Abstract
After the initiation of Human Microbiome Project in 2008, various biostatistic and bioinformatic tools for data analysis and computational methods have been developed and applied to microbiome studies. In this review and perspective, we discuss the research and statistical hypotheses in gut microbiome studies, focusing on mechanistic concepts that underlie the complex relationships among host, microbiome, and environment. We review the current available statistic tools and highlight recent progress of newly developed statistical methods and models. Given the current challenges and limitations in biostatistic approaches and tools, we discuss the future direction in developing statistical methods and models for the microbiome studies.
Keywords: Bioinformatics, Biostatistics, Cancer, Hypothesis testing, IBD, Microbiome, Statistical methods and models, Vitamin D receptor
Introduction
The gut microbiome plays fundamental roles in the human health. It can be considered as a newly identified organ that interacts with other organs and influences the development of disease.1, 2 Human Microbiome Project (HMP) was initiatively funded in 2008 by the National Institutes of Health Roadmap for Biomedical Research and constructed as a large, genome-scale community research project.3 The HMP project needs data analysis, computational methods development, and the public availability of tools and data.4
Human gut microbiome study is to understand not only the microbiome community composition, but also the dynamic interactions among microbiome, host, environment, and disease intervention. The microbiome studies require a multi-disciplinary team effort, involving basic, translational, and clinical investigators. The next phase of research investigation of the gut microbiome should be guided by specific biological questions relevant to the clinical aspects and natural history of the disease, utilizing the full spectrum of ‘omic’ technologies, bioinformatic analysis, and experimental models.5 However, the significant roles of biostatisticians and bioinformatic and biostatistical methods in gut microbiome studies are underestimated, especially the appropriate use of biostatistic tests is largely ignored.
Here, we discuss the statistical hypothesis tests in both community composition and microbiome-host interactions. We review the utility of various statistical approaches for assessing the diversity of microbiome communities and analyzing and modeling the association between community composition of the microbiome and host. We summarize the current available statistic tools for microbiome studies. We highlight the recent progress in new statistical methods and models. In doing so, we provide specific examples of these methods and discuss how to appropriately apply them into microbiome study. In the meantime, we bring up the limitations and daunting challenges ahead of us that must be overcome in order to move the field forward. Furthermore, we discuss the development of statistical methods, the limits, and future direction.
Research and statistical hypotheses in human microbiome studies
In the current microbiome studies, there are mainly two themes: 1) to characterize the relationship between microbiome features and biological, genetic, clinical or experimental conditions; and 2) to identify potential biological and environmental factors that are associated with microbiome composition. The goal of these studies is to understand mechanisms of host genetic and environmental factors that shape our microbiome. Insights gained from the studies potentially contribute to the development of therapeutic strategies in modulating the microbiome composition in human diseases.6, 7
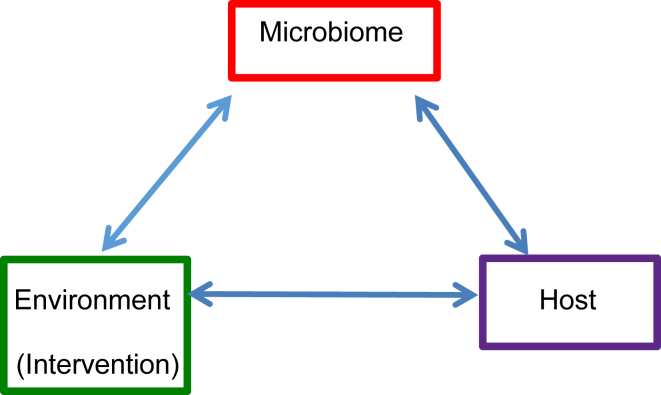
Dynamic interactions exist among environment, microbiome and host (Fig. 1). To study the complicated interactions among these factors, three general research hypotheses have been developed and used in the field: hypothesis 1 is to test the association between environment and host. There are no specific features for this hypothesis compared to other biomedical sciences. To test the hypothesis 1, we can use the standard statistical methods and models, which are commonly used in other biomedical sciences. For the microbiome studies, the focus is on the following research hypothesis 2 and 3:
Figure 1.
Dynamic Interactions among environment, microbiome and host for the research hypotheses in microbiome studies.
The research hypothesis 2 is to test the association between microbiome and host: whether the composition of the microbiome or “dysbiotic” microbiome is linked to the health or disease of host. For example, in inflammatory bowel diseases (IBD) research,8, 9 dysbiosis is associated with the progression of the diseases. Lack of vitamin D receptor (VDR) causes dysbiosis and changes the functions of the murine intestinal microbiome.10 Altered bacterial community is associated with different intestinal epithelial VDR status.11
The research hypothesis 3 is to test whether microbiome is associated with environmental or biological covariates,12 the impact of environmental factors on microbiome,13 or whether there is an effect of intervention on a specific microbiome composition (diversity) in health and disease. The examples include testing whether dietary interventions shape gut microbiota,8, 14 the impact of a probiotic intervention on the composition of the human microbiota.15 The longitudinal studies have tested antibiotics and diet effects on gut microbial community structure,9 analyzed whether nutrition influence gut microbiome composition at the level of bacterial species,16 or hypothesized that antibiotic treatments affect the diversity of strains of gut bacteria.13 In a recent paper, Bokulich et al showed that antibiotic exposure and delivery mode alter bacterial diversity and delay microbiota maturation and infant diet affects diversity of intestinal microbiome.17
Statisticians usually develop their statistical hypotheses based on the research hypotheses. Based on the research hypotheses, the null statistical hypothesis is developed as “there is no difference of microbiome composition in health and disease (or experimental groups or genetic conditions)” or “there is no difference (change) of microbiome composition in different environmental factors (or intervention). Although these statistical hypotheses have the core theme that explores impacts of environmental or external factors (e.g. interventions) on composition and/or richness of microbiota, they could focus on various topics, including, alpha diversity (species diversity in each individual sample), bacterial richness, total number of unique operational taxonomic units (OTUs), phylogenetic diversity (the relative amount of diverse phylogenetic lineages), and species evenness in each sample.17
The statistical hypothesis could be alpha diversity. For example, for antibiotic studies, we can hypothesize that antibiotic treatment does not decrease microbial diversity or specifically antibiotic-treated children have same diverse gut microbiota13; antibiotic treatment decreases microbial diversity,13, 18, 19, 20, 21 so the antibiotic-treated children have a less diverse gut microbiota.13 The statistical hypothesis can also be beta diversity, such as, Jaccard index of species or strains13 or UniFrac phylogenetic distance.17
The statistical hypotheses could be even temporal microbiome community. For example, we can hypothesize that all strains are similar,13 the microbiome community is stable (not change over time), or compared to non-antibiotic users, antibiotic treatment make the strains less similar and less stable.13
Statistical methods and models for microbiome studies
An appropriate statistical method is needed to prove a scientific hypothesis. In this section, we review the classical statistical tests, multivariate statistical tools, and some newly developed models and methods in analyzing microbiome data.
