Abstract
The TP53 tumor-suppressor gene is mutated in >50% of human tumors and Li-Fraumeni patients with germ line inactivation are predisposed to developing cancer. Here, we generated tp53 deleted zebrafish that spontaneously develop malignant peripheral nerve-sheath tumors, angiosarcomas, germ cell tumors, and an aggressive Natural Killer cell-like leukemia for which no animal model has been developed. Because the tp53 deletion was generated in syngeneic zebrafish, engraftment of fluorescent-labeled tumors could be dynamically visualized over time. Importantly, engrafted tumors shared gene expression signatures with predicted cells of origin in human tissue. Finally, we showed that tp53del/del enhanced invasion and metastasis in kRASG12D-induced embryonal rhabdomyosarcoma (ERMS), but did not alter the overall frequency of cancer stem cells, suggesting novel pro-metastatic roles for TP53 loss-of-function in human muscle tumors. In summary, we have developed a Li-Fraumeni zebrafish model that is amenable to large-scale transplantation and direct visualization of tumor growth in live animals.
Research organism: Zebrafish
Introduction
TP53 is a tumor suppressor protein that is mutated or functionally disrupted in more than 50% of all human tumors (Kastenhuber and Lowe, 2017; Muller and Vousden, 2014). Moreover, genetic mutation of TP53 in Li-Fraumeni patients leads to cancer predisposition early in life and is associated with transformation in a broad range of target tissues (Malkin, 2011). TP53 is commonly inactivated by single amino acid mutations that create dominant-negative forms of the protein that inhibit efficient tetramer formation and block transcriptional activity (Muller and Vousden, 2014). In this setting, TP53 alleles likely alter transcriptional activity of TP53 and its related transcription factor family members, TP63 and TP73 (Lang et al., 2004; Olive et al., 2004). By contrast, TP53 deletion is expected to have less wide-ranging transcriptional effects that are confined to tetrameric transcription factor function. Regardless of the genetic alteration, TP53 transcriptional inactivation can lead to genomic instability and impaired apoptotic responses that often are predisposing to a wide array of cancers (Kastenhuber and Lowe, 2017; Muller and Vousden, 2014).
To date, several murine genetic models have been developed to assess the effects of both loss- and gain-of-function Tp53 mutations in cancer (Donehower et al., 1992; Harvey et al., 1993; Jacks et al., 1994; Lang et al., 2004; Lavigueur et al., 1989; Lee et al., 1994; Olive et al., 2004). Both Tp53 mutant and null alleles spontaneously develop cancer. However, similar to human Li-Fraumeni patients, the spectrum varies among different alleles, suggesting that the mode of Tp53 inactivation has important implications in regulating the types of cancer that develop, the time to onset, and the overall propensity for tumor progression (Lavigueur et al., 1989; Lee et al., 1994). For example, mice heterozygous for the 172His point mutation are predisposed to developing osteosarcoma while animals harboring the 270His mutation develop hemangiosarcoma and carcinoma (Olive et al., 2004). By contrast, mice with homozygous Tp53 deletion mainly develop lymphoma, with rare cases of angiosarcoma, undifferentiated sarcoma, osteosarcoma, rhabdomyosarcoma, testicular tumors, nervous system tumors, teratoma, and mammary carcinoma being reported (Donehower et al., 1992; Harvey et al., 1993; Jacks et al., 1994). Together, these data suggest that differences in gain- and loss-of-function alleles have profound effects on tumor onset and spectrum in genetically engineered mice and yet, largely recapitulate the wide array of cancers observed in Li-Fraumeni patients. Importantly, a small subset of Li-Fraumeni syndrome patients harbor genomic deletions in the TP53 locus and cancers that develop in dominant-negative, heterozygous point-mutation carriers often display deletion of the second TP53 allele (Malkin, 2011). Thus, modeling complete TP53 loss-of-function in different animal models will likely provide novel insights into human disease.
TP53 is also commonly mutated in human sarcomas and is predictive of poor outcome (Taubert et al., 1996). For example, the TP53 locus is mutated in 16% of human embryonal rhabdomyosarcoma (ERMS), a common pediatric cancer of muscle and transcriptional activity is altered in >30% of human ERMS through TP53 locus disruption or MDM2 amplification (Taylor et al., 2000). Interestingly, TP53 mutations are also acquired at ERMS relapse (Chen et al., 2013), suggesting a likely role for TP53 in ERMS progression and therapy resistance. Finally, Li-Fraumeni patients with germline TP53 mutations commonly develop ERMS (Malkin, 2011), suggesting important roles for TP53 loss in the genesis of this disease. Yet, to date, the effect of TP53 pathway inactivation on cancer stem cell number, tumor progression, and metastasis in ERMS is not fully understood. Moreover, because genetically engineered mouse and human xenograft models of ERMS do not metastasize in vivo, assessing TP53 loss-of-function in the context of rhabdomyosarcoma metastasis has not been possible. Finally, to date, no tp53 deletion models have been generated in syngeneic zebrafish, precluding large-scale transplantation studies to assess how deletion regulates cancer stem cells and tumor invasion in vivo for a wide array of cancers.
To better study tp53 biology in vivo, we generated a complete loss-of-function tp53 deletion allele in syngeneic CG1-strain zebrafish using TALEN endonucleases. tp53del/del animals spontaneously developed a wide range of tumors including malignant peripheral nerve-sheath tumors (MPNSTs), angiosarcomas, germ cell tumors, and an aggressive natural killer cell-like leukemia not previously described in any animal model. This model contrasts with currently available point-mutation alleles for zebrafish tp53 that predominantly develop MPNSTs (Berghmans et al., 2005). Moreover, because the tp53del/del mutant was generated in CG1-strain syngeneic zebrafish (Mizgireuv and Revskoy, 2006), tumors efficiently transplanted into recipient fish enabling expansion of unlabeled and GFP-labeled tumors, dynamic live animal imaging of metastatic progression, and analysis of transcriptional differences between tumors using RNA sequencing approaches. Roles for tp53 were also assessed in kRASG12D-induced ERMS using large-scale cell transplantation assays and live fluorescent imaging over time. Using these approaches, we showed that the overall frequency of ERMS self-renewing cancer stem cells was unaffected by loss of tp53. In contrast, tp53del/del ERMS were more invasive, providing a potential explanation for increased aggression associated with TP53 disruption in human ERMS (Seki et al., 2015). Taken together, our work has uncovered novel roles for Tp53 loss in the onset of a wide array of cancers and has provided new insights into how tp53 affects ERMS progression in vivo.
Results and discussion
tp53 deletion mutants spontaneously develop MPNSTs, angiosarcoma, germ cell tumors, and leukemia
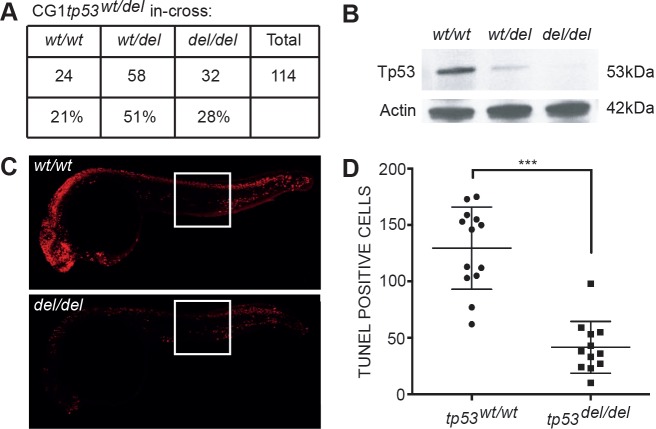
Given the critical function of Tp53 as a tumor suppressor and the absence of a complete null allele in zebrafish, we created a tp53 deletion mutant using two pairs of TALENs (Transcription Activator-Like Effector Nucleases) that cleaved at the 5’ and 3’ end of the tp53 locus (Figure 1A). One-cell stage CG1 syngeneic embryos (Mizgireuv and Revskoy, 2006) were microinjected with mRNA encoding each TALEN pair and raised to adulthood. F1 embryos were screened by genomic PCR to identify a single founder line with deletion of 12.1 kb tp53 genomic sequence (Figure 1A). CG1 tp53wt/del heterozygous fish were in-crossed and progeny assessed for Tp53 protein loss and the ability to undergo apoptosis following ionizing irradiation. As expected, homozygous tp53del/del embryos lacked protein expression and were resistant to radiation-induced apoptosis (Figure 1—figure supplement 1B–D).
