Abstract
Recent evidence demonstrates that epigenetic regulation of gene transcription is critically involved in learning and memory. Here, we discuss the role of histone acetylation and DNA methylation, which are two best understood epigenetic processes in memory processes. More specifically, we focus on learning-strength-dependent changes in chromatin on the fibroblast growth factor 1 (Fgf1) gene and on the molecular events that modulate regulation of Fgf1 transcription, required for memory enhancement, with the specific focus on CREB-regulated transcription coactivator 1 (CRTC1).
Keywords: Memory formation, Memory enhancement, Epigenetics, Gene transcription, FGF1, CRTC1
1. Introduction
Activity-dependent changes in gene transcription and de novo protein synthesis are required for memory processes (Alberini, 2009; Klann and Dever, 2004; Mayford et al., 2012). On the other hand, a deficiency in activity-dependent gene transcription is involved in cognitive decline prominent in many neuropsychiatric disorders, such as Alzheimer’s disease and depression, as well as in memory loss during healthy ageing (Greer and Greenberg, 2008; West and Greenberg, 2011). Epigenetic modifications have recently emerged as one of the central mechanisms regulating gene transcription in the brain (Day and Sweatt, 2010; Graff and Tsai, 2013; Peixoto and Abel, 2013).
The cAMP-responsive element-binding protein (CREB)-dependent gene expression is essential for synaptic plasticity, learning and memory (Barco et al., 2002; Barco et al., 2005; Bito et al., 1996; Bourtchuladze et al., 1994; Deisseroth et al., 1996; Impey et al., 1998; Josselyn et al., 2004; Kida et al., 2002; Kida and Serita, 2014; Mayford et al., 2012; Silva et al., 1998; Suzuki et al., 2011). The CREB-regulated transcriptional coactivators or cAMP-responsive transcriptional coactivators (CRTCs, also referred to as TORCs) may potentiate the interaction of CREB with CBP/p300 (Xu et al., 2007) and significantly increase CREB transcriptional activity independently of Ser133 phosphorylation (Conkright et al., 2003; Iourgenko et al., 2003) (Fig. 1). CRTC1 is translocated from the synapses/dendrites to the nucleus in response to neural activity and learning (Ch'ng et al., 2012; Kovacs et al., 2007; Li et al., 2009; Nonaka et al., 2014; Parra-Damas et al., 2017; Uchida et al., 2017a). Some reports have shown recently that CRTC1 plays a key role in synaptic plasticity and memory formation in rodents (Nonaka et al., 2014; Sekeres et al., 2012; Uchida et al., 2017a; Zhou et al., 2006a). Moreover, CRTC1 is associated with memory enhancement and memory maintenance via epigenetic regulation of gene transcription (Hirano et al., 2016; Uchida and Shumyatsky, 2017; Uchida et al., 2017a).
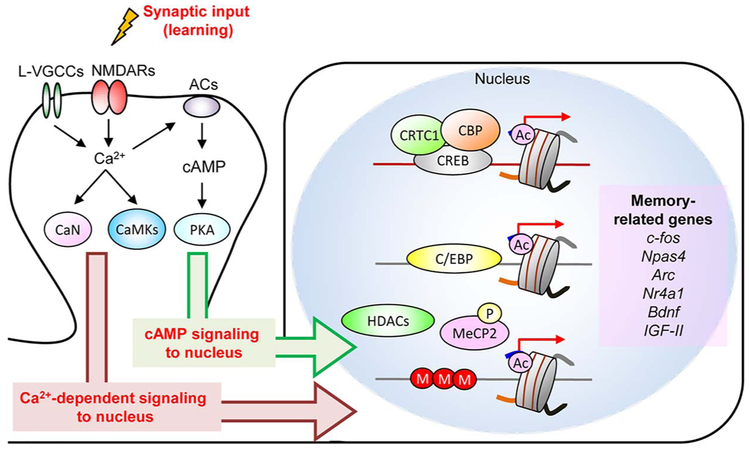
Fig. 1.
Learning-dependent gene expression program required for memory formation.
Activation of L-type voltage-sensitive calcium channel (L-VGCCs) and NMDA receptors (NMDARs) triggers calcium influx and induce calcium-dependent signaling molecules such as calcineurin (CaN) and Ca2+/cal-modulin-dependent protein kinases (CaMKs). Calcium influx also activates cAMP signaling pathway such as protein kinase (PKA) via Ca2+-sensitive adenylate cyclase (ACs). These molecules regulate the activity of transcription modulators (CREB, CBP, HDACs, CRTC1, and MeCP2) via phosphorylation and dephosphorylation. These transcriptional modulators contribute to the control of activity-dependent gene transcription which is required for synaptic plasticity and memory formation. Ac: acetylation: P: phosphorylation; M: DNA methylation.
In this review, we will begin by describing previous studies and recent progress demonstrating that histone acetylation and DNA methylation are importnat for memory. We will then describe the role of CRTC1-mediated epigenetic regulation of the fibroblast growth factor 1 (Fgf1) gene transcription in memory enhancement. We will also address how CRTC1 and FGF1 pathways may contribute to the development of memory-related disorders.
2. Epigenetic mechanisms in memory formation
An increasing evidence has indicated that epigenetic modifications of histones in neuronal cells constitute a powerful mechanism of memory processing (Day and Sweatt, 2010; Graff and Tsai, 2013; Peixoto and Abel, 2013).