Classic statistical tests
Many classic statistical tests are available to analyze gut microbiome. A hypothesis testing in microbial taxa can be conducted by comparing alpha and beta diversity indices. Depending on whether the data are normally or non-normally distributed, number of experimental groups, or experimental conditions, we can use a t-test, analysis of variance, or corresponding non-parametric test.
Standard t-test was used to compare alpha diversity between two groups22 or population abundance23, 24, 25, 26 between two sets of relative abundance data. Standard t-test was used even to compare the relative abundances of different phyla and genera between healthy volunteers and colorectal cancer (CRC) patients in a gut microbiota study in CRC.27, 28 The non-parametric analogous Wilcoxon rank-sum test (also called Mann–Whitney test) was conducted to compare alpha diversity29: Shannon diversity across groups,22 two clusters as defined by the bacterial taxonomic composition.9 It was used to identify the statistically significant differences in microbial taxa or OTUs, and other nonparametric measures,29 or the relative abundances of different phyla and genera.28 In summary, two-sample t-test and its nonparametric counterpart Wilcoxon rank-sum test were widely used in microbiome studies to comparing continuous variables between two groups.
When comparing more than two groups, the one-way analysis of variance (ANOVA) or its non-parametric equivalent of the Kruskal–Wallis test are appropriate, depending on that the variables are normally or not normally distributed. ANOVA was reviewed to analyze taxonomic diversity data including intergroup and intragroup beta diversity,30 to compare proportional abundance,23, 31 or to assess the performance of risk model of the gut microbiome on BMI or lipids24, 31 and taxonomic and functional-specific biases.32
ANOVA test was also used to compare the functional capacity of microbiome among intestinal locations.33 Kruskal–Wallis one-way ANOVA was used to compare normalized z-score of the bacterial and fungal proportions for samples, and unequal variances of microbiome data.34
When comparing categorical microbiome data, for example testing a single a priori specified taxon is present at different rates across groups, a chi-square test was usually used.22
Clearly, the classical statistical methods are still widely used and will be used in gut microbiome studies. However, it is important that the appropriate statistical tests and methods should be carefully chosen to analyze microbiome data.
Overdispersion and zero-inflated models in microbiome studies
Taxa count data in microbiome studies, such as microbiome taxonomy reads or OTU counts from amplicon sequencing experiments or differential expression data from RNA-Seq experiments are often overdispersed and have many zeros. In order to fit the microbiome count data with overdispersion and excess zeros, typically, the negative binomial (NB), and zero inflated models are often applied. For example, a NB35 model was fitted in microbiome abundance data analysis,36 used to analyze gut microbiome in Parkinson's disease.29 A NB model developed by30 was used to test for assessing differences in sequence tag abundance and used for detecting differentially abundant features in clinical metagenomic samples.37
The abundance of bacteria in the human gut is characterized by an increasing number of zeros at lower taxonomic levels and right screwed. In order to capture the characteristic of excess zeros and model the screwed microbiome data, a zero-inflated model, such as Zero-Inflated Poisson (ZIP), Zero-Inflated Negative Binomial (ZINB) or hurdle model, is needed to be chosen for modeling the excess zeros. The appropriateness of using zero-inflated model in gut microbiome study was assessed by extensive simulations and applied in a real human microbiome study.38 Recently, for the same reason: capture the excess zeros and model the screwed microbiome data, Wang et al used the hurdle model with a negative binomial distribution to analyze the species of bacteria (97% similarity threshold OTUs).39
In order to identify the environmental or biological covariates that are associated with different bacterial taxa while accounting for overdispersion and many zeros, Xia et al proposed to apply an additive logistic normal multinomial regression model40 to link covariates to bacterial composition (counts)40 and applied the model to a study of the association between diet and stool microbiome composition.41
Motivated by the observed strong correlation between the number of OTUs detected in a sample and the corresponding sequencing depth in high-throughput 16S rDNA studies, in 2013, Paulson et al proposed Zero-inflated Gaussian (ZIG) mixture model.42 The mixture model has two components: a novel cumulative sum scaling (CSS) normalization technique to correct the bias in the assessment of differential abundance introduced by total-sum normalization (TSS), and a zero-inflated Gaussian distribution mixture model to account for biases in differential abundance testing resulting from under-sampling of the microbial community. The model seeks to directly estimate the probability that an observed zero is generated from the detection distribution due to under-sampling or from the count distribution (absence of the taxonomic feature in the microbial community). ZIG is implemented in the metagenomeSeq Bioconductor package. The authors evaluated metagenomeSeq using simulated data and compared ZIG to existing tools, such as Kruskal–Wallis test using oral microbiota data from the Human Microbiome Project and concluded that ZIG outperforms approaches that are widely used in the field and yields a more precise biological interpretation of the data.42 ZIG methods may have broader applicability for other differential abundance analyses such as gut microbiome. However, the methods need be evaluated with sufficient amount of studies.
Multivariate statistical tools in microbiome studies
Microbiome communities in an environmental context can be analyzed by multivariate statistical methods or models. Many statistical models and methods exist for analyzing the association of microbiome community composition and environmental covariates and outcomes. Most multivariate statistical tools used in microbiome study mainly adopted from well-developed ecological research fields and environmental sciences.
Due to high dimensionality, non-normality and phylogenetic structure of the data, it is difficult to test the association of microbiome composition directly with potential environmental factors using OTUs or taxa abundances. Thus, multivariate analyses generally first need to choose one distance measure method and then conduct analysis of the estimated distances, in which a distance measure is defined between any of two microbiome samples.
Several tests of among-group differences are available in analyzing microbiome data: multivariate analysis of variance with permutation (PERMANOVA), and analysis of group similarities (ANOSIM), multi-response permutation procedures (MRPP), and Mantel's test (MANTEL).
