Abstract
A key public health need is to identify individuals at high risk for a given disease to enable enhanced screening or preventive therapies. Because most common diseases have a genetic component, one important approach is to stratify individuals based on inherited DNA variation.1 Proposed clinical applications have largely focused on finding carriers of rare monogenic mutations at several-fold increased risk. Although most disease risk is polygenic in nature,2–5 it has not yet been possible to use polygenic predictors to identify individuals at risk comparable to monogenic mutations. Here, we develop and validate genome-wide polygenic scores for five common diseases. The approach identifies 8.0%, 6.1%, 3.5%, 3.2% and 1.5% of the population at greater than three-fold increased risk for coronary artery disease (CAD), atrial fibrillation, type 2 diabetes, inflammatory bowel disease, and breast cancer, respectively. For CAD, this prevalence is 20-fold higher than the carrier frequency of rare monogenic mutations conferring comparable risk.6 We propose that it is time to contemplate the inclusion of polygenic risk prediction in clinical care and discuss relevant issues.
For various common diseases, genes have been identified in which rare mutations confer several-fold increased risk in heterozygous carriers. An important example is the presence of a familial hypercholesterolemia mutation in 0.4% of the population, which confers an up to 3-fold increased risk for coronary artery disease (CAD).6 Aggressive treatment to lower circulating cholesterol levels among such carriers can significantly reduce risk.7 Another example is the p.E508K missense mutation in HNF1A, with carrier frequency of 0.1% of the general population and 0.7% of Latinos,8 which confers up to 5-fold increased risk for type 2 diabetes.9 Although ascertainment of monogenic mutations can be highly relevant for carriers and their families, the vast majority of disease occurs in those without such mutations.
For most common diseases, polygenic inheritance, involving many common genetic variants of small effect, plays a greater role than rare monogenic mutations.2–5 However, it has been unclear whether it is possible to create a genome-wide polygenic score (GPS) to identify individuals at clinically significantly increased risk—for example, comparable to levels conferred by rare monogenic mutations.10-11
Previous studies to create GPS had only limited success, providing insufficient risk stratification for clinical utility (for example, identifying 20% of a population at 1.4-fold increased risk relative to the rest of the population).12 These initial efforts were hampered by three challenges: (i) the small size of initial genome-wide association studies (GWAS), which affected the precision of the estimated impact of individual variants on disease risk; (ii) limited computational methods for creating GPS; and (iii) lack of large datasets needed to validate and test GPS.
Using much larger studies and improved algorithms, we set out to revisit the question of whether a GPS can identify subgroups of the population with risk approaching or exceeding that of a monogenic mutation. We studied five common diseases with major public health impact – CAD, atrial fibrillation, type 2 diabetes, inflammatory bowel disease, and breast cancer.
For each of the diseases, we created several candidate GPS based on summary statistics and imputation from recent large GWAS in participants of primarily European ancestry (Table 1). Specifically, we derived 24 predictors based on a pruning and thresholding method and 7 additional predictors using the recently described LDPred algorithm13 (online Methods; Figure 1; Supplementary Tables 1–6). The UK Biobank has genotype data and extensive phenotypic information on 409,258 participants of British ancestry (average age 57 years; 55% female).14,15
Table 1.
GPS derivation and testing for five common, complex diseases
| Disease | Discovery GWAS ( n ) |
Prevalence in validation dataset |
Prevalence in testing dataset |
Polymorphisms in GPS |
Tuning parameter | AUC (95% CI) in validation dataset |
AUC (95% CI) in testing dataset |
|---|---|---|---|---|---|---|---|
| CAD | 60,801 cases; 123,504 controls16 | 3,963/120,280 (3.4%) | 8,676/288,978 (3.0%) | 6,630,150 | LDPred (ρ = 0.001) | 0.81 (0.80–0.81) | 0.81 (0.81–0.81) |
| Atrial fibrillation |
17,931 cases; 115,142 controls30 | 2,024/120,280 (1.7%) | 4,576/288,978 (1.6%) | 6,730,541 | LDPred (ρ = 0.003) | 0.77 (0.76–0.78) | 0.77 (0.76–0.77) |
| Type 2 diabetes |
26,676 cases; 132,532 controls31 | 2,785/120,280 (2.4%) | 5,853/288,978 (2.0%) | 6,917,436 | LDPred (ρ = 0.01) | 0.72 (0.72–0.73) | 0.73 (0.72–0.73) |
| Inflammatory bowel disease |
12,882 cases; 21,770 controls32 | 1,360/120,280 (1.1%) | 3,102/288,978 (1.1%) | 6,907,112 | LDPred (ρ = 0.1) | 0.63 (0.62–0.65) | 0.63 (0.62–0.64) |
| Breast cancer |
122,977 cases; 105,974 controls33 | 2,576/63,347 (4.1%) | 6,586/157,895 (4.2%) | 5,218 | Pruning and thresholding (r/2 < 0.2; P < 5 × 10−4) | 0.68 (0.67–0.69) | 0.69 (0.68–0.69) |
AUC was determined using a logistic regression model adjusted for age, sex, genotyping array, and the first four principal components of ancestry. The breast cancer analysis was restricted to female participants. For the LDPred algorithm, the tuning parameter ρ reflects the proportion of polymorphisms assumed to be causal for the disease. For the pruning and thresholding strategy, r2 reflects the degree of independence from other variants in the linkage disequilibrium reference panel, and P reflects the P value noted for a given variant in the discovery GWAS. CI, confidence interval.
