Abstract
The wild species of chickpea have tremendous potential for enhancing genetic gains of cultigen and have resistant accessions against major biotic and abiotic stresses. In the present study, two wild annual accessions, one each of C. reticulatum Ladiz. (ILWC 229) and C. echinospermum Davis (ILWC 246) were assessed for their agro-morphological features and hybridized with different cultivated varieties (BGD 72, PBG 5, ICKG 96029, Pusa 372 and JG 11) of chickpea. Fertile F1 plants were developed as revealed by their normal meiotic chromosomal configuration including high pollen stainability percentage and seed set. The effect of genetic and non-genetic factors on crossability performance with respect to pod and seed set was also evident under two growing conditions of North-Western Indian Himalayas. The segregation analysis using F2 phenotypic ratio of some distinct morphological (plant growth habit, stem pigmentation at seedling stage and testa texture) characters indicated their monogenic inheritance pattern. The study would also be useful to chickpea breeders to identify true to type interspecific plants. Further, the F1, F2 and F3 generations of all seven crosses along with parents were evaluated under natural field condition to determine the extent of variability created into the cultivated background of chickpea. There was a wide range of variation in F3 population against cold stress, suggesting selection of tolerant recombinant lines at an early stage. We also studied fruitful heterosis (%) as a useful approach, instead of residual heterosis to identify better performing transgressive segregants. The values of most of the interspecific crosses for important traits assessed in F2 and F3 generations were higher than that of better parent, suggesting isolation of inbred vigour for pod numbers and earliness. The results indicated that wild Cicer annual accessions of C. reticulatum and C. echinospermum species can be exploited after proper screening for traits of interest for diversification of cultivated gene pool and subsequent use in chickpea improvement.
Introduction
Chickpea (Cicer arietinum L.), is a true diploid (2n = 2x = 16) annual grain legume having genome size of ~738 Mbp [1]. It ameliorates soil fertility through biological nitrogen fixation and also plays a significant role in human diet as a useful source of protein, vitamins and minerals for ever increasing populations in the developing world [2]. As far as chickpea acreage is concerned, it is the second largest pulse crop after dry bean with 14 million hectare area under cultivation [3]. However, the world average yield is 982 kg/ha, which is far below than the actual potential yield of crop [3] because it is generally sown in marginal land with less inputs under rainfed agricultural production system [4]. The traditional breeding methods have not developed much high yielding varieties with large-scale impact on production and productivity of chickpea [5, 6]. Till date, more than 350 improved and 25 mutant varieties have been released by several crop improvement institutions worldwide [7, 8] and the crop has also been classified into desi and kabuli types characterized by its seed size, shape and color [9]. Further, the narrow genetic base of chickpea varieties does not provide much contrasting features for developing improved cultivars [10]. During the process of crop domestication, certain useful productivity genes and alleles were lost, which resulted in narrow genetic base of domesticated species [1, 2, 11–18]. Therefore, chickpea breeders are looking at crop wild relatives (CWRs) as an alternative source of variation for tailoring new germplasm [2, 19]. The genus Cicer comprises of 49 taxa [20, 21] and only two wild Cicer species, C. reticulatum Ladiz. and C. echinospermum Davis are crossable with cultivated gene pool [7]. The wild Cicer species consists of useful characters for distinct morphological, agronomical, protein content, and resistance against major biotic and abiotic stresses [22–25]. The present study was, therefore, undertaken to characterize, identify and introgress wild Cicer species and appearance of transgression for important quantitative traits, thus exhibiting the favorable effects to determine the extent of variability created into the background of different cultivated chickpea varieties.
Materials and methods
Morphological characterization and meiotic study
A total of 88 global wild annual Cicer accessions comprising 20 of C. reticulatum Ladizinsky, 10 of C. echinospermum P.H. Davis, 25 each of C. judaicum Boiss. and C. pinnatifidum Jarb. & Spach, 6 of C. bijugum Rech. f., and 2 of C. yamashitae Kitam. were characterized for distinct morphological characters viz. plant pigmentation, plant hairiness, number of leaflets leaf-1, seed shape, testa texture and seed color. These were also evaluated for important agro-morphological traits (days to flowering, days to maturity, plant height (cm), number of branches plant-1, number of pods plant-1, number of seeds pod-1, 100-seed weight (g), seed yield plant-1 (g) and biological yield plant-1 (g) in two cropping seasons at two different locations. The wild species were introduced from Biodiversity and Integrated Gene Management (BIGM) Unit at ICARDA, then in Aleppo, Syria during 2010–2011. Further, the characterization and evaluation study has helped us in identifying certain useful accessions including ILWC 229 of C. reticulatum (resistant to ascochyta blight and desirable pod numbers) and ILWC of C. echinospermum (resistant to botrytis grey mold and desirable pod numbers) found useful for their introgression into the background of cultivated varieties. Meiotic analysis was carried out by taking appropriate flower buds from the growing plants in pots under green house condition. The flower buds were taken from 25 randomly selected plants of each accession and fixed in Carnoy’s fixative solution (6:3:1 ethanol/chloroform/acetic acid) for 24 h [26]. The collected flower buds were washed properly and preserved in 70% ethanol at 4°C until its use for cytological study. Further, smears of appropriate buds were made in acetocarmine solution [27]. About 25–30 fresh slides of each accession were prepared from different flower buds to draw the valid conclusions. To count chromosome number, about 40 pollen mother cells (PMCs) were observed in different stages at diakinesis/ metaphase-I/anaphase-I and II. Pollen stainability was also assessed by mounting mature pollen grains (PGs) in glycero–acetocarmine (1:1) mixture. About 350–400 pollen grains were analyzed in each case for assessing pollen viability including pollen size. Filled pollen with stained nuclei was recorded as fertile grains, while shrivelled and unstained grains were taken as infertile. Photomicrographs of pollen mother cells and pollen grains were also made from freshly prepared slides using Nikon 80i eclipse Digital Imaging Microscope [26].
Interspecific hybridization of chickpea
The interspecific hybridization experiments were undertaken at the ICAR-National Bureau of Plant Genetic Resources (NBPGR) Pusa, New Delhi (28◦ 35′ N′, 70◦ 18′ E, 226m amsl) and the Mountain Agricultural Research and Extension Centre (MAREC) of CSKHPKV, Sangla (31◦ 55′ and 32◦ 20′ N and 77◦ 00′ and 79◦ 50′ E, 2,758m amsl) Himachal Pradesh, India. Wild Cicer accessions, ILWC 229 (C. reticulatum Ladiz.) and ILWC 246 (C. echinospermum Davis) were intercrossed with five released varieties of chickpea viz. BGD 72, PBG 5, ICKG 96029, Pusa 372 and JG 11. Total seven interspecific cross-combinations of BGD 72 x ILWC 229, PBG 5 x ILWC 229, BGD 72 x ILWC 246, PBG 5 x ILWC 246, ICKG 96029 x ILWC 246, Pusa 372 x ILWC 246 and JG 11 x ILWC 246 were successfully obtained. To accomplish wide hybridization studies, the emasculation was carried out between 3 to 5 P.M. at both the centers. Pollination was done on next day morning between 8:30 to 10:00 A.M. with mature pollen grains of the male parents as per standard procedure [28, 29]. A solution of growth hormones (GA3-120 ppm + NAA-30 ppm + Kinetin-15 ppm) was also applied to the base of peduncle and pollinated flower buds after pollination to prevent premature pod abscission [29, 30]. The resulted F1 seeds of all plants were grown at NBPGR, New Delhi, to obtain F2 seeds of each cross-combination during winter season of 2013–2014, and pollen fertility (% stainable pollen) of F1 plants was also determined by staining mature pollen grains with 2% acetocarmine solution. However, F2 seeds of all crosses were advanced to produce F3 seeds and evaluated for important agro-morphological characters in the summer season of 2014 at Sangla.
