., 2014). More recently a related effort has been established for GBM (Sunkin et al., 2013). Altogether these ongoing efforts should eventually lead to comprehensive understanding of the gene expression signatures that define the full inventory of distinct neural progenitors.
Pluripotent stem cells (PSCs) – embryonic stem cells (ESCs) and induced PSCs (iPSCs) – are phenotypically similar to the early pre-gastrulation stage human embryo, and therefore provide a valuable tool to explore early human development. Importantly, they also have practical value as a means to produce human neural cell types in the laboratory (Dolmetsch and Geschwind, 2011; Pourquié et al., 2015), providing a potentially unlimited source of neurons and glia that can be utilized in chemical and genetic screening.
Our knowledge of neural development has been useful to guide approaches to generate, expand and differentiate neural stem cells in vitro (Aboody et al., 2011). Neuroepithelial cells emerge early during ES and iPS cell differentiation – mirroring the primitive ectoderm to neural ectoderm developmental transition; these then transit into radial glia/apical progenitors that lose epithelial features such as expression of the cell-cell tight junction marker ZO-1 and acquire a ‘rosette’-like appearance in culture (Elkabetz et al., 2008). These in turn go on to differentiate into neurons, and then a later wave of glial differentiation (astrocytes and oligodendrocytes).
It has proven difficult to capture the more primitive neuroepithelial cells and expand them long term. However, mouse or human radial glia-like apical progenitors, whether derived from PSC differentiation, or freshly isolated foetal/adult CNS tissue, can be expanded long-term in culture using the growth factors EGF and FGF-2. These neural stem cells – herein termed ‘NS cells’ – can be propagated either in suspension culture as ‘neurospheres’, or using adherent monolayer. The advantages and disadvantages of these in vitro models have been discussed previously elsewhere (Pastrana et al., 2011). NS cells are somewhat restricted in their differentiation capacity and are glial biased, with features more akin to proliferative adult SVZ neural stem cells. It remains unclear to what extent distinct positional and temporal identities are permanently erased by the culture environment, or if some epigenetic memory of their original identity persists. The rest of this article focuses on these NS cell cultures. This cell state most closely corresponds to the glioma stem cells in their patterns of marker expression, glial differentiation bias and requirement for EGFR signaling. Comparisons of NS cells with their malignant GNS cell counterparts can identify tumour-associated pathways.
3. Neural stem cells and brain cancer
Around 10 years ago there was increased interest in the relationship between neural development, neural stem cells and cancer biology. It became clear that many neural stem cell markers were frequently expressed in and required for growth of gliomas, such as OLIG2 (Ligon et al., 2004, 2007). This raised a related issue of whether CNS derived tumours might arise from stem cells gone awry, and whether these putative cancer stem cells are critical to sustaining tumour growth (Stiles et al., 2008). Functional data supporting a hierarchy of tumour cell malignancy came via improved methods for fractionating tumour populations based on neural stem cell markers and interrogating their potency in immunocompromised mice (Singh et al., 2004). Brain tumours may therefore be a ‘caricature’ of the normal tissue stem cell, as was proposed in the 1960s for tertatocarcinoma (Pierce and Speers, 1988). Thus, gliomas might not be viewed simply as corrupted proliferative astrocytes, but instead may depend upon, and exploit, the core apparatus used by radial glia-like neural stem cells.
There has been much debate and discussion regarding the significance of cancer stem cell-based models for understanding cancer biology and guiding therapeutic strategies (Kreso and Dick, 2014). Arguably the most critical question for GBM is whether the putative cancer stem cells are hijacking and exploiting the self-renewal pathways that underpin normal neural stem cell self-renewal, as this information could eventually be exploited to halt tumour growth and relapse after therapy. Indeed, many of the essential transcriptional and epigenetic regulators of neural stem cells are highly expressed in gliomas and have clear functional importance in sustaining tumour growth (Gallo et al., 2013; Gangemi et al., 2009; Ligon et al., 2007; Verginelli et al., 2013).
Importantly, those culture conditions widely deployed for the propagation of neural stem cells (serum-free media with EGF and FGF-2) proved extremely well-suited to expansion of patient-derived putative glioma stem cells (Galli et al., 2004; Lee et al., 2006; Singh et al., 2003). Thus, these tumour-initiating cells can be expanded readily while retaining tumour-initiating capacity. They can also be propagated in adherent monolayer, simplifying chemical and genetic screening (Hubert et al., 2013; Pollard et al., 2009; Wurdak et al., 2010). Gliomas are therefore one of the few human cancers for which we can isolate, culture, and manipulate primary cancer stem cells, as well as their ‘normal’ tissue stem cell counterparts (NSCs). Comparing and contrasting glioma stem cells with their genetically and epigenetically normal NS cell counterparts will therefore help to identify potential cancer-specific vulnerabilities.
4. Patient-derived cellular models and matched controls: a unique opportunity for gliomas
Recurrent genetic and epigenetic perturbations in glioma have now been extensively catalogued (Brennan et al., 2013; Sturm et al., 2012). Functional genetic analyses in relevant human preclinical models are now needed. There have been recent reports of the exploitation of iPS cells to engineer oncogenic events prior to neural differentiation, thereby providing a model to interrogate glioma associated mutations across a range of regional and temporal human progenitors (Funato et al., 2014; Sancho-Martinez et al., 2016).
