Abstract
Focal adhesion kinase (FAK) mediates tumor cell–intrinsic behaviors that promote tumor growth and metastasis. We previously showed that FAK also induces the expression of inflammatory genes that inhibit antitumor immunity in the microenvironment. Here, we identified a crucial, previously unknown role for the dual-function cytokine IL-33 in FAK-dependent immune evasion. In murine squamous cell carcinoma (SCC) cells, specifically nuclear FAK enhanced the expression of the genes encoding IL-33, the chemokine CCL5, and the soluble, secreted form of the IL-33 receptor sST2. The abundance of IL-33 and CCL5 was increased in FAK-positive SCC cells but not in normal keratinocytes. IL-33 associated with FAK in the nucleus, and the FAK–IL-33 complex interacted with a network of chromatin modifiers and transcriptional regulators, including TAF9, WDR82 and BRD4, which promote the activity of nuclear factor κB (NF-κB) and its induction of genes encoding chemokines, including CCL5. We did not detect secretion of IL-33 from FAK-positive SCC cells; thus, we propose that the increased production and secretion of sST2 likely sequesters IL-33 secreted by other cell types within the tumor environment, thus blocking its stimulatory effects on infiltrating host immune cells. Depleting FAK, IL-33, or sST2 from SCC cells before implantation induced tumor regression in syngeneic mice, except when CD8+ T cells were co-depleted. Our data provide mechanistic insight into how FAK controls the tumour immune environment, namely through a transcriptional regulatory network mediated by nuclear IL-33. Targeting this axis may boost antitumor immunity in patients.
Introduction
Reprogramming the immuno-suppressive tumor environment to promote anti-tumor immunity is a major objective of immuno-modulatory therapies currently in clinical use or development. Cancer cells contribute to orchestrating the composition of this environment through driving enrichment of immune cell populations with intrinsic immuno-suppressive function, thereby evading the anti-tumor activity of cytotoxic CD8 T-cells. Identification and characterization of key molecular pathways that regulate cancer cell expression of immune modulators, such as chemokines and cytokines, may therefore provide new therapeutic strategies for use in combination immunotherapy.
Focal Adhesion Kinase (FAK) is a non-receptor tyrosine kinase that signals downstream of integrins and growth factor receptors to control the malignant phenotype in multiple ways, including by regulating adhesion, migration, proliferation, and survival (1). FAK is frequently increased in abundance in human cancers (2–4), and contributes to skin, mammary, intestinal and prostate tumorigenesis in mouse models (5–8). A number of small-molecule FAK kinase inhibitors are now in early-phase clinical trials. In addition to its role at the plasma membrane, FAK can also translocate to the nucleus where it can regulate gene expression (9–11). In a mouse model of skin squamous cell carcinoma (SCC) (12), we demonstrated that nuclear FAK controls expression of chemokines and cytokines, including Ccl5 and Tgfb2, that drive increased numbers of regulatory T (Treg) cells in the tumor environment, resulting in protection from an anti-tumor CD8+ T-cell response (9).
IL33 is a member of the IL1 family of cytokines and is secreted by necrotic epithelial cells and activated innate immune cells, or held within the cell as a nuclear factor. Secreted IL33 binds to its cognate receptor, a heterodimeric complex comprised of ST2L (IL1RL1) and IL-1 receptor accessory protein (IL-1RAcP), to initiate activation of mitogen-activated protein kinase (MAPK) and nuclear factor -κB (NF-κB) (13), wherein IL33 has potent pro-inflammatory functions and is considered a “danger” signal. IL33 released into the tumor environment has been suggested to both inhibit (14) and promote tumor formation (15), indicating that IL33’s roles in cancer development and progression are unclear. IL33 can also localize to the nucleus, where it either activates or represses transcription through association with the transcription factor NF-κB or the NF-κB p65 promoter, respectively (16, 17). The precise mechanisms that underpin IL33’s regulation of transcription are not well understood.
ST2, a component of the IL33 receptor complex, can exist as a functional transmembrane receptor (ST2L) or as a shorter secreted decoy receptor (sST2) (13). ST2L is present on the cell surface of several hematopoietic cells, including T cells, macrophages, neutrophils and MDSCs, and activation of downstream signaling can alter cytokine production or immunosuppressive capacity (14, 18). ST2L of host immune cells is required for tumor clearance; hence, it has been linked to the anti-tumor properties of IL33 (14). Conversely, soluble ST2 (sST2) is proposed to function as a decoy receptor for IL33, suppressing its potent pro-inflammatory functions, and high serum levels of sST2 have been correlated with poor prognosis in estrogen receptor (ER)-positive breast cancer (19). Like IL33, the precise role of sST2 in cancer remains somewhat controversial.
Here we found that nuclear FAK critical for the expression of IL33 and ST2 in cancer cells. IL33 was restricted to the nucleus in murine SCC cells, where it acts downstream of FAK to promote Ccl5 expression and tumor growth. Mechanistic protein network analyses suggested that IL33 regulates gene expression by interacting with chromatin modifiers and transcriptional regulators. ST2 was secreted by SCC cells, and it suppressed CD8+ T cell-mediated tumor clearance. Our findings reveal new insight into the molecular mechanisms by which nuclear FAK regulates chemokine expression, placing nuclear IL33 at the heart of a complex transcriptional network that specifies the anti-tumor immune response.
Results
Nuclear FAK regulates expression of IL33 and its receptor ST2
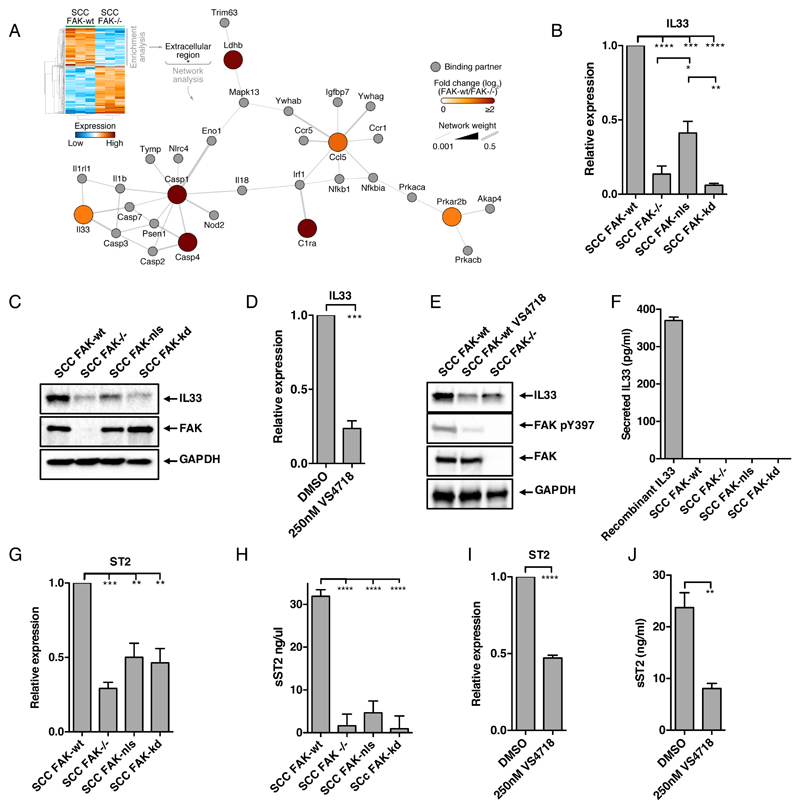
We have previously reported that nuclear FAK regulates the expression of chemokines, including Ccl5, and this is important for driving infiltration of regulatory T (Treg) cells into murine SCC tumors, enabling them to evade the anti-tumor immune response (9). To explore the mechanisms of FAK-dependent chemokine regulation, we analysed Affymetrix microarray data comparing murine FAK-/- SCC cells, with those re-expressing FAK-wt (herein referred to as FAK-wt) to identify genes that are regulated by FAK. In the set of genes significantly downregulated after FAK depletion, the only significantly enriched gene ontology term was “extracellular region” (p = 0.049). Using the genes contained within this category, we generated a protein interaction network based on direct physical interactions. The largest connected network was found to contain Ccl5 and the gene encoding the cytokine IL33 (Fig.1A). Given the link between IL33 and the regulation of gene expression (16, 17), we investigated whether, and if so how, IL33 contributed to FAK-dependent transcription of chemokines.
Figure 1. Nuclear FAK regulates expression of IL33 and its receptor ST2.
(A) Gene ontology enrichment analysis (cellular component terms) on the significantly down-regulated set of genes in the FAK-/- SCC transcriptome relative to the wild-type (wt) (predicated false positives (pfp)< 0.05). Genes annotated with the overrepresented term (extracellular region; Benjamini–Hochberg-corrected hypergeometric test) were used to seed a protein interaction network based on direct physical interactions (grey lines). Color of each node (circle) is proportional to the log-transformed fold change in gene expression. The largest connected graph component is displayed. (B and C) Abundance of IL33 at the mRNA level (B; by qRT-PCR) and protein level (C; by Western blot) in FAK-wt, FAK-/-, FAK-nls, and FAK-kd SCC cells. (D and E) Abundance of IL33 mRNA (D; by qRT-PCR) and IL33 protein (E; by Western blot) in FAK-wt SCC cells treated with control (DMSO) or VS4718 (250 nM, 24 hours). Western blot additionally assessed in FAK-/- SCC cells for reference. (F) Analysis of ELISA for IL33 in conditioned media from FAK-wt, FAK-/-, FAK-nls, and FAK-kd SCC cells. (G) qRT-PCR analysis of ST2 expression in FAK-wt, FAK-/-, FAK-nls, and FAK-kd SCC cells. (H) Analysis of ELISA for sST2 in conditioned media from FAK-wt, FAK-/-, FAK-nls, and FAK-kd SCC cells. (I) qRT-PCR analysis of ST2 expression in FAK-wt SCC cells treated with control (DMSO) or VS4718 (250 nM, 24 hours). (J) Analysis of ELISA for sST2 in conditioned media from FAK-wt SCC cells treated with control (DMSO) or VS4718 (250 nM, 24 hours). Data are mean ± S.E.M. n=3 experiments. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001; by Tukey corrected one-way ANOVA (B, G and H) or two-tailed unpaired t-test (D, I, and J).