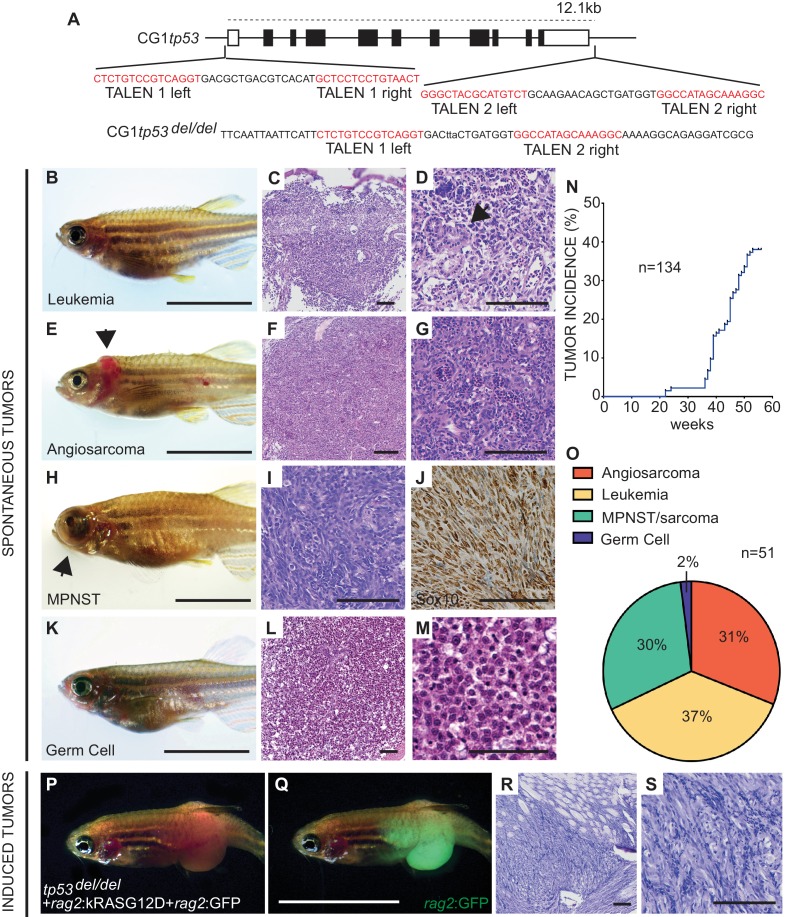
Figure 1. Homozygous tp53del/del zebrafish spontaneously develop a wide range of tumor types.
(A) tp53 genomic locus and CG1 tp53del/del allele. TALEN arms were designed to target the 5’ and 3’ genomic sequence of tp53 (red). (B–M) CG1 tp53del/del zebrafish develop leukemia (B–D), angiosarcoma (E–G), MPNSTs (H–J), and germ cell tumors (K–M). Whole animal images (B,E,H,K), hematoxylin/eosin (H and E) stained sections (C,D,F,G,I,L,M), and immunohistochemistry for Sox10 (J). Blast-like leukemia cells predominate in the kidney marrow and efface the renal tubules (black arrow, (D). (N) Tumor incidence in CG1 tp53del/del zebrafish (n = 134). (O) Quantitation of tumor types that form in CG1 tp53del/del mutant zebrafish by 55 weeks of life based on histology review (n = 51). (P–S) kRASG12D-induced embryonal rhabdomyosarcoma (ERMS) generated in CG1 tp53del/del zebrafish. Whole animal bright field and GFP-epifluorescence overlap images (P and Q, respectively). H and E stained sections revealed features consistent with human ERMS (R,S). Scale bars equal 12.5 mm in whole animal images and 100 μm in histology images.
Figure 1—figure supplement 1. tp53del/del zebrafish survive at expected Mendelian ratios, lack Tp53 protein expression, and are resistant to irradiation-induced cell death.
Figure 1—figure supplement 2. Tumor latency in tp53del/del zebrafish.

Progeny from tp53wt/del crosses were also raised to adulthood and assessed for viability and tumor onset over time. Both heterozygous tp53 wt/del and homozygous tp53del/del fish survived until adulthood at expected ratios (Figure 1—figure supplement 1A). By 4 months of age, tp53del/del zebrafish began to spontaneously develop tumors. Phenotypes of the earliest malignant tp53del/del cohort were consistent with loss of osmoregulation and kidney damage. Histopathological analysis of these animals revealed features consistent with leukemia, including blast-like cells predominating in the kidney marrow and loss of kidney stromal architecture, including effacement of the renal tubules (Figure 1B–D, Figure 1—figure supplement 2). Beginning by 7 months of age, a subset of tp53del/del animals developed externally visible tumors and histology consistent with angiosarcoma and malignant peripheral nerve sheath tumors (MPNSTs) (Figure 1E–J, Figure 1—figure supplement 2) (Berghmans et al., 2005; Choorapoikayil et al., 2012; Parant et al., 2010). A small subset of tp53del/del fish also developed prominent externally visible abdominal masses that were diagnosed as germ cell tumors following histopathological analysis (n = 2, Figure 1K–M, Figure 1—figure supplement 2) (Neumann et al., 2011). MPNST assignment was validated using IHC staining for sox10, which is a well-established clinical marker for this tumor type (Figure 1J) (Shin et al., 2012). As expected, tp53wt/del zebrafish infrequently developed tumors, which is consistent with studies in other tp53 deficiency mouse models (Donehower et al., 1992; Harvey et al., 1993; Jacks et al., 1994).
In total, 37% of tp53del/del animals developed externally visible tumors by 12 months of age with a wider tumor spectrum than previously reported in homozygous tp53M214K and tp53I166T mutant zebrafish (Figure 1N,O) (Berghmans et al., 2005; Parant et al., 2010). For example, these point mutation models predominantly developed MPNSTs with only a rare, single melanoma being detected in homozygous animals (Berghmans et al., 2005). Remarkably, the spectrum in tp53del/del zebrafish was more similar to that reported in Tp53-null mice, with angiosarcomas and germ cell tumors occurring in both models (Donehower et al., 1992; Harvey et al., 1993; Jacks et al., 1994). However, the predominance of T cell lymphomas seen in Tp53-null mice was not observed in tp53del/del zebrafish, likely reflecting species differences in sensitivity to Tp53 loss in target cells. It is also possible that tumor types seen in Tp53-null mice exhibit longer latency in our model and would not manifest in the short-lived CG1 strain zebrafish. Finally, we also generated ERMS in tp53del/del fish by microinjecting linearized human kRASG12D oncogene and GFP under the control of rag2 promoter (Figure 1P,Q) (Langenau et al., 2007). Histopathological analysis of transgene-induced tp53del/del ERMS showed consistent morphology with the spindle-variant of human ERMS (Figure 1R,S) (Langenau et al., 2007).