PERMANOVA was proposed by Anderson and McArdle to apply the powerful ANOVA to multivariate ecological datasets.43, 44 PERMANOVA is one of most widely used nonparametric methods to fit multivariate models to microbiome data. It is a multivariate analysis of variance based on distance matrices and permutation.44 Similarly as MRPP, and other multivariate analyses, PERMANOVA is generally used with one of distance measure methods. For example, a PERMANOVA using unweighted UniFrac distance measure was conducted to show the composition of the gut microbiota in omnivore versus vegans,45 to assess the association with beta diversity measures,46 to test for microbial divergence among populations,47 and Bray–Curtis dissimilarity matrix.48, 49
Similar as PERMANOVA, ANOSIM is one of most widely used multivariate methods in microbiome studies. ANOSIM was used to compare within- and between-group similarity50 through a distance measure, to test the null hypothesis that the average rank similarity between samples within a group is the same as the average rank similarity between samples belonging to different groups.51 For example, Kelly et al use weighted and unweighted UniFrac distances to test the strength of association with microbiome composition between treatments and among time points within treatments.52
In microbiome literature, the MRPP on the pairwise weighted UniFrac distance matrix was conducted to confirm the significance of the clustering,53 to test the factors influencing microbial communities,54 and to compare community dissimilarities with Bray–Curtis distances.49
Like a correlation analysis, Mantel's test was used to test association between environmental factors and host microbiome. For example, to test whether microbiome variation explains microbiome variation in host,47 the association between the host genetic distance and the variance in community beta-diversity,55 donor microbiome and BMI,56 and even to identify the predictors of microbiome composition.48
Newly developed statistical methods
In order to specifically fit multivariate data, especially microbiome data, recently, the researchers and statisticians have developed several parametric and non-parametric models.
Dirichlet-multinomial model
Among these parametric probability models, the multinomial and Dirichlet-multinomial distributions57, 58, 59 are the most popular ones. For the purposes of hypothesis testing and power calculations of taxonomic-based human microbiome data, La Rosa and colleagues proposed a multivariate statistic method,57 based on Dirichlet multinomial mixtures models.59 The authors reparametrize the Dirichlet multinomial model to the Dirichlet multinomial mixtures to make it suitable to perform hypothesis testing across groups, based on difference between location (mean comparison) and scales (variance comparison/dispersion).58 It is implemented in R statistical software package “HMP”60 via using the data from the NIH Human Microbiome Project (HMP).3 Its capability of performing power calculations is also attractive for researchers and statisticians when they design their microbiome study and prepare their grant application.
UniFrac distance metric family
We have reviewed above, to compare microbial communities, multivariate analyses first need to choose one distance measure method. Numerous distance measures have been proposed.61, 62 Among them, phylogenetic distance measures, which account for the phylogenetic relationship among the taxa, are very powerful toolboxes because they exploit the degree of divergence between different sequences.
In order to capture phylogenetic information when computing differences between microbial communities, in 2005, Lozupone and Knight proposed the UniFrac distance metric. UniFrac measures the phylogenetic distance between sets of taxa in a phylogenetic tree.63 The goal of the UniFrac distance metric was to enable objective comparison between microbiome samples from different conditions. In 2007, Lozupone et al added a proportional weighting to the original UniFrac and differentiated them as unweighted UniFrac and weighted UniFrac.64, 65 Since then, two versions of UniFrac were available in the microbiome literature and have been applied in thousands of research publications covering almost everything from human disease to general ecology.65, 66 Unweighted UniFrac distance considers only species presence and absence information and counts the fraction of branch length unique to either community, and weighted UniFrac distance uses species abundance information and weights the branch length with abundance difference.
Although these two UniFrac distances have been become the most widely used phylogenetic distance measures, their limitations also have been noticed: These two measures are evaluated assign too much weight either to rare lineages (unweighted UniFrac distance) or to most abundant lineages (weighted UniFrac distances), thus, may not be very powerful in detecting change in moderately abundant lineages.12 Based on a variance adjusted weighted UniFrac distance (VAWUniFrac),67 Chen et al in 2012 develop generalized UniFrac distances that extend the weighted and unweighted UniFrac distances for detecting a much wider range of biologically relevant changes in microbiome composition.12 Thus, the UniFrac toolbox family has been expanded from UniFrac distances to generalized UniFrac distances. The generalized UniFrac distances were demonstrated in detecting the microbiome differences by analysis of two real human gut microbiome data sets related to linking human gut microbiome composition to long-term diet41 and testing upper respiratory tract microbiome difference between smokers and non-smokers68 using PERMANOVA. Thus, through incorporating UniFrac distances and PERMANOVA, generalized UniFrac distance measure provides a statistical approach to test the association between microbiome composition and environmental covariates.
Two newly developed UniFrac tools were added to the UniFrac toolboxes: one is micropower R package contributed by Kelly et al69 and another is Wong et al's UniFrac R programs.70 In the micropower package, Kelly et al incorporated the measures of unweighted and weighted UniFrac distances into analyses of pairwise distances and PERMANOVA to power and sample-size estimation. Under the compositional data analysis setting, Wong et al introduced two new weightings: information UniFrac and ratio UniFrac that are not as sensitive to rarefaction and allow greater separation of outliers than classic unweighted and weighted UniFrac. The goal is to address the limitations of unweighted UniFrac's highly sensitive to rarefaction instance and to sequencing depth in uniform data sets with no clear structure or separation between groups.
To our knowledge, no formal manuals are available for micropower R package and Wong et al's UniFrac R programs, although Gregory B Gloor lab hosted UniFrac workshop on February 29, 2016 to illustrate the uses of information UniFrac and ratio UniFrac.
Compositional analysis for microbiome data
Much earlier in 1897, Pearson already warned that “spurious correlation” may be formed when use the ratio of two absolute measurements in the measurement of organs.71 Since the second half of the twentieth century, researchers in the field of geology have awakened the fact that use the standard statistical approaches to analyze composition data may make the results uninterpretable. Aitchison in the 1980s and particularly in his 1986 seminal work72, 73, 74, 75, 76 realized that every statement about a composition can be stated in terms of ratios of components and developed a set of fundamental principles, a variety of methods, operations, and tools for compositional data analysis. Of those, the logratio transformation methodology was widely accepted by statisticians and researchers in geology, ecology and other fields73, 77, 78, 79 since with logratio transformations, the problem of a constrained sample space (the simplex) of the compositional data will be removed, and data are projected into multivariate real space. Therefore, all available standard multivariate techniques can be used again79 to analyze compositional data. However, although a series of publications have shown that the existing tools for compositional data analysis in geology, ecology and other fields are readily adapted and also a valid approach to analyze microbiome high-throughput sequencing data,76, 77, 78, 79, 80, 81 the development of methods and tools for microbiome compositional data analysis are most recent. The representative works are Gloor team's ANOVA-like differential express (ALDEx and ALDEx2)80, 81, 82 and Mandal et al's analysis of composition of microbiomes (ANCOM).83 Fundamentally, both approaches use the logratio transformation techniques to convert microbiome data, thus remove the constraints to make the standard multivariate techniques suitable for analysis.