Field evaluation of parents and their advanced progenies
The genetic materials comprising of F1, F2 and F3 progenies alongwith parental lines were sown in the Experimental Farm of NBPGR, Regional Station Shimla (31◦ 05′ 53′ N and 77◦ 09′ 35′ E 1924 m amsl) during winter season of 2015–16. The seeds were planted in 3 m long rows spaced at 10 cm (plant to plant) and 40 cm (row to row) apart. One pre-sowing irrigation was applied to ensure satisfactory seed germination. Recommended cultural practices were followed for raising the genetic materials. During the whole cropping season, one rain and two snow falls was experienced and necessity of manual irrigation was not felt. Five plants of each parental line and F1 plants, and all available plants of F2 and F3 populations were selected for recording data on days to flowering, days to maturity, plant height (cm), number of branches plant-1, number of pods plant-1, 100-seed weight (g), seed yield plant-1 (g), and biological yield plant-1 (g). Fruitful heterosis was also calculated following Koseoglu et al. [31] in both F2 and F3 generations as HF (%) = [(F2, F3-BP)/BP] × 100, where, F2 and F3 are generations of interspecific populations and BP is the mean of better parent of a cross.
Screening of F3 progeny against cold tolerance
All F3 interspecific plant populations of seven crosses were screened against cold tolerance under natural field condition at Shimla centre. The experiment was conducted in Augmented Block Design [32] in which the best cold tolerant accession, IC31649 was used as standard check repeated after every 25th rows. For visual screening of interspecific plant populations against cold tolerance, a 1–9 rating scale was used [33] where 1- No visible expression of damage; 2- Highly tolerant, up to 10% leaflets shows drying no killing; 3- Tolerant, 11–20% leaflets shows withering, but no killing; 4- Moderately tolerant, 21–40% leaflets shows withering and drying; 5- Intermediate, 41–60% leaflets show withering and drying leaflets, along with 5% plant killing; 6- Moderately susceptible, 61–80% leaflets shows withering and drying symptoms and 6–25% plant killing; 7- Susceptible, 81–99% leaflets shows withering and drying along with 26–50% plant killing; 8- Highly susceptible, 100% leaflets shows withering and drying symptoms, and 51–99% plant killing; 9–100% plants were killed from cold.
Further, observations were recorded three times during the cropping season and all plants were also covered in fresh snow for three days, during winter season of 2016–2017. The lowest average mean temperature (2.8°C) was recorded during flowering stage. The average weather conditions (from October 2016 to April 2017) of cropping period, total rainfall, maximum, minimum temperatures and relative humidity (%) is given in Table 1.
Table 1. Total rainfall, maximum, minimum temperatures and relative humidity during cropping period for screening of F3 progeny against cold tolerance.
| Months | Rainfall (mm) | Temperature (0C) | Relative humidity (%) | ||
|---|---|---|---|---|---|
| Max | Min | Max | Min | ||
| October | 2.30 | 22.90 | 12.90 | 60.39 | 50.32 |
| November | 0.00 | 21.00 | 10.56 | 45.70 | 34.20 |
| December | 5.50 | 19.00 | 8.38 | 48.97 | 36.97 |
| January | 148.10 | 12.50 | 2.80 | 69.52 | 58.00 |
| February | 19.60 | 17.38 | 6.47 | 55.32 | 42.86 |
| March | 36.10 | 25.05 | 7.77 | 59.23 | 48.32 |
| April | 85.20 | 24.78 | 13.18 | 55.00 | 43.10 |
Statistical analysis
The genetic materials comprised parental lines and advanced progenies, which were studied for segregation analysis of important distinct morphological characters. Each F2 plant was observed for contrasting traits and the chi-square test for goodness of fit was also estimated. The X2 value is calculated from observed (O) and expected (E) results from total populations using following formula:
Where, ∑ refers to sum of values of X2 over the classes of an experiment. Yate’s factor was also used, where the population size was small. Further, the means were adjusted using online software package for augmented block design (ABD) developed by Rathore et al. [32]. The quantitative characters were further analyzed for various statistical parameters viz. range, mean, coefficient of variation, and fruitful heterosis (%) using the statistical software SYSTAT-12. For screening chickpea F3 plant populations against cold tolerance, data were taken using 1–9 rating scale and further analysis following MS Office Excel and SAS software (SAS 2011).
Results
Morphological characterization and meiotic study
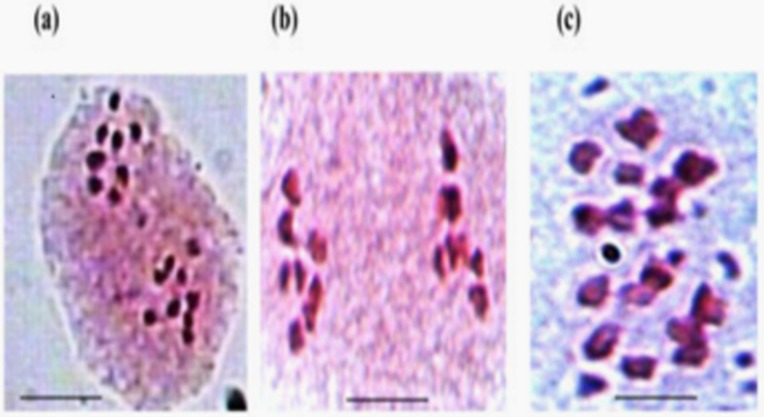
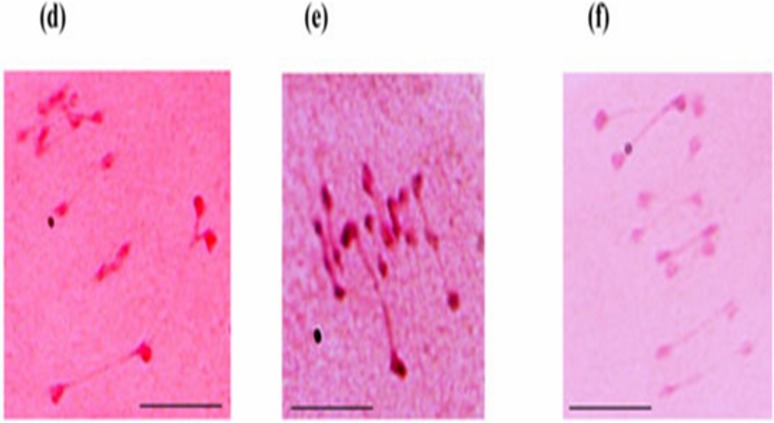
Both the wild annual accessions of C. reticulatum Ladiz. (ILWC 229) and C. echinospermum Davis (ILWC 246) were studied for important morphological characters viz. plant pigmentation, plant hairiness, number of leaflets leaf-1, seed shape, testa texture, flower colour and seed colour. The plant pigmentation showed low anthocyanin in ILWC 229 and no anthocyanin in ILWC 246. Light pubescence on leaves was observed in both the species. The number of leaflets leaf-1 was 9–11 in ILWC 229 and 11–13 in ILWC 246. Both the species expressed angular shape of seeds. The texture of testa was rough in ILWC 229 and it was tuberculated in ILWC 246. An accession ILWC 229 produced brown seed, while ILWC 246 black in color. Both the wild species showed normal chromosome pairing producing 8 bivalents (2n = 2x = 16) along with high percentage of pollen stainability. However, its pollen viability percentage ranged from 81.76 (pollen size 22.23 × 17.23 μm) in ILWC 246 to 100 (pollen size 24.23 × 18.56 μm) in ILWC 229 (Fig 1). The F1’s of all interspecific cross-combinations showed normal meiosis along with high pollen viability percentage (Fig 2).
Fig 1.
Pollen mother cells (PMCs) of wild annual Cicer species indicates normal breeding behavior where (a) accession ILWC 229 (C. reticulatum) showing usual separation of chromosomes at anaphase I, (b) accession ILWC 246 (C. echinospermum) indicates stable late anaphase, (c) accession ILWC 246 (C. echinospermum) chromosomes arranged in metaphase. Scale = 10 μm.
Fig 2.
Meiotic analysis of F1s of (d) C. arietinum x C. reticulatum showing normal separation of chromosomes at anaphase II and (e and f) of C. arietinum x C. echinospermum exhibiting normal division of chromosomes at metaphase stage. Scale = 10 μm.