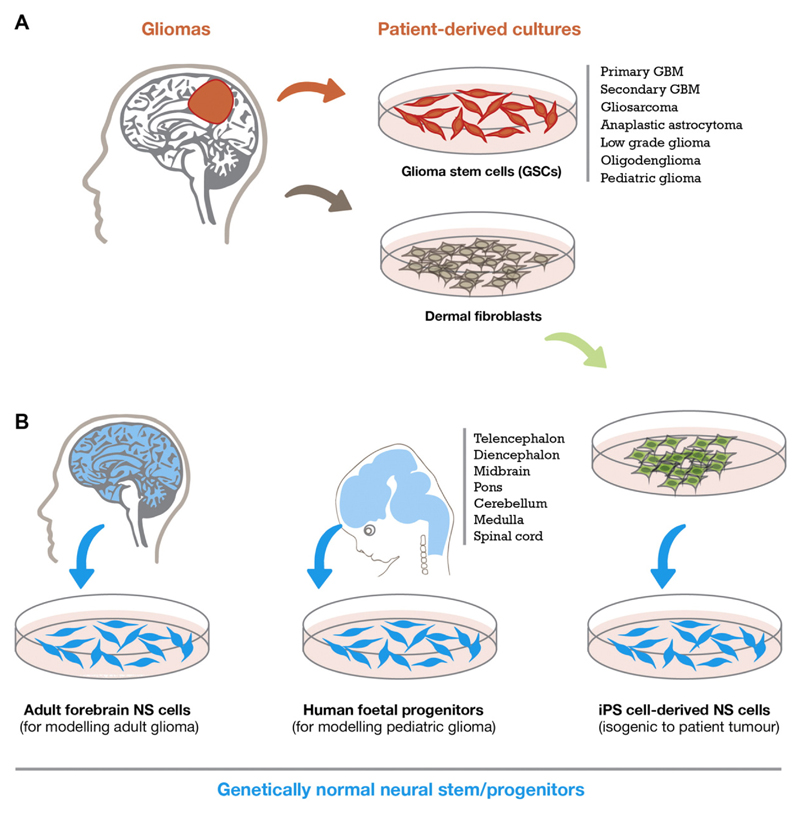
A complementary approach to iPSC engineering is to work with primary foetal and adult derived NS cell lines. Indeed, many groups have now developed large collections of GSC and NS cell lines that can be used in both basic and translational projects (Xie et al., 2015). Primary human NS cells provide the genetically normal controls for functional studies and comparison with glioma stem cell cultures (Fig. 1). Primary NS cells from foetal forebrain, midbrain and hindbrain tissues (typically from week 7 to 14 human embryos) will likely be very useful for modelling paediatric GBM and assessing how regional identity influences the competence to respond to particular glioma mutations. Adult human NS cell lines, derived from essentially healthy donors (CNS material from epilepsy surgery), are more difficult to establish, but there have been reports of successful cultures established from adult forebrain SVZ (Sanai et al., 2004). In the coming years these cellular models will help us to study the origins of GBM and how their spatial and temporal diversity influences the response to oncogenic insults.
Fig. 1.
Sources and diversity of primary cell lines. Cells in red are cancerous, blue cells are neural stem cells free from disease. Each cell type gives an orthogonal view of neural stem cells in development and disease. All resulting cell lines can be cultured for screening purposes under identical growth conditions allowing direct comparisons of screening results.
One of the limitations of functional cellular genetic studies in human iPS cells has been cell line variations, in part due to their diverse genetic backgrounds. Isogenic matched cell line pairs, where the only genetic difference is at candidate gene/locus of interest provide the most rigorous controls to draw meaningful conclusions. Similarly, for GSCs it would be valuable to generate NS cells that are isogenic to original patient tumour cell of origin. There is in fact a good opportunity to do this, as iPS cells can be differentiated into NS cells in vitro, and compared to the patient tumour-derived GSCs. Such isogenic matched cell lines will be essential as high quality genetic controls (Fig. 1). These sets of patient-derived models and their normal genetically matched controls should also be amenable to genome editing, opening up opportunities to engineer glioma mutations.
5. Genome editing to engineer normal and glioma-derived stem cells
Engineering precise genetic changes into the mouse genome of PSCs using gene targeting has been the gold standard approach to explore cancer gene function and creation of genetically engineered mouse models (GEMMs). Standard gene targeting approaches involve generation of a targeting vector and delivery to PSCs by in vitro transfection and subsequent selection of rare clonal lines with the desired change. However, this is often time-consuming and requires significant expertise. CRISPR/Cas technologies have caused tremendous excitement across many areas of life sciences and medicine, as these designer nucleases enable site-specific engineering of the genome in a much more efficient manner – including efficient editing of the human genome in iPS cells.
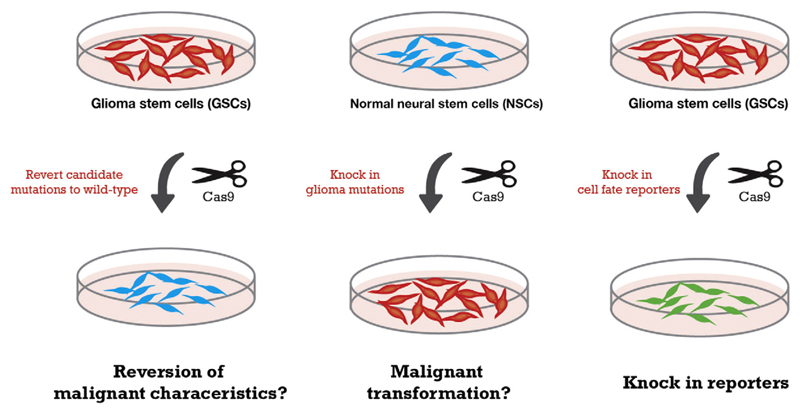
A unique opportunity has arisen to deploy these technologies for new drug development for GBM, across several areas: first, identification of new molecular targets through genome-wide CRISPR screens (Hart et al., 2015; Munoz et al., 2016; Toledo et al., 2015); second, engineering of candidate driver mutations into normal NS cells; third, reversion of genetic drivers to wild-type in GSCs; fourth, creation of useful live cell reporters (epitope tags or fluorescent proteins) or biosensors for cell based phenotypic screening.
NSCs and GSCs are easily transfected, expandable and clonogenic and should be well suited to genome editing. We have recently found that CRISPR/Cas-based genetic editing is straightforward in NSC and GSC lines (Pollard lab, submitted). It is therefore now possible to efficiently introduce targeted and sophisticated genetic changes such as: specific insertion/deletions; introduction of somatic point mutations; gene targeting to introduce conditional alleles, replace exons or modify cis-regulatory elements; knock-in of fluorescent marker gene reporters and biochemical tags; and engineering of large-scale chromosome structural engineering (e.g. large focal amplifications or deletions) (Fig. 2). Thus, it will be possible to construct a whole suite of useful isogenic human cellular model and reporter cell lines.
Fig. 2.
A genome editing ‘toolkit’ for functional genetic studies and novel engineered cellular models. Reversion of candidate drivers to wild-type (left) and/or introduction of key drivers into normal NSCs (middle) using CRISPR/Cas9 provides both a means for proving mutation causality, and matched cell lines as perfect isogenic controls for drug screening. Creation of a variety of useful engineered alleles such as live cell reporters and safe-harbours (right) removes need for fluorescent staining. Co-culture of fluorescently tagged cell lines carrying potentially druggable driver mutations with isogenic non-tagged controls provides an ideal internally controlled cell assay.