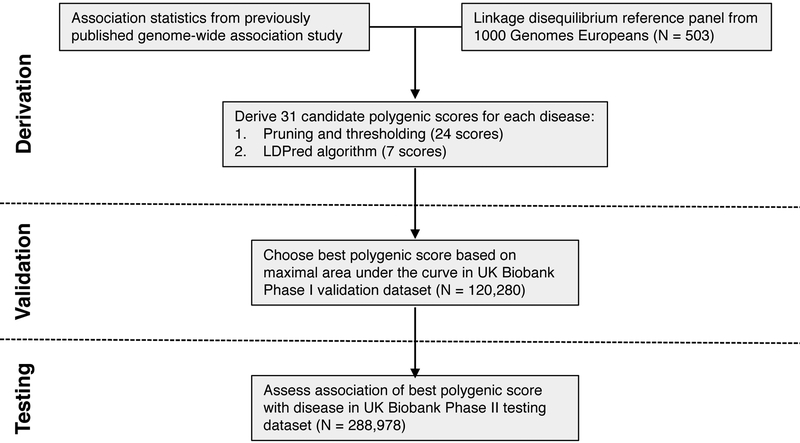
Figure 1. Study design and workflow.
A genome-wide polygenic score (GPS) for each disease was derived by combining summary association statistics from a recent large GWAS and a linkage disequilibrium reference panel of 503 Europeans.34 31 candidate GPS were derived using two strategies: 1. ‘pruning and thresholding’ – aggregation of independent polymorphisms that exceed a specified level of significance in the discovery GWAS and 2. LDPred computational algorithm,13 a Bayesian approach to calculate a posterior mean effect for all variants based on a prior (effect size in the prior GWAS) and subsequent shrinkage based on linkage disequilibrium. The seven candidate LDPred scores vary with respect to the tuning parameter ρ, the proportion of variants assumed to be causal, as previously recommended.13 The optimal GPS for each disease was chosen based on area under the receiver-operator curve (AUC) in the UK Biobank Phase I validation dataset (N=120,280 Europeans) and subsequently calculated in an independent UK Biobank Phase II testing dataset (N=288,978 Europeans).
We used an initial validation dataset of the 120,280 participants in the UK Biobank Phase 1 genotype data release to select the GPS with the best performance, defined as the maximum area under the receiver-operator curve (AUC). We then assessed the performance in an independent testing set comprised of the 288,978 participants in the UK Biobank Phase 2 genotype data release. For each disease, the discriminative capacity within the testing dataset was nearly identical to that observed in the validation dataset.
Taking CAD as an example, our polygenic predictors were derived from a GWAS involving 184,305 participants16 and evaluated based on their ability to detect the participants in the UK Biobank validation dataset diagnosed with CAD (Table 1). The predictors had AUC ranging from 0.79 – 0.81 in the validation set, with the best predictor (GPSCAD) involving 6,630,150 variants (Supplementary Table 1). This predictor performed equivalently well in the testing dataset, with AUC of 0.81.
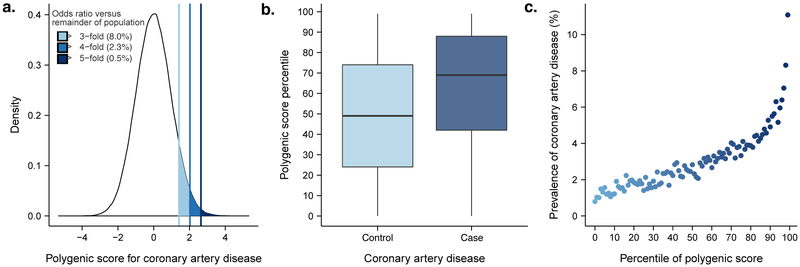
We then investigated whether our polygenic predictor, GPSCAD, could identify individuals at similar risk to the 3-fold increased risk conferred by a familial hypercholesterolemia mutation.6 Across the population, GPSCAD is normally distributed with the empirical risk of CAD rising sharply in the right tail of the distribution, from 0.8% in the lowest percentile to 11.1% in the highest percentile (Figure 2). The median GPSCAD percentile score was 69 for individuals with CAD vs. 49 for individuals without CAD. By analogy to the traditional analytic strategy for monogenic mutations, we defined ‘carriers’ as individuals with GPSCAD above a given threshold and ‘non-carriers’ as all others.
Figure 2. Risk for coronary artery disease according to genome-wide polygenic score.