2.1. Histone acetylation
Among the various types of histone modifications (acetylation, phosphorylation, methylation, ubiquitylation, sumoylation, ADP-ribosylation, deamination, proline isomerization), histone acetylation is one of the most well studied. In histone acetylation, a negatively charged acetyl group is added to lysin (K) residues of histone proteins (Graff and Tsai, 2013). Histone deacetylase (HDAC) inhibitors including trichostatin A, suberoylanilide, valproic acid, and sodium butyrate ameliorate cognitive deficits and improve learning and memory (Alarcon et al., 2004; Bredy et al., 2007; Guan et al., 2009; Korzus et al., 2004; Levenson et al., 2004; McQuown et al., 2011; Peleg et al., 2010; Wood et al., 2005). The enzymes primarily responsible for reversible histone acetylation that control memory are histone acetyltransferase (HAT) CBP/p300 and histone deacetylase HDAC2 (Table 1). These two molecules have opposite effects on memory. CBP loss-of-function mutation in mice shows decreased fear memory (Alarcon et al., 2004; Korzus et al., 2004; Wood et al., 2006). Also, p300 is required for long-term recognition memory and fear memory (Oliveira et al., 2007). Conversely, HDAC2 knockout mice show increased fear memory, whereas HDAC2 overexpression reduces memory (Guan et al., 2009). In addition, there is considerable evidence indicating the role of class 1 HDAC family (HDAC1, HDAC2, HDAC3, and HDAC8) in memory formation (Table 1). Viral-mediated overexpression of HDAC1 in the mouse hippocampus increases fear extinction, whereas pharmacological blockade of HDAC1 leads to impaired extinction (Bahari-Javan et al., 2012). The same paper also reported that HDAC1 regulates an activity-dependent gene (c-fos), suggesting a key role of HDAC1 in transcriptional regulation of memory-related genes. Both the focal deletion of HDAC3 in the CA subregion of hippocampus as well as HDAC3 inhibition via RGFP136 significantly enhances long-term memory (McQuown et al., 2011). Similarly, viral-mediated acute knockdown of HDAC3 in the CA subregion of the hippocampus or intra-hippocampal injection of an HDAC3 selective inhibitor leads to enhanced contextual fear memory (Uchida et al., 2017a). However, a prolonged HDAC3 depletion reduces memory (Nott et al., 2016). The discrepancy between these studies may be due to the duration of HDAC3 deficiency, but these reports support at least the contribution of HDACs to memory formation. In addition to class I HDAC family, HDAC4, belonging to class Ha HDACs family (HDAC4, −5, −7 and −9), regulates memory formation (Table 1). Conditional brain-specific HDAC4 knockout mice showed significant impairments in contextual fear and spatial memory (Kim et al., 2012). Given that HDAC4 synapse-to-nucleus shuttling is regulated in response to neuronal activity (Sando et al., 2012; Uchida and Shumyatsky, 2017), learning-dependent relocation of HDACs may have an important role for synaptic plasticity and memory.
Table 1.
Brief summary of the role of HDACs/DNMTs/TET1 in memory formation.
| Molecules | Findings | References | |
|---|---|---|---|
| HDACs | HDAC1 | Hippocampal HDAC1 is required for extinction learning via H3K9 deacetylation. | Bahari-Javan et al. (2012) |
| HDAC2 | HDAC2 deficiency causes increased synapse number and memory facilitation. HDAC2 overexpression decreases dendritic spine density, synaptic plasticity, and memory formation. S-nitrosylation of HDAC2 is involved in recent memory updating. |
Guan et al. (2009)
Graff et al. (2014) |
|
| HDAC3 | Focal deletion of HDAC3 in hippocampal CA1 region of adult mice enhances long-term memory. A prolonged HDAC3 depletion in forebrain reduces memory. HDAC3 knockdown in hippocampal CA region of adult mice enhances long-term memory. Enzymatic activity of HDAC3 is required for long-term memory formation |
McQuown et al. (2011)
Nott et al. (2016) Uchida et al., (2017a) |
|
| HDAC4 | HDAC4 regulates synaptic transmission and memory without deacetylating histones. Selective loss of HDAC4 in brain results in impairments in hippocampal-dependent memory and long-term synaptic plasticity. |
Sando et al. (2012)
Kim et al. (2012) |
|
| HDAC5 | Loss of HDAC5 does not impact learning and memory. HDAC5 deficiency leads to spatial memory impairment. |
Kim et al. (2012)
Agis-Balboa et al. (2013) |
|
| HDAC7 | HDAC7 in the hippocampus is involved selectively in the consolidation of contextual fear memory. | Jing et al. (2017) | |
| HATs | CBP/p300 | CBP +/− mice show impairments of chromatin acetylation, synaptic plasticity, and memory. HAT activity of CBP is required for memory consolidation. CREB-binding domain of CBP is required for memory formation. p300 is required for the formation of long-term memory. HAT activity of p300 is required for memory formation. |
Alarcon et al. (2004)
Korzus et al. (2004) Wood et al. (2006) Oliveira et al. (2011) Oliveira et al. (2007) |
| PCAF | PCAF KO animals show memory deficits. PCAF activator treatment enhances memory for fear extinction and prevents fear renewal. |
Maurice et al. (2008)
Wei et al. (2012) |
|
| KAT5 (Tip60) | KAT5 is required for H4K12 acetylation, synaptic plasticity, and memory enhancement. KAT5 is required for long-term memory maintenance via H4K16 acetylation. |
Uchida et al. (2017a)
Hirano et al. (2016) |
|
| DNMTs | DNMT1 | DNMT1 knockout mice show normal memory. | Morris et al. (2014) |
| DNMT1/3a | Double knockout mice show abnormal long-term plasticity in the hippocampal CA1 region together with deficits in learning and memory. | Feng et al. (2010) | |
| DNMT3a | DNMT3a knockout mice show reduced memory and abnormal synaptic plasticity. | Morris et al. (2014) | |
| DNMT3a2 | Reducing hippocampal Dnmt3a2 levels in young adult mice impairs memory formation. Restoring hippocampal Dnmt3a2 levels in aged mice rescues cognitive ability. |
Oliveira et al. (2012) | |
| TETs | TET1 | TET1 deficiency leads to abnormal hippocampal long-term depression and impaired memory extinction Hippocampal TET1 overexpression leads to impairment of contextual fear memory. |
Rudenko et al. (2013)
Kaas et al. (2013) |
We apologize to the authors whose articles were not cited here due to space limitation.
While we know a lot about the role of HDACs in memory formation, there is scarce evidence supporting the role of HATs except for CBP/p300 (Table 1). A recent paper has shown that KAT5 (also referred to as Tip60) plays a key role in learning-induced histone H4K12 acetylation, a potentially important lysine residue of histone H4 for memory (Guan et al., 2009; Peleg et al., 2010), and this mechanism is important for regulation of memory enhancement (Uchida et al., 2017a).
A very recent paper has shown that acetyl coenzyme A (acetyl-CoA) synthetase (ACSS2) regulates histone acetylation and hippocampal memory (Mews et al., 2017). As mentioned above, histones can be modified by acetyl groups, leading to the modulation of gene expression. Such acetylation requires a nuclear pool of the metabolite acetyl-CoA (Wellen et al., 2009). In mammalian cells, there are two principal enzymes that generate acetyl-CoA for histone acetylation: ACSS2 and citrate-dependent ATP-citrate lyase (ACL) (Pietrocola et al., 2015). ACSS2 is highly expressed in the mouse hippocampus (Lein et al., 2007). Mews et al. demonstrated that ACSS2 is required in the mouse hippocampus for the induction of immediate early genes and for long-term spatial memory through the epigenetic regulation of transcription dynamics (Mews et al., 2017). Thus, this report provides first evidence for metabolic signaling to be involved in chromatin regulation in the brain and show that this signaling has a crucial role in memory consolidation.