For comparison of microbial composition, it is inappropriate to draw inferences regarding the total abundance in the ecosystem from the abundance of OTUs in the specimen. Instead, the inferences are drawn regarding the relative abundance of a taxon in the ecosystem, using its relative abundance in the specimen. Thus, it exists a compositional constraint: all microbial relative abundances within a specimen sum to one, which results in compositional data residing in a simplex73, 76 rather than the Euclidean space.
Recently, Mandal et al83 developed a novel statistical framework called ANCOM to account for the compositional constraints to reduce false discoveries in detecting differences in microbial mean taxa abundance at an ecosystem level. It is based on compositional log-ratios. The authors compared ANCOM with zero inflated Gaussian (ZIG) and t-test with simulation studies and real data. They concluded ANCOM outperforms ZIG method by substantially reducing the FDR and increasing power.
The attractive features of ANCOM are: it makes no distributional assumptions and can be implemented in a linear model framework to adjust for covariates as well as model longitudinal data.
Compared to ANCOM, ALDEx and ALDEx2 are more comprehensive: they are applicable to nearly any type of data that is generated by high-throughput sequencing and are suitable for the comparison of many different experimental designs. The statistical analyses include both two-sample and paired t-test, ANOVA, and non-parametric test, such as, Welch's t test, Wilcoxon test, Kruskal–Wallace test. They also have option to adjust p-values using Benjamin-Hochberg method.
Statistical packages that implement hypothesis testing and statistical analysis
In the developing statistical methods and models for hypothesis testing and statistical analysis, bioinformatics pipelines and R packages play a very important role. The two bioinformatics pipelines are QIIME84 and mothur.85 Both QIIME and mother are all self-contained pipelines that can be used to analyze 16S rRNA gene sequencing data. Due to their comprehensive features and support documentation, QIIME and mother were reviewed as the two outstanding pipelines.86, 87 QIIME and mothur have the capability to perform microbiome composition and some statistical analyses such as alpha and beta diversities, ANOVA, paired t test, two sample t-test, adonis, ANOSIM, MRPP, PERMANOVA, PERMDISP, db-RDA, and Mantel test.88, 89
Vegan is a very important and most widely used R package.90 It was initially designed for community ecologists. Vegan is not self-contained, it depends on many other R packages, and must be run under R statistical environment. However, vegan contains the most popular methods of multivariate analysis and tools for diversity analysis, and other potentially useful functions. Therefore, it is commonly used in analyzing ecological communities, and has been adapted to analyze microbiome data.
Other R packages which are useful for hypothesis testing and statistical analysis include DESeq,91 DESeq2,92 edgeR,93 limma,94 metagenomeSeq,95 microbiome96 and phyloseq.97 All these packages have their specific capabilities to conduct hypothesis testing and statistical analysis. Both DESeq and DESeq2 use the negative binomial distribution to test for differential expression; edgeR package implements original statistical methodology described in93, 98, 99, 100; limma is to detect the differential abundance of the species in the samples95, 101; metagenomeSeq includes a non-parametric permutation test on t-statistics, a non-parametric Kruskal–Wallis test, and a mixture model that implements a zero-inflated Gaussian (ZIG) distribution of mean group abundance for each taxonomic feature.95
Among these packages, microbiome and phyloseq are more comprehensive statistical tools.
First of all, microbiome and phyloseq have integrated other available statistical packages to perform statistical hypothesis testing and analysis. For example, the microbiome package contains general-purpose tools for microarray-based analysis of microbiome profiling data sets in R. This package also conducts statistical analysis based on the phyloseq class. Additionally, phyloseq has integrated with or extended to other R packages, such as, DESeq, DESeq2, edgeR, to facilitate taxanomic diversity analysis and statistical modeling.
Second, they have tools to manage microbiome data sets. For example, phyloseq package has capability of importing and exporting data from other packages, even from bioinformatics pipelines, such as QIIME and mother.
Third, phyloseq also has capability to perform various diversity metrics analyses and sophisticated analyses. For example, after importing data into the R, one may easily perform beta diversity analysis using any or all of over 40 different ecological distance metrics; implement alpha diversity metrics; perform more sophisticated analyses, such as k-tables analysis102 and differential analysis of microbiome data. The microbiome package adds extra functionality for microbiome data sets to perform microbiota composition analysis, bistability analysis, calculate diversity indices and also to fit linear models, do pairwise comparisons, and association studies, et al.
Fourth, both microbiome and phyloseq packages have functions and tools to visualize microbiome data via barplots, boxplots, density plots, heatmaps, motion charts, and networks, and ordination and clustering.
Longitudinal microbiome data analysis and causal inference
The microbiome is inherently dynamic, driven by interactions with the host and the environment, and varies over time. Thus, longitudinal microbiome data analysis provides more information on the profile of microbiome with host and environment interactions.
Several computational methods for analyzing longitudinal microbiome data have been developed and applied into microbiome studies. A regression-based time series model can be used to analyze a series of observations (dependent variables) including relative abundances of an OUT over time, ecological diversity of the gut microbiota over time, and a function of time and other covariates (independent variables). For example, a regression model was conducted to evaluate the dependence of the human vaginal microbiome on time in the menstrual cycle and other covariates103, 104; an autoregressive (AR) model was used to assess the tendency of the different taxonomic groups of bacteria.105 Gerber and his colleagues developed the microbiome counts trajectories infinite mixture model engine,106 a time-series clustering method for analyzing microbiome data that automatically infers the number of temporal patterns from the data.104 However, current approach of longitudinal microbiome data analysis cannot provide appropriate statistical tools to model dynamic and complicated microbiome data.
First, microbiome may have causative effects on host. It was evidenced by the following factors of both human and animal studies: 1) studies in wild type mice107, 108 and zebrafish109, 110 have found a number of similarities in their microbiotic function and host interactions; 2) the microbiota have played a role in maturation of the host immune system and even anatomical development of the intestine.111, 112
Second, the bacterial composition (species members and abundance) of the gut microbiota is personalized.113, 114 Most microbiomes are strikingly divergent between distinct host species.115, 116 During the lifespan, our microbiome varies systematically across body habitats and time, can be dramatically altered, either transiently or long term, by diseases such as infections117 or medical interventions such as antibiotics,19, 118, 119 such trends may ultimately reveal how changes of microbiome cause or prevent diseases.120 For example, reduced species diversity has been observed in obese humans107, 116; the abundance of phylum Fusobacteria increased significantly in the colon of colorectal cancer patients.121, 122 Thus, researchers in the microbiome field need understand not only the association, but also the causative functions of bacteria in human diseases.123, 124, 125
Third, the mutual relationship between microbiome and its host suggests a causal inference model, or mediation analysis and longitudinal analysis may be granted. Currently, microbiome researchers shift their emphasis from correlation to causality. The distinguishing feature of longitudinal studies is that the subjects are measured repeatedly during the study, permitting the direct assessment of changes in response variable over time.126, 127 Thereby, longitudinal study captures both between-individual differences (heterogeneity among individuals) and within subject dynamics. It offers the opportunity to study complex biological, psychological, and behavioral hypotheses, especially those involving changes over time.128 The advantage of longitudinal analysis is also suitable for microbiome data. It will enhance our understanding of short-and long-term trends of microbiome by intervention, such as diet, and the development and persistence of disease caused by microbiome.