Seed set in F1 hybrids
The seed set in F1 hybrids under two agro-ecological conditions was recorded with respect to number of cross-pollinations attempted, pod set, seed set, number of seeds pod-1 and pollen viability (Table 2). The observations recorded in first set of wide hybridization experiment resulted the production of 54 healthy F1 seeds, of 907 cross-pollinations attempted. The maximum seed set (8.03%) was recorded for JG 11 x ILWC 246. The seed set percentage ranged from 4.48 (ICKG 96029 x ILWC 246) to 8.03 (JG 11 x ILWC 246) with an average seed set of 6.44%. Number of seeds pod-1 was observed more than one in all F1 crosses and pollen viability percentage ranged from 88.12 (ICKG 96029 x ILWC 246) to 93.10 (BGD 72 x ILWC 229). An average maximum/minimum temperature was recorded as 31.4/19.2°C. Identical observations were also reported in the second set of experiment, which resulted in production of 66 F1 seeds of 813 cross-pollinations. The highest seed set (10.67%) was observed for PBG 5 x ILWC 229. Pollen stainability percentage ranged from 87.10 (BGD 72 x ILWC 246) to 92.12 (BGD 72 x ILWC 229). The maximum/minimum summer season Himalayan nursery temperature was observed as 26.3/12.5°C. It is summarized that influence of genetic (species/accession groups) and non-genetic factors (day length, growing conditions and temperature) on seed set played a vital role under varied growing conditions.
Table 2. Number of cross-pollinations attempted, pod set, seed set, seeds pod-1 and pollen viability (%) during winter 2012–13 at New Delhi and summer Himalayan nursery 2013 at Sangla.
| No. of cross-pollinations attempted | Pod set | Seed set | Number of seeds crossed pod-1 | Pollen viability (%) of F1 plants | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| S.N. | Cross-combination | Delhi | Sangla | Delhi | Sangla | Delhi | Sangla | Delhi | Sangla | Delhi | Sangla |
| 1 | BGD 72 x ILWC 229 | 150 | 147 | 10 (6.66) μ | 12 (8.16) | 10 (6.66) | 12 (8.16) | 1.00 | 1.00 | 93.10 | 92.12 |
| 2 | PBG 5 X ILWC 229 | 114 | 103 | 09 (7.89) | 10 (9.70) | 08 (7.01) | 11 (10.67) | 1.12 | 1.10 | 91.10 | 90.11 |
| 3 | BGD 72 X ILWC 246 | 112 | 105 | 06 (5.35) | 10 (9.52) | 06 (5.35) | 07 (6.66) | 1.00 | 0.70 | 90.11 | 87.10 |
| 4 | PBG 5 X ILWC 246 | 111 | 110 | 06 (5.40) | 10 (9.09) | 07 (6.30) | 09 (8.18) | 1.16 | 1.11 | 89.10 | 89.12 |
| 5 | ICKG 96029 X ILWC 246 | 156 | 113 | 08 (5.12) | 09 (7.96) | 07 (4.48) | 09 (7.96) | 1.14 | 1.00 | 88.12 | 90.00 |
| 6 | PUSA 372 X ILWC 246 | 152 | 119 | 09 (5.92) | 11 (9.24) | 07 (4.60) | 09 (7.56) | 1.28 | 1.22 | 91.11 | 88.88 |
| 7 | JG 11 X ILWC 246 | 112 | 116 | 09 (8.03) | 11 (9.82) | 09 (8.03) | 09 (7.75) | 1.00 | 1.22 | 90.12 | 90.10 |
| Average performance | 129.57 | 116.14 | 8.14 (6.42) | 10.42 (9.15) | 7.71 (6.44) | 9.42 (8.60) | 1.09 | 1.02 | 90.03 | 89.63 | |
*μ Percentage of cross-pollinations attempted in parenthesis; Average max/min winter temperature during whole crossing period 31.4/ 19.2°C at Delhi; Average max/min summer temperature during whole crossing period 26.3/12.5°C at Sangla
Inheritance of distinct morphological traits
Plant growth habit
Two interspecific crosses were made between PBG 5 (erect growth habit) and ILWC 229 and ILWC 246 having spreading growth habit. The F1 plants showed erect growth indicating that erect plant growth is dominant over spreading habit. In F2 generation, 373 plants showed erect plant type and 144 plants showed spreading habit for cross PBG 5 x ILWC 229, while 181 plants showed erect plant type and 78 showed spreading growth habit for cross PBG 5 x ILWC 246 (Table 3).
Table 3. Inheritance pattern of distinct morphological traits in cultivated x wild chickpea crosses.
| Trait | Cross | Generation | Observed progeny | Expected ratio | Chi-square | P value |
|---|---|---|---|---|---|---|
| Plant growth habit | PBG 5 × ILWC 229 | F1 | 18 (erect) | - | - | - |
| F2 | 373 (erect) : 144 (spreading) | 3:1 | 2.24 | 0.2–0.3 | ||
| PBG 5 × ILWC 246 | F1 | 20 (erect) | - | - | - | |
| F2 | 181(erect) : 78 (spreading) | 3:1 | 2.61 | 0.3–0.5 | ||
| Stem pigmentation at seedling stage | Pusa 372 x ILWC 229 | F1 | 18 (pigmented) | - | - | - |
| F2 | 172 (pigmented) : 114 (non-pigmented) | 9:7 | 1.76 | 0.3–0.5 | ||
| PBG 5 x ILWC 229 | F1 | 16 (pigmented) | - | - | - | |
| F2 | 276 (pigmented) : 241 (non-pigmented) | 9:7 | 1.72 | 0.2–0.5 | ||
| Testa Texture | Pusa 256 x ILWC 229 | F1 | 20 (rough) | - | - | - |
| F2 | 94 (rough) : 33 (smooth) | 3:1 | 0.07 | 0.3–0.5 | ||
| BGD 72 x ILWC 229 | F1 | 18 (rough) | - | - | - | |
| F2 | 82 (rough) : 34 (smooth) | 3:1 | 1.15 | 0.2–0.3 |
Stem pigmentation at seedling stage
The stem pigmentation at seedling stage was studied in two interspecific crosses of Pusa 372 x ILWC 229 and PBG 5 x ILWC 229. The F1 plants expressed pigmented stems at seedling stage indicating that the trait is dominant over normal or non-pigmented, and in F2 generation of both crosses, the trait segregated into 9 pigmented: 7 non-pigmented ratio (Table 3).
Testa texture
The trait testa texture was studied in two interspecific crosses of Pusa 256 x ILWC 229 and BGD 72 x ILWC 229. The F1 hybrids of each cross exhibited rough texture, indicating that it is dominant over smooth (Table 3) and F2 generation of both crosses segregated into rough and smooth seeds fitting well into the ratio of 3 rough : 1 smooth texture pattern.
Field evaluation of parental lines and their advanced progenies
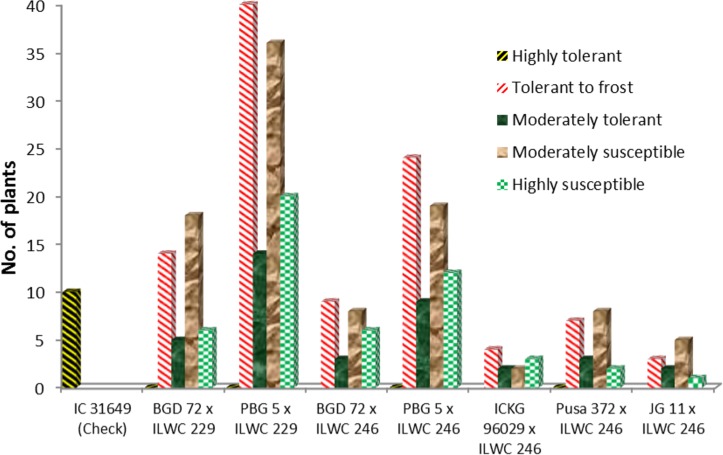
The parental lines, their F1s, F2s and F3s were evaluated under natural field conditions to assess the variability created through transgressive segregants of various interspecific crosses (Table 4). The F1s, involving early flowering and maturing cultivated parents (BGD 72, PBG 5, ICKG 96029, Pusa 372 and JG 11) and late flowering wild accessions (ILWC 229 and ILWC 246) showed interesting comparability. For the trait plant height, all F1 plants were tall for majority of crosses, and in F2 and F3 generations, it revealed variability through segregations towards dwarf to taller plants. However, number of pods plant-1 also manifested desirable performance for majority of F1 crosses, and in F2 and F3 generations, a wide range of variation occurred in all wide cross-combinations such as plant height, number of branches-1, number of pods-1 and seed yield plant-1. The seed yield plant-1, exhibited desirability in F1 hybrids, while in F2, and F3 derivatives, a substantial variation did appear in majority of interspecific cross-combinations of BGD 72 x ILWC 229, PBG 5 x ILWC 229, BGD 72 x ILWC 246, PBG 5 x ILWC 246, ICKG 96029 x ILWC 246 and Pusa 372 x ILWC 246. Likewise, for biological yield plant-1, F1 hybrids of almost all crosses manifested high biomass score as compared to their parents, and in F2 and F3 generations, variability appeared for this trait from low to high biomass yield. Furthermore, the F3 plant populations of all interspecific crosses were screened against cold tolerance and results showed that in each cross-combination, some interspecific recombinant lines exhibited complete tolerance against cold stress. The maximum number of plants were observed tolerant against cold stress in cross- combination of PBG 5 x ILWC 229 followed by PBG 5 x ILWC 246 and BGD 72 x ILWC 229. None of the plants were recorded highly tolerant against cold stress under natural field condition. An average mean temperature for the whole cropping season is presented in Table 1 and the distribution of plant populations of each interspecific cross in different resection against cold is given in Fig 3.