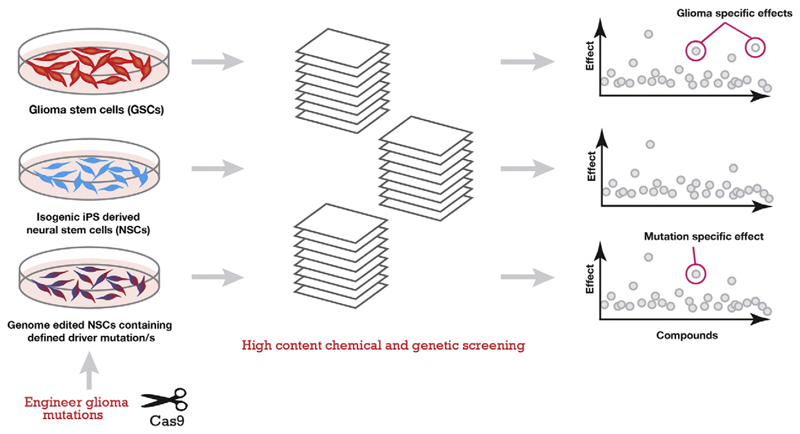
The combination of improved methods for isolating and propagating normal human NS cell lines as well as their glioma-derived malignant counterparts, alongside these remarkable developments in genome editing, is opening up prospects for development of new therapeutics for GBM. The gene editing pipeline would allow identification of drivers that could be targeted via existing or newly developed small molecules. Moreover, GBM stem cell cultures and matched controls – along with associated knock in fluorescent markers of stem cell and differentiation state – could be used directly in cell based phenotypic screens. The gene editing pipeline described above would also be instrumental for validation of drug specificity (Fig. 3). Thus, we can now envisage moving into a new era of sophisticated genetic analysis in primary human cells – both normal NS and glioma-derived NS cells. Some specific examples are described below.
Fig. 3.
High Content Screening approach and hit interpretation. Cell lines can be co-cultured or screened in parallel. Compound effects common to all cell lines are nonspecific while effects found in only one line are cancer or mutation specific. ‘Effects’ can be found in a wide variety of cell and nuclear morphologies at selected time points given the multiparametric nature of high content screening. Iterative rounds of genome engineering can be used to verify drug targets and determine mechanism of action. These phenotypic screens provide richer information; but are more difficult to implement than biochemical assays used in target based drug discovery.
5.1. Engineering of oncogenes and tumour suppressors
Tumours often develop progressively through preneoplastic growth, to full malignant transformation and undergo further evolution under the selective pressure associated with treatments. Deciphering what are true drivers from passenger mutations is critical to define the best therapeutic targets. Thus, to make rapid progress towards novel therapies, efforts should be focused on deletion of the most frequently amplified oncogenes thought to drive the disease (EGFR, PDGFR, CDK4). Complementary studies would aim to revert mutant to wild-type alleles for TP53, H3F3A, TERT (promoter) and IDH1/2. Additionally, using the matched genetically normal isogenic controls from the same patient (iPS-derived NSCs) glioma mutations can be engineered stepwise and in combination (Fig. 2). Together, this strategy should provide the gold standard genetically defined cellular models for chemical screens.
5.2. Knock in reporter cell lines
In parallel to exploring driver mutations it should be possible to perform knock in of useful reporters or other cargos to the safe-harbour AAVS1 locus (the AAVS1 is equivalent to the ROSA26 locus in mouse). For example, in a simple scenario, knock in of a constitutive eGFP and Luciferase labelled expression cassette would be extremely useful for tracking xenograft growth in live animals using small animal imaging with a bioluminescence or intravitral imaging system. This overcomes the issue of stable transfection via lentivirus, and removes issues of silencing: a persistent problem when using random insertion transgenesis. Introduction of eGFP, mCherry or BFP2, fluorescent reporters under the control of a variety of endogenous gene promoters (SOX2, FOXG1, OLIG2) also provides useful live cell reporters to monitor ‘stermness’ and conversely differentiation (AQP4, CNPase, NFM; for astrocyte, oligodendrocyte and neuron, respectively). These lineage reporters have great utility in vitro for high content phenotypic screening.
6. Cell-based phenotypic screening
Phenotypic screening can be defined as the quantification of functional biological endpoints from physiological-based model systems following exposure to libraries of small molecule chemicals, gene-targeting perturbations or proteins/antibodies (Lee and Berg, 2013; Yarrow et al., 2003; Carragher, 2008). This is particularly useful in stem cell based models where cell heterogeneity and shifts in differentiation status mean individual cell behavior must be tracked.
Phenotypic screening is typically performed with cell-based assays or model organisms amenable to automated medium- to high-throughput screening platforms. Smaller focused phenotypic screens using high quality tool compound libraries (see Box 1) can also be employed to determine which pathways are involved in a particular process.
Box 1. High quality commercially available and open source tool compound libraries.
(Brown and Müller, 2015; Drewry et al., 2014; Elkins et al., 2015; Mei et al., 2014; Moisan et al., 2015)
Published Kinase Inhibitor set available from GSK
An open access tool of 367 annotated small molecule kinase inhibitors. [Drewry et al., 2014 and Elkins et al., 2016]
Bioactive Compound Library from Selleck
Commercially available customizable library of 1902 bioactive compounds. [Mei et al., 2014 and Moisan et al., 2015]
http://www.selleckchem.com/screening/chemical-library.html
StemSelect
Library of 303 pharmacologically active, structurally diverse small molecules targeting a variety of pathways involved in proliferation, migration and differentiation. Available from Merck Millipore.
InhibitorSelect I, II & III
243 well-characterized protein kinase inhibitors spread over three 96 well plates. Also available from Merck Millipore.
Phenotypic toolbox from BioAscent
Composed of FDA approved drugs, reference compounds and diverse lead-like molecules in a small combined library.
http://www.bioascent.com/phenotypic/
Epigenetic chemical probes from the Structural Genomics Consortium (SGC)
More than 30 open access high quality tool compounds targeting a variety of epigenetic regulators. [Brown et al., 2015]
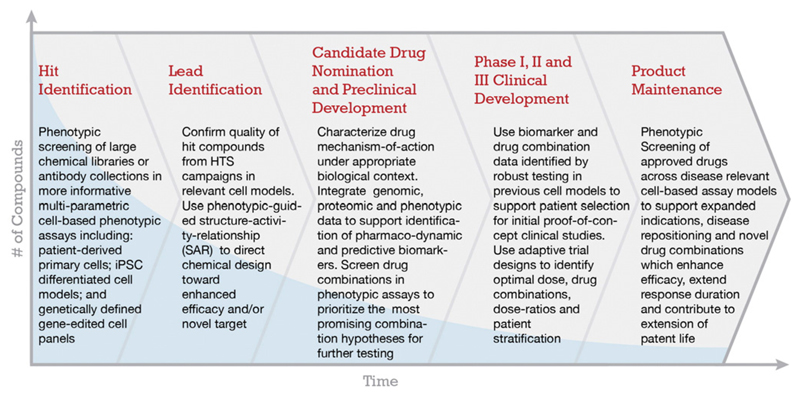
In contrast to the widely adopted target-directed drug discovery (TDD) model, in which screening is directed upon a pre-nominated protein target, phenotypic drug discovery (PDD) does not depend on prior knowledge of molecular targets (Fig. 4). PDD can also be applied to the discovery of novel drug combinations, or to support disease repositioning by screening approved drug libraries (Dawson and Carragher, 2014; Reaume, 2012). In contemporary drug discovery projects phenotypic screens are typically placed as secondary screening assays to confirm the quality and physiological relevance of novel antagonists or agonists derived from high throughput biochemical or structure-based screening. By placing phenotypic screening at the beginning of the drug discovery paradigm however it is possible to identify new targets or novel small molecules for further investigation.