(a) Distribution of genome-wide polygenic score for CAD (GPSCAD) in the UK biobank testing dataset (N=288,978). The x-axis represents GPSCAD, with values scaled to a mean of 0 and standard deviation of 1 to facilitate interpretation. Shading reflects proportion of population with 3, 4, and 5-fold increased risk versus remainder of the population. Odds ratio assessed in a logistic regression model adjusted for age, sex, genotyping array, and the first four principal components of ancestry; (b) GPSCAD percentile among CAD cases versus controls in the UK biobank validation cohort. Within each boxplot, the horizontal lines reflect the median, the top and bottom of the box reflects the interquartile range, and the whiskers reflect the maximum and minimum value within each grouping; (c) prevalence of CAD according to 100 groups of the validation cohort binned according to percentile of the GPSCAD.
We found that 8% of the population had inherited a genetic predisposition that conferred ≥3-fold increased risk for CAD (Table 2). Strikingly, the polygenic score identified 20-fold more people than found by familial hypercholesterolemia mutations in previous studies,6,7 at comparable or greater risk. Moreover, 2.3% of the population (‘carriers’) inherited ≥4-fold increased risk for CAD and 0.5% (‘carriers’) had inherited ≥5-fold increased risk. GPSCAD performed substantially better than two previously published polygenic scores for coronary artery disease that included 50 and 49,310 variants, respectively (Supplementary Table 7 and Supplementary Fig. 1).17,18
Table 2.
Proportion of the population at three-, four- and fivefold increased risk for each of the five common diseases
| High GPS definition | Individuals in testing dataset ( n ) | % of individuals |
|---|---|---|
| Odds ratio ≥3.0 | ||
| CAD | 23,119/288,978 | 8.0 |
| Atrial fibrillation | 17,627/288,978 | 6.1 |
| Type 2 diabetes | 10,099 288,978 | 3.5 |
| Inflammatory bowel disease | 9,209 288,978 | 3.2 |
| Breast cancer | 2,369/157,895 | 1.5 |
| Any of the five diseases | 57,115/288,978 | 19.8 |
| Odds ratio ≥4.0 | ||
| CAD | 6,631/288,978 | 2.3 |
| Atrial fibrillation | 4,335/288,978 | 1.5 |
| Type 2 diabetes | 578/288,978 | 0.2 |
| Inflammatory bowel disease | 2,297/288,978 | 0.8 |
| Breast cancer | 474/157,895 | 0.3 |
| Any of the five diseases | 14,029/288,978 | 4.9 |
| Odds ratio ≥5.0 | ||
| CAD | 1,443/288,978 | 0.5 |
| Atrial fibrillation | 2,020 288,978 | 0.7 |
| Type 2 diabetes | 144/288,978 | 0.05 |
| Inflammatory bowel disease | 571/288,978 | 0.2 |
| Breast cancer | 158/157,895 | 0.1 |
| Any of the five diseases | 4,305/288,978 | 1.5 |
For each disease, progressively more extreme tails of the GPS distribution were compared with the remainder of the population in a logistic regression model with disease status as the outcome, and age, sex, the first four principal components of ancestry, and genotyping array as predictors. The breast cancer analysis was restricted to female participants.
GPSCAD has the advantage that it can be assessed from the time of birth, well before the discriminative capacity emerges for risk factors (for example, hypertension or type 2 diabetes) used in clinical practice to predict CAD. Moreover, even for our middle-aged study population, practicing clinicians could not identify the 8% of individuals at ≥3-fold risk based on GPSCAD in the absence of genotype information (Supplementary Table 8). For example, conventional risk factors such as hypercholesterolemia was present in 20% of those with ≥3-fold risk based on GPSCAD versus 13% of those in the remainder of the distribution, hypertension in 32% versus 28%, and family history of heart disease in 44% versus 35%. Making high GPSCAD individuals aware of their inherited susceptibility may facilitate intensive prevention efforts. For example, we previously showed that a high polygenic risk for CAD may be offset by either of two interventions: adherence to a healthy lifestyle or cholesterol-lowering therapy with statin medications.19–21
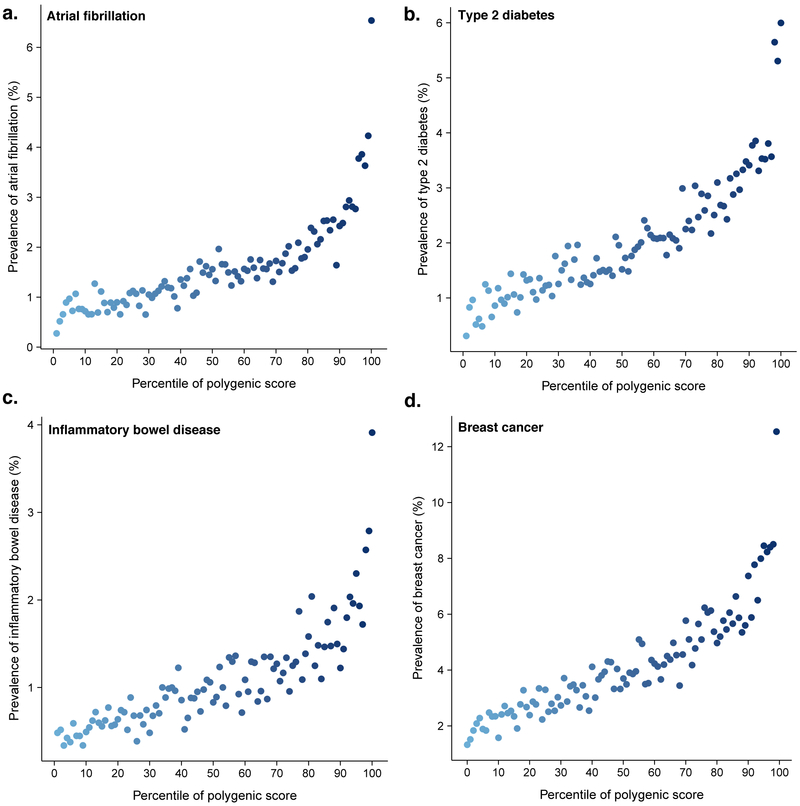
Our results for CAD generalized to four other diseases: risk increased sharply in the right tail of the GPS distribution (Figure 3). For each disease, the shape of the observed risk gradient was consistent with predicted risk based only on the GPS (Supplementary Figs. 2–3).