tp53del/del tumors are transplantable
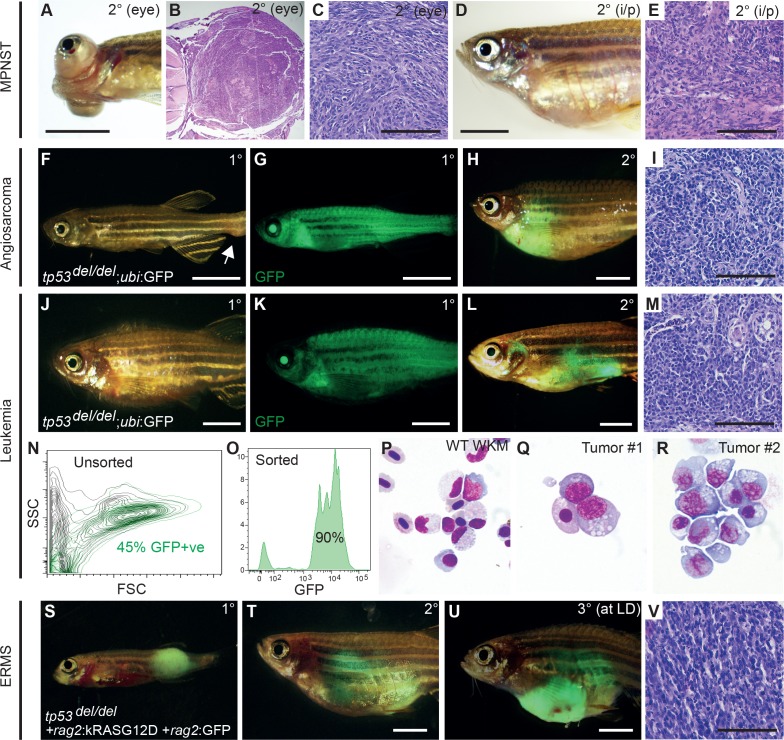
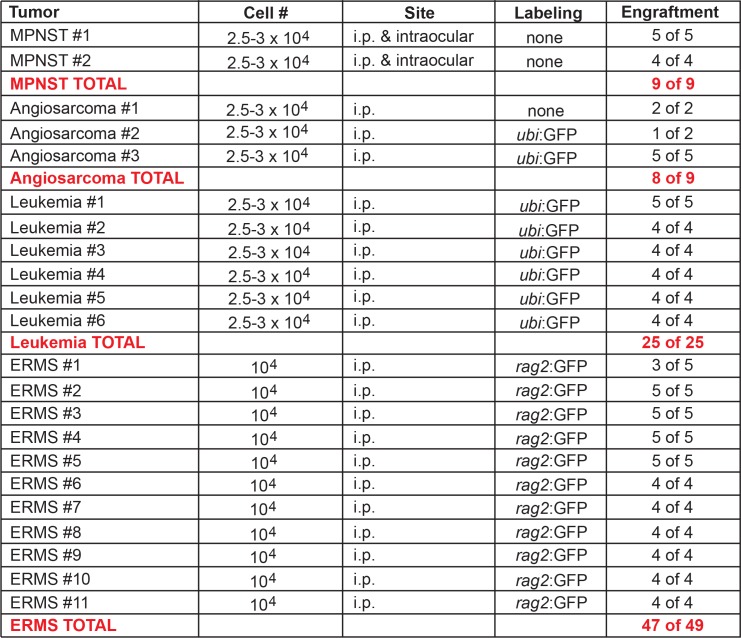
One major advantage of generating tp53del/del mutations in the CG1 syngeneic stain of zebrafish is the ease with which tumors can be used in cell transplantation assays. Primary MPNSTs that arose in the eye were dissected from euthanized tp53del/del animals and orthotopically transplanted into the equivalent periocular space or into the peritoneum of CG1-strain recipient fish (n = 2 primary tumors, n = 9 recipient fish total, 2 × 104 cells/fish). All recipient animals engrafted tumor with histology similar to the primary disease (Figure 2A–E, Figure 2—figure supplement 1). To more easily track tumor cells in host animals, we also crossed tp53del/del animals into CG1 syngeneic ubi:GFP transgenic zebrafish. We successfully engrafted tp53del/del angiosarcomas into CG1-recipient animals and ubi:GFP+ tumor cells were easily traceable in non-fluorescent recipients (n = 3 primary tumors, n = 8 of 9 transplant fish developed tumors, Figure 2F–I, Figure 2—figure supplement 1). Finally, ubi:GFP+ leukemias were also assessed by cell transplantation. Specifically, blood cells were engrafted into non-irradiated CG1 strain fish (2.5 × 104 cells/intraperitoneal injection). In total, five of five primary leukemias engrafted into recipient fish with GFP+ cells disseminating widely throughout the animal by 60 days post-transplantation (n = 5 primary leukemias, n = 25 of 25 animals engrafted leukemia, Figure 2J–M, Figure 2—figure supplement 1). Whole animal imaging and flow cytometric analysis revealed that GFP+ cells also invaded the recipient kidney marrow, the site of hematopoiesis in adult zebrafish (Figure 2M,N). Leukemic cells consisted of as much as 45% of the reconstituted marrow in transplanted fish (Figure 2N, n = 3 independent primary leukemias analyzed). To more closely observe leukemia cell morphology, FACs sorted GFP+ leukemia cells were assessed by cytospin and Wright/Giemsa staining (Figure 2O–R). The leukemic cells were large with prominent nucleoli and abundant, vacuolated cytoplasm, consistent with a rare, high-grade aggressive NK cell leukemia. In the context of gene expression profiling (outlined below), these leukemias were also similarly classified as aggressive NK cell-like leukemia, suggesting important roles for Tp53 in initiation of leukemias of NK cell origin (Figure 2Q,R). Finally, GFP-labeled kRASG12D-induced tp53del/del ERMS were readily transplantable when engrafted into syngeneic recipient fish (n = 11 primary tumors analyzed, n = 47/49 fish engrafted, Figure 2S–V, Figure 2—figure supplement 1). Histology was similar between primary and transplanted MPNSTs, angiosarcomas, leukemias and ERMS (Figure 2B,C,E,I,M,V).
Figure 2. tp53del/del tumors efficiently transplant into syngeneic CG1 strain zebrafish.
(A–E) A primary tp53del/del MPNSTs that formed in the eye transplanted orthotopically into the periocular space (A–C) or into the peritoneum of CG1-strain recipient fish (D–E). Intraperitoneal injection (i/p). (F–I) tp53del/del Tg(ubi:GFP)-positive angiosarcoma. Primary tumor-bearing fish (F–G) and transplanted animal (H–I). (J–R) tp53del/del Tg(ubi:GFP)-positive leukemia. Primary leukemia (J–K) and transplanted leukemia shown at 20 days post-transplantation (L–R). Whole kidney marrow was isolated from leukemia-engrafted fish and analyzed by FACS (N–O). (N) Forward and side scatter plot of whole kidney marrow of unlabeled CG1 host animal to assess ubi:GFP-positive tp53del/del leukemia cells following transplantation. (O) Analysis of GFP+ ubi:GFP-positive tp53del/del leukemia cells following FACS. Purity was ≥90%. (P–R) Cytospins and Wright/Giemsa staining of whole kidney marrow cells isolated from wildtype fish (P) compared with FACS sorted GFP+ cells from two representative aggressive NK cell-like leukemias, showing large blastic cells with abundant basophilic, vacuolated cytoplasm (Q–R). (S–V) Embryonal rhabdomyosarcoma arising in tp53del/del fish micro-injected at the one-cell stage with linearized rag2:kRASG12D + rag2:GFP. Primary (S), transplanted (2°) (T), and serially transplanted ERMS (3°) (U,V). Whole animal bright-field images (A,D,F,J) and merged GFP-fluorescence images (G,H,K,L,S–U). Hematoxylin and eosin stained sections of engrafted tumors (B–C,E, I, M,V). Scale bars are 5 mm in whole animal images and 100 μm for histology images.
Figure 2—figure supplement 1. Engraftment of tp53del/del tumors into CG1 recipient zebrafish.
Gene expression analysis of tp53del/del tumors arising in transplant recipient fish
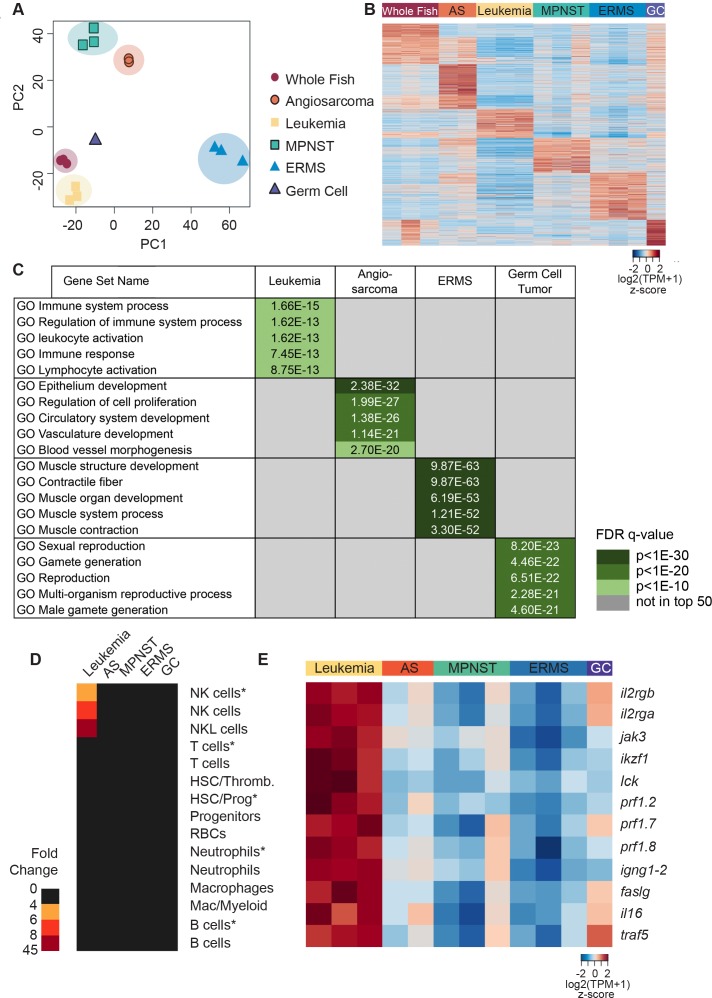
We next profiled the transcriptome of tumor cells isolated from fish transplanted with tp53del/del MPNSTs, angiosarcomas, leukemias, a germ cell tumor and kRASG12D-induced ERMS by Poly(A)+ RNA-sequencing (RNAseq). MPNSTs and the germ cell tumor were analyzed from bulk tumor isolated from non-GFP labeled animals, whereas angiosarcomas, leukemias, and ERMS were FACS sorted from engrafted GFP-labeled tumors (purity and viability >85%). Bulk mRNA from three independent wild-type CG1 strain fish was also sequenced and used as a control. Principal component analysis identified six distinct clusters corresponding to whole CG1 syngeneic fish, leukemia, MPNSTs, angiosarcomas, germ cell tumor and ERMS (Figure 3A). By comparing gene expression among different tumor types, unique tumor-specific expression profiles were identified and each assessed for overlap with gene sets found in the Molecular Signatures Database (MSigDB, Figure 3B,C, Figure 3—source datas 1–3). For example, the upregulated leukemia gene set identified in zebrafish was significantly enriched for GO terms associated with immune system processes, leukocyte activation, immune response and lymphocyte activation. By contrast, angiosarcomas were enriched in GO gene sets associated with vasculature, blood vessel morphogenesis and cellular proliferation. The germ cell tumor showed enrichment of GO gene sets associated with sexual reproduction and gamete formation. As expected, ERMS shared significant overlap of gene signatures with muscle structure, muscle contraction and muscle development.