Mediational analysis provides the researcher with a story about a sequence of effects that leads to something.129, 130, 131 It allows us to conduct scientific investigations to explain how something comes about. Detecting the dynamic causation132 among microbiome, intervention and the host is very critical. However, to our knowledge, there are limited applications of causal inferences and mediation analysis in microbiome studies.
Limitations of existing methods
The existing statistical approaches have their limitations. First, the existing statistical methods for analyzing microbiome proportional data do not solve constraint problem, and some researchers even do not know it exists. Most standard statistical methods, such as the Pearson correlation coefficient, t-test, ANOVA are still widely used or exist in current literature on the analysis of microbiome data23, 24, 25, 26 without testing the data distribution and transformation. One assumption of standard statistical methods is that the compared data are independent. Since the sum of the relative abundances is unity, it indicates the data are not independent with the unity or any constant constrain. Thus it is not appropriate to directly use these methods for analyzing microbiome relative abundance data.83 Actually, this constraint problem is not limited to standard statistical methods; even the family of probability models requires all pairs of OTUs to be negatively correlated.83 Thus, it may not be appropriate for microbiome data. When we examine univariate or multivariate differences, correlations, and methods that depend on correlation, any inference made on proportional data has the potential to be very misleading80 if the statistical methods do not consider constraint problem.
The focus of microbiome research shifts from correlation to causality. Although longitudinal microbiome data analyses are available,104 suitable longitudinal and causal inference models are very limited in microbiome studies. The available statistical tools for longitudinal data analysis are far away from meeting the needs of modeling the dynamic microbiome data.
Conclusion and future direction
In summary, three resources of statistical tools are available for analyzing microbiome data. First and convenient way is to use standard tests and models because of its familiarity; second is to borrow statistical methods and models from other relevant research fields such as multivariate methods and techniques from ecology; third is to develop new statistical methods for fitting in specific microbiome data. The progress has been made from choosing standard statistical methods, borrowing them from other fields to develop its own unique methods. Some available statistical approaches for microbiome data seem feasible.
There are two challenges in developing statistical methods and models in microbiome study: one is to fully consider the constraint feature of microbiome data to model and analyze compositional data; another is to develop longitudinal and causal models to fit the dynamic and complicated association among microbiome, environments and host.
For hypothesis testing and statistical analysis of microbiome data, further work is needed to develop methods and models that are more suitable for analyzing microbiome compositional data. Specially, the efforts should be focused on three approaches: 1) to analyze microbiome data as compositional and further develop statistical tools to capture compositional feature of microbiome data; 2) to develop statistical tools for longitudinal microbiome data analysis, and 3) to shift correlation analysis to causation analysis. Currently, both longitudinal and mediation analyses are very limited. To model causative effects of microbiome data, both appropriate design and suitable statistical models are needed. Because microbiome data are very complicated, it is really a challenge for statisticians and microbiome researchers to develop statistical tools to conduct longitudinal and mediation analyses of microbiome data.
The future studies need a team effort involving biomedical researcher, physicians, bioinformatic experts, and biostatisticians. More mechanism-driven studies should be based on appropriate statistical design and perform analysis using the experimental models, human samples, ‘omic’ technologies, bioinformatic analysis, and statistic modeling.
Conflicts of interest
The authors declare no competing financial interests.
Acknowledgement
We would like to acknowledge the NIDDK/National Institutes of Health grant R01 DK105118 to Jun Sun and UIC Cancer Center for supporting her research.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Contributor Information
Yinglin Xia, Email: yxia@uic.edu.
Jun Sun, Email: junsun7@uic.edu.
References
- 1.Clemente J.C., Ursell L.K., Parfrey L.W., Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148(6):1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhernakova A., Kurilshikov A., Bonder M.J. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352(6285):565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peterson J., Garges S., Giovanni M. The NIH human microbiome project. Genome Res. 2009;19:2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gevers D., Pop M., Schloss P.D., Huttenhower C. Bioinformatics for the human microbiome project. PLoS Comput Biol. 2012;8(11):e1002779. doi: 10.1371/journal.pcbi.1002779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun J., Chang E.B. Exploring gut microbes in human health and disease: pushing the envelope. Genes Dis. 2014;1(2):132–139. doi: 10.1016/j.gendis.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Virgin H.W., Todd J.A. Metagenomics and personalized medicine. Cell. 2011;147(1):44–56. doi: 10.1016/j.cell.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spor A., Koren O., Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9(4):279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 8.Albenberg L.G., Lewis J.D., Wu G.D. Food and the gut microbiota in IBD: a critical connection. Curr Opin Gastroenterol. 2012;28(4):314–320. doi: 10.1097/MOG.0b013e328354586f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis J.D., Chen E.Z., Baldassano R.N. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn's disease. Cell Host Microbe. 2015;18(4):489–500. doi: 10.1016/j.chom.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin D., Wu S., Zhang Y.G. Lack of vitamin D receptor causes dysbiosis and changes the functions of the murine intestinal microbiome. Clin Ther. 2015;37(5):996–1009.e7. doi: 10.1016/j.clinthera.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Wu S., Zhang Y.G., Lu R. Intestinal epithelial vitamin D receptor deletion leads to defective autophagy in colitis. Gut. 2015;64(7):1082–1094. doi: 10.1136/gutjnl-2014-307436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J., Bittinger K., Charlson E.S. Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics. 2012;28(16):2106–2113. doi: 10.1093/bioinformatics/bts342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yassour M., Vatanen T., Siljander H. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci Transl Med. 2016;8(343):343ra81. doi: 10.1126/scitranslmed.aad0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Albenberg L.G., Wu G.D. Diet and the intestinal microbiome: associations, functions, and implications for health and disease. Gastroenterology. 2014;146(6):1564–1572. doi: 10.1053/j.gastro.2014.