We first used q(RT)-PCR to compare IL33 expression in SCC cells expressing FAK-wt, FAK-/-, FAK-nls (a mutant that is largely excluded from the nucleus), and FAK-kd (a kinase deficient mutant), and found that regulation of IL33 mRNA was dependent on both FAK kinase activity and its nuclear localisation (Fig. 1B). Western blotting for IL33 abundance in whole cell lysates revealed similar findings at the protein level (Fig. 1C and fig. S1A). We have shown previously that mutation of the nuclear localization signal in FAK does not completely abolish FAK nuclear localization. We believe this explains why we observed slightly increased abundance of IL33 in cells expressing this mutant when compared to FAK-/- or FAK-kd SCC cells. Similarly, analysis of SCC cells expressing FAK-Y397F, an autophosphorylation-defective mutant of FAK that is kinase deficient, showed reduced IL33 expression similar to that observed in FAK-/- SCC cells (fig. S1B). Treatment of FAK-wt cells with the FAK catalytic inhibitor VS4718 inhibited IL33 abundance on both mRNA and protein levels (Fig. 1, D and E), even with a low (50 nM) concentration of VS4718 (fig. S1, C and D). IL33 can function both as a nuclear cytokine and a secreted alarmin (20). Using both an anti-IL33 ELISA (Fig. 1F) and Western blotting (fig. S1E), we could not detect IL33 in SCC cell-conditioned media, implying that IL33 predominantly functions as a nuclear cytokine in SCC cells.
As an extracellular cytokine, IL33 mediates signalling by binding to the IL-33R complex, comprised of ST2 (also termed interleukin 1 receptor-like 1, IL1RL1), and the IL-1 receptor accessory protein (IL-1RAcP) (21). Furthermore, both IL33 signalling, and nuclear IL33 have been shown to regulate ST2 expression (22, 23). The protein encoded by ST2 exists in two forms: (i) as ST2L, a membrane-anchored receptor that activates downstream signalling upon IL33 engagement, or (ii) as sST2, a secreted soluble decoy receptor that inhibits IL33 signalling (13). Using q(RT)-PCR with a primer set that would detect cumulative amounts of mRNA encoding both ST2L and sST2, we found that ST2 expression was greater in FAK-wt cells when compared to FAK-/-, FAK-nls, or FAK-kd cells (Fig. 1G). Thus, FAK’s regulation of ST2 expression is also dependent on both nuclear localization and kinase activity. Using an anti-ST2 ELISA, we found abundant secreted amounts of sST2 in FAK-wt cell-conditioned medium, and this was reduced in media conditioned by FAK-/-, FAK-nls, and FAK-kd cells (Fig. 1H). However, flow cytometry analysis did not detect the presence of ST2L on the surface of FAK-wt and FAK-/- SCC cells (fig. S1F), indicating that sST2 is the predominantly produced isoform upon ST2 induction in SCC cells. Treatment with the FAK inhibitor VS4718 reduced its expression and secretion (Fig. 1, I and J). Collectively, these results indicate that the kinase activity of FAK in the nucleus is a key regulator of the abundance of both nuclear IL33 and sST2, thereby influencing both the nuclear and extracellular/alarmin functions of IL33 signaling.
We next investigated the mechanism by which FAK controls IL33 expression and sST2 abundance in SCC cells. Using an experimentally derived nuclear FAK interactome from FAK-wt cells (9), we used Ingenuity Pathway Analysis (IPA) to identify direct experimentally observed relationships between nuclear FAK-interacting proteins and transcription factors that regulate the expression of IL33 and ST2 (obtained from Qiagen’s DECODE database) (fig. S1G). This identified associations with several IL33- and ST2-regulatory transcription factors. We noticed that 4 of these associations, RUNX1, SP1, NCOA2 and NR3C1, were linked to transcription factors associated with regulating the expression of both IL33 and ST2. We know that FAK associates with SP1 (confirmed in fig. S1, H and I) and RUNX1 (24) and is involved in regulating RUNX1-containing protein complexes, posttranslational modification, and ultimately transcription factor function. Small interfering RNA (siRNA)-mediated depletion of both RUNX1 and SP1 suggested that these transcription factors acted together to regulate IL33 abundance (fig. S1, J and K). The precise details of their coordinated activities require further investigation, but nonetheless our findings identify several connections between FAK-interacting proteins and transcription factors that regulate IL33 and ST2.
IL33 is required for FAK-dependent expression of a subset of chemokines
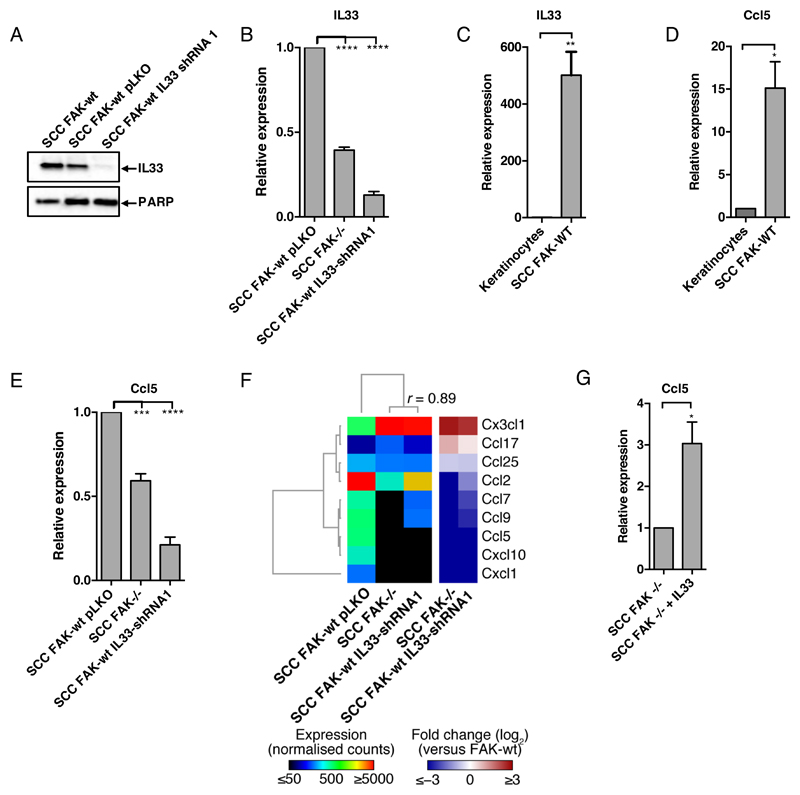
Nuclear IL33 has been linked to regulation of gene expression in several model systems (16, 17). To determine whether nuclear IL33 was required for FAK-dependent chemokine expression, we depleted IL33 in FAK-wt SCC cells using both short hairpin RNA (shRNA) (Fig. 2, A and B, and fig. S2A) and CRISPR/Cas9 (fig. S2B). IL33 has previously been linked to the regulation of Ccl5 (23), and we have shown that FAK-dependent Ccl5 expression regulates the anti-tumor immune response (9). We found that IL33 and Ccl5 expression was increased in SCC cells when compared to primary keratinocytes (Fig. 2, C and D), correlating with nuclear FAK in SCC cells, and that IL33 was required for Ccl5 expression in FAK-wt SCC cells (Fig. 2E and fig. S2C). A previous study in endothelial cells (23) demonstrated that IL33 silencing increased sST2 expression, here we found that IL33 silencing reduced sST2 abundance (fig. S2D). Although we have used different readouts (mRNA vs protein), it is likely that the regulation of ST2 by IL33 may be context dependent. To investigate further the requirement for IL33 in FAK-dependent chemokine expression, we performed Nanostring PanCancer Immune profiling of FAK-wt/pLKO, FAK-/-, and FAK-wt/IL33-shRNA cells. Hierarchical clustering of log-transformed fold changes, relative to control cells, identified a subset of chemokines co-regulated by FAK and IL33 (Fig. 2F), including CCL5. Furthermore, overexpression of IL33 in FAK-/- SCC cells increased Ccl5 expression (Fig. 2G), suggesting IL33 is sufficient to promote Ccl5 expression. Therefore, FAK regulates IL33, which in turn can mediate FAK-dependent chemokine expression.
Figure 2. Nuclear FAK and IL33 cooperate to regulate chemokine expression.
(A) Representative Western blot of IL33 abundance in FAK-wt, FAK-wt pLKO, and FAK-wt IL33 shRNA1 SCC cells. (B) qRT-PCR analysis of IL33 expression in FAK-wt pLKO, FAK-/-, and FAK-wt IL33-shRNA1 SCC cells. (C and D) qRT-PCR analysis of IL33 (C) and Ccl5 (D) expression in primary keratinocytes and FAK-wt SCC cells. (E) qRT-PCR analysis of Ccl5 expression in FAK-wt pLKO, FAK-/-, and FAK-wt IL33-shRNA1 cells. (F) NanoString analysis of chemokine expression in FAK-wt pLKO, FAK-/- and FAK-wt IL33-shRNA1 SCC cells. Log10-transformed expression levels for chemokines with at least 50 counts were hierarchically clustered and displayed as a heatmap. Pearson correlation coefficient between expression profiles in FAK-/- and FAK-wt IL33-shRNA1 SCC cells is shown. Log2-transformed fold changes (over SCC FAK-wt pLKO) are also displayed. (G) qRT-PCR analysis of Ccl5 expression in SCC FAK-/- and FAK-/-+IL33 cells. Data are mean ± S.E.M. n = 3 for all experiments. *p≤ 0.05, **p ≤ 0.01,***p ≤ 0.001, ****p ≤ 0.0001; by Tukey corrected one-way ANOVA (B and E) or two-tailed unpaired t-test (C, D and G).