Figure 3. Gene expression analysis of tp53del/del tumors.
(A) Principal component analysis (PCA) of gene expression profiles from whole CG1 syngeneic fish, MPNSTs, a germ cell tumor, and FACS sorted GFP+ leukemia, angiosarcomas, and ERMS. All tumor samples were obtained following engraftment in CG1 syngeneic recipient fish. (B) Heat map of genes differentially expressed with respect to controls identifies molecularly defined tumor groups. (C) Upregulated genes identified within each tumor type are enriched for Molecular Signature Database (MSigDB) signatures consistent with the expected tissue of origin. (D) NK cell leukemias are enriched for gene signatures identified from normal NK and NKL cells in the kidney marrow and NK cells isolated from rag1-/-, tg(lck:GFP) transgenic fish (NK cells*). For each analysis, enrichment is shown for the top 30 lineage-restricted genes identified from single-cell transcriptional profiling of transgenic cells using SMARTseq2 (denoted by asterisks) or unsorted cells using InDrops single-cell RNA sequencing approaches. (E) Heat map highlighting NK lineage genes significantly upregulated in tp53del/del leukemias when compared to all other tumor types analyzed. [log2(fold-change)]. Angiosarcoma (AS) and germ cell tumor (GC).
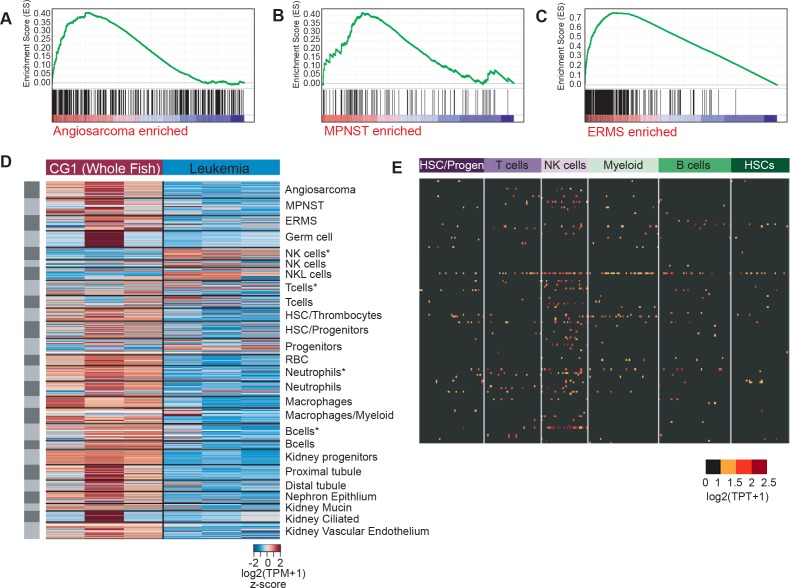
Figure 3—figure supplement 1. Zebrafish cancers share common gene expression with human tumors and confirmation of NK-cell linage derivation for tp53del/del leukemias.
To assess similarities between tp53del/del tumors and human, we next assessed if zebrafish tumors express tumor-specific gene signatures identified from human angiosarcoma (Andersen et al., 2013), MPNST (Kolberg et al., 2015) and ERMS (experimentally determined using GEO:GSE108022) (Figure 3—figure supplement 1—source data 1). Using Gene set enrichment analysis (GSEA) (Mootha et al., 2003; Subramanian et al., 2005), we identified significant enrichment of signatures associated with human angiosarcoma (FDR q-value = 0.001, Figure 3—figure supplement 1A), MPNST (FDR q-value = 0.00433526, Figure 3—figure supplement 1B), and ERMS (FDR q-value = 0, Figure 3—figure supplement 1C) in the corresponding zebrafish tp53del/del tumors but not other tumor types (Figure 3—figure supplement 1—source data 1). Taken together, these data reveal conserved gene expression programs associated with both the predicted cells of origin and the corresponding human cancer counterpart.
Given that GSEAsig analysis failed to assign leukemias to a specific lineage, we next assessed if these tumors were enriched for signatures associated with normal blood cell lineages identified previously by our group using single-cell RNA sequencing of the zebrafish marrow (Tang et al., 2017). Using these lineage-specific gene sets, we found that tp53del/del leukemias expressed markers indicative of NK and NK-like cells but largely failed to express genes associated with other hematopoietic cell lineages (Figure 3D and Figure 3—figure supplement 1D,E). To independently confirm our results, we next identified the top 200 most differentially regulated genes in leukemias compared to all other tumor types and assessed if these genes were differentially expressed in each zebrafish blood lineage. Significant enrichment was only observed in NK cells (Figure 3—figure supplement 1E, p=0.015, one-sided binomial test), supporting a NK cell origin of tp53del/del leukemias. Importantly, tp53del/del NK cell-like leukemias also expressed well-known genes commonly associated with human NK cells, including il2ga and b, jak3, perforins 2,7, and 8, and these genes were highly up-regulated when compared to all other tumor types in our analysis (Figure 3E).
In humans, aggressive NK cell leukemias (ANKLs) have a very poor prognosis and often express perforins but lack markers of mature T- and B- cell lineages (Liang and Graham, 2008; Suzuki and Nakamura, 1999). In human disease, ANKLs are associated with Epstein-Barr virus infection, however, CD3-/CD4-/CD56+/CD13-/CD33- leukemias without EBV infection and intact germline configured T-cell receptor and immunoglobulin have been reported (Liang and Graham, 2008). Interestingly, both TP53 point mutations and deletions have been identified in human ANK cell leukemias, suggesting a role for TP53 in pathogenesis of this disease (Soliman et al., 2014; Yagita et al., 2000). Yet, to date no in vivo animal models of ANKL have been reported precluding direct functional assessment of TP53 loss in eliciting transformation of NK cells.
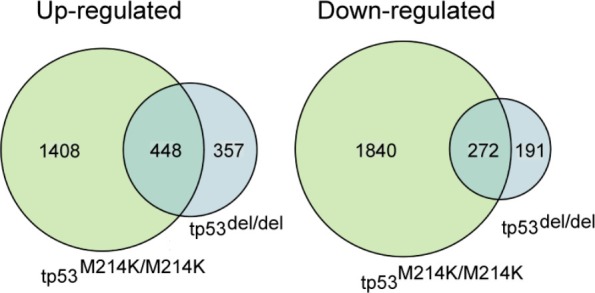
Given that tumor onset and spectrum differ based on the nature of Tp53 mutation or deletion in mice, we next compared gene expression between MPNSTs arising in tp53del/del and tp53M214K/M214K mutant zebrafish (Figure 3—source data 4). As may be expected, we found significant overlap in expression between homozygous tp53 deletion and point-mutant MPNSTs when compared to whole fish (p=4e-321 for up-regulated genes and p=5e-182 for down-regulated genes, one-sided Fisher’s exact test (Figure 3—source data 4), confirming the previously described loss-of-function activity for tp53M214K (Berghmans et al., 2005). Interestingly, differences in gene expression were also noted when comparing these tumors, likely arising from differences in the underlying mutations (Figure 3—source data 4), which are known to differentially affect Tp53 function and tumor etiology in genetically engineered mouse models.
tp53del/del ERMS display increased metastasis but did not alter cancer stem cell number
TP53 has roles in regulating self-renewal of normal stem cells and human cancer cells, including acute myeloid leukemia and breast cancer (Cicalese et al., 2009; Meletis et al., 2006; TeKippe et al., 2003; Zhao et al., 2010). Thus, we predicted that tp53 loss may affect the overall frequency of self-renewing cancer stem cells in zebrafish ERMS. To test this hypothesis, GFP+ ERMS cells were isolated by FACS and injected at limiting dilution into the peritoneum of CG1 recipients (1 × 104–10 cells/recipient, Figure 2T). Animals were followed for 90 days for engraftment using whole animal epi-fluorescent imaging. Unexpectedly, kRASG12D-induced tumors harboring wild-type tp53 had similar frequency of tumor-propagating stem cells when compared with those of tp53del/del ERMS (n ≥ 3 tumors analyzed per genotype, p=0.647 EDLA analysis, Table 1). We concluded that Tp53 loss-of-function does not alter the overall frequency of tumor-sustaining, cancer stem cells in ERMS, which contrasts with previous studies that defined major roles for NOTCH1, MYF5/MYOD, and WNT signaling in regulating self-renewal and the overall number of tumor sustaining cell types in rhabdomyosarcoma (Chen et al., 2014; Hayes et al., 2018; Ignatius et al., 2017; Tenente et al., 2017).