Fig. 4.
Proposal for a streamlined drug discovery process based on phenotypic screening. From the beginning and at each step during PDD assay quality and biological relevance is emphasised. This is in contrast to the brute force massively high quantity screening in traditional TDD pipelines.
The emergence of new molecular biology techniques, such as genome sequencing and more in-depth understanding of genetic linkage with human disease, has led to TDD being preferred drug discovery strategy by both the biopharmaceutical industry and core academic drug discovery groups. However, the anticipated increase in clinical approval rates of novel first-in-class therapies has not followed, indeed, the exponential increase in investment in reductionist TDD strategies has coincided with an overall decline in R&D productivity (Paul et al., 2010; Scannell et al., 2012). Cancer has been highlighted as a fruitful area for modern TDD strategies (Hoelder et al., 2012), however, cancer subtypes which do not fall into well-defined molecular subtypes remain intractable to new treatments derived from both strategies. This is particularly apparent in glioma where there have been no advances in effective targeted molecular therapies. The minimal impact of TDD strategies on the most serious cancers, including gliomas is of serious concern.
The rapid evolution and convergence of new technologies, including advances in image-based phenotypic screening, iPSC technologies and gene editing, are well placed to advance a new era of modern phenotypic screening in cell-based models of disease. The widespread use of basic phenotypic screening assays applied to established cell lines, which poorly model disease, most likely underestimates the true impact that PDD strategies will have. Furthermore, the development of more informative phenotypic screening across more disease relevant cell models plays a major role in enhancing TDD by supporting more robust target validation and secondary screening assay cascades. We propose that further development and increased adoption of the latest advances in image-based phenotypic screening, patient-derived primary cell models, iPSC and genome editing should have a significant impact in identifying new hits and leads for GBM. Clearly TDD and PDD represent complementary approaches and both have their value in the drug discovery effort. PDD involves increased effort and costs (labour and reagent costs) in acquiring large amounts of imaging data and lower throughput compared to TDD. However, for primary GBM clear-cut therapeutic target hypothesis and robust validation of novel molecular targets, which could be drivers across all the disease subtypes, have not emerged. Consequently an unbiased PDD approach is therefore particularly appealing, as unexpected pathways and biology can emerge, as recently exemplified by the study of Kitambi et al. (Kitambi et al., 2014).
7. Advances in image-based phenotypic screening
Microscopy has a long established role in the history of scientific research and represents a cornerstone of cell biology and pathology. Traditionally, microscopic imaging has been applied manually across small sample numbers to provide subjective evaluation of cell behavior and in vivo physiology. Robotic automation of microscope operation and associated phenotypic assays (e.g. immunocytochemistry protocols) is currently transforming the field of microscopy towards higher throughput and more quantitative applications (see Box 2 for an overview of commercially available high content imaging systems). Significant advances have also been made in optical performance, fluorescent reporter molecules, image-analysis and image-informatics, High content analysis of multiparametric features extracted from fluorescent or brightfield images of cells, tissues or small-model organisms is now possible (Taylor et al., 2001) providing quantitative readouts of phenotypic traits.
Box 2. Overview of available image based phenotypic screening technologies.
| Platform | Vendor |
|---|---|
| Epifluorescent High content platforms | |
| ImageXpress MicroXLS | Molecular Devices |
| CellInsight | Thermo Fisher |
| ArrayScan | Thermo Fisher |
| IN Cell Analyzer 2200 | GE Healthcare |
| ScanR | Olympus |
| WiScan | IDEA Bio-Medical Ltd |
| Cytation 5 | BioTek |
| Cellavista | SynenTec Bio Services |
| Laser Scanning imaging cytometer | |
| Acumen Cellista | TTP Labtech |
| Confocal High content platforms | |
| Operetta | Perkin Elmer |
| Opera Phenix | Perkin Elmer |
| ImageXpress Ultra | Molecular devices |
| IN Cell Analyzer 6000 | GE Healthcare |
| Yokogawa CQ1 | Wako Automation |
| Yokogawa CV700 | Wako Automation |
| “Live-cell” high-content imaging platforms tailored for long-term kinetic studies | |
| IncuCyte-ZoomTM | Essen Bioscience |
| Cell-IQ | CM Technologies |
| BioStation-CT | Nikon Instruments |
These developments have also supported higher throughput application of phenotypic screening to large chemical libraries, whole genome arrayed si/shRNA libraries, dose-matrix combination screening and comparison of phenotypic response across genetically distinct cell panels (Caie et al., 2010; Dawson and Carragher, 2014; Neumann et al., 2006; Yarrow et al., 2003). A significant advantage in high-content imaging over other high throughput phenotypic screening platforms is the extraction of functional data points with high spatial resolution across X, Y and Z dimensions, allowing meaningful screening assays to be conducted in more complex multicellular and 3D models (Wenzel et al., 2014).
8. Live-cell kinetic imaging
Integration of robust atmospheric and temperature control of biological samples mounted within high-content screening platforms, together with the development of novel optical biosensors and fluorescent reporter constructs, have increased the scope of phenotypic screening to include live-cell kinetic imaging (Isherwood et al., 2011). These approaches inevitably push resource limits in terms of assay development timelines and computational power required for image storage and analysis. So they are difficult to scale to high throughput screening (tens of thousands of compounds). Nevertheless, they provide extremely rich phenotypic information, and critical kinetic and spatial information, which may have greater value in hit selection and validation of compounds or the dose-response assays, for example to capture cellular transitions during differentiation; differentiation therapy is one potential strategy to limit cancer stem cell self-renewal which is being explored. Developments in the design of compact automated imaging platforms, which can be placed inside an environmental chamber (IncuCyte-Zoom™; Cell-IQ® and Biostation CT, see Box 2) and integrated with external and internal plate handling robotics further supports longer-term kinetic cellular assays across multiwell plates within a stably controlled microenvironment at scale. Long-term kinetic imaging and phenotypic screening can thus be performed under defined environmental conditions, for example normoxia or hypoxia, which may more accurately reflect the physiology and pathophysiology of tissues in vivo.