So far, we summarized the role of HDACs and HATs in epigenetic regulation of gene transcription in memory formation, whereas it should be noted that HDACs and HATs have multiple functions and substrates instead of histone proteins. For instance, an integrated database of Compendium of Protein Lysine Modifications reported that there are 7151 lysine acetylation sites in 3311 proteins (Liu et al., 2014). This is supported by experimentally that CBP/p300, PCAF, and HDACs have non-histone target proteins, such as transcription factors, transcription cofactors, and cytoskeleton (Brochier et al., 2013; Chen et al., 2005; Dompierre et al., 2007; Federman et al., 2013; Gaub et al., 2010). Thus, there is a possibility that HATs and HDACs modulate directly the acetylation/deacetylation of transcription modulators, which then lead to the regulation of gene transcription without affecting histone acetylation. Indeed, HDAC4 regulates synaptic transmission and memory without deacetylating histones (Sando et al., 2012). Further studies will be necessary to understand the role of epigenetic regulation of gene transcription by HATs and HDACs in memory formation and will be required to identify a clear distinction between cause and effect and between epigenetics and classical transcription machinery (see (Lopez-Atalaya and Barco, 2014) for extensive review).
2.2. DNA methylation
DNA methylation is another major epigenetic modification which leads to the addition of a methyl group to the 5′ position of a cytosine pyrimidine ring by DNA methyltransferases (DNMTs) to form 5-methylcytosine, thereby often modifying the chromatin state and gene expression. The vast majority of cytosine methylation occurs as part of a CG dinucleotide sequence. Changes in DNA methylation can result from various alterations in cell status, including after neuronal activity. Treatment with inhibitor for DNMTs inhibits synaptic plasticity and memory formation (Levenson et al., 2006). Contextual fear conditioning rapidly (i.e., within 30 min) increases the methylation level of memory suppressor gene protein phosphatase 1, while concurrently demethylating the promoter region of the plasticity-related gene reelm (Miller and Sweatt, 2007). Contextual fear conditioning transiently induces demethylation of the Bdnf exon III and exon IV promoters in the hippocampus, and these effects are blocked by application of the NMDA receptor antagonist MK801 (Lubin et al., 2008; Mizuno et al., 2012). DNA methylation of the memory suppressor gene calcmeurin is increased in the prefrontal cortex seven days following fear conditioning (Miller et al., 2010). Infusion of DNMT inhibitors into the anterior cingulate cortex prevents memory retrieval 30 days after training. These findings indicate that cortical DNA methylation is triggered by a learning experience and is a perpetuating signal used by the brain to help preserve remote memories (Miller et al., 2010). In addition, double knockout mice that lack Dnmt1 and Dnmt3a exclusively in forebrain excitatory neurons shows abnormal long-term plasticity in the hippocampal CA1 region together with deficits in learning and memory (Feng et al., 2010) (Table 1). Moreover, the reduced expression of DNA methyltransferase Dnmt3a2 is associated with age-related memory loss, and rescuing DNMT3a2 levels in the hippocampus of aged mice restore cognitive function (Oliveira et al., 2012). The authors also show that Dnmt3a2 is an immediate-early gene, activity of which is partially dependent upon nuclear calcium signaling. These findings suggest that activity-dependent DNA methylation may be associated with neurode-generative memory loss.
More recent work identified DNA methylation changes that are associated with contextual fear memory consolidation and maintenance (Haider et al., 2016). They charted an unbiased genome-wide profile of DNA methylation, brain region (hippocampal CA1 and anterior cingulate cortex) and cell type specificity (neuron and non-neuron), over time (1 h and 4 weeks after learning), and found that substantial changes in DNA methylation during memory consolidation and maintenance are present at specific inter- and intragenic regions. In neurons, differentially methylated regions were preferentially located in inter-genic (64%) and intronic (30%) regions. This evidence is similar to the distribution of differentially methylated regions in activity-induced dentate gyrus neurons (Guo et al., 2011a). Moreover, associative memory–induced differentially methylated regions significantly colocalized with the acetylation of H3K27-positive regions, indicating that 21–29% of the differentially methylated regions reside in functional cis-regulatory regions, many of which are intronic. These results suggest that a substantial proportion of differentially methylated regions might regulate transcription factor binding in cis-regulatory regions. Fischer and Bonn groups also demonstrated directly lasting memory-associated changes in DNA methylation in the cortex (Haider et al., 2016). Their comprehensive genome-wide assessments strongly support the hypothesis that DNA cytosine methylation contributes to long-term memory stabilization and storage in vivo. Thus, active methylation of cytosine bases in DNA is required for memory formation and maintenance, and neuronal activity and behavioral experiences lead to site-specific reorganization of DNA methylation dynamics (Day et al., 2013; Guo et al., 2011a; Haider et al., 2016; Miller et al., 2010).
A novel epigenetic mark, 5-hydroxymethylcytosine, has been recently shown to be important for neuronal function. 5-hydroxymethylcytosine is enriched in the brain (Szulwach et al., 2011) and is regulated by neuronal activity through ten-eleven translocation proteins (TETs) (Feng et al., 2015; Kaas et al., 2013; Rudenko et al., 2013). TETs can convert 5-methylcytosine to 5-hydroxymethylcytosine in mammals. Tet1 knockout mice show downregulation of neuronal activity-regulated genes (e.g., Npas4, c-fos) in the cortex and hippocampus (Rudenko et al., 2013). Tet1 knockout mice also have abnormal hippocampal LTD and impaired memory extinction (Rudenko et al., 2013). Another study showed that Tet1 itself can be regulated by neuronal activity and the enhancement of Tet1 function in the hippocampus leads to contextual fear memory impairment (Kaas et al., 2013). However the exact roles of 5-hydroxymethylcytosine and TETs in synaptic function and memory formation are still insufficient.
Methyl-CpG binding protein 2 (MeCP2) binds to methylated cytosines in DNA and acts epigenetically as a transcriptional repressor. Neuronal activity induces the phosphorylation of MeCP2 at Ser421 (Chen et al., 2003; Deng et al., 2010; Zhou et al., 2006b) across the genome, suggesting that activity-dependent phosphorylation of MeCP2 mediates a genome-wide chromatin response to neuronal activity (Cohen et al., 2011) (Fig. 1). MeCP2 knock-in mice were generated with Ser421 converted to alanine (Ser421Ala), preventing phosphorylation (Cohen et al., 2011). These knock-in mutants showed increased dendritic complexity and increased inhibitory synaptic strength in the cortex (Cohen et al., 2011). The knock-in mice showed abnormal responses to inanimate objects or conspecifics (Cohen et al., 2011). These findings suggest that activity-dependent phosphorylation of MeCP2 regulates synapse development and behavioral responses to environmental stimuli. However, MeCP2 Ser421Ala knock-in neurons did not have any changes in activity-dependent target gene transcription (Cohen et al., 2011). Thus, it remains to be determined whether activity-dependent phosphorylation of MeCP2 affects gene transcription and chromatin structure. More recently, phosphorylation of MeCP2 at Thr308 was reported to be induced by neuronal activity and MeCP2 T308 knock-in mice showed altered activity-dependent gene expression (e.g., Bdnf, Arc, Fos, Npas4) (Ebert et al., 2013). Another study with knock-in mice that carry point mutations in the endogenous Mecp2 gene locus that abolish phosphorylation at both S421 and S424 (Mecp2f421A:S424A/y mice) showed enhanced hippocampus-dependent contextual fear and spatial memory (Li et al., 2011). These mutants also display enhanced synaptic plasticity (long-term potentiation (LTP)) and synaptogenesis. Moreover these knock-in mice have increased Bdnf transcription. Mice expressing a truncated Mecp2 variant, which lacks the carboxy-terminal region, exhibit deficiency in learning and memory, as well as synaptic plasticity (Moretti et al., 2006). These mice showed deficits in hippocampus-dependent spatial memory, contextual fear and social memory (Moretti et al., 2006).