Table 1. Results from limiting dilution cell transplantation experiments comparing engraftment potential of tp53wt/wt and tp53del/del kRASG12D-induced ERMS.
| tp53wt/wt + rag2:kRASG12D ERMS | ||||
|---|---|---|---|---|
| Cell # | Tumor 1 | Tumor 2 | Tumor 3 | Tumor 3 |
| 10000 | 7 of 7 | 5 of 6 | 6 of 6 | 6 of 6 |
| 1000 | 2 of 6 | 2 of 7 | 6 of 8 | 1 of 8 |
| 100 | 0 of 9 | 0 of 8 | 1 of 8 | 0 of 8 |
| TPC# | 1 in 2832 | 1 in 4810 | 1 in 726 | 1 in 7388 |
| 1 in 3495 (2291–5333) | ||||
| tp53del/del + rag2:kRASG12D ERMS | ||||
| Cell # | Tumor 1 | Tumor 2 | Tumor 3 | |
| 10000 | 3 of 5 | 5 of 6 | 6 of 6 | |
| 1000 | 3 of 4 | 3 of 5 | 0 of 7 | |
| 100 | 3 of 9 | 3 of 7 | 1 of 8 | |
| TPC# | 1 in 3546 | 1 in 2228 | 1 in 3640 | |
| 1 in 3038 (1739–5307), p=0.647 | ||||
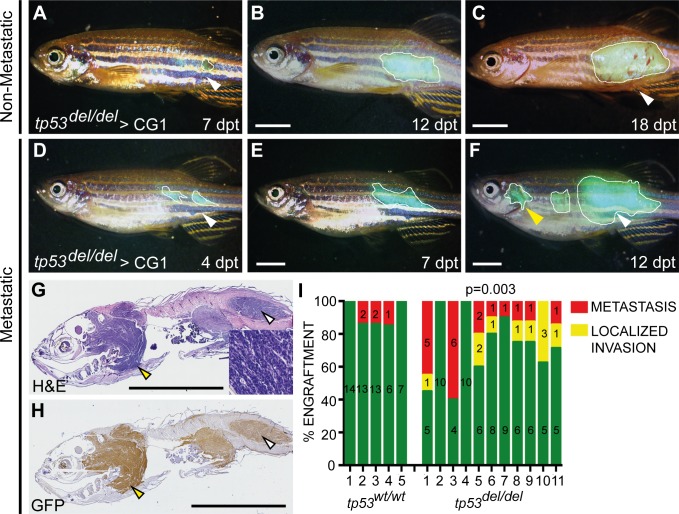
TP53 loss is predictive of poor outcome in human ERMS (Seki et al., 2015); however, given that tp53 loss did not regulate the overall frequency of ERMS stem cells in zebrafish, we reasoned that loss of Tp53 might rather affect tumor invasion and metastasis. To test this hypothesis, we undertook tumor cell transplantation experiments whereby GFP-labeled tumor cells were injected into the dorsolateral musculature of recipient fish and animals monitored for spread into the viscera using epifluoresence whole animal imaging (n = 5 tp53wt/wt and n = 11 tp53del/del ERMS, n = 7–15 recipient fish per tumor,>2×104 cells/recipient) (Tang et al., 2016). As expected, all animals developed GFP+ masses at the site of primary injection (Figure 4A,D, n = 160). Tumor growth was followed for up to 30 days after cell transplantation and animals were assessed for 1) local infiltrative disease defined by growth well beyond but contiguous with the primary site, or 2) metastasis defined by growth at sites unconnected to the primary lesion and/or associated with infiltration into organs within the peritoneal cavity (Tang et al., 2016). Through serial imaging of engrafted fish over time, we identified only rare metastatic lesions in zebrafish engrafted with tp53wt/wt ERMS (n = 5 of 58 engrafted animals, Figure 4A–F). By contrast, tp53del/del ERMS were highly aggressive and displayed elevated local invasion and disseminated metastatic disease (n = 28 of 102 engrafted fish had metastatic ERMS, p=0.003, one-sided Fisher’s exact test, Figure 4I). These metastatic lesions were confirmed on paraffin-embedded sections using both hematoxylin/eosin staining and anti-GFP antibody IHC (Figure 4G,H). Thus, our in vivo experiments demonstrate an important consequence for tp53 loss in stimulating local infiltration and metastasis, revealing a property that may account for poor outcome in RMS patients with TP53 pathway deregulation.
Figure 4. tp53del/del kRASG12D-induced ERMS have increased invasion and metastasis.
(A–F) Whole animal fluorescent images of CG1-strain fish engrafted into the dorsolateral musculature with non-disseminated (A–C) and disseminated ERMS (D–F). Days post transplantation (dpt). White lines demarcate GFP+ tumor area. White arrowheads show site of injection and yellow arrowheads denote metastatic lesions. (G) H and E and (H) GFP immunohistological staining of fish engrafted with metastatic tp53del/del kRASG12D-induced ERMS. (I) Quantification of growth confined to site of injection (green bars) and compared with animals that exhibited local invasion or metastatic ERMS following tumor engraftment until fish were moribund. X-axis identifies 5 tp53wt/wt and 11 tp53del/del ERMS primary tumors that were transplanted into wild-type CG1 syngeneic host zebrafish. p=0.003, one-sided Fisher’s exact test. Scale bars denote 5 mm.
Taken together, our work has defined syngeneic zebrafish as a novel model to assess Tp53 loss-of-function phenotypes and has generated a wide array of cancer types now available for study by the community. This is particularly important for modeling angiosarcoma and aggressive NK cell leukemias for which readily available tp53-deficient zebrafish models are lacking. To date, our tp53del/del zebrafish is the first description of any animal model of aggressive NK cell-like leukemia, highlighting the importance of Tp53 loss in the genesis of these leukemias and opening exciting new avenues of future study. Finally, our work in embryonal rhabdomyosarcoma revealed that Tp53 loss likely has major impacts on regulating ERMS invasion and metastasis, without altering the overall frequency of relapse driving cancer stem cells. Such findings likely account for why human RMS are more aggressive following TP53 pathway disruption (Seki et al., 2015). This work is important, because unlike available genetically engineered mouse models and human ERMS xenografts, zebrafish ERMS are metastatic, which can be readily quantified and visualized in vivo. Future experiments will likely utilize the tp53del/del model to study cancer stem cell self-renewal pathways and metastatic progression in a wider array of tumor types including angiosarcoma, MPNSTs, and available transgenic models that require tp53 loss.
Methods and materials
Key resources table.
| Reagent type (species) or resource |
Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (danio rerio) |
CG1 | Mizgireuv and Revskoy (2006) | ||
| Genetic reagent (danio rerio) |
rag2E450fs/E450fs | Tang et al. (2014) | ||
| Antibody | anti-tp53 (zebrafish) | Abcam | ab77813, RRID: AB_10864112 |
WB 5 ug/mL |
| Antibody | anti-Actin | Sigma | A2066, RRID: AB_476693 |
WB 1:200 |
| Antibody | anti-mouse HRP | Sigma | NA931, RRID: AB_772210 |
WB 1:1000 |
| Antibody | anti-rabbit HRP | Cell Signalling Technology |
7074, RRID: AB2099233 |
WB 1:1000 |
| Recombinant DNA construct |
rag2:kRASG12D | Langenau et al. (2007) | ||
| Recombinant DNA construct |
rag2:GFP |
Langenau et al. (2007)
Langenau et al. (2008) |
||
| Recombinant DNA construct |
ubi:GFP | Mosimann et al. (2011) | ||
| Commercial assay or kit |
In situ Cell Death Detection Kit, TMR red |
Sigma | 12156792910 |
Animals
Zebrafish used in this work included: CG1 strain zebrafish (Mizgireuv and Revskoy, 2006), CG1-strain Tg(ubi:GFP) animals that were generated using Tol2-mediated transgenesis (Kawakami et al., 2000; Mosimann et al., 2011), and rag2E450fs/E450fs;casper strain zebrafish that were used in a subset of ERMS metastasis assays. All animal studies were approved by the Massachusetts General Hospital Subcommittee on Research Animal Care under the protocol #2011 N-000127.