Kinetic analysis of cell phenotype following exposure to chemicals can reveal unique insights into cellular pharmacology, which are not apparent from traditional fixed endpoint assays. For example, cell physiological processes that operate under precise temporal and spatial control such as cell-cycle transition, motility, cell death, cell signaling and subcellular trafficking of proteins including oscillatory nuclear-to-cytoplasmic translocations can only reliably be recorded by kinetic analysis of live cells.
Long-term image-based kinetic analysis of cell phenotypes facilitates quantification of stem cell differentiation and cell fate. Such image-based live cell phenotypic screening studies can be applied to discover modulators which accelerate or delay stem cell differentiation. Our groups have utilized such live cell imaging to probe small chemical libraries for effects on both NS and GNS cells (Danovi et al., 2010, 2013; Pollard et al., 2009). Kinetic analysis of phenotypic response further supports the selection of the most informative and appropriate time-points for higher throughput endpoint assays. This may include the selection of the most optimal time-point or time-points to measure transient phenotypic response or selection of earliest time-point/phenotypic measure predictive of long-term stem cell fate to accelerate stem cell screening studies. Thus, while the throughput is reduced, the potential for obtaining rich biological information is immense. This will guide assay development, hit selection and inform subsequent drug discovery and development strategy.
9. Multiparametric phenotypic profiling
Traditional cell-based phenotypic screening assays depend upon simple cell viability or genetic reporter readouts, and therefore provide limited information on drug mechanism-of-action to guide hit selection. Such basic phenotypic assays also provide limited opportunity to direct structural activity relationships (SAR) and guide chemical design towards precise phenotypes representing enhanced efficacy and safety features. These are important issues to address.
The evolution of multiparametric phenotypic assays combined with multivariate statistics and a variety of new image-informatics methods have produced the discipline of phenotypic profiling. Phenotypic profiling enables the classification of phenotypic response and thus compound mechanism-of-action based upon similar multiparametric phenotypic fingerprints (Carpenter et al., 2006; Feng et al., 2009; Perlman et al., 2004; Tanaka et al., 2005a). The application of phenotypic profiling and image-informatics methods incorporating machine learning and artificial neural networks has steadily evolved to support robust phenotypic profiling across distinct cell types (Caie et al., 2010; Ljosa et al., 2013; Smith and Horvath, 2014). A key requirement of image informatics methods applied to multiparametric high content data sets is a minimal number of measured features with retention of maximal phenotypic information. Feature reduction methods including principal component analysis, factor analysis and support vector machines have been commonly used to distil multiparametric high content data to single endpoints, which can be used to rank compound performance and integrate phenotypic response with orthogonal datasets (Ljosa et al., 2013; Tanaka et al., 2005b). Phenotypic data reduction to single endpoints supports the association of biological similarity with chemical similarity to guide chemical design and direct SAR from phenotypic response (Young et al., 2007).
10. Cheminformatics and chemical library design for phenotypic screening
Phenotypic profiling methods have also been combined with cheminformatics approaches to further guide chemical design based on phenotypic data and development of the most appropriate chemical libraries for phenotypic screening applications (Wagner and Clemons, 2009; Wawer et al., 2014). While the methods for defining chemical similarity and linking chemical structure to phenotypic response data are still evolving, chemical features associated with frequent ‘hits’ in multiple diverse cell-based assays representing ‘non-specific’ phenotypic outcomes can be identified and interpreted appropriately (Tanikawa et al., 2009). Alternatively, compounds with distinct chemical features which produce similar biological phenotypes might be considered starting points for new library synthesis directed at specific phenotypic outcomes.
The development of publicly available cheminformatics databases, combined with commercially available chemically diverse libraries and annotated compounds, supports further advancement of phenotypic screening (see Box 1 for links to further information on these resources). Specifically, custom-designed phenotypic toolbox compounds, which provide a reference library for classifying the mechanism-of-action of unknown compounds, continue to be developed and expanded. Such development supports the expansion of phenotypic screening beyond large pharma into small biotech and academic translational research groups.
11. The road ahead: linking phenotype with genotype to advance pharmacogenomics studies in glioma
Application of high content screening across genetically distinct cell lines can help elucidate drug mechanism-of-action and guide clinical positioning by linking phenotype to genotype (Johannessen et al., 2015). Transcriptional profiling of well-characterized cancer cell-line panels such as the NCI-60 collection has supported such studies. Correlation of the NCI-60 transcriptional profiles with phenotypic response to compound treatments using existing cell viability assay results produced over 200 gene expression based signatures for compound sensitivity (Staunton et al., 2001). These early results provided the impetus for expansion of high throughput pharmacogenomics studies across larger cancer cell line panels including a study led by the Sanger Institute across 639 cancer cell lines and a separate study led by the Broad Institute in collaboration with Novartis across 949 cell lines (Barretina et al., 2012; Garnett et al., 2012). Both studies used a simple cell viability assay readout for recording phenotypic response and quantification of drug sensitivity.
Comparative analysis of both studies revealed inconsistencies (Haibe-Kains et al., 2013; Stransky et al., 2015). Some of this may be attributed to the use of transformed cell lines, which are often genetically unstable and no longer represent the primary cells and tissues from which they were originally derived. Also the simple whole well cell viability endpoint employed in these studies lacks precise mechanistic information. The advances in precise gene editing technologies such as CRISPR/Cas can provide genetically defined models representative of a wider variety of glioma subtypes. These cell lines can be used to precisely quantify and classify cell phenotype (Shalem et al., 2014). The convergence of gene-editing with iPSC and high content phenotypic screening further enable the expansion of high throughput pharmacogenomics studies from simple cell viability endpoints to more complex phenotypes and assay formats which can help define new targets for glioblastoma.
In summary, frontloading cell based phenotypic models of glioma at the earliest stages of the drug discovery using disease relevant cellular models will fast track the identification of the most promising therapeutic targets and candidate drugs. The rationale outlined here represents our perspectives on how these new technologies are opening up drug pipelines that will hopefully lead to new drugs and improved clinical outcomes for glioma patients.
Acknowledgements
SMP is supported by a Cancer Research UK Senior Research Fellowship (A19778). We thank Faye Robertson, Gillian Morrison and Paul Brennan for helpful comments on the manuscript.