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lahti L., Salonen A., Kekkonen R.A. Associations between the human intestinal microbiota, Lactobacillus rhamnosus GG and serum lipids indicated by integrated analysis of high-throughput profiling data. PeerJ. 2013;1:e32. doi: 10.7717/peerj.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Backhed F., Roswall J., Peng Y. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17(6):852. doi: 10.1016/j.chom.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 17.Bokulich N.A., Chung J., Battaglia T. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8(343):343ra82. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jakobsson H.E., Jernberg C., Andersson A.F., Sjolund-Karlsson M., Jansson J.K., Engstrand L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One. 2010;5(3):e9836. doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dethlefsen L., Relman D.A. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dethlefsen L., Huse S., Sogin M.L., Relman D.A. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6(11):e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nobel Y.R., Cox L.M., Kirigin F.F. Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat Commun. 2015;6:7486. doi: 10.1038/ncomms8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.La Rosa P.S., Zhou Y., Sodergren E., Weinstock G., Shannon W.D. Hypothesis Testing of Metagenomic Data. In: Izard J., Rivera M.C., editors. Metagenomics for Microbiology. Academic Press; Waltham, MA, USA: 2015. pp. 81–96. [Google Scholar]
- 23.Chen W., Liu F., Ling Z., Tong X., Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PloS One. 2012;7(6):e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim K.A., Jung I.H., Park S.H., Ahn Y.T., Huh C.S., Kim D.H. Comparative analysis of the gut microbiota in people with different levels of ginsenoside Rb1 degradation to compound K. PLoS One. 2013;8(4):e62409. doi: 10.1371/journal.pone.0062409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwai S., Fei M., Huang D. Oral and airway microbiota in HIV-infected pneumonia patients. J Clin Microbiol. 2012;50(9):2995–3002. doi: 10.1128/JCM.00278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsiao E.Y., McBride S.W., Hsien S. The microbiota modulates gut physiology and behavioral abnormalities associated with autism. Cell. 2013;155(7):1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao Z., Guo B., Gao R., Zhu Q., Qin H. Microbiota disbiosis is associated with colorectal cancer. Front Microbiol. 2015;6:20. doi: 10.3389/fmicb.2015.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang T., Cai G., Qiu Y. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6(2):320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yin X., Peng J., Zhao L. Structural changes of gut microbiota in a rat non-alcoholic fatty liver disease model treated with a Chinese herbal formula. Syst Appl Microbiol. 2013;36(3):188–196. doi: 10.1016/j.syapm.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 30.Alekseyenko A.V., Perez-Perez G.I., De Souza A. Community differentiation of the cutaneous microbiota in psoriasis. Microbiome. 2013;1:31. doi: 10.1186/2049-2618-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tong M., McHardy I., Ruegger P. Reprograming of gut microbiome energy metabolism by the FUT2 Crohn's disease risk polymorphism. ISME J. 2014;8(11):2193–2206. doi: 10.1038/ismej.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voigt A.Y., Costea P.I., Kultima J.R. Temporal and technical variability of human gut metagenomes. Genome Biol. 2015;16:73. doi: 10.1186/s13059-015-0639-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang H., Huang X., Fang S., Xin W., Huang L., Chen C. Uncovering the composition of microbial community structure and metagenomics among three gut locations in pigs with distinct fatness. Sci Rep. 2016;6:27427. doi: 10.1038/srep27427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorzelak M.A., Gill S.K., Tasnim N., Ahmadi-Vand Z., Jay M., Gibson D.L. Methods for improving human gut microbiome data by reducing variability through sample processing and storage of stool. PloS One. 2015;10(8):e0134802. doi: 10.1371/journal.pone.0134802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Falkenhorst G., Simonsen J., Ceper T.H. Serological cross-sectional studies on salmonella incidence in eight European countries: no correlation with incidence of reported cases. BMC Public Health. 2012;12:523. doi: 10.1186/1471-2458-12-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McMurdie P.J., Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol. 2014;10(4):e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White J.R., Nagarajan N., Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol. 2009;5(4):e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu L., Paterson A.D., Turpin W., Xu W. Assessment and selection of competing models for zero-inflated microbiome data. PLoS One. 2015;10(7):e0129606. doi: 10.1371/journal.pone.0129606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J., Thingholm L.B., Skieceviciene J. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat Genet. 2016;48(11):1396–1406. doi: 10.1038/ng.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia F., Chen J., Fung W.K., Li H. A logistic normal multinomial regression model for microbiome compositional data analysis. Biometrics. 2013;69(4):1053–1063. doi: 10.1111/biom.12079. [DOI] [PubMed] [Google Scholar]
- 41.Wu G.D., Chen J., Hoffmann C. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paulson J.N., Stine O.C., Bravo H.C., Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat Methods. 2013;10(12):1200–1202. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson M.J. A new method for non-parametric multivariate analysis of variance. Aust Ecol. 2001;26:32–46. [Google Scholar]
- 44.McArdle B.H., Anderson M.J. Fitting multivariate models to community data: a comment on distance based redundancy analysis. Ecology. 2001;82:290–297. [Google Scholar]
- 45.Wu G.D., Compher C., Chen E.Z. Comparative metabolomics in vegans and omnivores reveal constraints on diet-dependent gut microbiota metabolite production. Gut. 2016;65(1):63–72. doi: 10.1136/gutjnl-2014-308209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen J., Ryu E., Hathcock M. Impact of demographics on human gut microbial diversity in a US Midwest population. PeerJ. 2016;4:e1514. doi: 10.7717/peerj.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith C.C., Snowberg L.K., Gregory Caporaso J., Knight R., Bolnick D.I. Dietary input of microbes and host genetic variation shape among-population differences in stickleback gut microbiota. ISME J. 2015;9(11):2515–2526. doi: 10.1038/ismej.2015.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tung J., Barreiro L.B., Burns M.B. Social networks predict gut microbiome composition in wild baboons. Elife. 2015;4 doi: 10.7554/eLife.05224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan Q., Li J., Yu Y. Environmental filtering decreases with fish development for the assembly of gut microbiota. Environ Microbiol. 2016;18(12):4739–4754. doi: 10.1111/1462-2920.13365. [DOI] [PubMed] [Google Scholar]