Generation of tp53del/del zebrafish using TALENS
Four TALEN pairs (two pairs each flanking the tp53 gene locus in D. rerio) were designed to generate a ~12.1 kb deletion encompassing the entire tp53 coding sequence. Mojo Hand (http://talendesign.org) was used to design eight 15-mer repeat variable di-residue (RVD) TALENs with a 15 to 18 bp spacer (Ma et al., 2013; Neff et al., 2013). Each TALEN pair was designed to target a unique restriction site that could be used to determine TALEN cutting efficacy by restriction fragment length polymorphism (RFLP) analysis. All TALEN constructs were synthesized with the Golden Gate method using the RCIscript-GoldyTALEN scaffold (Addgene, https://www.addgene.org/Stephen_Ekker/, ID# 38142) (Ma et al., 2013; Neff et al., 2013). The RVDs NI, HD, NG and NN (recognizing A, C, T and G bases, respectively) were used to construct TALENs. Intermediate constructs containing RVDs for positions 1 to 10 were synthesized in the pFUS_A receiver plasmid in the first reaction. Pre-synthesized pFUS_B4 plasmids were then selected based on the target sequence. The library of 256 pFUS_B4 plasmids is available through Addgene (https://www.addgene.org/Stephen_Ekker/, Kit # 1000000038). The completed pFUS_A and pFUS_B4 as well as the last half-repeat plasmid (pLR-NI, -HD, -NN or -NG) were combined in the second Golden Gate reaction in the RCIscript-GoldyTALEN expression vector that has T3 promoter. The completed constructs were linearized using SacI, and mRNA was in vitro transcribed using the mMESSAGE mMACHINE T3 Transcription Kit (Thermo Fisher Scientific, cat. no. AM1348). Large deletions encompassing the tp53 locus were engineered through co-injection of TALEN pairs targeting the tp53 5’UTR and 3’UTR. Genotyping was performed using standard PCR: tp53 forward 5’-CACAGCAAGGACACATCTGC-3’, tp53del reverse 5’-AGATCAGTGCTTGTATTGTATCAGTTT-3’, tp53wt reverse 5’-GATCGCTCAGAGTCGCAAA-3’
Embryonic protein extraction and western blotting
24 hpf embryos of the respective genotypes were dissociated in PBS, spun down at 1000xg to de-yolk samples, and lysed in 10% SDS buffer. Western blot was performed using anti-tp53 (ab77813, Abcam) and anti-actin (A2066, Sigma) antibodies.
Apoptosis assay
Embryos were raised at 28°C and gamma-irradiated at 24 hpf. At 30 hpf embryos were fixed overnight in 4% paraformaldehyde followed by staining using the In Situ Cell Death Detection Kit, TMR Red (Roche Applied Bioscience) as per manufacturer protocol.
Histology and immunohistochemistry
Paraffin embedding, sectioning and immunohistochemical analysis of zebrafish sections were performed as previously described (Chen et al., 2014; Ignatius et al., 2012). Anti-human SOX10 was performed at the MGH and BWH DF/HCC Research Pathology Cores. Slides were imaged using a transmitted light Olympus BX41 microscope and a Motic Easy Scan Pro slide scanner. Pathology review and staging were completed by board-certified sarcoma (G.P.N and E.Y.C) and hematology pathologist (R.P.H).
Micro-injection and ERMS generation
rag2:kRASG12D and rag2:GFP constructs were described previously (Langenau et al., 2008; Langenau et al., 2007). DNA plasmids were linearized with Xho1, phenol:chloroform-extracted, ethanol-precipitated, resuspended in 0.5 × Tris EDTA + 0.1 M KCl, and injected into one-cell CG1 strain embryos.
FACS and tumor cell transplantation
FACS analysis and RMS cell transplantation were completed essentially as previously described (Chen et al., 2014; Ignatius et al., 2012; Langenau et al., 2007; Smith et al., 2010). tp53del/del angiosarcomas, leukemias and ERMS tumor cells were stained with DAPI to exclude dead cells and sorted twice using a Laser BD FACSAria II Cell Sorter. Sort purity and viability were assessed after two rounds of sorting, exceeding 85% and 90%, respectively. GFP+ ERMS tumors were transplanted at limiting dilution and monitored for tumor engraftment under a fluorescent dissecting microscope from 10 to 90 days post-transplantation. Tumor-propagating cell frequency was quantified using the Extreme Limiting Dilution Analysis software package (http://bioinf.wehi.edu.au/software/elda/). GFP+ tumor cells were isolated by FACS from a subset of transplanted fish and RNA isolated for RNA sequencing. Subsets of tumors were fixed in 4% PFA and embedded in paraffin blocks, sectioned and stained with Hematoxylin and Eosin. Sorted GFP+ tp53 del/del leukemia cells were spun down onto a cytospin slide and processed by Wright/Giemsa staining.
RNA sequencing and analysis
Paired-end reads from poly(A)+ RNA-seq were aligned to the GRCz10 reference zebrafish genome with STAR v2.4.0 (Dobin et al., 2013) using GRCz10v85 Ensembl annotations. PCR duplicates were removed with Picard v1.95 [http://broadinstitute.github.io/picard/] and reads aligning to ribosomal RNA were removed with RSeQC (Wang et al., 2012) Gene counts were obtained from reads with an alignment quality of at least 10 using featureCounts (Liao et al., 2014) and transformed to transcript per million (TPM) units. Human orthologues of zebrafish genes were obtained from the Beagle database (Tang et al., 2017; available at: http://chgr.mgh.harvard.edu/genomesrus/). Differential expression analysis was performed with DESeq2 (Love et al., 2014), requiring log2(FC) ≥2 and an FDR < 0.05 was required. Each tumor type was individually compared to the control samples. Clustering of differentially expressed genes used the partitioning around medoids (PAM) method in the cluster R package and the Pearson correlation was used as distance. The number of clusters was optimized with the silhouette function from the same cluster R package.
Gene set enrichment analysis
Human tumor-specific gene signatures were assessed for enrichment in tp53del/del tumor types using GSEA 3.0. (Mootha et al., 2003; Subramanian et al., 2005). Gene signatures were assessed for anigosarcoma (Andersen et al., 2013), MPNST (Kolberg et al., 2015), and ERMS (GEO:GSE108022). The ERMS signature was defined by genes up-regulated in both human and zebrafish kRASG12D-induced tp53wt/wt ERMS when compared with normal muscle (log2(FC) ≥2). GSEA was completed in comparing individual tumor types to all other tumors using the default parameters and 1000 permutations of the data.
Comparison of tp53del/del and tp53M214K/M214K MPNST
RNA sequencing data from four tp53M214K/M214K homozygous mutant MPNST samples were processed as described above. Differential expression analysis was performed as described above, comparing tp53del/del and tp53M214K/M214K MPNSTs to whole CG1 controls. Statistical significance was assessed with a one-sided Fisher’s exact test on a background of genes that were expressed in at least 4 out of 15 samples.
Molecular signature database (MSigDB) analysis and leukemia similarities with NK cells
The top 500 most differentially regulated genes within each tumor type were identified and assigned human gene IDs using the Beagle database. These humanized gene lists were then queried for overlaps with molecular signatures from MSigDB. Only the top 50 enriched gene sets were analyzed and representative examples of enriched data sets are shown in Figure 3C. For blood cell analysis in Figure 3D,a 30 gene signature was defined for each of the major blood cell lineages and then cumulative gene expression analyzed across tumor types as described by Tang et al. (2017). The top 200 genes up-regulated in tp53del/del leukemias compared to all other tumors where assessed relative to gene sets generated using SMARTseq as described in Tang et al. (2017).
Acknowledgements
We thank Dr. Jim Amatruda for sharing zebrafish MPNST RNAseq sample data.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
David M Langenau, Email: dlangenau@mgh.harvard.edu.
Marianne E Bronner, California Institute of Technology, United States.
Richard M White, Memorial Sloan Kettering Cancer Center, United States.
Funding Information
This paper was supported by the following grants:
National Cancer Institute R24OD016761 to Myron S Ignatius, Madeline N Hayes, Finola E Moore, Qin Tang, Sara P Garcia, Alexander Jin, Ashwin Ramakrishnan, Sophia Reeder, Gunnlaugur Petur Nielsen, Eleanor Y Chen, Robert P Hasserjian, David M Langenau.
Alex's Lemonade Stand Foundation for Childhood Cancer to Myron S Ignatius, Madeline N Hayes, David M Langenau.
National Cancer Institute R00CA175184 to Myron S Ignatius, Kunal Baxi, Long Wang, Yidong Chen.
National Cancer Center RR160062 to Myron S Ignatius, Kunal Baxi, Long Wang.