IL33/ST2 axis supports tumor growth by suppressing the immune response
IL33 is expressed by several cell types within the tumour environment, and secreted IL33 can have both pro and anti-tumour effects (14, 15). ST2L, the IL33 receptor, is present on various immune cells, including Treg cells, macrophages, and CD8+ T cells, and activation of ST2L signalling can alter their phenotype and/or function. For example, ST2L-positive Treg cells exhibit a more potent immunosuppressive function than ST2L-negative Treg cells (25), implying that IL33-ST2L signalling could enhance the suppressive activity of Treg cells, thereby promoting tumor growth. In contrast, activation of ST2L on cytotoxic CD8+ T cells enhances the cells’ cytotoxic function (18), resulting in an improved anti-tumor immune response. Therefore, it is possible that FAK-dependent regulation of the decoy receptor sST2 could have pro- or anti-tumor effects.
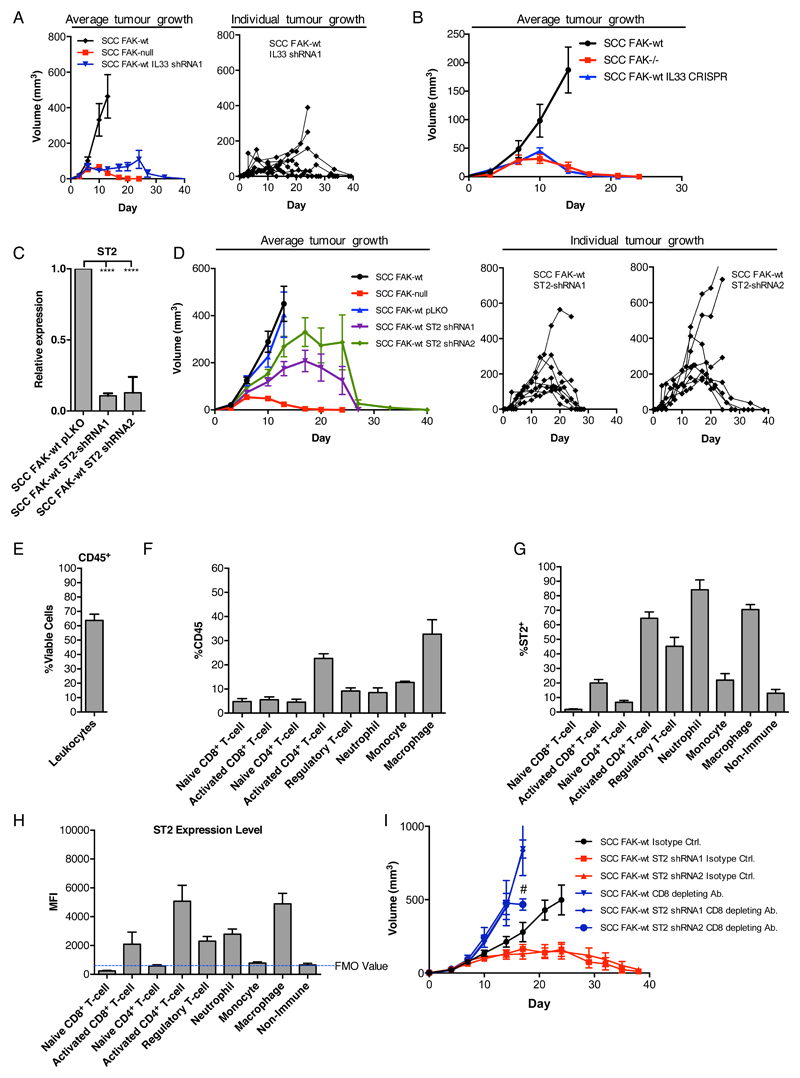
Having established that nuclear IL33 regulates chemokine expression, including Ccl5, we next assessed the effects of IL33 depletion on SCC tumor growth. Therefore, 1 x 106 FAK-wt, FAK-/-, and FAK-wt/IL33-shRNA1 cells were injected subcutaneously into FVB mice (the syngeneic host strain), and tumor growth was monitored. FAK-wt tumors exhibited exponential growth until they reached defined endpoints by which time the mice had to be sacrificed (see Methods section). In contrast, FAK-/- SCC tumours grew until approximately day 7, after which they stalled and underwent complete regression (Fig. 3A, left graph), as we have also reported previously (9). IL33 depletion from FAK-wt cells resulted in 6 out of 8 tumors exhibiting a period of growth followed by complete regression, in a similar manner to FAK-/- tumors, whereas 2 out of 8 tumours showed a growth delay (Fig. 3A). Similar studies using CRISPR/Cas9 to deplete IL33 from FAK-wt cells confirmed the requirement for IL33 in supporting SCC tumor growth (Fig. 3B). Thus, IL33 is required to support tumor growth and permit immune evasion, likely by regulating transcription of vital chemokines, including Ccl5.
Figure 3. IL33 and sST2 support SCC tumour growth through suppressing the anti-tumour immune response.
(A and B) Representative growth of FAK-wt, FAK-/-, and FAK-wt IL33-shRNA1 (A) or FAK-wt IL33-CRISPR (B) SCC tumors after orthotopic subcutaneous implantation. (C) qRT-PCR analysis of ST2 expression in FAK-wt pLKO, FAK-wt ST2-shRNA1, and FAK-wt ST2-shRNA2 SCC cells. (D) Growth of FAK-wt ST2-shRNA (pools 1 and 2) SCC tumors after orthotopic subcutaneous implantation. (E) CD45+ cells as a percentage of live cells isolated from FAK-wt SCC tumors 12 days after implantation. (F) Abundance of different immune cell populations as a percentage of CD45+ cells. Marker sets used to identify cell populations are listed in fig. S3. (G) Percentage ST2+ cells in different immune cell populations. (H) Mean fluorescent intensity of ST2 expression in different immune cell populations. (I) Representative growth of FAK-wt, FAK-wt ST2-shRNA1, and FAK-wt ST2-shRNA2 SCC tumors receiving treatment with either a CD8-depleting antibody or isotype control antibody. Statistics in C, ****p ≤ 0.0001 by Tukey corrected one-way ANOVA. Data are mean ± S.E.M, n = 3 for q(RT)-PCR, n = 6-8 tumors.
To next probe the role of sST2 in tumor growth, we generated sST2-depleted FAK-wt SCC cell lines using shRNA (Fig. 3C) and injected 1 x 106 FAK-wt, FAK-wt/pLKO (shRNA control), FAK-wt/ST2-shRNA1 and FAK-wt/ST2-shRNA2 SCC cells into syngeneic FVB mice. We found that most tumours (6 out of 8 for shRNA1 and 5 out of 8 for shRNA2) exhibited a period of growth followed by complete regression (Fig. 3D), like FAK-/- tumors, albeit with different kinetics. To address how sST2 depletion might result in tumor regression, we profiled the immune cells in FAK-wt tumors to determine the quantity of ST2L on the cell surface. We found that after 12 days of growth, FAK-wt tumors have an extensive immune cell infiltrate accounting for approximately 60% of the viable cell population (Fig. 3, E and F). Using a range of surface and intracellular markers (table S1), we identified activated CD8+ T cells, activated CD4+ T-cells, Treg cells, neutrophils, and macrophages as the major immune cell populations that displayed surface-bound ST2L (Fig. 3, G and H). Because IL33 engagement with ST2L expressed on activated CD8+ T cells can enhance cytotoxic function and drive increased expression of the effector cytokine interferon-gamma (IFNγ) (14), we hypothesized that tumor regression in response to depletion of sST2 from SCC cancer cells was likely CD8+ T-cell dependent. To test this, we used CD8-depleting antibodies and found that depletion of CD8+ T cells was sufficient to completely rescue the growth of FAK-wt/ST2-shRNA SCC tumors (Fig. 3I), implying that sST2 from the tumor cells plays an important role in supressing CD8+ T cell-mediated anti-tumour immunity. Consistent with previous results (9), we also observed enhanced growth of FAK-wt tumors upon depletion of CD8+ T cells, implying that even the FAK-wt cells remain under negative pressure from the CD8+ T-cell mediated immune response.