Figure 3. Risk gradient for disease according to genome-wide polygenic score percentile.
100 groups of the validation cohort were derived according to percentile of the disease-specific GPS. Prevalence of disease displayed for risk of (a) atrial fibrillation, (b) type 2 diabetes, (c) inflammatory bowel disease, and (d) breast cancer according to GPS percentile.
Atrial fibrillation is an underdiagnosed and often asymptomatic disorder in which an irregular heart rhythm predisposes to blood clots and is a leading cause of ischemic stroke.22 The polygenic predictor identified 6.1% of the population at ≥3-fold risk and the top 1% had 4.63-fold risk (Tables 2 & 3). Screening for atrial fibrillation has become increasingly feasible owing to the development of ‘wearable’ device technology; these efforts to increase detection may have maximal utility in those with high GPSAF.
Table 3.
Prevalence and clinical impact of a high GPS
| High GPS definition | Reference group | Odds ratio | 95% CI | P value |
|---|---|---|---|---|
| CAD | ||||
| Top 20% of distribution | Remaining 80% | 2.55 | 2.43–2.67 | <1 × 10−300 |
| Top 10% of distribution | Remaining 90% | 2.89 | 2.74–3.05 | <1 × 10−300 |
| Top 5% of distribution | Remaining 95% | 3.34 | 3.12–3.58 | 6.5 × 10−264 |
| Top 1% of distribution | Remaining 99% | 4.83 | 4.25–5.46 | 1.0 × 10−132 |
| Top 0.5% of distribution | Remaining 99.5% | 5.17 | 4.34–6.12 | 7.9 × 10−78 |
| Atrial fibrillation | ||||
| Top 20% of distribution | Remaining 80% | 2.43 | 2.29–2.59 | 2.1 × 10−177 |
| Top 10% of distribution | Remaining 90% | 2.74 | 2.55–2.94 | 7.0 × 10−169 |
| Top 5% of distribution | Remaining 95% | 3.22 | 2.95–3.51 | 1.1 × 10−152 |
| Top 1% of distribution | Remaining 99% | 4.63 | 3.96–5.39 | 2.9 × 10−84 |
| Top 0.5% of distribution | Remaining 99.5% | 5.23 | 4.24–6.39 | 3.5 × 10−56 |
| Type 2 diabetes | ||||
| Top 20% of distribution | Remaining 80% | 2.33 | 2.20–2.46 | 3.1 × 10−201 |
| Top 10% of distribution | Remaining 90% | 2.49 | 2.34–2.66 | 1.2 × 10−167 |
| Top 5% of distribution | Remaining 95% | 2.75 | 2.53–2.98 | 1.7 × 10−130 |
| Top 1% of distribution | Remaining 99% | 3.30 | 2.81–3.85 | 1.4 × 10−49 |
| Top 0.5% of distribution | Remaining 99.5% | 3.48 | 2.79–4.29 | 4.3 × 10−30 |
| Inflammatory bowel disease | ||||
| Top 20% of distribution | Remaining 80% | 2.19 | 2.03–2.36 | 7.7 × 10−95 |
| Top 10% of distribution | Remaining 90% | 2.43 | 2.22–2.65 | 8.8 × 10−88 |
| Top 5% of distribution | Remaining 95% | 2.66 | 2.38–2.96 | 3.0 × 10−68 |
| Top 1% of distribution | Remaining 99% | 3.87 | 3.18–4.66 | 1.4 × 10−43 |
| Top 0.5% of distribution | Remaining 99.5% | 4.81 | 3.74–6.08 | 9.0 × 10−37 |
| Breast cancer | ||||
| Top 20% of distribution | Remaining 80% | 2.07 | 1.97–2.19 | 3.4 × 10−159 |
| Top 10% of distribution | Remaining 90% | 2.32 | 2.18–2.48 | 2.3 × 10−148 |
| Top 5% of distribution | Remaining 95% | 2.55 | 2.35–2.76 | 2.1 × 10−112 |
| Top 1% of distribution | Remaining 99% | 3.36 | 2.88–3.91 | 1.3 × 10−54 |
| Top 0.5% of distribution | Remaining 99.5% | 3.83 | 3.11–4.68 | 8.2 × 10−38 |
Odds ratios were calculated by comparing those with high GPS with the remainder of the population in a logistic regression model adjusted for age, sex, genotyping array, and the first four principal components of ancestry. The breast cancer analysis was restricted to female participants. CI, confidence interval.