- 50.McCord A.I., Chapman C.A., Weny G. Fecal microbiomes of non-human primates in Western Uganda reveal species-specific communities largely resistant to habitat perturbation. Am J Primatol. 2014;76(4):347–354. doi: 10.1002/ajp.22238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giatsis C., Sipkema D., Smidt H., Verreth J., Verdegem M. The colonization dynamics of the gut microbiota in tilapia larvae. PLoS One. 2014;9(7):e103641. doi: 10.1371/journal.pone.0103641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kelley S.T., Skarra D.V., Rivera A.J., Thackray V.G. The gut microbiome is altered in a letrozole-induced mouse model of polycystic ovary syndrome. PLoS One. 2016;11(1):e0146509. doi: 10.1371/journal.pone.0146509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Narrowe A.B., Albuthi-Lantz M., Smith E.P. Perturbation and restoration of the fathead minnow gut microbiome after low-level triclosan exposure. Microbiome. 2015;3:6. doi: 10.1186/s40168-015-0069-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Degnan P.H., Pusey A.E., Lonsdorf E.V. Factors associated with the diversification of the gut microbial communities within chimpanzees from Gombe National Park. Proc Natl Acad Sci U S A. 2012;109(32):13034–13039. doi: 10.1073/pnas.1110994109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanders J.G., Powell S., Kronauer D.J., Vasconcelos H.L., Frederickson M.E., Pierce N.E. Stability and phylogenetic correlation in gut microbiota: lessons from ants and apes. Mol Ecol. 2014;23(6):1268–1283. doi: 10.1111/mec.12611. [DOI] [PubMed] [Google Scholar]
- 56.Ridaura V.K., Faith J.J., Rey F.E. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341(6150):1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.La Rosa P.S., Shands B., Deych E. Statistical object data analysis of taxonomic trees from human microbiome data. PLoS One. 2012;7(11):e48996. doi: 10.1371/journal.pone.0048996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.La Rosa P.S., Brooks J.P., Deych E. Hypothesis testing and power calculations for taxonomic-based human microbiome data. PLoS One. 2012;7(12):e52078. doi: 10.1371/journal.pone.0052078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Holmes I., Harris K., Quince C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS One. 2012;7(2):e30126. doi: 10.1371/journal.pone.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.LaRosa P.S., Deych E., Shands B., Shannon W.D. 2016. Hypothesis Testing and Power Calculations for Comparing Metagenomic Samples from HMP. R-package. [Google Scholar]
- 61.Kuczynski J., Liu Z., Lozupone C., McDonald D., Fierer N., Knight R. Microbial community resemblance methods differ in their ability to detect biologically relevant patterns. Nat Meth. 2010;7(10):813–819. doi: 10.1038/nmeth.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Swenson N.G. Phylogenetic beta diversity metrics, trait evolution and inferring the functional beta diversity of communities. PLoS One. 2011;6(6):e21264. doi: 10.1371/journal.pone.0021264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lozupone C.A., Knight R. UnifFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71 doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lozupone C.A., Hamady M., Kelley S.T., Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73(5):1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lozupone C., Lladser M.E., Knights D., Stombaugh J., Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2011;5(2):169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith M.I., Yatsunenko T., Manary M.J. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339(6119):548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chang Q., Luan Y., Sun F. Variance adjusted weighted UniFrac: a powerful beta diversity measure for comparing communities based on phylogeny. BMC Bioinformatics. 2011;12:118. doi: 10.1186/1471-2105-12-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Charlson E.S., Chen J., Custers-Allen R. Disordered microbial communities in the upper respiratory tract of cigarette smokers. PloS one. 2010;5(12):e15216. doi: 10.1371/journal.pone.0015216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kelly B.J., Gross R., Bittinger K. Power and sample-size estimation for microbiome studies using pairwise distances and PERMANOVA. Bioinformatics. 2015;31(15):2461–2468. doi: 10.1093/bioinformatics/btv183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wong R.G., Wu J.R., Gloor G.B. Expanding the UniFrac toolbox. PLoS One. 2016;11(9) doi: 10.1371/journal.pone.0161196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pearson K. Mathematical contributions to the theory of evolution. On a form of spurious correlation which may arise when indices are used in the measurement of organs. Proc R Soc Lond. 1897:489–502. [Google Scholar]
- 72.Aitchison J. A new approach to null correlations of proportions. Math Geol. 1981;13(2):175–189. [Google Scholar]
- 73.Aitchison J. The statistical analysis of compositional data (with discussion) J R Stat Soc Ser B Stat Methodol. 1982;44(2):139–177. [Google Scholar]
- 74.Aitchison J. Principal component analysis of compositional data. Biometrika. 1983;70(1):57–65. [Google Scholar]
- 75.Aitchison J. Reducing the dimensionality of compositional data sets. J Int Assoc Math Geol. 1984;16(6):617–635. [Google Scholar]
- 76.Aitchison J. Chapman and Hall Ltd; London: 1986. The Statistical Analysis of Compositional Data. Monographs on Statistics and Applied Probability. Reprinted in 2003 with additional material by The Blackburn Press. [Google Scholar]
- 77.Pawlowsky-Glahn V., Buccianti A. John Wiley & Sons, Ltd; Chichester, UK: 2011. Compositional Data Analysis: Theory and Applications. [Google Scholar]
- 78.van den Boogaart G.K., Tolosana-Delgado R. Springer; Heidelberg: 2013. Analyzing Compositional Data with R. [Google Scholar]
- 79.Pawlowsky-Glahn V., Egozcue J.J., Tolosana-Delgado R. John Wiley & Sons; London, UK.: 2015. Modeling and Analysis of Compositional Data. Springer. [Google Scholar]
- 80.Gloor G.B., Wu J.R., Pawlowsky-Glahn V., Egozcue J.J. It's all relative: analyzing microbiome data as compositions. Ann Epidemiol. 2016;26(5):322–329. doi: 10.1016/j.annepidem.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 81.Gloor G.B., Reid G. Compositional analysis: a valid approach to analyze microbiome high-throughput sequencing data. Can J Microbiol. 2016;62(8):692–703. doi: 10.1139/cjm-2015-0821. [DOI] [PubMed] [Google Scholar]
- 82.Fernandes A.D., Macklaim J.M., Linn T.G., Reid G., Gloor G.B. ANOVA-like differential expression (ALDEx) analysis for mixed population RNA-seq. PLoS One. 2013;8(7):e67019. doi: 10.1371/journal.pone.0067019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mandal S., Van Treuren W., White R.A., Eggesbo M., Knight R., Peddada S.D. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb Ecol Health Dis. 2015;26:27663. doi: 10.3402/mehd.v26.27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Caporaso J.G., Kuczynski J., Stombaugh J. QIIME allows analysis of high-throughput community sequencing data. Nat Meth. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schloss P.D., Westcott S.L., Ryabin T. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nilakanta H., Drews K.L., Firrell S., Foulkes M.A., Jablonski K.A. A review of software for analyzing molecular sequences. BMC Res Notes. 2014;7(1):830. doi: 10.1186/1756-0500-7-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Plummer E., Twin J., Bulach D.M., Garland S.M., Tabrizi S.N. A comparison of three bioinformatics pipelines for the analysis of preterm gut microbiota using 16S rRNA gene sequencing data. J Proteomics Bioinform. 2015;8:283–291. [Google Scholar]