Table 4. Range, mean, standard error and coefficient of variation for agro-morphological traits in different generations of crosses involving cultivated x wild chickpea.
| Character | P1 | P2 | F1 | F2 | F3 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Range | Mean±SE | CV (%) | Range | Mean±SE | CV (%) | Range | Mean±SE | CV (%) | Range | Mean±SE | CV (%) | Range | Mean±SE | CV (%) | |
| Days to flowering | |||||||||||||||
| BGD 72 x ILWC 229 | 62.0–72.0 | 67.3±2.9 | 7.4 | 75–80 | 77.5±2.5 | 4.5 | 78.1–89 | 84.0±0.2 | 13.7 | 68–91 | 76.1±2.7 | 10.3 | 83.0–92.0 | 87.1±0.41 | 2.13 |
| PBG 5 x ILWC 229 | 67.0–71.0 | 69.0±1.1 | 2.8 | 75–80 | 77.5±2.5 | 4.5 | 72.0–86.0 | 80.3±2.4 | 8.6 | 55–117 | 75.0±1.8 | 17.9 | 81.0–89.0 | 84.2±0.25 | 2.15 |
| BGD 72 x ILWC 246 | 62.0–72.0 | 67.3±2.9 | 7.4 | 65–70 | 67.3±2.1 | 3.7 | 80.0–89.0 | 86.8±3.4 | 7.9 | 67–98 | 79.8±4.5 | 14.9 | 83.0–88.0 | 85.7±0.41 | 1.73 |
| PBG 5 x ILWC 246 | 67.0–71.0 | 69.0±1.1 | 2.8 | 65–70 | 67.3±2.1 | 3.7 | 90.0–96.0 | 93.7±3.6 | 6.6 | 60–115 | 83.4±3.1 | 20.3 | 89–99.0 | 96.3±0.40 | 2.43 |
| ICKG 96029 x ILWC 246 | 57.0–66.0 | 60.6±2.7 | 7.7 | 65–70 | 67.3±2.1 | 3.7 | 95.0–102 | 99.0±0.5 | 0.9 | 57–108 | 72.5±6.8 | 24.8 | 92.0–104 | 102.8±1.55 | 1.60 |
| PUSA 372 x ILWC 246 | 64.0–67.0 | 65.3±0.8 | 2.3 | 65–70 | 67.3±2.1 | 3.7 | 89.0–96.0 | 93.0±0.5 | 0.9 | 75.0–96.0 | 84.5±5.0 | 15.6 | 85.8–93.6 | 89.6±0.70 | 2.52 |
| JG 11 x ILWC 246 | 56.0–67.0 | 60.6±3.2 | 9.3 | 65–70 | 67.3±2.1 | 3.7 | 87.0–99.9 | 93.0±0.5 | 0.7 | 65.0–93.0 | 80.3±7.4 | 22.5 | 82.0–97.2 | 94.5±0.50 | 1.39 |
| Days to maturity | |||||||||||||||
| BGD 72 x ILWC 229 | 114.0–122.0 | 117.6±2.3 | 2.9 | 125–135 | 130.0±2.0 | 8.2 | 110–152 | 146.6±0.6 | 0.6 | 115–132 | 124.1±6.2 | 9.1 | 114–125 | 119.1 ± 0.57 | 1.84 |
| PBG 5 x ILWC 229 | 125.0–130.0 | 122.6±1.4 | 1.4 | 125–135 | 130.0±2.0 | 8.2 | 116–126 | 112.0±1.5 | 4.1 | 116–140 | 132.9±3.2 | 15.2 | 123–135 | 129.3 ± 0.29 | 1.55 |
| BGD 72 x ILWC 246 | 114.0–122.0 | 117.6±2.3 | 2.9 | 115–131 | 127.6±1.4 | 1.4 | 140–154 | 151.2±0.7 | 1.0 | 135–142 | 139.1±2.1 | 3.2 | 128–150 | 143.8 ± 0.60 | 1.58 |
| PBG 5 x ILWC 246 | 125.0–130.0 | 117.6±1.4 | 1.4 | 115–131 | 127.6±1.4 | 1.4 | 110–126 | 121.1±0.7 | 1.3 | 119–128 | 122.0±3.1 | 12.1 | 125–132 | 119.1 ± 0.26 | 1.01 |
| ICKG 96029 x ILWC 246 | 143.0–148.0 | 145.3±1.4 | 1.7 | 115–131 | 127.6±1.4 | 1.4 | 119–125 | 120.0±0.5 | 0.5 | 114–147 | 134.7±5.4 | 10.6 | 113–141 | 136.9 ± 0.83 | 1.25 |
| PUSA 372 x ILWC 246 | 145.0–149.0 | 146.6±1.2 | 1.4 | 115–131 | 127.6±1.4 | 1.4 | 120–127 | 123.0±1.7 | 1.7 | 121–138 | 133.2±9.5 | 16.4 | 115–139 | 123.2 ± 0.91 | 2.1 |
| JG 11 x ILWC 246 | 131.0–175.0 | 146.0±14.5 | 17.1 | 115–131 | 127.6±1.4 | 1.4 | 120–127 | 121.3±0.8 | 0.8 | 112–131 | 117.0±5.3 | 9.4 | 124–146 | 137.3± 0.82 | 1.44 |
| Plant height (cm) | |||||||||||||||
| BGD 72 x ILWC 229 | 40.0–52.0 | 44.6±3.7 | 14.3 | 29.0–51.0 | 40.0±11.0 | 38.8 | 68.0–76.0 | 71.3±2.4 | 5.8 | 30.0–63.0 | 39.6±3.6 | 25.8 | 34.8–65.2 | 52.9 ±1.28 | 15.5 |
| PBG 5 x ILWC 229 | 17.0–22.0 | 19.6±1.4 | 12.7 | 29.0–51.0 | 40.0±11.0 | 38.8 | 53.0–84.0 | 64.7±1.7 | 11.4 | 8.0–87.0 | 56.8±2.2 | 28.6 | 34.2–72.4 | 52.6 ±0.92 | 18.4 |
| BGD 72 x ILWC 246 | 40.0–52.0 | 44.6±3.7 | 14.3 | 41.0–50.0 | 44.6±2.7 | 10.5 | 57.0–65.0 | 60.8±1.3 | 4.8 | 32.0–70.0 | 51.5±5.6 | 28.8 | 38.4–69.4 | 53.4±1.63 | 15.1 |
| PBG 5 x ILWC 246 | 17.0–22.0 | 19.6±1.4 | 12.7 | 41.0–50.0 | 44.6±2.7 | 10.5 | 42.0–84.0 | 68.5±5.6 | 23.2 | 36.0–81.0 | 55.8±3.1 | 30.7 | 38.3–78.4 | 58.5 ±1.26 | 17.2 |
| ICKG 96029 x ILWC 246 | 57.0–63.0 | 60.0±1.7 | 5 | 41.0–50.0 | 44.6±2.7 | 10.5 | 48.0–52.0 | 50.0±1.1 | 4.0 | 28.0–83.0 | 54.0±8.4 | 41.5 | 37.8–70.8 | 56.1±3.30 | 19.5 |
| PUSA 372 x ILWC 246 | 28.0–41.0 | 34.6±3.7 | 18.7 | 41.0–50.0 | 44.6±2.7 | 10.5 | 51.0–54.0 | 52.3±0.8 | 2.9 | 43.0–82.0 | 55.4±5.2 | 25.2 | 32.4–59.5 | 40.1±1.31 | 14.7 |
| JG 11 x ILWC 246 | 65.0–71.0 | 68.3±1.7 | 4.4 | 41.0–50.0 | 44.6±2.7 | 10.5 | 56.0–63.0 | 59.0±2.0 | 6.1 | 45.0–67.0 | 53.8±3.8 | 17.2 | 34.4–53.8 | 42.3±1.74 | 14.3 |
| No. of branches plant-1 | |||||||||||||||
| BGD 72 x ILWC 229 | 4.0–7.0 | 5.6±0.8 | 26.8 | 9.0–12.0 | 10.5±1.5 | 20.1 | 10.0–15.0 | 12.6±1.4 | 19.8 | 2.0–12.0 | 7.0±1.2 | 48.8 | 7.0–23.0 | 15.4±0.57 | 23.6 |
| PBG 5 x ILWC 229 | 7.0–11.0 | 9.0±1.1 | 22.2 | 9.0–12.0 | 10.5±1.5 | 20.1 | 6.0–19.0 | 12.0±0.8 | 29.4 | 3.0–37.0 | 13.3±0.9 | 50.2 | 7.0–25.0 | 15.8±0.35 | 22.9 |
| BGD 72 x ILWC 246 | 4.0–7.0 | 5.6±0.8 | 26.8 | 8.0–14.0 | 11.0±1.7 | 27.2 | 13.0–30.0 | 19.6±2.8 | 32.9 | 8.0–16.0 | 11.4±1.3 | 31.8 | 7.0–19.0 | 12.3±0.51 | 21.2 |
| PBG 5 x ILWC 246 | 7.0–11.0 | 9.0±1.1 | 22.2 | 8.0–14.0 | 11.0±1.7 | 27.2 | 6.0–17.0 | 10.8±1.2 | 32.3 | 3.0–23.0 | 9.4±0.8 | 47.2 | 7.0–24.0 | 14.2±0.48 | 26.9 |