National Cancer Institute R01CA154923 to Myron S Ignatius, Madeline N Hayes, Finola E Moore, Qin Tang, Sara P Garcia, Alexander Jin, Ashwin Ramakrishnan, Sophia Reeder, Gunnlaugur Petur Nielsen, Eleanor Y Chen, Robert P Hasserjian, David M Langenau.
National Cancer Institute U54CA168512 to Myron S Ignatius, Madeline N Hayes, Finola E Moore, Qin Tang, Sara P Garcia, Alexander Jin, Ashwin Ramakrishnan, Sophia Reeder, Gunnlaugur Petur Nielsen, Eleanor Y Chen, Robert P Hasserjian, David M Langenau.
Rally Foundation Amanda Riley Foundation Research Fellowship to Madeline N Hayes.
National Cancer Center R01CA211734 to Madeline N Hayes, Sara P Garcia, Alexander Jin, David M Langenau.
National Cancer Center R01CA215118 to Madeline N Hayes, Sara P Garcia, Alexander Jin, David M Langenau.
National Cancer Institute UL1RR024150 to Patrick R Blackburn.
National Cancer Institute GM63904 to Stephen C Ekker.
St. Baldrick's Foundation to David M Langenau.
Massachusetts General Hospital Research Scholars Program to David M Langenau.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Writing—original draft, Writing—review and editing.
Data curation, Formal analysis, Investigation, Writing—original draft, Writing—review and editing.
Conceptualization, Data curation, Formal analysis, Investigation, Writing—review and editing.
Data curation, Formal analysis, Investigation, Writing—review and editing.
Formal analysis, Writing—review and editing.
Resources, Methodology, Writing—review and editing.
Formal analysis, Investigation, Methodology, Writing—review and editing.
Resources.
Formal analysis.
Investigation.
Data curation.
Resources.
Formal analysis, Writing—review and editing.
Formal analysis.
Formal analysis, Writing—review and editing.
Data curation.
Resources, Methodology, Writing—review and editing.
Supervision, Funding acquisition, Writing—original draft, Writing—review and editing.
Ethics
Animal experimentation: Animal studies were approved by the Massachusetts General Hospital Subcommittee on Research Animal Care under the protocol #2011-N-000127.
Additional files
Data availability
Sequencing data has been deposited in GEO under accession code GSE109581
The following dataset was generated:
Myron S Ignatius, author; Madeline N Hayes, author; David M Langenau, author. tp53 deficiency causes a wide tumor spectrum and increases embryonal rhabdomyosarcoma metastasis in zebrafish. 2018 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE109581 Publicly available at the NCBI Gene Expression Omnibus (accession no. GSE109581)
The following previously published datasets were used:
Qin Tang, author; David M Langenau, author. Dissecting hematopoietic and renal cell heterogeneity in adult zebrafish at single cell resolution using RNA sequencing [Smart-seq] 2017 http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE100911 Publicly available at the NCBI Gene Expression Omnibus (accession no. GSE100911)
Qin Tang, author; David M Langenau, author. Dissecting hematopoietic and renal cell heterogeneity in adult zebrafish at single cell resolution using RNA sequencing [inDrops] 2017 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE100910 Publicly available at the NCBI Gene Expression Omnibus (accession no. GSE100910)
Qin Tang, author; David M Langenau, author. Dissecting hematopoietic and renal cell heterogeneity in adult zebrafish at single cell resolution using RNA sequencing [bulk RNA-seq] 2017 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE100912 Publicly available at the NCBI Gene Expression Omnibus (accession no. GSE100912)
References
- Andersen NJ, Nickoloff BJ, Dykema KJ, Boguslawski EA, Krivochenitser RI, Froman RE, Dawes MJ, Baker LH, Thomas DG, Kamstock DA, Kitchell BE, Furge KA, Duesbery NS. Pharmacologic inhibition of MEK signaling prevents growth of canine hemangiosarcoma. Molecular Cancer Therapeutics. 2013;12:1701–1714. doi: 10.1158/1535-7163.MCT-12-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghmans S, Murphey RD, Wienholds E, Neuberg D, Kutok JL, Fletcher CD, Morris JP, Liu TX, Schulte-Merker S, Kanki JP, Plasterk R, Zon LI, Look AT. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. PNAS. 2005;102:407–412. doi: 10.1073/pnas.0406252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, DeRan MT, Ignatius MS, Grandinetti KB, Clagg R, McCarthy KM, Lobbardi RM, Brockmann J, Keller C, Wu X, Langenau DM. Glycogen synthase kinase 3 inhibitors induce the canonical WNT/β-catenin pathway to suppress growth and self-renewal in embryonal Rhabdomyosarcoma. PNAS. 2014;111:5349–5354. doi: 10.1073/pnas.1317731111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Stewart E, Shelat AA, Qu C, Bahrami A, Hatley M, Wu G, Bradley C, McEvoy J, Pappo A, Spunt S, Valentine MB, Valentine V, Krafcik F, Lang WH, Wierdl M, Tsurkan L, Tolleman V, Federico SM, Morton C, Lu C, Ding L, Easton J, Rusch M, Nagahawatte P, Wang J, Parker M, Wei L, Hedlund E, Finkelstein D, Edmonson M, Shurtleff S, Boggs K, Mulder H, Yergeau D, Skapek S, Hawkins DS, Ramirez N, Potter PM, Sandoval JA, Davidoff AM, Mardis ER, Wilson RK, Zhang J, Downing JR, Dyer MA, St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project Targeting oxidative stress in embryonal Rhabdomyosarcoma. Cancer Cell. 2013;24:710–724. doi: 10.1016/j.ccr.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choorapoikayil S, Kuiper RV, de Bruin A, den Hertog J. Haploinsufficiency of the genes encoding the tumor suppressor pten predisposes zebrafish to hemangiosarcoma. Disease Models & Mechanisms. 2012;5:241–247. doi: 10.1242/dmm.008326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicalese A, Bonizzi G, Pasi CE, Faretta M, Ronzoni S, Giulini B, Brisken C, Minucci S, Di Fiore PP, Pelicci PG. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell. 2009;138:1083–1095. doi: 10.1016/j.cell.2009.06.048. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Butel JS, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- Harvey M, McArthur MJ, Montgomery CA, Butel JS, Bradley A, Donehower LA. Spontaneous and carcinogen-induced tumorigenesis in p53-deficient mice. Nature Genetics. 1993;5:225–229. doi: 10.1038/ng1193-225. [DOI] [PubMed] [Google Scholar]
- Hayes MN, McCarthy KM, Jin A, Oliveira ML, Iyer S, Garcia SP, Sindiri S, Gryder B, Motala Z, Nielsen GP, Borg J-P, van de Rijn M, Malkin D, Khan J, Ignatius MS, Langenau DM. Vangl2 regulates stem cell self-renewal programs and growth in Rhabdomyosarcoma through a wnt/PCP/RhoA signaling Axis. Cell Stem Cell. 2018;22:414–427. doi: 10.1016/j.stem.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatius MS, Chen E, Elpek NM, Fuller AZ, Tenente IM, Clagg R, Liu S, Blackburn JS, Linardic CM, Rosenberg AE, Nielsen PG, Mempel TR, Langenau DM. In vivo imaging of tumor-propagating cells, regional tumor heterogeneity, and dynamic cell movements in embryonal Rhabdomyosarcoma. Cancer Cell. 2012;21:680–693. doi: 10.1016/j.ccr.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatius MS, Hayes MN, Lobbardi R, Chen EY, McCarthy KM, Sreenivas P, Motala Z, Durbin AD, Molodtsov A, Reeder S, Jin A, Sindiri S, Beleyea BC, Bhere D, Alexander MS, Shah K, Keller C, Linardic CM, Nielsen PG, Malkin D, Khan J, Langenau DM. The NOTCH1/SNAIL1/MEF2C pathway regulates growth and Self-Renewal in embryonal Rhabdomyosarcoma. Cell Reports. 2017;19:2304–2318. doi: 10.1016/j.celrep.2017.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Current Biology. 1994;4:1–7. doi: 10.1016/S0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Kastenhuber ER, Lowe SW. Putting p53 in Context. Cell. 2017;170:1062–1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Shima A, Kawakami N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the japanese medaka fish, and its transposition in the zebrafish germ lineage. PNAS. 2000;97:11403–11408. doi: 10.1073/pnas.97.21.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolberg M, Høland M, Lind GE, Ågesen TH, Skotheim RI, Hall KS, Mandahl N, Smeland S, Mertens F, Davidson B, Lothe RA. Protein expression of BIRC5, TK1, and TOP2A in malignant peripheral nerve sheath tumours--A prognostic test after surgical resection. Molecular Oncology. 