Recent evidence suggests that MeCP2 acts not only as a transcription repressor but also as transcription activator in a complex with CREB in vitro (Chahrour et al., 2008) and in vivo (Uchida et al., 2011), regulating a complex transcription program following neuronal activation. Indeed, MeCP2 can interact not only with transcriptional (co)-activators (e.g., CREB, CBP) but also transcriptional (co)-repressors (e.g., REST, HDACs). Furthermore, MeCP2 can bind to 5-methylcytosine- and 5-hydroxymethylcytosine-containing DNA with similar affinity in the mouse brain (Mellen et al., 2012). Thus, MeCP2 regulation is activity-dependent and complex, which might explain inconsistent results between the studies. In addition to CpG methylation, a recent report has shown that adult mouse dentate gyrus neurons consist of both CpG and CpH (H = A/C/T) methylation (Guo et al., 2014). This neuronal CpH methylation is conserved in human brain, enriched in regions of low CpG density and negatively correlates with gene expression. MeCP2 binding is greatly enhanced when CpG and CpH methylation are adjacent to each other. Although it is still unclear whether CpH methylation is involved in synaptic plasticity and memory formation, CpH methylation is a new layer of epigenetic modulation of the neuronal genome.
3. CRTC1-CREB signaling in memory enhancement
Transcription factors such as CREB and C/EBP are known to regulate gene transcription essential for synaptic plasticity and memory formation (Alberini and Chen, 2012) (Fig. 1). More recently, transcription cofactor CRTC1 has emerged as novel transcriptional regulators of essential biological functions, including brain plasticity and memory formation (Escoubas et al., 2017; Saura and Cardinaux, 2017; Uchida and Shumyatsky, 2017). Nuclear-cytoplasmic redistribution of CRTCs is dependent on their activity-regulated phosphorylation status (Altarejos and Montminy, 2011). Calcium signals promote nuclear translocation of CRTC1 via activation of calcineurin, which directly dephosphorylates CRTC1 at Ser151 (Bittinger et al., 2004; Ch'ng et al., 2012; Screaton et al., 2004). A recent report has shown that two phosphorylation sites (S151 and S245) contribute to nuclear import of CRTC1 (Nonaka et al., 2014). The nuclear transport of CRTC1 is observed in the CA1 and CA3 pyramidal neurons, but not in the dentate gyrus granule neurons, of the hippocampus following multiple behavioral tasks, including contextual fear conditioning (CFC), novel object location and Morris water maze (Parra-Damas et al., 2017; Uchida and Shumyatsky, 2017; Uchida et al., 2017a). Importantly, strong training of CFC induces much greater CRTC1 nuclear accumulation than weak CFC training (Uchida et al., 2017a), suggesting an association of CRTC1 nuclear translocation with memory strength. Moreover, CRTC1 nuclear accumulation is specific for learning, but as this accumulation is not observed in context-only or immediate-shock exposure (Uchida et al., 2017a). This learning-dependent nuclear accumulation of CRTC1 also occurs in the basolateral amygdala following CFC (Nonaka et al., 2014). Viral-mediated enhancement of CRTC1 function in the CA area of hippocampus increases CFM (Nonaka et al., 2014; Uchida et al., 2017a). Conversely, shRNA-mediated knockdown of CRTC1 in the CA region of hippocampus (Uchida et al., 2017a) or basolateral amygdala (Nonaka et al., 2014) leads to decreased CFM along with decreased CREB-mediated gene transcription. Furthermore, increased CRTC1 function promoted spine enlargement in the CA1 pyramidal cells (Nonaka et al., 2014) and the loss-of-function CRTC1 disrupted LTP in CA1 (Uchida et al., 2017a; Zhou et al., 2006a), clearly suggesting that CRTC1 is involved in structural and synaptic plasticity.
One step downstream of the activation of CRTC1 is regulation of the expression of its target genes. In the nucleus, CRTC1 increases the expression of a subset of CREB target genes, including c-fos, Arc, Nr4a1, Bdnf (Ch'ng et al., 2012; Fukuchi et al., 2015; Nonaka et al., 2014; Parra-Damas et al., 2017; Parra-Damas et al., 2014; Zhou et al., 2006a) (Fig. 1). Although all of these activity-regulated genes have been involved in synaptic plasticity and memory, the molecular mechanism underlying CRTC1-mediated memory enhancement has not been completely investigated. One of CRTC1-CREB target genes, brain-specific isoform B of Fgf1 (Fgf1b), was recently identified (Uchida et al., 2017a).
4. Fgf1b, a CRTC1-CREB target gene required for memory consolidation, promotes memory enhancement
Fgf1b, has been recently identified as a target gene of CRTC1/CREB-mediated transcription during memory consolidation in mice (Uchida et al., 2017a). In addition to neurotrophic factors (e.g., BDNF) and growth factors (e.g., IGF-II) (Chen et al., 2011; Finsterwald and Alberini, 2014; Tyler et al., 2002) (Fig. 1), the FGF signaling has recently emerged as a new key player in synaptic plasticity and memory (Bookout et al., 2013; Kang and Hebert, 2015; Owen et al., 2013; Turner et al., 2012; Wu et al., 2012). In mammals, the FGF family consists of 22 members, of which FGF1 is predominantly expressed in neurons (Elde et al., 1991). It was earlier reported that Fgf1b is induced in the mouse hippocampus immediately following electroconvulsive stimulation (Ma et al., 2009), suggesting that FGF1 signaling can be regulated by activity and possibly involved in synaptic plasticity.