| ICKG 96029 x ILWC 246 | 3.0–6.0 | 4.3±0.8 | 35.1 | 8.0–14.0 | 11.0±1.7 | 27.2 | 16.0–21.0 | 19.0±1.5 | 13.8 | 4.0–12.0 | 8.0±1.0 | 34.5 | 7.0–18.0 | 10.9±1.05 | 31.9 |
| PUSA 372 x ILWC 246 | 8.0–9.0 | 8.6±0.3 | 6.5 | 8.0–14.0 | 11.0±1.7 | 27.2 | 23.0–27.0 | 25.3±1.2 | 8.2 | 9.0–30.0 | 15.5±2.6 | 44.7 | 9.0–19.0 | 14.2±0.66 | 20.7 |
| JG 11 x ILWC 246 | 10.0–15.0 | 12.3±1.4 | 20.3 | 8.0–14.0 | 11.0±1.7 | 27.2 | 19.0–24.0 | 22.0±1.5 | 12 | 4.0–23.0 | 12.6±2.8 | 55.5 | 7.0–21.0 | 15.1±1.13 | 25.7 |
| No. of pods plant-1 | |||||||||||||||
| BGD 72 x ILWC 229 | 13.0–16.0 | 14.3±0.8 | 10.6 | 7.0–9.0 | 8.0±1.0 | 17.6 | 25.0–51.0 | 33.6±8.6 | 44.5 | 10.0–35.0 | 21.2±2.9 | 39.7 | 50–104 | 71.3± 2.27 | 20.38 |
| PBG 5 x ILWC 229 | 22.0–24.0 | 23.0±0.5 | 4.3 | 7.0–9.0 | 8.0±1.0 | 17.6 | 4.0–58.0 | 24.9±3.0 | 54 | 5.0–165.0 | 25.8±4.1 | 113.2 | 41–112 | 78.0± 1.45 | 19.53 |
| BGD 72 x ILWC 246 | 13.0–16.0 | 14.3±0.8 | 10.6 | 3.0–7.0 | 4.6±1.2 | 44.6 | 27.5–75.0 | 42.4±8.7 | 45.8 | 2.0–27.0 | 13.2±3.9 | 78.9 | 43–80 | 59.1± 1.83 | 15.81 |
| PBG 5 x ILWC 246 | 22.0–24.0 | 23.0±0.5 | 4.3 | 3.0–7.0 | 4.6±1.2 | 44.6 | 1.5–40.0 | 25.7±5.4 | 60.1 | 2.0–74.0 | 17.0±3.3 | 106.5 | 50–135 | 71.2± 2.16 | 24.26 |
| ICKG 96029 x ILWC 246 | 16.0–19.0 | 17.6±0.8 | 8.6 | 3.0–7.0 | 4.6±1.2 | 44.6 | 32.0–35.0 | 34.0±1.0 | 5.0 | 4.0–86.0 | 26.8±10.5 | 103.3 | 24–79 | 51.2± 4.25 | 27.49 |
| PUSA 372 x ILWC 246 | 16.0–25.0 | 19.6±2.7 | 24 | 3.0–7.0 | 4.6±1.2 | 44.6 | 37.0–39.0 | 38.0±0.5 | 2.6 | 2.0–105.0 | 37.5±13.8 | 97.5 | 39–91 | 64.9±3.04 | 20.94 |
| JG 11 x ILWC 246 | 47.0–94.0 | 69.6±13.6 | 33.7 | 3.0–7.0 | 4.6±1.2 | 44.6 | 30.0–32.0 | 31.0±0.5 | 3.2 | 5.0–25.0 | 11.8±2.9 | 61.2 | 46–79 | 58.2±2.77 | 16.5 |
| 100-seed weight (g) | |||||||||||||||
| BGD 72 x ILWC 229 | 26.0–28.0 | 27.0±0.5 | 3.7 | 11.0–12.0 | 11.5±0.4 | 6.0 | 18.4–24.2 | 20.3±1.9 | 16.4 | 16.0–45.0 | 23.1±3.4 | 41.6 | 11.1–28.7 | 17.8±0.92 | 32.87 |
| PBG 5 x ILWC 229 | 13.5–17.5 | 15.5±1.1 | 12.9 | 11.0–12.0 | 11.5±0.4 | 6.0 | 2.0–18.6 | 14.9±1.0 | 28.7 | 9.0–24.0 | 14.4±0.4 | 20.4 | 10.6–23.6 | 17.6±0.25 | 14.68 |
| BGD 72 x ILWC 246 | 26.0–28.0 | 27.0±0.5 | 3.7 | 7.1–7.8 | 7.4±0.2 | 0.1 | 12.8–21.2 | 17.6±1.9 | 25.0 | 15.0–20.8 | 18.2±0.9 | 13.2 | 12.6–22.6 | 17.8±0.41 | 11.86 |
| PBG 5 x ILWC 246 | 13.5–17.5 | 15.5±1.1 | 12.9 | 7.1–7.8 | 7.4±0.2 | 0.0 | 7.6–19.0 | 12.9±2.1 | 39.7 | 12.0–20.5 | 15.8±0.4 | 15.9 | 12.4–27.8 | 18.4±0.38 | 16.36 |
| ICKG 96029 x ILWC 246 | 13.0–28.0 | 21.6±4.4 | 35.8 | 7.1–7.8 | 7.4±0.2 | 0.0 | 16.3–16.4 | 16.3±0.1 | 0.3 | 8.0–18.4 | 13.4±1.7 | 34.1 | 13.6–21.6 | 18.0±0.87 | 13.7 |
| PUSA 372 x ILWC 246 | 12.0–14.8 | 12.9±0.9 | 12.4 | 7.10–7.8 | 7.46±0.2 | 0.02 | 17.6–17.7 | 17.6±0.1 | 0.2 | 13.6–15.0 | 14.4±0.2 | 3.6 | 13.8–19.8 | 16.5±0.38 | 10.28 |
| JG 11 x ILWC 246 | 19.2–21.2 | 20.0±0.6 | 5.2 | 7.1–7.8 | 7.4±0.2 | 0.1 | 18.4–18.5 | 18.4±0.1 | 0.2 | 1.8–20.0 | 4.2±2.6 | 45.8 | 11.0–18.2 | 14.7±0.65 | 15.2 |
| Seed-yield plant-1 (g) | |||||||||||||||
| BGD 72 x ILWC 229 | 2.7–3.0 | 2.8±0.2 | 5.3 | 0.8–2.7 | 1.7±0.9 | 76.5 | 4.6–15.7 | 8.3±3.6 | 77.1 | 1.8–6.0 | 3.6±0.5 | 41.8 | 5.6–19.9 | 12.9 ± 0.56 | 27.94 |
| PBG 5 x ILWC 229 | 3.5–4.1 | 3.8±0.1 | 7.8 | 0.8–2.7 | 1.7±0.9 | 76.5 | 0.1–6.4 | 4.3±0.4 | 42.5. | 0.6–25.4 | 4.2±0.6 | 108.9 | 6.1–27.4 | 13.9 ± 0.42 | 31.77 |
| BGD 72 x ILWC 246 | 2.7–3.0 | 2.8±0.4 | 5.3 | 0.3–1.0 | 0.5±0.2 | 66 | 4.6–11.4 | 7.1±1.2 | 38.9 | 0.2–6.7 | 2.5±1.0 | 103.8 | 6.1–16.5 | 10.9 ± 0.57 | 26.68 |
| PBG 5 x ILWC 246 | 3.5–4.1 | 3.8±0.1 | 7.8 | 0.3–1.0 | 0.5±0.2 | 66 | 0.1–7.7 | 5.5±2.0 | 46.7 | 0.2–17.4 | 3.8±0.8 | 117.8 | 6.2–26.4 | 13.3 ± 0.53 | 31.67 |
| ICKG 96029 x ILWC 246 | 3.0–3.6 | 3.2±0.1 | 9.2 | 0.3–1.0 | 0.5±0.2 | 66 | 4.5–4.6 | 4.5±0.1 | 1.0 | 0.4–11.4 | 3.7±1.3 | 97 | 3.4–16.3 | 10.0±1.37 | 38.68 |
| PUSA 372 x ILWC 246 | 2.6–4.6 | 3.6±0.5 | 27.5 | 0.3–1.0 | 0.5±0.2 | 66 | 5.2–5.3 | 5.2±0.1 | 0.8 | 0.2–14.9 | 5.9±2.1 | 93.7 | 7.9–14.8 | 11.3 ± 0.43 | 16.89 |
| JG 11 x ILWC 246 | 13.2–21.5 | 16.7±2.4 | 25.4 | 0.3–1.0 | 0.5±0.2 | 66 | 5.0–5.1 | 5.0±0.1 | 0.3 | 1.0–3.8 | 1.9±0.4 | 58.8 | 6.1–13.4 | 8.7 ±0.56 | 22.16 |
| Biological yield plant-1 (g) | |||||||||||||||
| BGD 72 x ILWC 229 | 3.9–4.9 | 4.3±0.3 | 12 | 2.2–4.10 | 3.1±0.95 | 42.5 | 13.4–34.2 | 20.3±6.9 | 59 | 4.2–27.9 | 12.4±2.5 | 56.9 | 16.6–51.8 | 39.6±1.17 | 18.9 |
| PBG 5 x ILWC 229 | 18.8–21.0 | 20.1±0.6 | 5.7 | 2.2–4.10 | 3.1±0.95 | 42.5 | 10.4–22.1 | 15.2±0.6 | 19.9 | 2.4–53.6 | 16.2±1.3 | 60.4 | 24.6–65.6 | 40.0±0.69 | 18.1 |
| BGD 72 x ILWC 246 | 3.9–4.9 | 4.3±0.3 | 12 | 10.1–12.9 | 11.7±0.8 | 12.3 | 18.8–43.3 | 24.7±4.6 | 42.2 | 8.9–22.8 | 15.5±2.1 | 37 | 27.9–51.2 | 37.8±0.97 | 13.1 |
| PBG 5 x ILWC 246 | 18.8–21.0 | 20.1±0.6 | 5.7 | 10.1–12.9 | 11.7±0.8 | 12.3 | 9.8–20.7 | 14.2±1.6 | 32 | 2.7–43.6 | 15.3±2.0 | 70.5 | 25.9–64.6 | 41.8±1.06 | 20.3 |