Nuclear IL33 interacts with an extensive network of transcriptional regulators
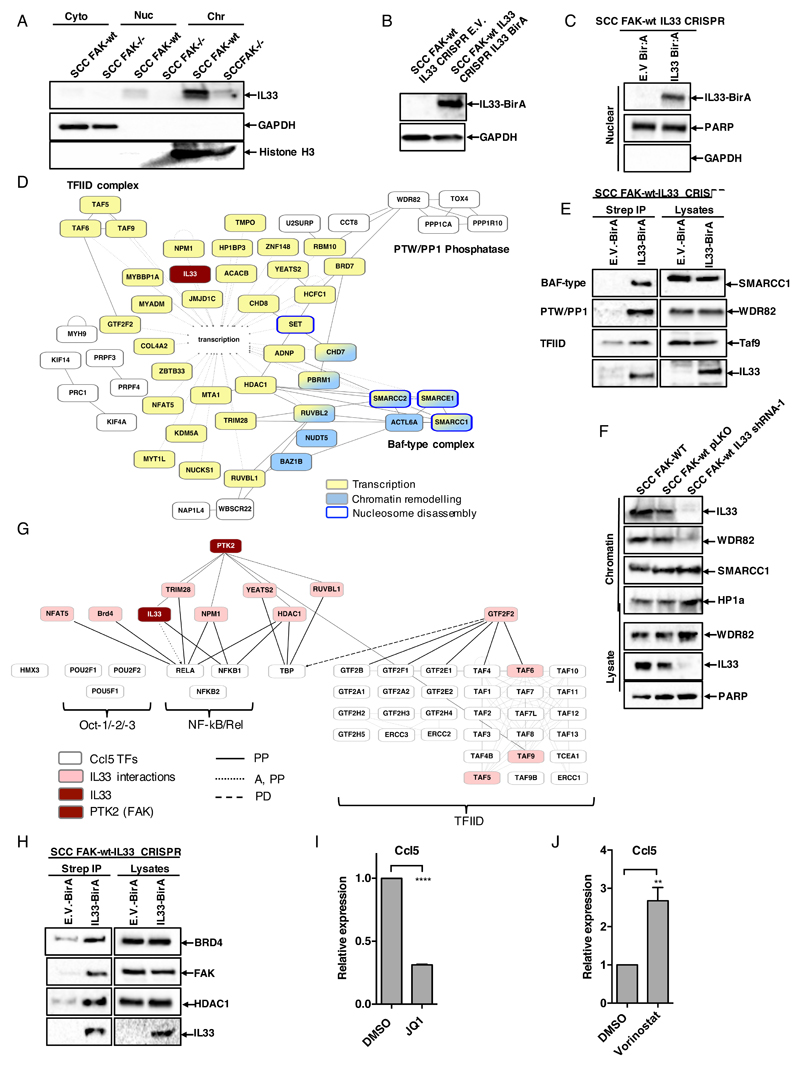
Having established an important role for nuclear IL33 in the regulation of chemokine expression and tumour growth, we addressed the molecular mechanisms that may underpin IL33’s regulation of chemokine transcription. Using cellular fractionation, we prepared cytoplasmic, nuclear, and chromatin extracts from FAK-wt and FAK-/- SCC cells as we have done before (9). Our results show that IL33 is largely chromatin-associated (Fig. 4A), in agreement with previous studies that identified IL33 as a histone H2A/2B binding protein (26). To investigate the functional significance of IL33 association with chromatin, we made use of a proteomics technique called “BioID” (27). Firstly, we generated an IL33 protein fused to the 35KDa E. coli biotin protein ligase BirA (IL33-BirA). Next, we expressed either IL33-BirA, or BirA alone, in FAK-wt cells from which endogenous IL33 was deleted (FAK-wt/IL33-CRISPR SCC cells; Fig. 4, B and C), and cultured these in the presence of biotin for 24 hours. This resulted in the biotinylation of proximal interacting proteins, enabling their purification and identification by mass spectrometry. To ensure identification of robust interactions, we applied stringent criteria, including that: (i) proteins must be present in all 3 biological replicates, (ii) proteins must have greater than 3-fold enrichment when compared to BirA only control, and (iii) fold enrichment must be statistically significant (p <0.05). With these criteria, we identified 105 proteins that associated with IL33. Gene ontology analysis of both biological and cellular processes (DAVID bioinformatics database) represented within the dataset identified significantly enriched terms associated with chromatin organisation and transcriptional regulation (fig. S3, A and B). Further analysis of the nuclear IL33 interactome using IPA identified a connected network of proteins that were implicated in regulation of transcription, chromatin remodelling and nucleosome disassembly (Fig. 4D). There were several members of the Baf-type complex (also known as SWI/SNF), the PTW/PP1 phosphatase complex, and the transcription factor TFIID complex, establishing a novel link between nuclear IL33 and the core transcriptional machinery. We extracted a complete list of genes belonging to these complexes from the gene ontology database AmiGO, and used IPA to reconstruct all three complexes based on known physical interactions (fig. S3, C to E). We next contextualised the BioID proteomics-informed IL33 nuclear interactome onto these networks (highlighted in pink in fig. S3, C to E), and identified potential interactions linking IL33 to key members of these complexes. Using streptavidin pulldown following incubation of IL33-BirA fusion protein expressing FAK-wt/IL33-CRISPR SCC cells in the presence of biotin, we confirmed the interactions of IL33 with WDR82, SMARCC1, and TAF9 (Fig. 4E). We found that IL33 depletion in FAK-wt cells resulted in loss of WDR82 from the chromatin fraction (Fig. 4F), implying that IL33 is required to stabilize the association of WDR82 with chromatin. We find this interesting because WDR82 is a key component of complexes associated with chromatin modification, such as the PTW/PP1 phosphatase complex (28) and the Set1A/Set1B histone H3-Lys4 (H3K4) methyltransferase complex (29). Therefore, IL33 likely plays a key role in scaffolding complexes required to modulate chromatin structure and permit transcription.
Figure 4. Nuclear IL33 interacts with an extensive network of transcriptional regulators.
(A) Representative Western blot of IL33 abundance in cytoplasmic (cyto), nuclear (Nuc), and chromatin (Chr) fractions from FAK-wt and FAK-/- SCC cells. (B) Representative Western blot of IL33 abundance in FAK-wt IL33-CRISPR/BirA-E.V. and FAK-wt IL33-CRISPR/IL33-BirA SCC cells. (C) Western blot analysis of lysates from nuclear fractionations of IL33-BirA fusion protein expression in FAK-wt IL33-CRISPR/E.V. or FAK-wt IL33-CRISPR/IL33-BirA SCC cells. (D) Functional interaction network analysis of the IL33 interactome. Direct physical interactions (solid grey lines) and functional association with transcription (dashed grey lines) are shown. Node style indicates association with additional relevant overrepresented functions (p<0.0001, Benjamini–Hochberg-corrected hypergeometric test). Components of the PTW/PP1 phosphatase, Baf-type and TFIID complexes are highlighted. (E) Western blot analysis of key network components using streptavidin pulldowns from biotinylated lysates of SCC FAK-wt IL33 CRISPR cells expressing either BirA empty vector or IL33-BirA fusion protein. (F) Representative Western blot of chromatin fractions and whole cell lysates from SCC FAK-wt, FAK-wt pLKO, and FAK-wt IL33 shRNA1 cells. (G) Interrogation of the IL33 BioID protein interaction network to identify potential upstream regulators of mCcl5 promoter associated transcription factors (taken from Qiagen ENCODE database). PP, protein-protein interaction; E, expression; PD, protein-DNA interaction; T, transactivation; A, activation. (H) Western blot analysis of IL33:BRD4, IL33:FAK, and IL33:HDAC1 associations using streptavidin pulldowns from biotinylated lysates of FAK-wt IL33-CRISPR SCC cells expressing either BirA empty vector or IL33-BirA fusion protein. (I) qRT-PCR analysis of Ccl5 expression in SCC FAK-wt cells treated with DMSO or JQ1 (200nM for 48 hours). (J) q(RT)-PCR analysis of Ccl5 expression in FAK-wt SCC cells treated with DMSO or vorinostat (10μM for 24 hours). Statistics in I and J, ****p ≤ 0.0001, ***p ≤ 0.001, ** ≤ 0.01. Two-tailed unpaired t-test. n = 3 for all experiments.
IL33 interacts with and enhances regulators of Ccl5 expression
As mentioned above, we previously showed that CCL5 secreted by FAK-wt SCC cells drives the infiltration of immuno-suppressive regulatory Treg cells into SCC tumors, shifting the CD8+ T-cell:Treg ratio in favor of tumor tolerance (9). Thus far, we have found that IL33 is both necessary and sufficient to drive Ccl5 expression downstream of FAK (Fig. 2, E and G). To define how IL33 contributes to regulation of Ccl5 expression we mapped the nuclear IL33 interactome onto a network of proteins associated with predicted Ccl5 transcription factors (Fig. 4G). This revealed that IL33 interacts with transcription factors and transcriptional regulators that impact on expression of Ccl5. Of note, three factors belonging to the transcription factor IID complex (TFIID) were found to interact with nuclear IL33, including TAF9, a protein we found previously to form complexes with FAK (9). To explore the connection between FAK, IL33, and Ccl5 transcription, we mapped the nuclear FAK interactome onto the set of Ccl5 regulatory proteins found to interact with nuclear IL33 (Fig. 4G). This identified proteins in common between the nuclear FAK and nuclear IL33 interactomes, suggesting that these proteins may be part of the same molecular complexes that regulate Ccl5 expression. Using streptavidin-coated beads, we isolated the IL33 BioID fusion protein, and confirmed that FAK and IL33 exist in complex under steady state conditions (Fig. 4H). Hence, we conclude that FAK binds to IL33, and together they complex with key Ccl5-regulatory transcription factors, to co-regulate chemokine gene expression.
Besides TFIID, many of the IL33 interacting partners converge on regulators of NF-κB, suggesting that NF-κB may be central to FAK and IL33s regulation of chemokine expression. Of note, we showed that IL33 interacts with the bromodomain protein BRD4, and the histone deacetylase, HDAC1 (Fig. 4H). BRD4 is a member of the bromodomain and extraterminal domain (BET) family of transcriptional coactivators and elongation factors that recruit chromatin remodelling factors, including the SWI/SNF complex (30), to the promoters of genes via recognition of polyacetylated histone tails (31). Because IL33 binds to both BRD4, and members of the Baf-type (SWI/SNF) complex, we hypothesize that IL33 plays a role in formation of this complex at actively transcribing genes. It is known that BRD4 directly binds to acetylated p65 NF-κB, which leads to enhanced NF-κB transactivation activity (32), together suggesting a role for BRD4 in inflammatory transcriptional signalling. To support these conclusions, we treated FAK-wt SCC cells with the BET family inhibitor JQ1. This resulted in reduced Ccl5 expression (Fig. 4I), implying a role for BET family proteins in the regulation of chemokine expression. In contrast, HDAC1 has been shown to negatively regulate NF-κB transcriptional activity via a direct interaction with p65 (RelA) (33). We used the HDAC inhibitor vorinostat, and identified a clear induction of Ccl5 expression upon HDAC inhibition (Fig. 4J). Collectively, our data support a model whereby FAK binds to IL33, which is a central component of a network of transcriptional regulators associated with the dynamic regulation of NF-κB-dependent chemokine transcription.
Discussion
Nuclear FAK is emerging as an important regulator of gene expression in cancer cells, controlling transcriptional networks that influence multiple cellular functions. For example, FAK is reported to interact with the transcription factors p53 and GATA4, resulting in their inactivation with effects on cell survival (34). We have shown that nuclear FAK regulates expression of chemokines and cytokines, including Ccl5, likely via interactions with transcription factors and transcriptional regulators (9). Here, we show for the first time that the both IL33 and sST2 are transcriptionally regulated by nuclear FAK in a kinase-dependent manner, and that FAK interacts with transcription factors and transcriptional regulators linked to control of expression of IL33 and ST2.