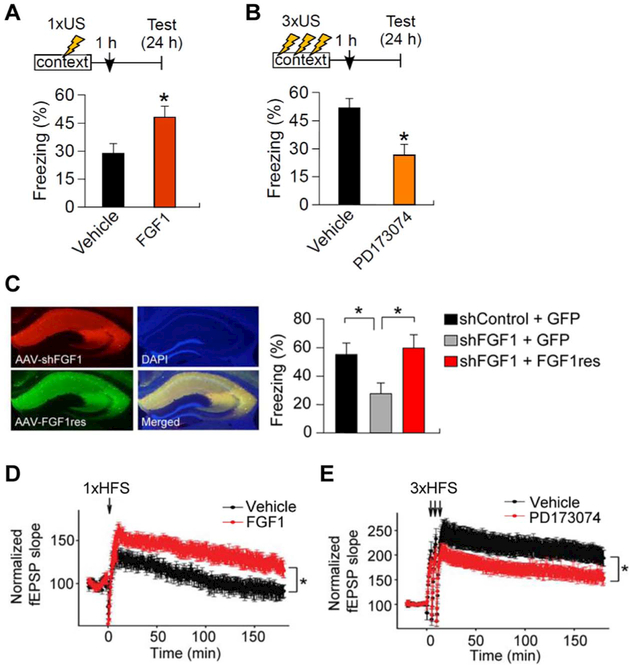
Confirming this possibility, Uchida et al. recently reported that persistent expression of the Fgf1b gene following learning within the CA region of the hippocampus is associated with an increase in memory strength (Uchida et al., 2017a). Intriguingly, Fgf1b expression is briefly increased within 1 h and returns to the baseline 2 h after “weak” (single-shock) training in CFC, while it is elevated 2h following “strong” (three-shock) training. There is no effect of learning on Fgf1b expression in the dentate gyrus of hippocampus. These data suggest that persistent expression of Fgf1b is CA-region-specific in the hippocampus and might be involved in establishing memory strength in the CA region. Injection of FGF1 increase memory after weak training (Fig. 2A). By contrast, an FGF receptor antagonist disrupted memory after strong training (Fig. 2B), suggesting that FGF1 signaling is required for memory consolidation. Moreover, FGF1 knockdown reduced long-term memory (Fig. 2C) following strong training. Similar to fear memory, object location memory (OLM), which is also hippocampus-dependent, is modulated by FGF1 (Uchida et al., 2017a). These results further confirm that FGF1 signaling in the CA subregion is required for memory consolidation. It should be noted that the FGF1 enhances the transient potentiation induced by weak high-frequency stimulation (1 × HFS) (Fig. 2D). Strong high-frequency stimulation (3 × HFS) elicits robust LTP and is attenuated by FGF1 antagonist (Fig. 2E). Thus, FGF1 appears necessary for the switch from transient plasticity to strong enduring plasticity, suggesting an interesting mechanism for memory consolidation.
Fig. 2.
FGF1 is essential for memory enhancement.
(A) Effect of recombinant FGF1 post-treatment on weak training of contextual fear conditioning. Mice were injected 1 h after 1-shock contextual fear conditioning and memory assessed 24 h later. *p < 0.05 vs. vehicle-treated group.
(B) Effect of PD173074 pretreatment on conditioned fear memory. Mice were injected with PD173074 into the hippocampus 1 h after 3-shock training, and memory was assessed 24 h later. *p < 0.05 vs. vehicle-treated group.
(C) Mice co-injected with AAV-shFGF1 and AAV-GFP showed decreased long-term (24 h) contextual fear memory following 3-shock training of contextual fear conditioning. This reduction was not observed in mice coinjected with AAV-shFGF1 together with shRNA-resistance Fgf1 (AAV-FGF1res). *p < 0.05.
(D) Effect of recombinant FGF1 on weak stimulus (1 × HFS)-evoked LTP. HFS: high frequency stimulation. *p < 0.05.
(E) Effect of the FGF receptor antagonist PD173074 on 3 × HFS-evoked LTP at CA3-CA1 synapses. *p < 0.05.
Adapted from Uchida et al. (2017a).
5. CRTC1-dependent switch of histone acetyltransferases on the Fgf1b promoter following learning
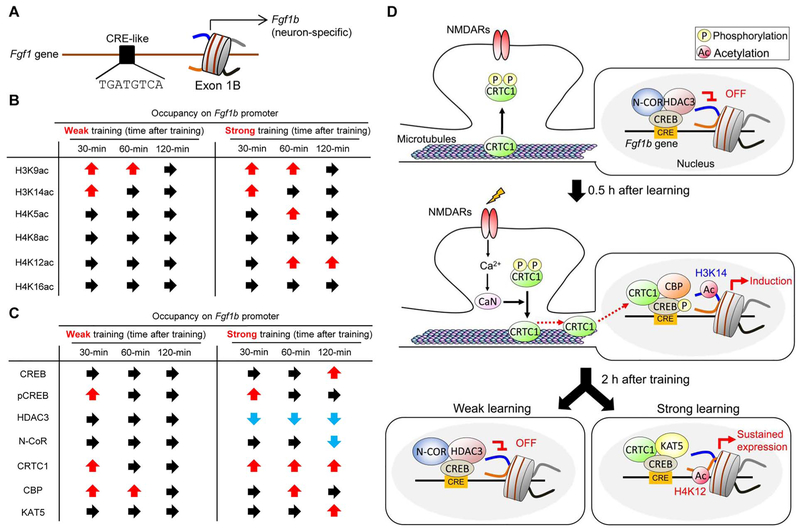
How is Fgf1b transcription involved in memory encoding? Recent work has shown that CRTC1 regulates two waves Fgf1b transcription and enhances memory strength (Uchida et al., 2017a). During the first wava, the acetylation levels of H3K9 and H3K14, but not of H4K8 or H4K16, are significantly increased on the Fgf1b gene promoter 0.5–1 h after both strong and weak fear conditioning (Fig. 3A and B). By contrast, acetylation of H4K5 and H4K12 is increased 1–2 h after strong training only, suggesting that these latter histone modifications are important for long term memory encoding and memory enhancement. Strong but not weak training induced progressive dissociation of HDAC3 and corepressor N-CoR from the Fgf1b promoter (Fig. 3C and D), suggesting that basal Fgf1b transcription is suppressed by recruitment of HDAC3-N-CoR to its promoter (Uchida et al., 2017a). This hypothesis is supported by pharmacological and genetic studies (Uchida et al., 2017a): mice injected bilaterally with T247, a potent and selective HDAC3 inhibitor (Suzuki et al., 2013), into the hippocampus show an increase in Fgf1b expression, H3K14 acetylation at the Fgf1b promoter and exhibit increased freezing 24 h after weak training. Similarly, mice with a HDAC3 knockdown in the CA subregion exhibit enhanced long-term memory (Uchida et al., 2017a). These results suggest that HDAC3-mediated regulation of Fgf1b transcription is involved in memory formation, in agreement with a previous report demonstrating that HDAC3 negatively regulates long-term memory (McQuown et al., 2011). Moreover, homozygous knock-in mice expressing mutant N-CoR lacking HDAC3 binding capacity show deficient long-term OLM (McQuown et al., 2011), again consistent with a role for HDAC3 in memory consolidation via its interaction with N-CoR. These results suggest that HDAC3 activity is important for epigenetic silencing of Fgf1b and HDAC is a negative regulator of long-term memory formation.