| ICKG 96029 x ILWC 246 | 3.5–4.2 | 3.8±0.2 | 9 | 10.1–12.9 | 11.7±0.8 | 12.3 | 18.0–18.6 | 18.3±0.1 | 1.6 | 3.6–20.7 | 10.4±2.6 | 65.9 | 23.6–50.9 | 35.8±2.58 | 20.4 |
| PUSA 372 x ILWC 246 | 7.0–12.4 | 9.5±1.5 | 28.6 | 10.1–12.9 | 11.7±0.8 | 12.3 | 34.1–34.9 | 34.5±0.2 | 1.1 | 15.7–44.1 | 29.2±3.9 | 35.2 | 29.8–46.6 | 38.3±1.06 | 12.3 |
| JG 11 x ILWC 246 | 17.3–26.3 | 21.3±2.6 | 21.3 | 10.1–12.9 | 11.7±0.8 | 12.3 | 21.0–22.1 | 21.4±0.3 | 2.7 | 4.5–20.8 | 13.3±2.7 | 49.9 | 31.1–44.6 | 35.2±1.11 | 10.9 |
Fig 3. Distribution of F3 interspecific populations of different crosses into various reactions against cold tolerance.
Estimates of fruitful heterosis (%)
The nature and magnitude of fruitful heterosis for inbred vigour was studied in F2 and F3 interspecific derivatives for seed yield plant-1 and its important component traits (Table 5). An extent of fruitful heterosis as a vigour was estimated as percent of deviation of enhanced progenies from the better performing parent. In F2 generation, the mean performance ranged from -400.70% for plant height (ICKG 96029 x ILWC 246) to 31.02% for number of pods plant-1 (BGD 72 x ILWC 229). Likewise, in F3 generation, the vigour ranged from -36.82% for plant height (Pusa 372 x LWC 246) to 45.38% for seed yield plant-1 (JG 11 x ILWC 246). There were wide range of variation with respect to fruitful heterosis (inbred vigour) for important traits of interest in both F2 and F3 generations. For majority of interspecific crosses in both the generations, number of pods plant-1 and seed yield plant-1 revealed positive vigour (Table 5).
Table 5. Estimates of fruitful heterosis (%) as inbred vigour for important traits of interest in F2 and F3 wide cross populations.
| Trait/Cross | Generation | |||
|---|---|---|---|---|
| F2 | F3 | |||
| Range | Mean±SE | Range | Mean±SE | |
| Pusa 372 x ILWC 246 | ||||
| Days to maturity | 1.14–6.29 | 4.26±0.34 | 5.71–13.71 | 10.40±0.52 |
| Plant height (cm) | -45.20-(-28.82) μ | -36.80±0.98 | -48.96-(-6.30) | -36.82±2.08 |
| No. of pods plant-1 | -88.33-(-75.00) | -81.25±0.75 | -35-(51.67) | 8.17±5.06 |
| Seed yield plant-1 (g) | -42.75-(-10.14) | -30.33±2.23 | -7.25-(2.75) | 13.88±3.10 |
| JG 11 x ILWC 246 | ||||
| Days to maturity | 5.06–8.43 | 6.84±0.28 | 8.99–14.04 | 10.86±0.46 |
| Plant height (cm) | -31.12-(-13.28) | -20.00±1.80 | -28.63-(+5.33) | -12.15±3.62 |
| No. of pods plant-1 | 11.54–30.77 | 20.67±2.09 | -38.67-(+5.33) | -22.33±3.70 |
| Seed yield plant-1 (g) | 2.44–20.73 | 10.26±1.67 | 29.35–85.87 | 45.38±4.25 |
| BGD 72 x ILWC 246 | ||||
| Days to maturity | -2.47-(+3.70) | 0.33±0.39 | 16.05–23.46 | 19.66±0.37 |
| Plant height (cm) | -25.56-(+13.69) | -8.58±2.06 | -29.93-(+26.64) | -2.52±2.97 |
| No. of pods plant-1 | -39.47-(+2.63) | -21.58±2.14 | 3.45–37.93 | 12.14±1.77 |
| Seed yield plant-1 (g) | -31.93-(+2.52) | -19.66±2.16 | 2.94–61.76 | 21.30±3.30 |
| ICKG 96029 x ILWC 246 | ||||
| Days to maturity | -6.71-(-0.61) | -4.01±0.93 | 11.59–16.46 | 13.97±0.51 |
| Plant height (cm) | -670.73-(-60.98) | -400.70±94.89 | -46.15-(+0.85) | -19.98±5.52 |
| No. of pods plant-1 | 7.41–31.48 | 17.20±3.91 | 0.01–46.30 | 11.11±4.50 |
| Seed yield plant-1 (g) | 0.01–5.49 | 3.30±0.68 | 11.10–79.12 | 27.87±6.74 |
| PBG 5 x ILWC 246 | ||||
| Days to maturity | -11.60-(-2.26) | -7.54±0.48 | 13.26–17.68 | 15.57±0.15 |
| Plant height (cm) | -37.21–3.16 | -13.51±1.96 | -36.38–30.22 | -2.74±2.10 |
| No. of pods plant-1 | 0.01–12.86 | 5.30±0.73 | 0.01–92.86 | 15.76±2.07 |
| Seed yield plant-1(g) | 0.01–16.96 | 5.93±0.89 | 0.89–135.71 | 29.74±3.50 |
| BGD 72 x ILWC229 | ||||
| Days to maturity | 1.18–1.18 | 4.33±1.69 | 7.2–13.3 | 10.03±0.32 |
| Plant height (cm) | -54.54-(-5.54) | 8.36±2.40 | -75.3–81.4 | 18.46±6.62 |
| No. of pods plant-1 | 1.96–1.96 | 31.02±5.83 | 0.01–57.6 | 15.78±2.24 |
| Seed yield plant-1 (g) | 2.25–2.25 | 25.94±4.27 | 2.00–97.0 | 34.68±4.02 |
| PBG 5 x ILWC 229 | ||||
| Days to maturity | -2.31–0.82 | 0.82±0.36 | 7.82–14.83 | 11.39±0.16 |
| Plant height (cm) | -32.19–7.37 | -7.37±1.63 | -34.73–38.17 | 0.54±1.77 |
| No. of pods plant-1 | 5.00–14.03 | 14.03±1.29 | -1.33–944.00 | 20.40±8.55 |
| Seed yield plant-1 (g) | 2.94–17.17 | 17.17±1.87 | -12.93–136.21 | 30.49±2.91 |
*μ negative range performance recorded in parentheses
Discussion