Type 2 diabetes is a key driver of cardiovascular and renal disease, with rapidly increasing global prevalence.23 The polygenic predictor identified 3.5% of the population at ≥3-fold risk and the top 1% had 3.30-fold risk. (Tables 2 & 3). Both medications and an intensive lifestyle intervention have been proven to prevent progression to type 2 diabetes,24 but widespread implementation has been limited by side effects and cost, respectively. Ascertainment of those with high GPST2D may provide an opportunity to target such interventions with increased precision.
Inflammatory bowel disease involves chronic intestinal inflammation and often requires lifelong anti-inflammatory medications or surgery to remove afflicted segments of the intestines.25 The polygenic predictor identified 3.2% of the population at ≥3-fold risk and the top 1% had 3.87-fold risk (Tables 2 & 3). Although no therapies to prevent inflammatory bowel disease are currently available, ascertainment of those with increased GPSIBD may enable enrichment of a clinical trial population to assess a novel preventive therapy.
Breast cancer is the leading cause of malignancy-related death in women. The polygenic predictor identified 1.5% of the population at ≥3-fold risk (Tables 2 & 3). Moreover, 0.1% of women had ≥5-fold risk of breast cancer—corresponding to a breast cancer prevalence of 19.0% in this group versus 4.2% in the remaining 99.9% of the distribution. The role of screening mammograms for asymptomatic middle-aged women has remained controversial owing to a low-incidence of breast cancer in this age group and a high false positive rate. Knowledge of GPSBC may inform clinical decision making about the appropriate age to recommend screening.26
The results above show that, for a number of common diseases, polygenic risk scores can now identify a substantially larger fraction of the population than found by rare monogenic mutations, at comparable or greater disease risk. Our validation and testing was performed in the UK Biobank population. Individuals who volunteered for the UK Biobank tended to be more healthy than the general population;27 although this nonrandom ascertainment is likely to deflate disease prevalence, we expect the relative impact of genetic risk strata to be generalizable across study populations. Additional studies are warranted to develop polygenic risk scores for many other common diseases with large GWAS data and validate risk estimates within population biobanks and clinical health systems.
Polygenic risk scores differ in important ways from the identification of rare monogenic risk factors. Whereas identifying carriers of rare monogenic mutations requires sequencing of specific genes and careful interpretation of the functional effects of mutations found, polygenic scores can be readily calculated for many diseases simultaneously, based on data from a single genotyping array. In our testing dataset, 19.8% of participants were at ≥3-fold increased risk for at least one of the five diseases studied (Table 2).
The potential to identify individuals at significantly higher genetic risk, across a wide range of common diseases and at any age, poses a number of opportunities and challenges for clinical medicine.
Where effective prevention or early detection strategies are available, key issues will include allocation of attention and resources across individuals with different levels of genetic risk and integration of genetic risk stratification with other risk factors—including rare monogenic mutations, clinical, and environmental factors. Where such strategies do not exist or are suboptimal, the identification of individuals at high risk should facilitate the design of efficient natural-history studies to discover early markers of disease onset and clinical trials to test prevention strategies. In both cases, it is important to recognize that the risk associated with a high polygenic score may not reflect a single underlying mechanism, but rather the combined influence of multiple pathways.28 Nonetheless, prevention and detection strategies may have utility regardless of underlying mechanism—as is the case for statin therapy for CAD, blood thinning-medications to prevent stroke in those with atrial fibrillation, or intensified mammography screening for breast cancer.
Risk communication will require serious consideration. While polygenic risk scores can be simultaneously calculated at birth for all common diseases, the usefulness of the knowledge and the potential harms to the individual may vary with the disease and stage of life—from juvenile diabetes to Alzheimer’s disease. Yet, it may not be feasible or appropriate to withhold information that can be readily calculated from genetic data. Moreover, it will be important to consider how to assess both absolute and relative risks and how to communicate these risks to best serve each patient—for example, to encourage the adoption of lifestyle modifications or disease screening.