Fig. 3.
CRTC1 modulates the epigenetic regulation of Fgf1b transcription.
(A) Putative CRE sites within the mouse Fgf1b promoter. Arrows indicate major transcription start sites.
(B) Summary of the data showing H3K9ac, H3K14ac, H4K5ac, H4K8ac, H4K12ac, and H4K16ac occupancy on the Fgf1b promoter following weak (single-shock) or strong (three-shock) CFC.
(C) Summary of the data showing the occupancies of transcription factor, transcription cofactors, HDACs, and HATs (CREB, pCREB, HDAC3, N-CoR, CRTC1, CBP, and KAT5) on the Fgf1b promoter following weak (single-shock) or strong (three-shock) CFC.
(D) Proposed model for memory enhancement. Under basal conditions, CRTC1 is phosphorylated and anchored to the synapses and dendrites. In the nucleus, HDAC3-N-CoR complex represses Fgf1b transcription. Upon learning, Ca2+ signals potentiate CRTC1 dephosphorylation via activation of calcineurin (CaN). Dephosphorylated CRTC1 translocates to the nucleus, where it binds to phosphorylated CREB (pCREB) and histone acetyltransferases (CBP) and enhances Fgf1b gene transcriptional activity by increasing the acetylation of H3K14 on its promoter. Strong training (e.g., three foot-shock CFC) maintains nuclear localization of CRTC1 and upregulates Fgf1b transcription independently of pCREB even 2 h after learning by enhancing H4K12 acetylation via KAT5 recruitment to its promoter region. Learning-induced KAT5 recruitment acetylates H4K12 on the Fgf1b promoter, thereby enhancing synaptic plasticity and memory formation. Ac: acetylation, P: phosphorylation.
Adapted from Uchida and Shumyatsky (2017).
The phosphorylated form of CREB at Seri 33 occupies the promoter of Fgf1b following both weak and strong training in contextual fear (Fig. 3C and D) and the recruitment of CRTC1 and CBP enhances the acetylation of H3K14, which is one of the substrates of CBP (Peixoto and Abel, 2013), leading to Fgf1b gene transcription. This molecular event is transient: CREB phosphorylation and CBP are not observed on the Fgf1b gene promoter 1 h after weak training. By contrast, strong training elicits prolonged (up to 2h) CRTC1 recruitment on the Fgf1b gene promoter and is associated with long-lasting upregulation of H4K12 acetylation, which is not observed following weak training. This sustained transcription following strong training is mediated at least in part by substitution of histone acetyltransferase KAT5 for CBP in a CRTC1-dependent manner (Fig. 3C and D). Strong training maintains upregulation of Fgf1b transcription 2h after learning by recruiting KAT5 to the promoter region independently of CREB phosphorylation, enhancing H4K12 acetylation. Given that increased H4K12 acetylation might be associated with memory enhancement (Guan et al., 2009; Peleg et al., 2010), nuclear translocation of CRTC1 and subsequent initiation of KAT5-dependent enhancement of H4K12 are critical for enduring synaptic plasticity and memory enhancement. This switching of histone acetyltransferases (CBP to KAT5) may be an important molecular mechanism underlying memory enhancement.
Importantly, CRTC1 knockdown suppresses KAT5 recruitment and subsequent H4K12 acetylation on Fgf1b promoter following strong training (Uchida et al., 2017a), indicating that CRTC1 is critical for sustained epigenetic regulation of Fgf1b transcription, required for memory enhancement. Given that deregulation of H4K12 acetylation is involved in age-associated memory loss (Peleg et al., 2010), it is possible that aberrant CRTC1-CREB-KAT5-mediated regulation of H4K12 acetylation may be involved in the pathophysiology of age-related cognitive disorders. Another study has also shown that the transition from memory formation to maintenance is mediated by H4K16 acetylation and shifting the transcriptional complex from CREB/CBP to CREB/CRTC1 in Drosophila (Hirano et al., 2016). This study also reported that KAT5-dependent histone acetylation is required for memory maintenance. These reports suggest that memory formation and maintenance are distinct processes, and involve a shifting array of transcription factors, coactivators and HATs. Thus, CRTC1 is a key molecule for memory enhancement and maintenance, and shows a sustained translocation to the nucleus in time-limited and learning-strength-dependent manner, which is associated with dynamic alteration of histone acetylation in the nucleus. Future studies aiming to understand these molecular mechanisms should lead to better insight into the involvement of CRTC1 in epigenetic regulation of gene transcription required for memory.
In addition to histone modification, DNA methylation is reported to be involved in Fgf1b transcription. Synchronous activation of adult hippocampal neurons in vivo by electroconvulsive stimulation leads to CpG demethylation of Fgf1b promoters in these neurons within 4 h after the activation (Ma et al., 2009). AAV-mediated overexpression of TET1 leads to significant decreases in the CpG methylation levels at Fgf1b in the hippocampus, and this is accompanied by a significant upregulation in transcription level (Guo et al., 2011b). Thus, endogenous TET1 is required for neuronal activity-induced active DNA demethylation and gene expression for Fgf1b in the adult hippocampus. Further studies will be necessary to clarify the role of DNA (de)methylation of Fgf1b in learning and memory.
6. Retrieval-induced reconsolidation for memory enhancement
In addition to pharmacological and genetic approaches, there are a number of behavioral manipulations found to be effective for memory enhancement (e.g., reconsolidation, exercise, and environmental enrichment). For example, memory can be enhanced by targeting retrieval-induced reconsolidation (Dudai and Eisenberg, 2004). Following retrieval, a once consolidated memory destabilizes and again requires gene transcription changes in order to re-stabilize (Nader et al., 2000; Suzuki et al., 2004). It should be noted that chromatin histone modifications are suggested to be involved not only in memory consolidation but also in memory reconsolidation (Hemstedt et al., 2017; Jarome and Lubin, 2014; Lattal and Wood, 2013). Importantly, CRTC1 overexpression enhanced both memory consolidation and reconsolidation (Sekeres et al., 2012). Increasing CRTC1 function is sufficient to enhance the strength of new, as well as established reactivated, memories. Similarly, CREB and CBP have shown as key modulators for memory reconsolidation (Kida et al., 2002; Maddox et al., 2013; Sekeres et al., 2012), suggesting that CRTC1/CREB/CBP complex-mediated epigenetic regulations of gene transcription also regulate memory reconsolidation. Indeed, recent evidence suggests that epigenetic histone modifications are associated with memory reconsolidation (Bredy and Barad, 2009; Graff et al., 2014; Lubin and Sweatt, 2007; Villain et al., 2016), but further studies are necessary to reveal how CRTC1 influences gene transcription during and after memory retrieval.