2015;9:1129–1139. doi: 10.1016/j.molonc.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, Terzian T, Caldwell LC, Strong LC, El-Naggar AK, Lozano G. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Langenau DM, Keefe MD, Storer NY, Guyon JR, Kutok JL, Le X, Goessling W, Neuberg DS, Kunkel LM, Zon LI. Effects of RAS on the genesis of embryonal Rhabdomyosarcoma. Genes & Development. 2007;21:1382–1395. doi: 10.1101/gad.1545007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenau DM, Keefe MD, Storer NY, Jette CA, Smith AC, Ceol CJ, Bourque C, Look AT, Zon LI. Co-injection strategies to modify radiation sensitivity and tumor initiation in transgenic zebrafish. Oncogene. 2008;27:4242–4248. doi: 10.1038/onc.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavigueur A, Maltby V, Mock D, Rossant J, Pawson T, Bernstein A. High incidence of lung, bone, and lymphoid tumors in transgenic mice overexpressing mutant alleles of the p53 oncogene. Molecular and Cellular Biology. 1989;9:3982–3991. doi: 10.1128/MCB.9.9.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Abrahamson JL, Kandel R, Donehower LA, Bernstein A. Susceptibility to radiation-carcinogenesis and accumulation of chromosomal breakage in p53 deficient mice. Oncogene. 1994;9:3731–3736. [PubMed] [Google Scholar]
- Liang X, Graham DK. Natural killer cell neoplasms. Cancer. 2008;112:1425–1436. doi: 10.1002/cncr.23316. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma AC, Lee HB, Clark KJ, Ekker SC. High efficiency in vivo genome engineering with a simplified 15-RVD GoldyTALEN design. PloS One. 2013;8:e65259. doi: 10.1371/journal.pone.0065259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkin D. Li-fraumeni syndrome. Genes & Cancer. 2011;2:475–484. doi: 10.1177/1947601911413466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meletis K, Wirta V, Hede SM, Nistér M, Lundeberg J, Frisén J. p53 suppresses the self-renewal of adult neural stem cells. Development. 2006;133:363–369. doi: 10.1242/dev.02208. [DOI] [PubMed] [Google Scholar]
- Mizgireuv IV, Revskoy SY. Transplantable tumor lines generated in clonal zebrafish. Cancer Research. 2006;66:3120–3125. doi: 10.1158/0008-5472.CAN-05-3800. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genetics. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Mosimann C, Kaufman CK, Li P, Pugach EK, Tamplin OJ, Zon LI. Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development. 2011;138:169–177. doi: 10.1242/dev.059345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA, Vousden KH. Mutant p53 in Cancer: new functions and therapeutic opportunities. Cancer Cell. 2014;25:304–317. doi: 10.1016/j.ccr.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff KL, Argue DP, Ma AC, Lee HB, Clark KJ, Ekker SC. Mojo hand, a TALEN design tool for genome editing applications. BMC Bioinformatics. 2013;14:1. doi: 10.1186/1471-2105-14-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann JC, Lillard K, Damoulis V, Amatruda JF. Zebrafish models of germ cell tumor. Methods in Cell Biology. 2011;105:1–24. doi: 10.1016/B978-0-12-381320-6.00001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, Jacks T. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Parant JM, George SA, Holden JA, Yost HJ. Genetic modeling of Li-Fraumeni syndrome in zebrafish. Disease Models & Mechanisms. 2010;3:45–56. doi: 10.1242/dmm.003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Nishimura R, Yoshida K, Shimamura T, Shiraishi Y, Sato Y, Kato M, Chiba K, Tanaka H, Hoshino N, Nagae G, Shiozawa Y, Okuno Y, Hosoi H, Tanaka Y, Okita H, Miyachi M, Souzaki R, Taguchi T, Koh K, Hanada R, Kato K, Nomura Y, Akiyama M, Oka A, Igarashi T, Miyano S, Aburatani H, Hayashi Y, Ogawa S, Takita J. Integrated genetic and epigenetic analysis defines novel molecular subgroups in Rhabdomyosarcoma. Nature Communications. 2015;6:8557. doi: 10.1038/ncomms8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Padmanabhan A, de Groh ED, Lee JS, Haidar S, Dahlberg S, Guo F, He S, Wolman MA, Granato M, Lawson ND, Wolfe SA, Kim SH, Solnica-Krezel L, Kanki JP, Ligon KL, Epstein JA, Look AT. Zebrafish neurofibromatosis type 1 genes have redundant functions in tumorigenesis and embryonic development. Disease Models & Mechanisms. 2012;5:881–894. doi: 10.1242/dmm.009779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AC, Raimondi AR, Salthouse CD, Ignatius MS, Blackburn JS, Mizgirev IV, Storer NY, de Jong JL, Chen AT, Zhou Y, Revskoy S, Zon LI, Langenau DM. High-throughput cell transplantation establishes that tumor-initiating cells are abundant in zebrafish T-cell acute lymphoblastic leukemia. Blood. 2010;115:3296–3303. doi: 10.1182/blood-2009-10-246488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman DS, Sabbagh AA, Omri HE, Ibrahim FA, Amer AM, Otazu IB. Rare aggressive natural killer cell leukemia presented with bone marrow fibrosis - a diagnostic challenge. SpringerPlus. 2014;3:390. doi: 10.1186/2193-1801-3-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. PNAS. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, Nakamura S. Malignancies of natural killer (NK) cell precursor: myeloid/NK cell precursor acute leukemia and blastic NK cell lymphoma/leukemia. Leukemia Research. 1999;23:615–624. doi: 10.1016/S0145-2126(98)00194-5. [DOI] [PubMed] [Google Scholar]
- Tang Q, Abdelfattah NS, Blackburn JS, Moore JC, Martinez SA, Moore FE, Lobbardi R, Tenente IM, Ignatius MS, Berman JN, Liwski RS, Houvras Y, Langenau DM. Optimized cell transplantation using adult rag2 mutant zebrafish. Nature Methods. 2014;11:821–824. doi: 10.1038/nmeth.3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Iyer S, Lobbardi R, Moore JC, Chen H, Lareau C, Hebert C, Shaw ML, Neftel C, Suva ML, Ceol CJ, Bernards A, Aryee M, Pinello L, Drummond IA, Langenau DM. Dissecting hematopoietic and renal cell heterogeneity in adult zebrafish at single-cell resolution using RNA sequencing. The Journal of Experimental Medicine. 2017;214:2875–2887. doi: 10.1084/jem.20170976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Moore JC, Ignatius MS, Tenente IM, Hayes MN, Garcia EG, Torres Yordán N, Bourque C, He S, Blackburn JS, Look AT, Houvras Y, Langenau DM. Imaging tumour cell heterogeneity following cell transplantation into optically clear immune-deficient zebrafish. Nature Communications. 2016;7:10358. doi: 10.1038/ncomms10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubert H, Meye A, Würl P. Prognosis is correlated with p53 mutation type for soft tissue sarcoma patients. Cancer Research. 1996;56:4134–4136. [PubMed] [Google Scholar]
- Taylor AC, Shu L, Danks MK, Poquette CA, Shetty S, Thayer MJ, Houghton PJ, Harris LC. P53 mutation and MDM2 amplification frequency in pediatric rhabdomyosarcoma tumors and cell lines. Medical and Pediatric Oncology. 2000;35:96–103. doi: 10.1002/1096-911X(200008)35:2<96::AID-MPO2>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- TeKippe M, Harrison DE, Chen J. Expansion of hematopoietic stem cell phenotype and activity in Trp53-null mice. Experimental Hematology. 2003;31:521–527. doi: 10.1016/S0301-472X(03)00072-9. [DOI] [PubMed] [Google Scholar]
- Tenente IM, Hayes MN, Ignatius MS, McCarthy K, Yohe M, Sindiri S, Gryder B, Oliveira ML, Ramakrishnan A, Tang Q, Chen EY, Petur Nielsen G, Khan J, Langenau DM. Myogenic regulatory transcription factors regulate growth in Rhabdomyosarcoma. eLife. 2017;6:e19214. doi: 10.7554/eLife.19214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang S, Li W, Wei L. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- Yagita M, Huang CL, Umehara H, Matsuo Y, Tabata R, Miyake M, Konaka Y, Takatsuki K. A novel natural killer cell line (KHYG-1) from a patient with aggressive natural killer cell leukemia carrying a p53 point mutation. Leukemia. 2000;14:922–930. doi: 10.1038/sj.leu.2401769. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Zuber J, Diaz-Flores E, Lintault L, Kogan SC, Shannon K, Lowe SW. p53 loss promotes acute myeloid leukemia by enabling aberrant self-renewal. Genes & Development. 2010;24:1389–1402. doi: 10.1101/gad.1940710. [DOI] [PMC free article] [PubMed] [Google Scholar]