References
- Aboody K, Capela A, Niazi N, Stern JH, Temple S. Translating stem cell studies to the clinic for CNS repair: current state of the art and the need for a Rosetta stone. Neuron. 2011;70:597–613. doi: 10.1016/j.neuron.2011.05.007. [DOI] [PubMed] [Google Scholar]
- Agrotis A, Ketteler R. A new age in functional genomics using CRISPR/Cas9 in arrayed library screening. Front Genet. 2015;6 doi: 10.3389/fgene.2015.00300. 1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Láhar J, Kryukov GV, Sonkin D, Reddy A, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–307. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond AM, Ming G-L, Song H. Adult mammalian neural stem cells and neurogenesis: five decades later. Cell Stem Cell. 2015;17:385–395. doi: 10.1016/j.stem.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CW, Verhaak RGW, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, Beroukhim R, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PJ, Müller S. Open access chemical probes for epigenetic targets. Future Med Chem. 2015;7:1901–1917. doi: 10.4155/fmc.15.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caie PD, Walls RE, Ingleston-Orme A, Daya S, Houslay T, Eagle R, Roberts ME, Carragher NO. High-content phenotypic profiling of drug response signatures across distinct cancer cells. Mol Cancer Ther. 2010;9:1913–1926. doi: 10.1158/1535-7163.MCT-09-1148. [DOI] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carragher NO. Profiling distinct mechanisms of tumour invasion for drug discovery: imaging adhesion, signalling and matrix turnover. Clin Exp Metastasis. 2008;26:381–397. doi: 10.1007/s10585-008-9222-y. [DOI] [PubMed] [Google Scholar]
- Danovi D, Falk A, Humphreys P, Vickers R, Tinsley J, Smith AG, Pollard SM. Imaging-based chemical screens using normal and glioma-derived neural stem cells. Biochem Soc Trans. 2010;38:1067–1071. doi: 10.1042/BST0381067. [DOI] [PubMed] [Google Scholar]
- Danovi D, Folarin A, Gogolok S, Ender C, Elbatsh AMO, Engström PG, Stricker SH, Gagrica S, Georgian A, Yu D, U KP, et al. A high-content small molecule screen identifies sensitivity of glioblastoma stem cells to inhibition of polo-like kinase 1. PLoS ONE. 2013;8:e77053. doi: 10.1371/journal.pone.0077053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson JC, Carragher NO. Quantitative phenotypic and pathway profiling guides rational drug combination strategies. Front Pharmacol. 2014;5:118. doi: 10.3389/fphar.2014.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolmetsch R, Geschwind DH. The human brain in a dish: the promise of iPSC-derived neurons. Cell. 2011;145:831–834. doi: 10.1016/j.cell.2011.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewry DH, Willson TM, Zuercher WJ. Seeding collaborations to advance kinase science with the GSK Published Kinase Inhibitor Set (PKIS) Curr Top Med Chem. 2014;14:340–342. doi: 10.2174/1568026613666131127160819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkabetz Y, Panagiotakos G, Al Shamy G, Socci ND, Tabar V, Studer L. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008;22:152–165. doi: 10.1101/gad.1616208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins JM, Fedele V, Szklarz M, Abdul Azeez KR, Salah E, Mikolajczyk J, Romanov S, Sepetov N, Huang X-P, Roth BL, Zen H, et al. Comprehensive characterization of the Published Kinase Inhibitor Set. Nat Biotechnol. 2015;34:95–103. doi: 10.1038/nbt.3374. [DOI] [PubMed] [Google Scholar]
- Feng Y, Mitchison TJ, Bender A, Young DW, Tallarico JA. Guide to drug discovery: multi-parameter phenotypic profiling: using cellular effects to characterize small-molecule compounds. Nat Rev Drug Discov. 2009;8:1–12. doi: 10.1038/nrd2876. [DOI] [PubMed] [Google Scholar]
- Florio M, Albert M, Taverna E, Namba T, Brandl H, Lewitus E, Haffner C, Sykes A, Wong FK, Peters J, Guhr E, et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science. 2015;347:1465–1470. doi: 10.1126/science.aaa1975. [DOI] [PubMed] [Google Scholar]
- Funato K, Major T, Lewis PW, Allis CD, Tabar V. Use of human embryonic stem cells to model pediatric gliomas with H3.3K27M histone mutation. Science. 2014;346:1529–1533. doi: 10.1126/science.1253799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage FH, Temple S. Neural stem cells: generating and regenerating the brain. Neuron. 2013;80:588–601. doi: 10.1016/j.neuron.2013.10.037. [DOI] [PubMed] [Google Scholar]
- Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, DiMeco F, Vescovi A. Isolation and characterization of tumorigenic, stemlike neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- Gallo M, Ho J, Coutinho FJ, Vanner R, Lee L, Head R, Ling EKM, Clarke ID, Dirks PB. A tumorigenic MLL-Homeobox network in human glioblastoma stem cells. Cancer Res. 2013;73:417–427. doi: 10.1158/0008-5472.CAN-12-1881. [DOI] [PubMed] [Google Scholar]
- Gangemi RMR, Griffero F, Marubbi D, Perera M, Capra MC, Malatesta P, Ravetti GL, Zona GL, Daga A, Corte G. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells. 2009;27:40–48. doi: 10.1634/stemcells.2008-0493. [DOI] [PubMed] [Google Scholar]
- Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J, Liu Q, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haibe-Kains B, El-Hachem N, Birkbak NJ, Jin AC, Beck AH, Aerts HJWL, Quackenbush J. Inconsistency in large pharmacogenomic studies. Nature. 2013;504:389–393. doi: 10.1038/nature12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Parker PRL, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, Mero P, et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- Hoelder S, Clarke PA, Workman P. Discovery of small molecule cancer drugs: successes, challenges and opportunities. Mol Oncol. 2012;6:155–176. doi: 10.1016/j.molonc.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert CG, Bradley RK, Ding Y, Toledo CM, Herman J, Skutt-Kakaria K, Girard EJ, Davison J, Berndt J, Corrin P, Hardcastle J, et al. Genome-wide RNAi screens in human brain tumor isolates reveal a novel viability requirement for PHF5A. Genes Dev. 2013;27:1032–1045. doi: 10.1101/gad.212548.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isherwood B, Timpson P, McGhee EJ, Anderson KI, Canel M, Serrels A, Brunton VG, Carragher NO. Live cell in vitro and in vivo imaging applications: accelerating drug discovery. Pharmaceutics. 2011;3:141–170. doi: 10.3390/pharmaceutics3020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen CM, Clemons PA, Wagner BK. Integrating phenotypic small-molecule profiling and human genetics: the next phase in drug discovery. Trends Genet. 2015;31:16–23. doi: 10.1016/j.tig.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MB, Wang PP, Atabay KD, Murphy EA, Doan RN, Hecht JL, Walsh CA. Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nat Neurosci. 2015;18:637–646. doi: 10.1038/nn.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitambi SS, Toledo EM, Usoskin D, Wee S, Harisankar A, Svensson R, Sigmundsson K, Kalderén C, Niklasson M, Kundu S, Aranda S, et al. Vulnerability of glioblastoma cells to catastrophic vacuolization and death induced by a small molecule. Cell. 2014;157:313–328. doi: 10.1016/j.cell.2014.02.021. [DOI] [PubMed] [Google Scholar]
- Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CLL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Berg EL. Neoclassic drug discovery: the case for lead generation using phenotypic and functional approaches. J Biomol Screen. 2013;18:1143–1155. doi: 10.1177/1087057113506118. [DOI] [PubMed] [Google Scholar]
- Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, Park JK, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Ligon KL, Alberta JA, Kho AT, Weiss J, Kwaan MR, Nutt CL, Louis DN, Stiles CD, Rowitch DH. The oligodendroglial lineage marker OLIG2 is universally expressed in diffuse gliomas. J Neuropathol Exp Neurol. 2004;63:499–509. doi: 10.1093/jnen/63.5.499. [DOI] [PubMed] [Google Scholar]
- Ligon KL, Huillard E, Mehta S, Kesari S, Liu H, Alberta JA, Bachoo RM, Kane M, Louis DN, DePinho RA, Anderson DJ, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53:503–517. doi: 10.1016/j.neuron.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljosa V, Caie PD, ter Horst R, Sokolnicki KL, Jenkins EL, Daya S, Roberts ME, Jones TR, Singh S, Genovesio A, Clemons PA, et al. Comparison of methods for image-based profiling of cellular morphological responses to small-molecule treatment. J Biomol Screen. 2013;18:1321–1329. doi: 10.1177/1087057113503553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell. 2011;146:18–36. doi: 10.1016/j.cell.2011.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JH, Nowakowski TJ, Pollen AA, Javaherian A, Kriegstein AR, Oldham MC. Radial glia require PDGFD-PDGFRβ signalling in human but not mouse neocortex. Nature. 2014;515:264–268. doi: 10.1038/nature13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei F, Fancy SPJ, Shen Y-AA, Niu J, Zhao C, Presley B, Miao E, Lee S, Mayoral SR, Redmond SA, Etxeberria A, et al. Micropillar arrays as a high-throughput screening platform for therapeutics in multiple sclerosis. Nat Med. 2014;20:954–960. doi: 10.1038/nm.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle FT, Tramontin AD, Garcia-Verdugo J-M, Alvarez-Buylla A. Radial glia give rise to adult neural stem cells in the subventricular zone. Proc Natl Acad Sci U S A. 2004;101:17528–17532. doi: 10.1073/pnas.0407893101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JA, Ding S-L, Sunkin SM, Smith KA, Ng L, Szafer A, Ebbert A, Riley ZL, Royall JJ, Aiona K, Arnold JM, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisan A, Lee Y-K, Zhang JD, Hudak CS, Meyer CA, Prummer M, Zoffmann S, Truong HH, Ebeling M, Kiialainen A, Gerard R, et al. White-to-brown metabolic conversion of human adipocytes by JAK inhibition. Nat Cell Biol. 2015;17:57–67. doi: 10.1038/ncb3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz DM, Cassiani PJ, Li L, Billy E, Korn JM, Jones MD, Golji J, Ruddy DA, Yu K, McAllister G, DeWeck A, et al. CRISPR screens provide a comprehensive assessment of cancer vulnerabilities but generate false-positive hits for highly amplified genomic regions. Cancer Discov. 2016;6:900–913. doi: 10.1158/2159-8290.CD-16-0178. [DOI] [PubMed] [Google Scholar]
- Neumann B, Held M, Liebel U, Erfle H, Rogers P, Pepperkok R, Ellenberg J. High-throughput RNAi screening by time-lapse imaging of live human cells. Nat Methods. 2006;3:385–390. doi: 10.1038/nmeth876. [DOI] [PubMed] [Google Scholar]
- Pastrana E, Silva-Vargas V, Doetsch F. Eyes wide open: a critical review of sphere-formation as an assay for stem cells. Stem Cells. 2011;8:486–498. doi: 10.1016/j.stem.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul SM, Mytelka DS, Dunwiddie CT, Persinger CC, Munos BH, Lindborg SR, Schacht AL. How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nat Rev Drug Discov. 2010:1–12. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- Perlman ZE, Slack MD, Feng Y, Mitchison TJ, Wu LF, Altschuler SJ. Multidimensional drug profiling by automated microscopy. Science. 2004;306:1194–1198. doi: 10.1126/science.1100709. [DOI] [PubMed] [Google Scholar]
- Pierce GB, Speers WC. Tumors as caricatures of the process of tissue renewal: prospects for therapy by directing differentiation. Cancer Res. 1988;48:1996–2004. [PubMed] [Google Scholar]
- Pilz G-A, Shitamukai A, Reillo I, Pacary E, Schwausch J, Stahl R, Ninkovic J, Snippert HJ, Clevers H, Godinho L, Guillemot F, et al. Amplification of progenitors in the mammalian telencephalon includes a new radial glial cell type. Nat Commun. 2013;4 doi: 10.1038/ncomms3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard SM, Yoshikawa K, Clarke ID, Danovi D, Stricker S, Russell R, Bayani J, Head R, Lee M, Bernstein M, Squire JA, et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4:568–580. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- Pourquié O, Bruneau B, Keller G, Smith A. Looking inwards: opening a window onto human development. Development. 2015;142:1–2. doi: 10.1242/dev.119727. [DOI] [PubMed] [Google Scholar]
- Reaume AG. Drug repurposing through nonhypothesis driven phenotypic screening. Drug Discov Today. 2012;8:85–88. doi: 10.1016/j.ddstr.2011.09.007. [DOI] [Google Scholar]
- Rowitch DH, Kriegstein AR. Developmental genetics of vertebrate glial-cell specification. Nature. 2010;468:214–222. doi: 10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- Sanai N, Tramontin AD, Quinones-Hinojosa A, Barbaro NM, Gupta N, Kunwar S, Lawton MT, McDermott MW, Parsa AT, Verdugo JM-G, Berger MS, et al. Unique astrocyte ribbon in adult human brain contains neural stem cells but lacks chain migration. Nature. 2004;427:740–744. doi: 10.1038/nature02301. [DOI] [PubMed] [Google Scholar]