7. Possible involvement of CRTC1 and FGF1 in memory-related disorders
Alzheimer’s disease is a neurodegenerative disorder, characterized by progressive decline in memory, cognitive functions, and changes in behavior and personality (Mattson, 2004; Reddy and Beal, 2008). There are two major features observed in postmortem brains from patients with Alzheimer’s disease: 1) intracellular neurofibrillary tangles and 2) extracellular amyloid beta (Aβ) deposits in the regions of the brain that are responsible for learning and memory. Alzheimer’s disease is also associated with the loss of synapses, synaptic function and neuronal loss. It is interesting to note that CREB/CRTC1-mediated regulation of gene expression in the hippocampus of a mouse model of Alzheimer’s disease is changed not only in naïve conditions but also in response to learning (Parra-Damas et al., 2014). Moreover, disrupted memory and diminished induction of activity-dependent genes Nr4a1 and Nr4a2 following learning in mice lacking the Alzheimer’s disease-linked presenilin genes (presenilin conditional double knockout mice) are rescued by CRTC1 overexpression in the dorsal hippocampus (Parra-Damas et al., 2017), suggesting an impairment in activity-dependent gene regulation in Alzheimer’s disease. Analysis of human patients shows a reduction of both total and phosphorylated CRTC1 in human hippocampus at Braak IV and V–VI pathological stages of Alzheimer’s disease (Parra-Damas et al., 2014). More recently, DNA methylation at the CRTC1 promoter was reported to be significantly lower in the hippocampus samples of patients with Alzheimer’s disease (Mendioroz et al., 2016), suggesting a potential role of DNA methylation in the pathophysiology. In addition to CRTC1, there are also reports suggesting a possible role of FGF1 in Alzheimer’s disease. Genetic studies indicate significant association of single nucleotide polymorphisms (SNPs) within the FGF1 gene (Tao et al., 2014; Yamagata et al., 2004). In addition, the number of FGF1-immunolabeled neurons is reduced in patients with Alzheimer’s disease (Thorns et al., 2001; Thorns and Masliah, 1999). Taken together, deficiency in the CRTC1-FGF1 pathway might be associated with the pathophysiology of Alzheimer’s disease.
Meta-analysis has revealed significant relationship between depression and memory impairment (Kizilbash et al., 2002). Also, symptoms of depression are present in a significant proportion of Alzheimer’s disease patients (Cerejeira et al., 2012). While, pathophysiology of depression remains poorly understood, aberrant epigenetic regulation of gene transcription has been suggested in patients with major depression as well as in animal models of depression (Consortium, 2015; Robison and Nestler, 2011; Sun et al., 2013; Uchida et al., 2017b). Several of the activity-dependent transcription factors and cofactors have been associated with depression, including CREB (Carlezon et al., 2005), MeCP2 (Hutchinson et al., 2012; Uchida et al., 2011) and CRTC1 (Meylan et al., 2016a; Meylan et al., 2016b). These findings suggest that aberrant epigenetic regulation of gene transcription mediated by CRTC1 pathway may also account at least in part for the pathophysiology of Alzheimer’s disease as well as depression.
8. Conclusion
In conclusion, this review shows that epigenetic mechanisms are involved in the memory consolidation and memory enhancement. The challenge for the translational field is to develop the therapeutic agents for treatment of memory-related conditions, such as Alzheimer’s disease. As recombinant FGFs have been applied in clinical settings to treat multiple disorders (Beenken and Mohammadi, 2009), FGF1 could be a novel target for cognitive enhancement. In addition, although it is still debated whether HDACs and HATs are directly modulate histone acetylation during memory formation, the development of drugs targeting transcriptional modulators, such as HDAC3 or KAT5, may have the potential as a treatment of memory-related symptoms.
Acknowledgements
This work was supported by the NIH R01MH107555, Whitehall Foundation (#2008-12-104), March of Dimes, NARSAD Independent Investigator Grant and the New Jersey Commission on Brain Injury Research (to G.P.S); JST-CREST, KAKENHI (15K09807 and 15H04895) and the NAITO Foundation (to S.U.).
Footnotes
Conflict of interest
The authors declare that there are no conflicts of interest.
References
- Agis-Balboa RC, Pavelka Z, Kerimoglu C, Fischer A, 2013. Loss of HDAC5 impairs memory function: implications for Alzheimer’s disease. J. Alzheimers Dis 33, 35–44. [DOI] [PubMed] [Google Scholar]
- Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A, 2004. Chromatin acetylation, memory, and LTP are impaired in CBP +/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron 42, 947–959. [DOI] [PubMed] [Google Scholar]
- Alberini CM, Chen DY, 2012. Memory enhancement: consolidation, reconsolidation and insulin-like growth factor 2. Trends Neurosci 35, 274–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberini CM, 2009. Transcription factors in long-term memory and synaptic plasticity. Physiol. Rev 89, 121–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altarejos JY, Montminy M, 2011. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat. Rev. Mol. Cell Biol 12, 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahari-Javan S, Maddalena A, Kerimoglu C, Wittnam J, Held T, Bahr M, Burkhardt S, Delalle I, Kugler S, Fischer A, et al. , 2012. HDAC1 regulates fear extinction in mice. J. Neurosci 32, 5062–5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barco A, Alarcon JM, Kandel ER, 2002. Expression of constitutively active CREB protein facilitates the late phase of long-term potentiation by enhancing synaptic capture. Cell 108, 689–703. [DOI] [PubMed] [Google Scholar]
- Barco A, Patterson SL, Alarcon JM, Gromova P, Mata-Roig M, Morozov A, Kandel ER, 2005. Gene expression profiling of facilitated L-LTP in VP16-CREB mice reveals that BDNF is critical for the maintenance of LTP and its synaptic capture. Neuron 48, 123–137. [DOI] [PubMed] [Google Scholar]
- Beenken A, Mohammadi M, 2009. The FGF family: biology, pathophysiology and therapy. Nat. Rev. Drug Discov 8, 235–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bito H, Deisseroth K, Tsien RW, 1996. CREB phosphorylation and dephosphorylation: a Ca(2+)- and stimulus duration-dependent switch for hippocampal gene expression. Cell 87, 1203–1214. [DOI] [PubMed] [Google Scholar]