Depletion of IL33 abundance in FAK-wt cells revealed that it is vital downstream of FAK in the regulation of chemokine expression, including that of Ccl5. We have previously identified a CCL5–CCR1/3/5 paracrine signalling axis between SCC FAK-wt cells and tumor infiltrating Treg cells, and have shown that CCL5 depletion results in FAK-wt tumor regression as a result of reduced tumor infiltrating Treg cells (9). Our tumor growth studies here revealed that IL33 depletion caused FAK-wt tumor regression, presumably due to IL33-dependent regulation of Ccl5 and other chemokines. Thus, IL33 regulates pro-inflammatory gene programs downstream of FAK that we have shown play an important role in defining the tumour immune environment, impacting on SCC tumor growth and survival.
sST2 functions as a decoy receptor that is secreted into the tumor environment, leading to competitive inhibition of IL33-ST2 autocrine and paracrine signalling (13). Although IL33 is not secreted by SCC cells, it can be secreted by macrophages and neutrophils (35), influencing the function of immune cell populations. ST2L is also present on activated CD8+ T-cells and Natural Killer (NK) cells, and IL33 stimulation can increase IFNγ expression (14) and cytotoxic activity (18). We identified ST2L on several immune cell types in the SCC tumor environment, including activated CD8+ T cells. Depletion of sST2 in FAK-wt tumors largely resulted in CD8+ T cell-dependent tumor regression, implying that sST2 contributes to inhibition of CD8+ T cell-mediated immunity. Studies using an ST2 knockout mouse have shown that host ST2 signalling is required for tumor regression to occur in response to overexpression of a secreted form of IL33 (14). Therefore, we conclude that the presence of sST2 enables FAK-abundant tumors to benefit from nuclear IL33 while counteracting the potential anti-tumor effects of secreted IL33 (Fig. 5).
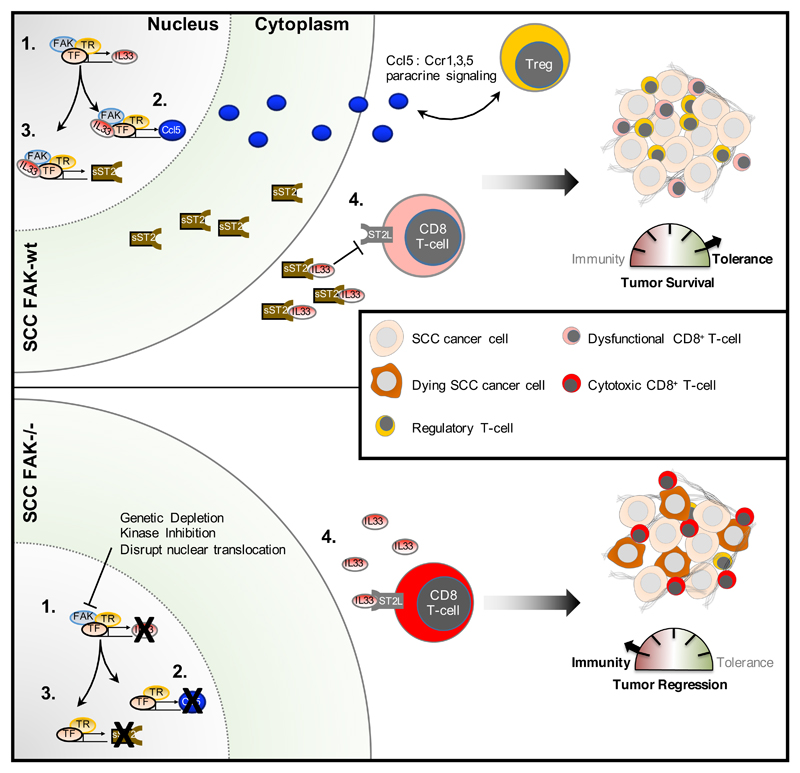
Figure 5. Nuclear FAK regulates IL33/ST2 signaling to control the anti-tumor immune response.
Model of the mechanism. Nuclear FAK regulates IL33 expression (“1”) through interaction with transcription factors (TF) and transcriptional regulators (TR). Nuclear FAK and IL33 cooperate to drive expression of Ccl5 and sST2 (“2” and “3”, respectively) through interaction with TFs and TRs. Ccl5 and sST2 are secreted from SCC cancer cells, promoting immune evasion. We have previously reported a CCL5-CCR1,3, and 5 paracrine signaling axis between FAK-wt SCC cells and tumor-infiltrating Treg cells that contributes to immune evasion. We propose that sST2 contributes to immune evasion through competitive inhibition of IL33/ST2 signaling on cytotoxic CD8+ T-cells (“4”), resulting in tumor tolerance.
Nuclear IL33 has been linked to regulation of transcription previously (16, 17), although the precise mechanisms underpinning this are unknown. We confirmed that nuclear IL33 interacts with chromatin, supporting its role in transcriptional regulation, and BioID proteomics identified proximal interactions with components of the Baf-type (SWI/SNF), PTW/PP1 phosphatase and TFIID complexes. This implies that nuclear IL33 functions in chromatin remodelling and transcriptional initiation. In support of this, we confirmed binding of WDR82, SMARCC1, TAF9, BRD4, and HDAC1, and showed that WDR82 was absent from chromatin after depletion of IL33 from FAK-wt cells, showing that IL33 is required to stabilize the WDR82 chromatin complex. WDR82 is a component of the PTW/PP1 phosphatase complex and is involved in regulating chromatin structure (28). It is also a core component of the mammalian Set1A/Set1B histone H3K4 methyltransferase complex that is associated with regulating H3K4 trimethylation, a key step in transcriptional activation (36).
Finally, we identified BRD4 and HDAC1 positive (37) and negative (33) regulators of NF-κB activity, respectively, as previously unknown nuclear IL33-interacting proteins. BRD4 is also required for recruitment of the SWI/SNF complex to active promoters (30), and we have shown that IL33 binds to several components of this complex. Therefore, IL33 likely binds to BRD4, HDAC1, and other chromatin modifiers to control the dynamic expression of NF-κB-target genes, such as Ccl5 and other chemokines. Collectively, our data suggest that IL33 acts to regulate chromatin organization and FAK-dependent transcription, promoting a pro-inflammatory gene program that enables evasion of the anti-tumor immune response.
Materials and Methods
Cell lines
Isolation and generation of the FAK SCC cell model is described in detail in Serrels et al. (12). Briefly, squamous cell carcinoma cells (SCCs) were induced in K14CreER FAKflox/flox mice on an FVB background using the DMBA/TPA two-stage skin chemical carcinogenesis protocol and cells isolated. Following treatment with 4-hydroxytamoxifen, a FAK-null (FAK-/-) cell clone was isolated, and retroviral transduction was used to stably re-express FAK-wt and FAK mutant proteins. Phoenix Ecotropic cells were transfected with pWZL (Hygro) FAK using Lipofectamine 2000 (Thermo Scientific) according to manufacturer’s instructions. Twenty-four hours post-transfection, cell culture supernatant was removed, filtered through a 0.45-μm Millex-HA filter (Millipore), diluted at a 1:1 ratio in normal SCC cell culture medium, supplemented with 5 μg/ml polybrene, and added to SCC FAK-/- cells for 24 hours. A total of two rounds of infection were performed to generate each cell line. Cells were cultured at 37°C in Glasgow minimum essential medium (MEM) (Sigma-Aldrich) supplemented with 2 mM L-glutamine, MEM vitamins, 1 mM sodium pyruvate (all Sigma-Aldrich), MEM amino acids, and 10% fetal bovine serum (FBS) (both Life Technologies), and maintained under selection using 0.25 mg/ml hygromycin. To overexpress IL33 in FAK-/- SCC cells, the cells were transfected with 1.5 μg of IL33 pcDNA3.1 (synthesized optimized sequence by Geneart - Life Technologies) using Lipofectamine 2000. Cells were selected in 400 μg/ml of G418, and overexpression of IL33 was confirmed by Western blotting.
shRNA-mediated ST2 and IL33 Knockdown
To generate lentiviral particles, 2x106 HEK293FT cells were transfected with a mixture of 2 μg shRNA [RMM4534-EG77125 (IL33), RMM4534-EG17082 (ST2/IL1l1R); GE Healthcare], 0.5 μg MDG2 and 1 μg PAX2 plasmid DNA using Lipofectamine 2000 (ThermoFisher Scientific) as per manufacturers guidelines. Forty-eight hours post transfection, media was removed and filtered through a 0.45μm Millex-AC filter (Millipore) and mixed at a 1:1 ratio with normal SCC growth medium, supplemented with polybrene (Millipore) to a final concentration of 5 μg/ml, and added to SCC cells for twenty-four hours. Cells were subjected to two rounds of lentiviral infection prior to selection in 2 μg/ml of puromycin.
siRNA mediated knockdown of SP1 and RUNX1
To knockdown SP1 and RUNX1, FAK-wt SCC cells were transfected 10nmol of non-targeting siRNA Smartpool, SP1 siRNA Smartpool or Runx1 siRNA smartpool (all Dharmacon, siGenome), or a combination of 10nmol SP1 and RUNX1 siRNA Smartpool using hyperfect transfection reagent (Qiagen as per manufacturers guidelines). Transfection was performed in serum free media for 24 hours, after which the media was replaced with normal growth media. Protein lysates were collected 48 hours after transfection.
IL33 CRISPR construct generation
Guide sequence oligonucleotides (Forward: TTTCTTGGCTTTATATATCTTGTG GAAAGGACGAAACACCGTTCCTAGAATCCCGTGGAT Reverse: GACTAGCCTTA TTTTAACTTGCTATTTCTAGCTCTAAAACATCCACGGGATTCTAGGAAC) including EcoR I restriction overhangs and a PAM sequence targeting exon three within IL33 were annealed using High Fidelity Phusion Polymerase (NEB) according to manufacturers guidelines. 10 μM of each oligonucleotide was mixed with 25mM MgCl2, 2.5mM dNTP, 5x High Fidelity Phusion Buffer, and Phusion Polymerase to a final volume of 50 μl. Cycling conditions were 98°C (30 seconds), 30 cycles of 98°C (10 seconds), 55°C (30 seconds) 72°C (20 seconds), followed by a final incubation at 72°C (20 seconds). The final PCR product was analysed by agarose gel electrophoresis. Annealed oligonucleotides were ligated into the gRNA_Cloning Vector (pCR-Blunt II-TOPO), a gift from George Church (Addgene plasmid # 41824). 1.5 μg of gRNA_cloning vector DNA was digested with 1.5 units of Aflii restriction enzyme (NEB) according to manufacturers guidelines, and incubated for 1 hour at 37°C. 4 ng of annealed oligonucleotides were ligated with 30 ng of linearized gRNA_cloning vector using Gibson assembly master mix (New England Biolabs) according to manufacturers guidelines, and incubated for 1 hour at 50°C. Constructs were transformed into DH5α chemically competent cells and selected on agar plates containing 50 μg/ml of kanamycin. To identify positive colonies, DNA was prepared and digested with EcoR I restriction enzyme (NEB).