The narrow crop genetic base is forcing plant breeders to search for new adaptive traits of interest. The introduction, characterization, evaluation, identification, and utilization of unadapted gene sources for useful traits are prerequisites conducting successful base broadening programme in annual crop plants [34–36]. The results pertaining to chickpea interspecific hybridization accomplished under two growing seasons help us to conclude those longer days during summer season and optimum temperature has pivotal role in pod and seed setting. Singh et al. [30] have also observed identical role of these factors in determining the onset of ontogenesis in chickpea. The remarkable variation assessed in morphological traits, which can help to distinguish phenotypic groups and their segregation analysis and genetic controls can be established using Mendelian genetic studies. It also permits us in the phenotypic identification of specific alleles for specific gene loci [37,38]. The normal breeding behaviour (meiotic chromosome configuration) of both wild Cicer annual accessions and their F1 hybrids was confirmed from complete pairing of chromosomes in bivalent forms and the production of fertile off-springs, consequently regular segregation and elite random selection [39,40]. The segregation of some important morphological traits suggested their monogenic inheritance for plant growth habit, stem pigmentation at seedling stage and testa texture. The study would be useful in the identification of true to type F1 hybrids through distinct morphological characters.
Further, field evaluation of parents and their advanced progenies using range, mean and coefficient of variation revealed creation of desired variability exhibiting inclusion of useful genes and alleles and possibly major role of complementary gene action [41]. Plant height manifested taller plants in majority of F1 hybrids, and in F2 and F3 generations, variation appears for dwarf to taller plants offering ample scope for selection of elite recombinant lines. Further, desirable plants were recorded in F1 generation for high number of pods plant -1 and in F2 and F3, the trait appear with wide range of variation from low to high pod number, suggesting that C. reticulatum accession ILWC 229 was better for number of pods plant-1, offering possibilities of recovering better plant types with high yield potential in subsequent advanced generations [5, 21, 30]. For seed yield plant-1, desirable performance appears for most of crosses in F1 generation, and in F2 and F3 interspecific populations, substantial range of variation was assessed for this trait, suggesting the scope of improvement through single plant selection from F2 onwards [5]. Considering linkage drag as a barrier in interspecific hybrid populations, the generation advancement and useful selection in segregating populations is very important to select promising recombinants and therefore, F2 population should be adequate in size [5] beside many other useful traits can also be taken well from the segregating plant populations. It further advocated that high yielding lines could be developed from interspecific hybridization following single plant selection [7,42]. The presence of elite transgression for seed yield and other agronomic traits indicating genetic complementarity between recipient and donor parental genotypes [43, 44]. This foster better hope for the recovery of desirable alleles [5, 7, 42]. Further, heterosis breeding has fastened the genetic improvement of crop plants. The consistency in the magnitude of fruitful heterosis in F2 and F3 generations might be due to accumulation of favorable additive alleles. Such segregants may be tackled as suggested by Redden and Jensen [45] for selecting elite recombinant lines for the development of suitable genotypes. Furthermore, the F3 interspecific derivatives also exhibit tolerance against cold stress, such tolerance could be useful selection criteria for developing suitable genotypes or breeding populations for colder areas. Here the prime aim of our study was to characterize, evaluate, and identify useful traits of interest from wild Cicer accessions and their introgression for diversification of genetic base of cultivated gene pool. The interspecific hybridization between C. arietinum L. x C. reticulatum and C. arietinum L. x C. echinospermum crosses showed substantially higher variation for important agro-morphological traits, which offers scope for isolation of potential transgressive segregants for developing high yielding lines or useful donors for further practical breeding purposes [46]. The study would also be useful to the chickpea researchers, while planning their experiments for introgressing useful traits of interest from wild Cicer species. The genetic materials are being advanced for further breeding and desirable selection.
Acknowledgments
The authors acknowledge the staff of Biodiversity and Integrated Gene Management (BIGM) Unit at International Centre for Agricultural Research in Dry Areas (ICARDA) Aleppo, Syria for providing the global wild annual Cicer accessions and Department of Agriculture Cooperation and Farmers Welfare, Govt. of India for grant in aid.
Data Availability
All relevant data are within the paper.
Funding Statement
This study was supported by the Department of Agriculture and Cooperation- Germplasm Evaluation Division- Manuscript Submission Report 10.