Finally, we highlight a crucial equity issue. The polygenic risk scores described here were derived and tested in individuals of primarily European ancestry, the group in which most genetic studies have been undertaken to date. Because allele frequencies, linkage disequilibrium patterns, and effect sizes of common polymorphisms vary with ancestry, the specific GPS here will not have optimal predictive power for other ethnic groups.29 It will be important for the biomedical community to ensure that all ethnic groups have access to genetic risk prediction of comparable quality, which will require undertaking or expanding GWAS in non-European ethnic groups.
Online Methods:
Polygenic score derivation
Polygenic scores provide a quantitative metric of an individuals inherited risk based on the cumulative impact of many common polymorphisms. Weights are generally assigned to each genetic variant according to the strength of their association with disease risk (effect estimate). Individuals are scored based on how many risk alleles they have for each variant (for example, 0, 1, or 2 copies) included in the polygenic score.
For our score derivation, we used summary statistics from recent GWAS studies conducted primarily among participants of European ancestry for five diseases16,30–33 and a linkage disequilibrium reference panel of 503 European samples from 1000 Genomes phase 3 version 5.34 UK Biobank samples were not included in any of the five discovery GWAS studies. DNA polymorphisms with ambiguous strand (A/T or C/G) were removed from the score derivation. For each disease, we computed a set of candidate genome-wide polygenic scores (GPS) using the LDPred algorithm and a pruning and threshold derivation strategies.
The LDPred computational algorithm was used to generate seven candidate GPSs for each disease.13 This Bayesian approach calculates a posterior mean effect size for each variant based on a prior and subsequent shrinkage based on the extent to which this variant is correlated with similarly associated variants in the reference population. The underlying Gaussian distribution additionally considers the fraction of causal (e.g. non-zero effect sizes) markers via a tuning parameter, ρ. Because ρ is unknown for any given disease, a range of ρ, the fraction of causal variants, was used – 1, 0.3, 0.1, 0.03, 0.01, 0.003, 0.001.
A second approach, pruning and thresholding, was used to build an additional 24 candidate GPSs. Pruning and thresholding scores were built using a p-value and LD-driven clumping procedure in PLINK version 1.90b (--clump).35 In brief, the algorithm forms clumps around SNPs with association p-values less than a provided threshold. Each clump contains all SNPs within 250kb of the index SNP that are also in LD with the index SNP as determined by a provided r2 threshold in the LD reference. The algorithm iteratively cycles through all index SNPs, beginning with the smallest p-value, only allowing each SNP to appear in one clump. The final output should contain the most significantly disease-associated SNP for each LD-based clump across the genome. A GPS was built containing the index SNPs of each clump with association estimate betas (log odds) as weights. GPSs were created over a range of p-value (1, 0.5, 0.05, 5×10−4, 5×10−6, 5×10−8) and r2 (0.2, 0.4, 0.6, 0.8) thresholds, for a total of 24 pruning and thresholding-based candidate scores for each disease. The resulting GPS for a p-value threshold of 5×10−8 and r2 of < 0.2 was denoted the ‘GWAS significant variant’ derivation strategy.
Polygenic score calculation in the validation dataset
For each disease, the thirty-one candidate GPSs were calculated in a validation dataset of 120,280 participants of European ancestry derived from the UK Biobank Phase I release. The UK Biobank is a large prospective cohort study that enrolled individuals from across the United Kingdom, aged 40–69 years at time of recruitment, starting in 2006.14 Individuals underwent a series of anthropometric measurements and surveys, including medical history review with a trained nurse.
Scores were generated by multiplying the genotype dosage of each risk allele for each variant by its respective weight, and then summing across all variants in the score using PLINK2 software.35 Incorporating genotype dosages accounts for uncertainty in genotype imputation. The vast majority of variants in the GPSs were available for scoring purposes in the validation dataset with sufficient imputation quality (INFO > 0.3); Supplementary Tables 1–6.
For each of the five diseases, the score with the best discriminative capacity was determined based on maximal area under the receiver-operator curve (AUC) in a logistic regression model with the disease as the outcome and the disease-specific candidate GPS, age, sex, first four principal components of ancestry, and an indicator variable for genotyping array used (Supplementary Tables 1–6). AUC confidence intervals were calculated using the ‘pROC’ package within R.
Testing cohort
The testing dataset was comprised of 288,978 UK Biobank Phase 2 participants distinct from those in the validation dataset described above. Individuals in the UK Biobank underwent genotyping with one of two closely related custom arrays (UK BiLEVE Axiom Array or UK Biobank Axiom Array) consisting of over 800,000 genetic markers scattered across the genome.15 Additional genotypes were imputed centrally using the Haplotype Reference Consortium resource, the UK10K panel, and the 1000 Genomes panel. In order to analyze individuals with a relatively homogenous ancestry and owing to small percentages of non-British individuals, the present analysis was restricted to the white British ancestry individuals. This subpopulation was constructed centrally using a combination of self-reported ancestry and genetically confirmed ancestry using principal components. Additional exclusion criteria included outliers for heterozygosity or genotype missing rates, discordant reported versus genotypic sex, putative sex chromosome aneuploidy, or withdrawal of informed consent, derived centrally as previously reported.15
For each of the five diseases, proportion of variance explained was calculated for each disease using the Nagelkerke’s pseudo-R2 metric (Supplementary Table 9). The R2 was calculated for the full model inclusive of the genome-wide polygenic score plus the covariates minus R2 for the covariates alone, thus yielding an estimate of the explained variance. Covariates in the model included age, gender, genotyping array, and the first four principal components of ancestry.