IL33 CRISPR/Cas9 transfection and single cell clone expansion
SCC-FAK-wt cells were seeded on a 100mm tissue culture dish and grown until ~70% confluent. Cells were co-transfected with 3 μg of hCas9, and 3 μg of gRNA vector containing IL33 specific guides, using Lipofectamine 2000 (Thermo Scientific) according to manufacturers guidelines. IL33 CRISPR knockout clones were isolated by dilution cloning; once cells reached confluence they were trypsinized and re-suspended in 10 mls of growth media. To ensure single cell suspension, cells were passed through a cell strainer (Thermo Scientific), and 5 μl of the cell suspension was plated into a 15cm plate. Several colonies were picked and expanded, and knockout of IL33 was confirmed by qPCR and western blot. IL33 primers used were: forward GGATCCGATTTTCGAGAGCTTAAACAT reverse: GCGGCCGCATGAGACCTAGAATGAAGT
Immunoblotting
Cells were washed twice in ice cold phosphate buffered saline (PBS) and lysed in RIPA lysis buffer (50mM Tris-HCL at pH7.4, 150mM Sodium Chloride, 5mM EGTA, 0.1% SDS, 1% NP40, 1% Deoxycholate) supplemented with a protease/phosphatase inhibitor cocktail (mini complete Ultra Protease tablet and phosSTOP tablet from Roche). Lysates were clarified by high-speed centrifugation (16,000g for 15 min at 4°C). Protein concentration was measured using a micro BCA protein assay (ThermoScientific), and 10-20 μg of total protein was supplemented with 2x SDS sample buffer (Tris pH6.8, 20% Glycerol, 5% SDS, β-mercaptoethanol, Bromo-phenol blue), and boiled at 95°C for 5 minutes. Samples were separated by polyacrylamide gel electrophoresis using 4-15% mini-protean TGX gels (BioRad), proteins transferred to nitrocellulose, blocked [5% bovine serum albumin in PBS–Tween 20 (BSA/PBS-T)], and probed with either anti-IL33 (R&D Biosystems), anti-GAPDH (Cell Signaling Technology), anti-phospho-FAK (Y397) (Cell Signaling Technology), anti-FAK (Cell Signaling), anti-WDR82 (Abcam), anti-TAF9 (Abcam), anti- SMARC C1 (Abcam), anti-BRD4 (Abcam), anti-HP1 aplha/beta (Cell Signaling Technology), anti-SP1(Abcam), anti-Runx1 (Cell Signalling Technology), and anti-HDAC1 Abs (Cell Signalling Technology) (all 1:1000 in 5% BSA/PBS-T). Bound antibody was detected by incubation with anti-rabbit, anti-mouse or streptavidin conjugated-HRP secondary antibody (Cell Signaling Technology), and visualized using the BioRad ChemiDoc MP Imaging System.
Chromatin preparation
1.5x106 cells were plated into 100mm dishes and after twenty-four hours, washed twice in cold PBS. Cells were lysed in 400 μl of extraction buffer (10mM HEPES, pH7.9, 10mM KCl, 1.5mM MgCl2, 0.34M sucrose, 10% glycerol, 0.2% NP-40 substitute) containing protease/phopshatase inhibitors (mini complete Ultra Protease tablet and phosSTOP tablet from Roche). Lysates were cleared at 6,500g for 5 minutes at 4°C. The resulting nuclear pellet was washed in extraction buffer without NP-40, and centrifuged at 6,500g for 5 minutes at 4°C. The pellet was re-suspended in 400 μl of no salt buffer (10mM HEPES pH7.9, 3mM EDTA, 0.2mM EGTA), incubated at 4°C for 30 minutes with agitation, and centrifuged at 6500g for 5 minutes at 4°C. The pellet was re-suspended in 160 μl of high salt solubilisation buffer (50mM Tris-HCl pH8.0,2.5M NaCl, 0.05% NP-40) vortexed briefly, incubated at 4°C for 30 minutes with agitation, and centrifuged at 16,000g for 10 minutes at 4°C. The supernatant containing chromatin fraction was collected, and TCA precipitation was performed. Trichloroacetic acid (TCA) was added to a final volume of 10%, and samples were incubated for 15 minutes on ice. Following centrifugation at 21,000g for 15 minutes the resulting pellet was washed twice in 500 μl of cold acetone, and then allowed to air dry. The pellet was then re-suspended in 20 μl 2x sample buffer (Tris pH6.8, 20% Glycerol, 5% SDS, β-mercaptoethanol, Bromo-phenol blue) and boiled at 75°C for 10 minutes. Samples were separated by polyacrylamide gel electrophoresis on a 12% Mini-Protean TGX gel (BioRad), transferred onto nitrocellulose membrane, blocked [5% bovine serum albumin in PBS–Tween 20 (BSA/PBS-T)], then incubated with primary and secondary antibodies as above.
sST2 and IL33 ELISA
2x106 cells were plated on a 100mm tissue culture dish and left to adhere overnight. Media was replaced with 4ml fresh complete growth media and conditioned for 24hrs, collected and spun at 1000rpm for 5mins to remove debris. Media samples were analysed for sST2 levels using a mouse ST2 Quantikine ELISA kit or mouse IL33 Quantikine Elisa kit (R and D Systems). Cells adhered to the tissue culture dish were washed 2x in ice cold PBS and lysed in RIPA lysis buffer as above. Protein concentration was measured using micro BCA protein assay and protein quantities were used for normalisation of ELISA values.
q-RT-PCR and Nanostring
RNA was prepared from cells using the Qiagen RNeasy mini kit as per manufacturer’s instructions, inclusive of DNase digestion. Final concentration of RNA was measured using a Nanodrop (Thermo Scientific). For q-RT-PCR, 5 μg of total RNA was converted to cDNA using the superscript II cDNA synthesis kit (Thermo Scientific). For quantitative real-time PCR, 62.5ng of cDNA was added to SYBR Green (Applied Biosystems), supplemented with 0.25 μl of 10μM qPCR primers to a final reaction volume of 10 μl. IL33 primers used were: forward GGATCCGATTTTCGAGAGCTTAAACAT reverse: GCGGCCGCATGAGACCTAGAATGAAGT, ST2 primers used were: GCGGAGAATGGAAGCAACTA reverse: AAGCAAGCTGAACAGGCAAT, Ccl5 primers used were: forward CCCTCACCATCATCCTCACT reverse CCTTCGAGTGACAAACACGA. Quantitative real time PCR was performed on a StepOne Plus qRT-PCR instrument (Applied Biosystems). PCR conditions were as follows: 95°C (3 minutes), followed by 40 cycles of 95°C (5 seconds), 60°C (10 seconds), and 72°C (10 seconds). Melt curve analysis was performed following each qPCR reaction. Data was analysed using the ddCT method, and expression calculated relative to GAPDH. For Nanostring analysis, 150ng of RNA was labelled with gene specific bar codes as per manufacturer’s instructions and quantified using a Nanostring ncounter DX. Nanostring analysis was carried out by the Newcastle Nanostring ncounter analysis service. Analysis was performed using nSolver analysis software (Nanostring).
Generation of IL33-BirA expressing SCC Cell Line
IL33 cDNA was amplified by PCR using gene specific primers (Forward: GCAGCAGCGGCCGCATGAGACCTAGAATGAAGTATTCCAAC Reverse: TGCTGCGGATCCGATTTTCGAGAGCTTAAACATAATATTG), and sub-cloned into the Not1/BamH1 sites of pQCXIN-BirA-Myc. Specifically, 20ng of pcDNA-3.1 IL33 DNA template was mixed with 10 μM of each primer in PfuUltra Hotstart master mix (Stratagene) and subjected to PCR with cycle conditions as follows: 98°C (30 seconds), 30 cycles; 98°C (10 seconds), 60°C (30 seconds), 72°C (1 minute), followed by a final incubation at 72°C (10 minutes). Following PCR, samples were gel purified (Qiagen Gel Purification Kit) and eluted in 30 μl of nuclease free water (NFW). Purified PCR product was incubated with 1 μl BamH1 restriction enzyme, 1 μl NOT1 restriction enzyme, 5 μl of Buffer 3.1(all New England Biolabs) to a total of 20 μl, and incubated at 37°C for 2 hours. 1 μg of pQCXIN-BirA-Myc was incubated with 1 μl BamH1 restriction enzyme, 1μl NOT1 restriction enzyme, 5 μl of Buffer 3.1 to a total of 20ul and incubated at 37°C for 1 hour. Following digestion, both the PCR product and digested vector were gel purified and eluted in 30 μl of nuclease free water. To ligate the digested IL33 PCR product into the BamH1/NOT1 sites of pQCXIN-BirA-Myc, a 1:4 ratio of digested vector to PCR product was used, and incubated with 0.5 μl of T4 DNA ligase (New England Biolabs), along with DNA ligase buffer in a final reaction volume of 10ul, and incubated for 2 hours at room temperature. 50 μl of DH5alpha chemically competent cells (Life Technologies) were transformed with 5 μl of ligation reaction according to manufacturers guidelines, and plated onto agar plates containing 100 μg/ml of ampicillin. Resulting colonies were checked for successful ligation using a diagnostic digest with BamH1 and NOT1 restriction enzymes, and if positive sequenced. Phoenix Ecotropic cells were transfected with empty vector-BirA or IL33-BirA using Lipofectamine 2000 (Thermo Scientific) according to manufacturer’s guidelines. Forty-eight hours post-transfection, cell culture supernatant was removed, filtered through a 0.45-μm Millex-HA filter (Millipore), diluted at a 1:1 ratio in normal SCC cell culture medium, supplemented with 5 μg/ml polybrene, and added to SCC FAK-wt IL33-CRISPR cells for 24 hours. A total of two rounds of infection were performed to generate each cell line prior to selection in 400 μg/ml of G418.