- Sancho-Martinez I, Nivet E, Xia Y, Hishida T, Aguirre A, Ocampo A, Ma L, Morey R, Krause MN, Zembrzycki A, Ansorge O, et al. Establishment of human iPSC-based models for the study and targeting of glioma initiating cells. Nat Commun. 2016;7 doi: 10.1038/ncomms10743. 10743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell JW, Blanckley A, Boldon H, Warrington B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat Rev Drug Discov. 2012;11:1–10. doi: 10.1038/nrd3681. [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- Singh SK, Singh SK, Hawkins C, Hawkins C, Clarke ID, Clarke ID, Squire JA, Squire JA, Bayani J, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Smith K, Horvath P. Active learning strategies for phenotypic profiling of high-content screens. J Biomol Screen. 2014;19:685–695. doi: 10.1177/1087057114527313. [DOI] [PubMed] [Google Scholar]
- Staunton JE, Slonim DK, Coller HA, Tamayo P, Angelo MJ, Park J, Scherf U, Lee JK, Reinhold WO, Weinstein JN, Mesirov JP, et al. Chemosensitivity prediction by transcriptional profiling. Proc Natl Acad Sci U S A. 2001;98:10787–10792. doi: 10.1073/pnas.191368598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles CD, Stiles CD, Rowitch DH, Rowitch DH. Glioma stem cells: a midterm exam. Neuron. 2008;58:832–846. doi: 10.1016/j.neuron.2008.05.031. [DOI] [PubMed] [Google Scholar]
- Stransky N, Ghandi M, Kryukov GV, Garraway LA, Lehár J, Liu M, Sonkin D, Kauffmann A, Venkatesan K, Edelman EJ, Riester M, et al. Pharmacogenomic agreement between two cancer cell line data sets. Nature. 2015:1–15. doi: 10.1038/nature15736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm D, Witt H, Hovestadt V, Khuong-Quang D-A, Jones DTW, Konermann C, Pfaff E, Tönjes M, Sill M, Bender S, Kool M, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- Sunkin SM, Ng L, Lau C, Dolbeare T, Gilbert TL, Thompson CL, Hawrylycz M, Dang C. Allen brain atlas: an integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res. 2013;41:D996–D1008. doi: 10.1093/nar/gks1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Bateman R, Rauh D, Vaisberg E, Ramachandani S, Zhang C, Hansen KC, Burlingame AL, Trautman JK, Shokat KM, Adams CL. An unbiased cell morphology-based screen for new, biologically active small molecules. PLoS Biol. 2005a;3:e128–13. doi: 10.1371/journal.pbio.0030128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Bateman R, Rauh D, Vaisberg E, Ramachandani S, Zhang C, Hansen KC, Burlingame AL, Trautman JK, Shokat KM, Adams CL. An unbiased cell morphology-based screen for new, biologically active small molecules. PLoS Biol. 2005b;3:e128–13. doi: 10.1371/journal.pbio.0030128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanikawa T, Fridman M, Zhu W, Faulk B, Joseph IC, Kahne D, Wagner BK, Clemons PA. Using biological performance similarity to inform disaccharide library design. J Am Chem Soc. 2009;131:5075–5083. doi: 10.1021/ja806583y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna E, Götz M, Huttner WB. The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annu Rev Cell Dev Biol. 2014;30:465–502. doi: 10.1146/annurev-cellbio-101011-155801. [DOI] [PubMed] [Google Scholar]
- Taylor DL, Woo ES, Giuliano KA. Real-time molecular and cellular analysis: the new frontier of drug discovery. Curr Opin Biotechnol. 2001;12:75–81. doi: 10.1016/s0958-1669(00)00180-4. [DOI] [PubMed] [Google Scholar]
- Toledo CM, Ding Y, Hoellerbauer P, Davis RJ, Basom R, Girard EJ, Lee E, Corrin P, Hart T, Bolouri H, Davison J, et al. Genome-wide CRISPR-Cas9 screens reveal loss of redundancy between PKMYT1 and WEE1 in glioblastoma stem-like cells. Cell Rep. 2015;13:2425–2439. doi: 10.1016/j.celrep.2015.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verginelli F, Perin A, Dali R, Fung KH, Lo R, Longatti P, Guiot M-C, Del Maestro RF, Rossi S, di Porzio U, Stechishin O, et al. Transcription factors FOXG1 and Groucho/TLE promote glioblastoma growth. Nat Commun. 2013;4:1–16. doi: 10.1038/ncomms3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner BK, Clemons PA. Connecting synthetic chemistry decisions to cell and genome biology using small-molecule phenotypic profiling. Curr Opin Chem Biol. 2009;13:539–548. doi: 10.1016/j.cbpa.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawer MJ, Li K, Gustafsdottir SM, Ljosa V, Bodycombe NE, Marton MA, Sokolnicki KL, Bray M-A, Kemp MM, Winchester E, Taylor B, et al. Toward performance-diverse small-molecule libraries for cell-based phenotypic screening using multiplexed high-dimensional profiling. PNAS. 2014;111:10911–10916. doi: 10.1073/pnas.1410933111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel C, Riefke B, Gründemann S, Krebs A, Christian S, Prinz F, Osterland M, Golfier S, Räse S, Ansari N, Esner M, et al. 3D high-content screening for the identification of compounds that target cells in dormant tumor spheroid regions. Exp Cell Res. 2014;323:131–143. doi: 10.1016/j.yexcr.2014.01.017. [DOI] [PubMed] [Google Scholar]
- Wurdak H, Zhu S, Romero A, Lorger M, Watson J, Chiang CY, Zhang J, Natu VS, Lairson LL, Walker JR, Trussell CM, et al. An RNAi screen identifies TRRAP as a regulator of brain tumor-initiating cell differentiation. Cell Stem Cell. 2010;6:37–47. doi: 10.1016/j.stem.2009.11.002. [DOI] [PubMed] [Google Scholar]
- Xie Y, Bergström T, Jiang Y, Johansson P, Marinescu VD, Lindberg N, Segerman A, Wicher G, Niklasson M, Baskaran S, Sreedharan S, et al. The human glioblastoma cell culture resource: validated cell models representing all molecular subtypes. EBioMedicine. 2015;2:1351–1363. doi: 10.1016/j.ebiom.2015.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarrow JC, Feng Y, Perlman ZE, Kirchhausen T, Mitchison TJ. Phenotypic screening of small molecule libraries by high throughput cell imaging. Comb Chem High Throughput Screen. 2003;6:279–286. doi: 10.2174/138620703106298527. [DOI] [PubMed] [Google Scholar]
- Young DW, Bender A, Hoyt J, McWhinnie E, Chirn G-W, Tao CY, Tallarico JA, Labow M, Jenkins JL, Mitchison TJ, Feng Y. Integrating high-content screening and ligand-target prediction to identify mechanism of action. Nat Chem Biol. 2007;4:59–68. doi: 10.1038/nchembio.2007.53. [DOI] [PubMed] [Google Scholar]