A sensitivity analysis was performed by removing one individual from each pair of related individuals (third-degree or closer; kinship coefficient > 0.0442), confirming similar results within this subpopulation comprised of 222,529 of the 288,978 (77%) testing dataset participants (Supplementary Table 10).
Diagnosis of prevalent disease was based on a composite of data from self-report in an interview with a trained nurse, electronic health record (EHR) information including inpatient International Classification of Disease (ICD-10) diagnosis codes and Office of Population and Censuses Surveys (OPCS-4) procedure codes.
Coronary artery disease ascertainment was based on a composite of myocardial infarction or coronary revascularization. Myocardial infarction was based on self-report or hospital admission diagnosis, as performed centrally. This included individuals with ICD-9 codes of 410.X, 411.0, 412.X, 429.79 or ICD-10 codes of I21.X, I22.X, I23.X, I24.1, I25.2 in hospitalization records. Coronary revascularization was assessed based on an OPCS-4 coded procedure for coronary artery bypass grafting (K40.1–40.4, K41.1–41.4, K45.1–45.5) or coronary angioplasty with or without stenting (K49.1–49.2, K49.8–49.9, K50.2, K75.1–75.4, K75.8–75.9).
Atrial fibrillation ascertainment was based on self-report of atrial fibrillation, atrial flutter, or cardioversion in an interview with a trained nurse, ICD-9 codes of 427.3 or ICD-10 codes of I48.X in hospitalization records, or history of a percutaneous ablation or cardioversion based on OPCS-4 coded procedure (K57.1, K62.1, K62.2, K62.3, K 62.4) as performed previously.30
Type 2 diabetes ascertainment was based on self-report in an interview with a trained nurse or ICD-10 codes of E11.X in hospitalization records. Inflammatory bowel disease ascertainment was based on report in an interview with a trained nurse, ICD-9 codes of 555.X or ICD-10 codes of K51.X in hospitalization records.
Breast cancer ascertainment was based on self-report in an interview with a trained nurse, ICD-9 codes (174, 174.9) or ICD-10 codes (C50.X) in hospitalization records, or a breast cancer diagnosis reported to the national registry prior to date of enrollment.
Statistical analysis within the testing dataset
For each disease, the GPS with the best discriminative capacity in the testing dataset was calculated in the testing dataset of 288,278 participants using genotyped and imputed variants using the Hail software package.36 The proportion of the population and of diseased individuals with a given magnitude of increased risk was determined by comparing progressively more extreme tails of the distribution to the remainder of the population in a logistic regression model predicting disease status and adjusted for age, gender, four principal components of ancestry, and genotyping array. Individuals were next binned into 100 groupings according to percentile of the GPS and unadjusted prevalence of disease within each bin determined. We next compared the observed risk gradient across percentile bins to that which would be predicted by the GPS. For each individual, the predicted probability of disease was calculated using a logistic regression model with only the genome-wide polygenic score (GPS) as a predictor. The predicted prevalence of disease within each percentile bin of the GPS distribution was calculated as the average predicted probability of all individuals within that bin. The shape of the predicted risk gradient was consistent with the empirically observed risk gradient for each of the five disease (Supplementary Fig. 2–3).
Statistical analyses were conducted using R version 3.4.3 software (The R Foundation).
A Life Sciences Reproducibility Summary for this paper is available.
Supplementary Material
Acknowledgments:
UK Biobank analyses were conducted via application 7089 using a protocol approved by the Partners HealthCare Institutional Review Board. The analysis was supported by a KL2/Catalyst Medical Research Investigator Training award from Harvard Catalyst funded by the National Institutes of Health (TR001100) (A.V.K.), a Junior Faculty Research Award from the National Lipid Association (A.V.K.), the National Heart, Lung, and Blood Institute of the US National Institutes of Health under award numbers T32 HL007208 (K.A.), K23HL114724 (S.L.), R01HL139731 (S.L.), RO1HL092577 (P.E.), R01HL128914 (P.E.), K24HL105780 (P.E.), and RO1 HL127564 (S.K.), the National Human Genome Research Institute of the US National Institutes of Health under award number 5UM1HG008895 (S.L., E.L., S.K), the Doris Duke Charitable Foundation under award number 2014105 (S.L.), the Foundation Leducq under award number 14CVD01 (P.E.), and the Ofer and Shelly Nemirovsky Research Scholar Award from Massachusetts General Hospital (S.K.)
The authors thank Dr. David Altshuler (Vertex Pharmaceuticals; Boston, MA) for comments on an earlier version of this manuscript.