Proteomic analysis of the IL33 interactome using BioID
Cells were incubated with 50 μM biotin (Sigma Aldridge) in complete cell culture medium at 37°C for 24hrs, washed 2x in ice cold PBS, and lysed in RIPA lysis buffer as above. Cell lysates were sonicated using a Bioruptor (Diagenode), 30 second pulses with 30 second intervals over 5mins, and cleared by high speed centrifugation (16,000g for 15min at 4°C). Protein concentration was measured using a micro BCA protein assay (Thermo Scientific) and 1 mg of total cell lysate was incubated with 50 μl of streptavidin-C1 dynabeads (Life Technologies) overnight at 4°C with agitation. Beads were washed using a magnetic tube rack 3x with ice cold RIPA buffer and 2x with ice cold PBS. Captured proteins (from experiments performed in biological triplicate) were subjected to on-bead proteolytic digestion, desalting and liquid chromatography-tandem mass spectrometry as described previously (38). Mean label-free MS intensities were calculated for each biological replicate. Peptide and protein false discovery rates were set to 1%. Proteins enriched from SCC-FAK-wt lL33-CRSIPR-IL33-BirA cells by at least 3-fold when compared to SCC-FAK-wt IL33-CRISPR-EV-BirA (p<0.05) were considered specific. Proteomics analysis of nuclear FAK protein complexes is described in (9). All other protein interaction network analysis was performed using Ingenuity Pathway Analysis (Qiagen). The IL33 mass spectrometery proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (39) with the dataset identifier PXD007698.
Interaction network analysis
Genes differentially expressed in FAK-/- SCC cells compared to FAK-wt SCC cells were extracted from microarray data (9) using rank product analysis. Significantly differentially expressed genes (p < 0.0005, percentage false positives (pfp) < 0.05) were subjected to gene ontology enrichment analysis using DAVID Bioinformatics Resources (version 6.8) (40).Genes annotated with overrepresented top-level cellular component terms as determined by Benjamini–Hochberg-corrected hypergeometric test (p < 0.05) were used to seed a protein interaction network based on direct physical interactions constructed using the GeneMANIA plugin (version 3.4.1; mouse interactions) in Cytoscape (version 3.3.0) (41). The largest connected graph component was clustered using the Allegro Spring-Electric force-directed algorithm (AllegroViva).
Hierarchical cluster analysis
Unsupervised agglomerative hierarchical clustering was performed on the basis on Euclidean distance or Pearson correlation computed with an average-linkage or complete-linkage matrix using R or Cluster 3.0 (C Clustering Library, version 1.50) (42). Clustering results were visualised using R or Java TreeView (version 1.1.6) (43) and MultiExperiment Viewer (version 4.8.1) (44).
Inhibitor treatment
FAK-wt SCC cells were treated with doses ranging from 50-250nM VS4718 for 24 hours, 10 μM vorinostat for 24 hours, or 200 nM JQ1 for 48 hours, after which RNA or protein lysates were collected for q-RT-PCR or western blot analysis as described above. All inhibitors were obtained from Selleckchem.
Subcutaneous tumour growth
All experiments involving animals were carried out in accordance with the UK Coordinating Committee on Cancer Research guidelines by approved protocol (Home Office Project License no. 60/4248). 1x106 cells SCC cells (defined earlier) were injected into both flanks of FVB mice and tumour growth was measured twice-weekly using calipers. Animals were sacrificed by cervical dislocation when tumours reached maximum allowed size (1.5cm diameter) or when signs of ulceration were evident. Group sizes ranged from 3-5 mice, each bearing two tumours, and tumour volume was calculated in Excel (Microsoft) using formula x-4/3πr3. Statistics and graphs were calculated using Prism (GraphPad).
CD8+T-cell Depletion
Anti-mouse CD8 (clone 53-6.7) and appropriate isotype control Abs (Rat IgG2a) were purchased from BioXcel. T-cell depletion was achieved following intra-peritoneal (IP) injection of 150 μg of depleting antibody (same for all antibodies) into female age-matched FVB mice for 3 consecutive days, and maintained by further IP injection at 7-day intervals until the study was terminated. 1 x 106 SCC FAK-wt or FAK-/- cells were injected subcutaneously into both flanks 6 days after initial antibody treatment, and tumor growth was measured twice-weekly as described above.
FACS Analysis of Immune Cell Populations
Tumors established following injection of 1 x 106 SCC cells into both flanks of FVB mice were removed at day 12 into RPMI (Sigma-Aldrich) supplemented with 10% FBS (Life Technologies). Tumor tissue was mashed into a pulp using a scalpel and re-suspended in DMEM (Sigma-Aldrich) supplemented with 2 mg/ml collagenase D and DNase1 (200U/ml) (Roche). Samples were incubated for 30 mins at 37°C with agitation, pelleted by centrifugation at 1600 rpm for 5 min at 4°C, resuspended in 5 ml of red blood cell lysis buffer (Pharm Lysis Buffer, Becton Dickinson) for 10 min at 37°C, pelleted by centrifugation at 1600 rpm for 5 min at 4°C, re-suspended in PBS and mashed through a 70-μm cell strainer. The resulting single cell suspension pellet was pelleted by centrifugation at 1600 rpm for 5 min at 4°C and re-suspended in PBS. This step was repeated a further time and the resulting cell pellets resuspended in 100 μl PBS containing e506 fixable viability dye (1/1000 dilution) and transferred into the well of a round bottom 96-well plate. Samples were incubated at 4°C for 30 minutes. Cells were then pelleted by centrifugation at 1600 rpm for 5 min at 4°C and re-suspended in FACS buffer (PBS containing 1% FBS and 0.1% sodium azide). This step was repeated a total of three times. Cell pellets were re-suspended in 100 μl of Fc block (1:200 dilution of Fc antibody (eBioscience) in FACS buffer) and incubated for 15 min at room temperature (RT). 100 μl of antibody mixture (1:200 dilution of antibodies except anti-FoxP3 which was used at 1:100 (listed in fig. S3) in FACS buffer) were added to each well and the samples incubated for 30 min in the dark at 4°C. The plate was then centrifuged at 1600 rpm for 5 min at 4°C and the cells re-suspended in FACS buffer. This step was repeated 3 times. Samples were analyzed using a BD Fortessa FACS Analyser. Data analysis was performed using FlowJo software. All antibodies were from eBioscience. Statistics and graphs were calculated using Prism (GraphPad).
Supplementary Material
One-sentence summary.
The kinase FAK coordinates chromatin modifiers and transcription factors to orchestrate evasion of antitumor immune responses.
Editor’s summary.
FAK directs tumor immune evasion
Tumors are adept at escaping the immune system’s surveillance or suppressing its activity. The kinase FAK is implicated in immune escape mechanisms. Serrels et al. found that FAK activates a transcriptional network that induces—and is then further mediated by—the nuclear abundance of interleukin 33 (IL-33) in tumor cells. In a mouse model of squamous cell carcinoma, FAK–IL-33 complexes boosted the production and secretion of two key factors in immunosuppression: the chemokine CCL5, which stimulates immunosuppressive regulatory T cells, and soluble ST2, a decoy receptor for cytotoxic T cell–stimulatory IL-33. Blocking these FAK-mediated signals may help the patient’s immune system find and kill tumors.
Acknowledgments
We would like to thank the FACS facility at the MRC-Centre for Inflammation Research, University of Edinburgh for their help with FACS analysis, and the CBS animal technicians at LF2, University of Edinburgh for their assistance with biological work.
Funding: This work was supported by Cancer Research UK (Grant no. C157/A15703 to M.C.F.), European Research Council (Grant no. 299440 Cancer Innovation to M.C.F.). A.v.K. is supported by the Cancer Research UK Edinburgh centre grant. N.Q is supported by the Welcome Trust-UoE ISSF2 programme. D.T. is supported by Cancer Research UK (Grant no. C54352/A22011 awarded to S.M.A.).
Footnotes
Author contributions: B.S., A.S., and M.C.F. devised and oversaw the project. B.S., A.S., N.M., A.B., S.M.A., and M.C.F. designed the experiments with contributions from M.C. and H.M. B.S., A.S., N.M., M.C., A.B., S.C.J., N.Q. D.T., performed the experiments with contributions from A.v.K., and B.S., A.S., N.M., M.C., A.B., N.Q., S.C.J., analysed the data. B.S., and A.B., performed bioinformatics analysis. B.S., A.S., and M.C.F., wrote the manuscript with contributions from A.B., and N.M.; all authors commented on and approved the final version.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The mass spectrometery proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository, dataset identifier PXD007698.