- 88.D'Argenio V., Casaburi G., Precone V., Salvatore F. Comparative metagenomic analysis of human gut microbiome composition using two different bioinformatic pipelines. BioMed Res Int. 2014:2014. doi: 10.1155/2014/325340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.He Y., Zhou B.-J., Deng G.-H., Jiang X.-T., Zhang H., Zhou H.-W. Comparison of microbial diversity determined with the same variable tag sequence extracted from two different PCR amplicons. BMC Microbiol. 2013;13(1):208. doi: 10.1186/1471-2180-13-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Oksanen Jari, Guillaume Blanchet F., Michael Friendly . 2016. Vegan: community Ecology Package. R package version 2.4-1. [Google Scholar]
- 91.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Smyth G. Limma: linear models for microarray data. In: Gentleman R., Carey V., Dudoit S., Irizarry R., Huber W., editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- 95.Paulson J.N., Stine O.C., Bravo H.C., Pop M. Robust methods for differential abundance analysis in marker gene surveys. Nat Methods. 2013;10(12):1200–1202. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lahti L., Salojarvi J. 2014–2016. Microbiome R Package. [Google Scholar]
- 97.McMurdie P.J., Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4) doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Robinson M.D., Smyth G.K. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics. 2007;23(21):2881–2887. doi: 10.1093/bioinformatics/btm453. [DOI] [PubMed] [Google Scholar]
- 99.Robinson M.D., Smyth G.K. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics. 2008;9(2):321–332. doi: 10.1093/biostatistics/kxm030. [DOI] [PubMed] [Google Scholar]
- 100.McCarthy D.J., Chen Y., Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40(10):4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Praveen P., Jordan F., Priami C., Morine M.J. The role of breast-feeding in infant immune system: a systems perspective on the intestinal microbiome. Microbiome. 2015;3(1):41. doi: 10.1186/s40168-015-0104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Thioulouse J. Simultaneous analysis of a sequence of paired ecological tables: a comparison of several methods. Ann Appl Stat. 2011:2300–2325. [Google Scholar]
- 103.Gajer P., Brotman R.M., Bai G. Temporal dynamics of the human vaginal microbiota. Sci Transl Med. 2012;4(132):3003605. doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gerber G.K. Longitudinal Microbiome Data Analysis. In: Izard J., Rivera M.C., editors. Metagenomics for Microbiology. Elsevier Inc.; London, UK: 2015. [Google Scholar]
- 105.Palmer C., Bik E.M., DiGiulio D.B., Relman D.A., Brown P.O. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5(7):26. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gerber G.K., Onderdonk A.B., Bry L. Inferring dynamic signatures of microbes in complex host ecosystems. PLoS Comput Biol. 2012;8(8):e1002624. doi: 10.1371/journal.pcbi.1002624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ley R.E., Backhed F., Turnbaugh P., Lozupone C.A., Knight R.D., Gordon J.I. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102(31):11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Samuel B.S., Gordon J.I. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci U S A. 2006;103(26):10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rawls J.F., Samuel B.S., Gordon J.I. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci U S A. 2004;101(13):4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rawls J.F., Mahowald M.A., Ley R.E., Gordon J.I. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127(2):423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ivanov, Atarashi K., Manel N. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ivanov, Littman D.R. Segmented filamentous bacteria take the stage. Mucosal Immunol. 2010;3(3):209–212. doi: 10.1038/mi.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Baxter N.T., Wan J.J., Schubert A.M., Jenior M.L., Myers P., Schloss P.D. Intra- and interindividual variations mask interspecies variation in the microbiota of sympatric peromyscus populations. Appl Environ Microbiol. 2015;81(1):396–404. doi: 10.1128/AEM.02303-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lozupone C.A., Stombaugh J.I., Gordon J.I., Jansson J.K., Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489(7415):220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Morgan X.C., Huttenhower C. Chapter 12: human microbiome analysis. PLoS Comput Biol. 2012;8(12):27. doi: 10.1371/journal.pcbi.1002808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ley R.E., Turnbaugh P.J., Klein S., Gordon J.I. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 117.Koenig J.E., Spor A., Scalfone N. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;1:4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Perez-Cobas A.E., Gosalbes M.J., Friedrichs A. Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut. 2013;62(11):1591–1601. doi: 10.1136/gutjnl-2012-303184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Peterfreund G.L., Vandivier L.E., Sinha R. Succession in the gut microbiome following antibiotic and antibody therapies for Clostridium difficile. PLoS One. 2012;7(10):10. doi: 10.1371/journal.pone.0046966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Costello E.K., Lauber C.L., Hamady M., Fierer N., Gordon J.I., Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326(5960):1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Castellarin M., Warren R.L., Freeman J.D. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kostic A.D., Gevers D., Pedamallu C.S. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22(2):292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhang C., Zhao L. Strain-level dissection of the contribution of the gut microbiome to human metabolic disease. Genome Med. 2016;8(1):016–0304. doi: 10.1186/s13073-016-0304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fei N., Zhao L. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. 2013;7(4):880–884. doi: 10.1038/ismej.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhao L. The gut microbiota and obesity: from correlation to causality. Nat Rev Microbiol. 2013;11(9):639–647. doi: 10.1038/nrmicro3089. [DOI] [PubMed] [Google Scholar]
- 126.Fitzmaurice G.M., Laird N.M., Ware J.H. Wiley; 2004. Applied Longitudinal Analysis. [Google Scholar]
- 127.Diggle P.J., Heagerty P., Liang K.-Y., Zeger S.L. 2nd ed. Oxford University Press; 2002. Analysis of Longitudinal Data. [Google Scholar]
- 128.Zhang H., Xia Y., Chen R., Gunzler D., Tang W., Tu X. Modeling longitudinal binomial responses: implications from two dueling paradigms. J Appl Stat. 2011;38(11):2373–2390. [Google Scholar]
- 129.MacKinnon D.P., Fairchild A.J., Fritz M.S. Mediation analysis. Annu Rev Psychol. 2007;58:593–614. doi: 10.1146/annurev.psych.58.110405.085542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.MacKinnon D.P. Erlbaum; Mahwah, NJ: 2008. Introduction to Statistical Mediation Analysis. [Google Scholar]
- 131.Xia Y., Lu N., Zhang H., Gunzler D., Zubenko G.S., Tu X.M. Statistical methods and issues in the study of suicide. In: Lavigne J., Kemp J., editors. Frontiers in Suicide Risk: Research, Treatment and Prevention. Nova Science; Hauppauge, New York: 2012. pp. 139–158. Science. [Google Scholar]
- 132.Segata N., Haake S.K., Mannon P. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13(6):2012–2013. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]