Competing financial interests:
Drs. Khera and Kathiresan are listed as co-inventors on a patent application for the use of genetic risk scores to determine risk and guide therapy. Drs. Kathiresan and Ellinor are supported by a grant from Bayer AG to the Broad Institute focused on the genetics and therapeutics of myocardial infarction and atrial fibrillation.
Footnotes
URLs:
1000 Genomes Phase 3, http://www.internationalgenome.org/category/phase-3/, UK Biobank, https://www.ukbiobank.ac.uk/; R statistical software, http://www.R-project.org/; PLINK 2.0, https://www.cog-genomics.org/plink/2.0/, Hail, https://github.com/hail-is/hail
Data availability statement
Genome-wide polygenic scores for each of the five diseases are available for research uses at: http://www.broadcvdi.org/informational/data.
References:
- 1.Green ED, Guyer MS; National Human Genome Research Institute. Charting a course for genomic medicine from base pairs to bedside. Nature. 470, 204–213 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Fisher RA The correlation between relatives on the supposition of Mendelian inheritance. Proc. Roy. Soc. Edinburgh 52, 99–433 (1918). [Google Scholar]
- 3.Gibson G Rare and common variants: twenty arguments. Nat Rev Genet. 18, 135–45 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Golan D, Lander ES, Rosset S. Measuring missing heritability: inferring the contribution of common variants. Proc Natl Acad Sci U S A. 111, E5272–81 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuchsberger C, et al. The genetic architecture of type 2 diabetes. Nature. 536, 41–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abul-Husn NS, et al. Genetic identification of familial hypercholesterolemia within a single U.S. health care system. Science. 354 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Nordestgaard BG, et al. Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: guidance for clinicians to prevent coronary heart disease: consensus statement of the European Atherosclerosis Society. Eur Heart J. 34, 3478–90a (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 536, 285–91 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Estrada K, et al. Association of a low-frequency variant in HNF1A with type 2 diabetes in a Latino population. JAMA. 311, 2305–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatterjee N et al. Projecting the performance of risk prediction based on polygenic analyses of genome-wide association studies. Nat Genet. 45, 400–405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, et al. Estimation of complex effect-size distributions using summary-level statistics from genome-wide association studies across 32 complex traits and implications for the future. Preprint at: https://www.biorxiv.org/content/early/2017/08/11/175406 (2017). [DOI] [PubMed]
- 12.Ripatti S, et al. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 327, 1393–400 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vilhjálmsson BJ et al. Modeling linkage disequilibrium increases accuracy of polygenic scores. Am J Hum Genet. 97, 576–592 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sudlow C et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bycroft C, et al. Genome-wide genetic data on ~500,000 UK Biobank participants. Preprint at: https://www.biorxiv.org/content/early/2017/07/20/166298 (2017).
- 16.Nikpay M et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 47,1121–1130 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tada H, et al. Risk prediction by genetic risk scores for coronary heart disease is independent of self-reported family history. Eur Heart J. 37, 561–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abraham G, et al. Genomic prediction of coronary heart disease. Eur Heart J. 37, 3267–3278 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khera AV, et al. Genetic risk, adherence to a healthy lifestyle, and coronary disease. N Engl J Med. 375, 2349–2358 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mega JL, et al. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. Lancet. 385, 2264–2271 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Natarajan P, et al. Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation. 135, 2091–2101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.January CT, et al. 2014 AHA/ACC/HRS guideline for the management of patients with atrial fibrillation: a report of the American College of Cardiology/American Heart Association Task Force on practice guidelines and the Heart Rhythm Society. Circulation. 130, e199–267 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.GBD 2015 Disease and Injury Incidence and Prevalence Collaborators. Global, regional, and national incidence, prevalence, and years live with disability for 310 diseases and injuries, 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 388, 1545–1602 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knowler WC, et al. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med. 346, 393–403 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abraham C & Cho JH Inflammatory bowel disease. N Engl J Med. 361, 2066–78 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pharoah PD, Antoniou AC, Easton DF, Ponder BA. Polygenes, risk prediction, and targeted prevention of breast cancer. N Engl J Med. 358, 2796–803 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Fry A, et al. Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population. Am J Epidemiol. 186, 1026–34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khera AV & Kathiresan S Is coronary atherosclerosis one disease or many? Setting realistic expectations for precision medicine. Circulation. 135, 1005–07 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin AR et al. Human demographic history impacts genetic risk prediction across diverse populations. Am J Hum Genet. 100, 635–649 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christophersen IE, et al. Large-scale analyses of common and rare variants identify 12 new loci associated with atrial fibrillation. Nat Genet. 49, 946–952 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott RA, et al. An Expanded Genome-Wide Association Study of Type 2 Diabetes in Europeans. Diabetes. 66, 2888–2902 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu JZ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 47, 979–986 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michailidou K, et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 551, 92–94 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang CC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ganna A, et al. Ultra-rare disruptive and damaging mutations influence educational attainment in the general population. Nat Neurosci. 19, 1563–65 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.