References
- 1.Sulzmaier FJ, Jean C, Schlaepfer DD. FAK in cancer: mechanistic findings and clinical applications. Nat Rev Cancer. 2014;14:598–610. doi: 10.1038/nrc3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lightfoot HM, Jr, Lark A, Livasy CA, Moore DT, Cowan D, Dressler L, Craven RJ, Cance WG. Upregulation of focal adhesion kinase (FAK) expression in ductal carcinoma in situ (DCIS) is an early event in breast tumorigenesis. Breast Cancer Res Treat. 2004;88:109–116. doi: 10.1007/s10549-004-1022-8. [DOI] [PubMed] [Google Scholar]
- 3.Lark AL, Livasy CA, Calvo B, Caskey L, Moore DT, Yang X, Cance WG. Overexpression of focal adhesion kinase in primary colorectal carcinomas and colorectal liver metastases: immunohistochemistry and real-time PCR analyses. Clin Cancer Res. 2003;9:215–222. [PubMed] [Google Scholar]
- 4.Agochiya M, Brunton VG, Owens DW, Parkinson EK, Paraskeva C, Keith WN, Frame MC. Increased dosage and amplification of the focal adhesion kinase gene in human cancer cells. Oncogene. 1999;18:5646–5653. doi: 10.1038/sj.onc.1202957. [DOI] [PubMed] [Google Scholar]
- 5.McLean GW, Komiyama NH, Serrels B, Asano H, Reynolds L, Conti F, Hodivala-Dilke K, Metzger D, Chambon P, Grant SG, Frame MC. Specific deletion of focal adhesion kinase suppresses tumor formation and blocks malignant progression. Genes Dev. 2004;18:2998–3003. doi: 10.1101/gad.316304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lahlou H, Sanguin-Gendreau V, Zuo D, Cardiff RD, McLean GW, Frame MC, Muller WJ. Mammary epithelial-specific disruption of the focal adhesion kinase blocks mammary tumor progression. Proc Natl Acad Sci U S A. 2007;104:20302–20307. doi: 10.1073/pnas.0710091104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashton GH, Morton JP, Myant K, Phesse TJ, Ridgway RA, Marsh V, Wilkins JA, Athineos D, Muncan V, Kemp R, Neufeld K, et al. Focal adhesion kinase is required for intestinal regeneration and tumorigenesis downstream of Wnt/c-Myc signaling. Dev Cell. 2010;19:259–269. doi: 10.1016/j.devcel.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slack-Davis JK, Hershey ED, Theodorescu D, Frierson HF, Parsons JT. Differential requirement for focal adhesion kinase signaling in cancer progression in the transgenic adenocarcinoma of mouse prostate model. Mol Cancer Ther. 2009;8:2470–2477. doi: 10.1158/1535-7163.MCT-09-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serrels A, Lund T, Serrels B, Byron A, McPherson RC, von Kriegsheim A, Gomez-Cuadrado L, Canel M, Muir M, Ring JE, Maniati E, et al. Nuclear FAK controls chemokine transcription, Tregs, and evasion of anti-tumor immunity. Cell. 2015;163:160–173. doi: 10.1016/j.cell.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cardoso AC, Pereira AH, Ambrosio AL, Consonni SR, Rocha de Oliveira R, Bajgelman MC, Dias SM, Franchini KG. FAK Forms a Complex with MEF2 to Couple Biomechanical Signaling to Transcription in Cardiomyocytes. Structure. 2016;24:1301–1310. doi: 10.1016/j.str.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Lim ST, Miller NL, Chen XL, Tancioni I, Walsh CT, Lawson C, Uryu S, Weis SM, Cheresh DA, Schlaepfer DD. Nuclear-localized focal adhesion kinase regulates inflammatory VCAM-1 expression. J Cell Biol. 2012;197:907–919. doi: 10.1083/jcb.201109067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Serrels A, McLeod K, Canel M, Kinnaird A, Graham K, Frame MC, Brunton VG. The role of focal adhesion kinase catalytic activity on the proliferation and migration of squamous cell carcinoma cells. Int J Cancer. 2012;131:287–297. doi: 10.1002/ijc.26351. [DOI] [PubMed] [Google Scholar]
- 13.Milovanovic M, Volarevic V, Radosavljevic G, Jovanovic I, Pejnovic N, Arsenijevic N, Lukic ML. IL-33/ST2 axis in inflammation and immunopathology. Immunol Res. 2012;52:89–99. doi: 10.1007/s12026-012-8283-9. [DOI] [PubMed] [Google Scholar]
- 14.Gao X, Wang X, Yang Q, Zhao X, Wen W, Li G, Lu J, Qin W, Qi Y, Xie F, Jiang J, et al. Tumoral expression of IL-33 inhibits tumor growth and modifies the tumor microenvironment through CD8+ T and NK cells. J Immunol. 2015;194:438–445. doi: 10.4049/jimmunol.1401344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Davis C, Shah S, Hughes D, Ryan JC, Altomare D, Pena MM. IL-33 promotes growth and liver metastasis of colorectal cancer in mice by remodeling the tumor microenvironment and inducing angiogenesis. Mol Carcinog. 2017;56:272–287. doi: 10.1002/mc.22491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ali S, Mohs A, Thomas M, Klare J, Ross R, Schmitz ML, Martin MU. The dual function cytokine IL-33 interacts with the transcription factor NF-kappaB to dampen NF-kappaB-stimulated gene transcription. J Immunol. 2011;187:1609–1616. doi: 10.4049/jimmunol.1003080. [DOI] [PubMed] [Google Scholar]
- 17.Choi YS, Park JA, Kim J, Rho SS, Park H, Kim YM, Kwon YG. Nuclear IL-33 is a transcriptional regulator of NF-kappaB p65 and induces endothelial cell activation. Biochem Biophys Res Commun. 2012;421:305–311. doi: 10.1016/j.bbrc.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 18.Yang Q, Li G, Zhu Y, Liu L, Chen E, Turnquist H, Zhang X, Finn OJ, Chen X, Lu B. IL-33 synergizes with TCR and IL-12 signaling to promote the effector function of CD8+ T cells. Eur J Immunol. 2011;41:3351–3360. doi: 10.1002/eji.201141629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu DP, Zhou XY, Yao LT, Liu CG, Ma W, Jin F, Wu YF. Serum soluble ST2 is associated with ER-positive breast cancer. BMC Cancer. 2014;14:198. doi: 10.1186/1471-2407-14-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villarreal DO, Weiner DB. Interleukin 33: a switch-hitting cytokine. Curr Opin Immunol. 2014;28:102–106. doi: 10.1016/j.coi.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chackerian AA, Oldham ER, Murphy EE, Schmitz J, Pflanz S, Kastelein RA. IL-1 receptor accessory protein and ST2 comprise the IL-33 receptor complex. J Immunol. 2007;179:2551–2555. doi: 10.4049/jimmunol.179.4.2551. [DOI] [PubMed] [Google Scholar]
- 22.Schmieder A, Multhoff G, Radons J. Interleukin-33 acts as a pro-inflammatory cytokine and modulates its receptor gene expression in highly metastatic human pancreatic carcinoma cells. Cytokine. 2012;60:514–521. doi: 10.1016/j.cyto.2012.06.286. [DOI] [PubMed] [Google Scholar]
- 23.Shao D, Perros F, Caramori G, Meng C, Dormuller P, Chou PC, Church C, Papi A, Casolari P, Welsh D, Peacock A, et al. Nuclear IL-33 regulates soluble ST2 receptor and IL-6 expression in primary human arterial endothelial cells and is decreased in idiopathic pulmonary arterial hypertension. Biochem Biophys Res Commun. 2014;451:8–14. doi: 10.1016/j.bbrc.2014.06.111. [DOI] [PubMed] [Google Scholar]
- 24.Canel M, Byron A, Sims AH, Cartier J, Patel H, Frame MC, Brunton VG, Serrels B, Serrels A. Nuclear FAK and Runx1 cooperate to regulate IGFBP3, cell cycle progression and tumor growth. Cancer Res. 2017 doi: 10.1158/0008-5472.CAN-17-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schiering C, Krausgruber T, Chomka A, Frohlich A, Adelmann K, Wohlfert EA, Pott J, Griseri T, Bollrath J, Hegazy AN, Harrison OJ, et al. The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature. 2014;513:564–568. doi: 10.1038/nature13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roussel L, Erard M, Cayrol C, Girard JP. Molecular mimicry between IL-33 and KSHV for attachment to chromatin through the H2A-H2B acidic pocket. EMBO Rep. 2008;9:1006–1012. doi: 10.1038/embor.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JH, You J, Dobrota E, Skalnik DG. Identification and characterization of a novel human PP1 phosphatase complex. J Biol Chem. 2010;285:24466–24476. doi: 10.1074/jbc.M110.109801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JH, Skalnik DG. Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A Histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol Cell Biol. 2008;28:609–618. doi: 10.1128/MCB.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi J, Whyte WA, Zepeda-Mendoza CJ, Milazzo JP, Shen C, Roe JS, Minder JL, Mercan F, Wang E, Eckersley-Maslin MA, Campbell AE, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 2013;27:2648–2662. doi: 10.1101/gad.232710.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mujtaba S, Zeng L, Zhou MM. Structure and acetyl-lysine recognition of the bromodomain. Oncogene. 2007;26:5521–5527. doi: 10.1038/sj.onc.1210618. [DOI] [PubMed] [Google Scholar]
- 32.Huang B, Yang XD, Zhou MM, Ozato K, Chen LF. Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol Cell Biol. 2009;29:1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashburner BP, Westerheide SD, Baldwin AS., Jr The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol Cell Biol. 2001;21:7065–7077. doi: 10.1128/MCB.21.20.7065-7077.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim ST, Chen XL, Lim Y, Hanson DA, Vo TT, Howerton K, Larocque N, Fisher SJ, Schlaepfer DD, Ilic D. Nuclear FAK promotes cell proliferation and survival through FERM-enhanced p53 degradation. Mol Cell. 2008;29:9–22. doi: 10.1016/j.molcel.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lefrancais E, Cayrol C. Mechanisms of IL-33 processing and secretion: differences and similarities between IL-1 family members. Eur Cytokine Netw. 2012;23:120–127. doi: 10.1684/ecn.2012.0320. [DOI] [PubMed] [Google Scholar]
- 36.Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol. 2008;28:7337–7344. doi: 10.1128/MCB.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zou Z, Huang B, Wu X, Zhang H, Qi J, Bradner J, Nair S, Chen LF. Brd4 maintains constitutively active NF-kappaB in cancer cells by binding to acetylated RelA. Oncogene. 2014;33:2395–2404. doi: 10.1038/onc.2013.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turriziani B, Garcia-Munoz A, Pilkington R, Raso C, Kolch W, von Kriegsheim A. On-beads digestion in conjunction with data-dependent mass spectrometry: a shortcut to quantitative and dynamic interaction proteomics. Biology (Basel) 2014;3:320–332. doi: 10.3390/biology3020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vizcaino JA, Csordas A, Del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu QW, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:11033. doi: 10.1093/nar/gkw880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 43.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 44.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.