References
- 1.Varshney RK, Song C, Saxena RK, Azam S, Yu S, Sharpe AG, et al. Draft genome sequence of chickpea provides a resource for trait improvement. Nat Biotechnol. 2012; 31: 240–248. [DOI] [PubMed] [Google Scholar]
- 2.Abbo S, Berger J, Turner NC. Viewpoint: evolution of cultivated chickpea: four bottlenecks limit diversity and constraint adaptation. Func. Plant Biol. 2003; 30: 1081–1087. [DOI] [PubMed] [Google Scholar]
- 3.FAOSTAT. Food and agriculture organization statistical databases. http://faostat3.fao.org/download/Q/QC/E. Accessed 15th July 2017
- 4.Berger JD, Speijers J, Sapra RL, Sood UC. Genotype by environment interaction and chickpea improvement In: Yadav SS, Redden B, Chen W, and Sharma B (eds.) Chickpea breeding and management. CAB International, Wallingford, 2007, pp 193–12. [Google Scholar]
- 5.Singh KB, Ocampo B. Exploitation of wild Cicer species for yield improvement in chickpea. Theor. Appl. Genet. 1997; 95: 418–23. [Google Scholar]
- 6.Salimath PM, Toker C, Sandhu JS, Kumar J, Suma B, Yadva SS, et al. Conventional breeding methods In: Yadav SS, Redden B, Chen W, and Sharma B (eds.) Chickpea breeding and management. CAB International, Wallingford, 2007, pp 369–390. [Google Scholar]
- 7.Gaur PM, Gowda CLL, Knights EJ, Warkentin TD, Acikgoz N. Breeding Achievements. In: Yadav SS, Redden B, Chen W and Sharma B (eds.), Chickpea Breeding and Management CABI, UK, 2007, 391–416. [Google Scholar]
- 8.Toker C. Mutagenesis for resistance to abiotic stresses: chickpea as model crop In: Tomlekova NB, Kozgar MI and Wani MR (eds.) Mutagenesis: exploring novel genes and pathways. Wageningen Academic Publishers, Wageningen, 2014, pp 215–238. [Google Scholar]
- 9.van der Maesen LJ. Cicer L., a monograph of the genus, with special reference to the chickpea (Cicer arietinum L.), its ecology and cultivation Mededelingen Landbouwhoge School (Communications Agricultural University; ), Wageningen, 1972, pp 97–100. [Google Scholar]
- 10.Thudi M, Chitikineni A, Liu X, He W, Roorkiwal M, Yang W, et al. Recent breeding programs enhanced genetic diversity in both desi and kabuli varieties of chickpea (Cicer arietinum L.). Sci. Rep. 2016; 6:38636 10.1038/srep38636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ladizinsky IG, Adler A. The origin of chickpea as indicated by seed protein electrophoresis. Israel J Bot. 1975; 24: 183–189. [Google Scholar]
- 12.Tuwafe IS, Kahler AL, Boe A, Ferguson M. Inheritance and geographical distribution of allozyme polymorphisms in chickpea. J Heredity. 1988; 79: 170–174. [Google Scholar]
- 13.Kazan IK, Muehlbauer FJ. Allozyme variation and phylogeny in annual species of Cicer. Plant Syst. Evol. 1991; 175: 11–21. [Google Scholar]
- 14.Ahmad F, Gaur PM, Slinkard AE. Isozyme polymorphism and phylogenetic interpretations in the genus Cicer. Theor. Appl. Genet. 1992; 83: 620–627. 10.1007/BF00226907 [DOI] [PubMed] [Google Scholar]
- 15.Ahmad F, Slinkard AE. Genetic relationships in the genus Cicer as revealed by polyacrylamide gel electrophoresis of seed storage proteins. Theor. Appl. Genet. 1992; 84: 688–692. 10.1007/BF00224169 [DOI] [PubMed] [Google Scholar]
- 16.Labdi M, Robertson ID, Singh KB, Charrier A. Genetic diversity and phylogenetic relationships among annual Cicer species as revealed by isozyme polymorphism. Euphytica 1996; 88: 181–188. [Google Scholar]
- 17.Tayyar RI, Waines JG. Genetic relationships among annual species of Cicer using isozyme variation. Theor. Appl. Genet. 1996; 92: 245–254. 10.1007/BF00223381 [DOI] [PubMed] [Google Scholar]
- 18.Simon CJ, Muehlbauer FJ. Construction of a chickpea linkage map and its comparison with maps of pea and lentil. J Heredity. 1997; 88: 115–119. [Google Scholar]
- 19.Stalker HT. Utilization of wild species for crop improvement. Adv. Agron. 1980; 33: 111–147. [Google Scholar]
- 20.Toker C, Berger J, Kahraman A, Ayogan Can A, Bukun C, Penmetsa B, et al. Cicer reticulatum Ladizinsky, progenitor of the cultivated chickpea (C. arietinum L.). Legume Prospect. 2014b; 5: 26–27. [Google Scholar]
- 21.Toker C, Uzum B, Ceylan FO, Ikten C Chickpea In: Pratap A and Kumar J (eds.) Alien gene transfer in crop plants, vol, Achievements and impacts, Springer, New York: 2014a, pp. 125–151. [Google Scholar]
- 22.Croser JS, Ahmad CF, Clarke HJ, Siddique KHM. Utilization of wild Cicer in chickpea improvement- progress, constraints, and prospects. Austr. J. Agric. Res. 2003; 54: 429–44. [Google Scholar]
- 23.Singh S, Gumber RK, Joshi N, Singh K. Introgression from wild Cicer reticulatum to cultivated chickpea for productivity and disease resistance. Plant Breed. 2005; 124: 477–80. [Google Scholar]
- 24.Sandhu JS, Gupta SK, Singh G, Sharma YR, Bains TS. Inter-specific hybridization between Cicer arietinum and Cicer pinnatifidum for improvement of yield and other traits. In: 4th International Food Legumes Research Conference, New Delhi, India. 2006, 192: 18–22.
- 25.Singh M, Bisht IS, Dutta M, Kumar K, Basandrai AK, Kaur L, et al. Characterization and evaluation of wild annual Cicer species for agro-morphological traits and major biotic stresses under North-western Indian conditions. Crop Sci. 2014; 54: 229–239. [Google Scholar]
- 26.Marks GE. Acetocarmine glycerol jelly for use in pollen fertility counts. Stain Technol. 1994; 29: 277. [DOI] [PubMed] [Google Scholar]
- 27.Belling J. On counting chromosomes in pollen mother cells. American Nature. 1921; 55: 573–574. [Google Scholar]
- 28.Malhotra RS, Balyan HS, Gupta PK. Crossing technique in lentils. LENS Newsl. 1978; 5: 7–8. [Google Scholar]
- 29.Kumar A, Singh DP. Hybridization technique in lentil under field conditions. LENS Newsl. 1998; 25: 1–3. [Google Scholar]
- 30.Singh M, Kumar K, Bisht IS, Dutta M, Rana MK, Rana JC, et al. Exploitation of wild annual Cicer species for widening the gene pool of chickpea cultivars. Plant Breed. 2015; 134: 186–192. [Google Scholar]
- 31.Koseoglu K, Adak A, Sari D, Sari H, Ceylan FO, Toker C. Transgressive segregations for yield criteria in reciprocal interspecific crosses between C. arietinum L. and C. reticulatum Ladiz. Euphyt. 2017; 10.1007/s10681-017-1903-7 [DOI] [Google Scholar]
- 32.Rathore A, Prasad R, Gupta VK. Computer aided construction and analysis of augmented designs. J Indian Soc. Agric. Stat. 2004; 57: 320–344. [Google Scholar]
- 33.Singh KB, Malhotra RS, Saxena MC. Chickpea evaluation for cold tolerance under field conditions. Crop Sci. 1989; 29: 282–285. [Google Scholar]
- 34.Duvick DN. Genetic diversity in major farm crops on the farm and in reverse. Econ Bot. 1984; 38: 151–178. [Google Scholar]
- 35.Lazaro A, Ruiz M, Rosa L, Martin I. Relationship between agro-morphological characters and climate parameters in Spanish landraces of lentil. Genet Resour and Crop Evol. 2001; 48: 239–249. [Google Scholar]
- 36.Naghavi MR, Johansouz MR. Variation in the agronomic and morphological traits of lentil accessions. J Integer Plant Biol. 2005; 47: 375–379. [Google Scholar]
- 37.Muehlbauer FJ, Singh KB. Genetics of chickpea In: Saxena MC and Singh KB (ed.), The Chickpea, AB International, Cambridge, 1987, 99–126. [Google Scholar]
- 38.Pundir RPS, Rao NK, Van der Maesen LPJ. Distribution of qualitative traits in the world germplasm of chickpea. Euphyt. 1985; 34: 697–703. [Google Scholar]
- 39.Gill KS, Gill BS, Endo TR, Mukai Y. Fine physical mapping of ph1, a chromosome pairing regulator gene in polyploid wheat. Genetics. 1993; 134: 1231–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luan L, Wang X, Long WB, Liu YH, Tu SB. A comparative cytogenetic study of the rice auto tetraploid restorers and hybrids. Russian J Genet. 2009; 45: 1074–1081. [PubMed] [Google Scholar]
- 41.Vega U, Frey KJ. Transgressive segregation in inter and intra-specific crosses of barley. Euphyt. 1980; 29: 585–694. [Google Scholar]
- 42.Gaur PM, Mallikarjuna N, Knights T, Beebe TS, Debouck D. Gene introgression in grain legumes In: Gupta S, Ali M and Singh BB (eds.), Grain Legumes: Genetic Improvement, Management and Trade, Indian Society of Pulses Research and Development, Indian Institute of Pulses Research, Kanpur, India: 2008, 1–17. [Google Scholar]
- 43.Singh M, Rana JC, Singh B, Kumar S, Saxena DR, Saxena A, et al. Comparative agronomic performance and reaction to Fusarium wilt of L. culinaris x L. orientalis and L. culinaris x L. ervoides derivatives. Front Plant Sci. 2017; 8: 1162 10.3389/fpls.2017.01162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adak A, Sari D, Sari H, Toker C. Gene effects of Cicer reticulatum Ladiz. on qualitative and quantitative traits in the cultivated chickpea. Plant Breed. 2017; 10.1111/pbr.12464 [DOI] [Google Scholar]
- 45.Redden RJ, Jensen NF. Mass selection and mating systems in cereals. Crop Sci. 1974; 14: 345–350. [Google Scholar]
- 46.Kahraman A, Pandey A, Khan MK, Lindsay D, Moenga S, Vance L, et al. Distinct Subgroups of Cicer echinospermum are associated with hybrid sterility and breakdown in interspecific crosses with cultivated chickpea. Crop Sci. 2017; 10.2135/cropsci2017.06 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.