- Bittinger MA, McWhinnie E, Meltzer J, Iourgenko V, Latario B, Liu X, Chen CH, Song C, Garza D, Labow M, 2004. Activation of cAMP response element-mediated gene expression by regulated nuclear transport of TORC proteins. Curr. Biol 14, 2156–2161. [DOI] [PubMed] [Google Scholar]
- Bookout AL, de Groot MH, Owen BM, Lee S, Gautron L, Lawrence HL, Ding X, Elmquist JK, Takahashi JS, Mangelsdorf DJ, et al. , 2013. FGF21 regulates metabolism and circadian behavior by acting on the nervous system. Nat. Med 19, 1147–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtchuladze R, Frenguelli B, Blendy J, Cioffi D, Schutz G, Silva AJ, 1994. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell 79, 59–68. [DOI] [PubMed] [Google Scholar]
- Bredy TW, Barad M, 2009. Social modulation of associative fear learning by pheromone communication. Learn. Mem 16, 12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredy TW, Wu H, Crego C, Zellhoefer J, Sun YE, Barad M, 2007. Histone modifications around individual BDNF gene promoters in prefrontal cortex are associated with extinction of conditioned fear. Learn. Mem 14, 268–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brochier C, Dennis G, Rivieccio MA, McLaughlin K, Coppola G, Ratan RR, Langley B, 2013. Specific acetylation of p53 by HDAC inhibition prevents DNA damage-induced apoptosis in neurons. J. Neurosci 33, 8621–8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlezon WA Jr., Duman RS, Nestler EJ, 2005. The many faces of CREB. Trends Neurosci 28, 436–445. [DOI] [PubMed] [Google Scholar]
- Cerejeira J, Lagarto L, Mukaetova-Ladinska EB, 2012. Behavioral and psychological symptoms of dementia. Front. Neurol 3, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ch'ng TH, Uzgil B, Lin P, Avliyakulov NK, O'Dell TJ, Martin KC, 2012. Activity-dependent transport of the transcriptional coactivator CRTC1 from synapse to nucleus. Cell 150, 207–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY, 2008. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science 320, 1224–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME, 2003. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science 302, 885–889. [DOI] [PubMed] [Google Scholar]
- Chen J, Zhou Y, Mueller-Steiner S, Chen LF, Kwon H, Yi S, Mucke L, Gan L, 2005. SIRT1 protects against microglia-dependent amyloid-beta toxicity through inhibiting NF-kappaB signaling. J. Biol. Chem 280, 40364–40374. [DOI] [PubMed] [Google Scholar]
- Chen DY, Stern SA, Garcia-Osta A, Saunier-Rebori B, Pollonini G, Bambah-Mukku D, Blitzer RD, Alberini CM, 2011. A critical role for IGF-II in memory consolidation and enhancement. Nature 469, 491–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Gabel HW, Hemberg M, Hutchinson AN, Sadacca LA, Ebert DH, Harmin DA, Greenberg RS, Verdine VK, Zhou Z, et al. , 2011. Genome-wide activity-dependent MeCP2 phosphorylation regulates nervous system development and function. Neuron 72, 72–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M, 2003. TORCs: transducers of regulated CREB activity. Mol. Cell 12, 413–423. [DOI] [PubMed] [Google Scholar]
- Consortium, T.N.a.P.A.S.o.t.P.G, 2015. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat. Neurosci 18, 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD, 2010. DNA methylation and memory formation. Nat. Neurosci 13, 1319–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Childs D, Guzman-Karlsson MC, Kibe M, Moulden J, Song E, Tahir A, Sweatt JD, 2013. DNA methylation regulates associative reward learning. Nat. Neurosci 16, 1445–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K, Bito H, Tsien RW, 1996. Signaling from synapse to nucleus: post-synaptic CREB phosphorylation during multiple forms of hippocampal synaptic plasticity. Neuron 16, 89–101. [DOI] [PubMed] [Google Scholar]
- Deng JV, Rodriguiz RM, Hutchinson AN, Kim IH, Wetsel WC, West AE, 2010. MeCP2 in the nucleus accumbens contributes to neural and behavioral responses to psychostimulants. Nat. Neurosci 13, 1128–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dompierre JP, Godin JD, Charrin BC, Cordelieres FP, King SJ, Humbert S, Saudou F, 2007. Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington's disease by increasing tubulin acetylation. J. Neurosci 27, 3571–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudai Y, Eisenberg M, 2004. Rites of passage of the engram: reconsolidation and the lingering consolidation hypothesis. Neuron 44, 93–100. [DOI] [PubMed] [Google Scholar]
- Ebert DH, Gabel HW, Robinson ND, Kastan NR, Hu LS, Cohen S, Navarro AJ, Lyst MJ, Ekiert R, Bird AP, et al. , 2013. Activity-dependent phosphorylation of MeCP2 threonine 308 regulates interaction with NCoR. Nature 499, 341–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elde R, Cao YH, Cintra A, Brelje TC, Pelto-Huikko M, Junttila T, Fuxe K, Pettersson RF, Hokfelt T, 1991. Prominent expression of acidic fibroblast growth factor in motor and sensory neurons. Neuron 7, 349–364. [DOI] [PubMed] [Google Scholar]
- Escoubas CC, Silva-Garcia CG, Mair WB, 2017. Deregulation of CRTCs in aging and age-related disease risk. Trends Genet 33, 303–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federman N, de la Fuente V, Zalcman G, Corbi N, Onori A, Passananti C, Romano A, 2013. Nuclear factor kappaB-dependent histone acetylation is specifically involved in persistent forms of memory. J. Neurosci 33, 7603–7614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G, 2010. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci 13, 423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Shao N, Szulwach KE, Vialou V, Huynh J, Zhong C, Le T, Ferguson D, Cahill ME, Li Y, et al. , 2015. Role of Tet1 and 5-hydroxymethylcytosine in cocaine action. Nat. Neurosci 18, 536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finsterwald C, Alberini CM, 2014. Stress and glucocorticoid receptor-dependent mechanisms in long-term memory: from adaptive responses to psychopathologies. Neurobiol. Learn Mem 112, 17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi M, Tabuchi A, Kuwana Y, Watanabe S, Inoue M, Takasaki I, Izumi H, Tanaka A, Inoue R, Mori H, et al. , 2015. Neuromodulatory effect of Galphas- or Galphaq-coupled G-protein-coupled receptor on NMDA receptor selectively activates the NMDA receptor/Ca2 + /calcineurin/cAMP response element-binding protein-regulated transcriptional coactivator 1 pathway to effectively induce brain-derived neurotrophic factor expression in neurons. J. Neurosci 35, 5606–5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaub P, Tedeschi A, Puttagunta R, Nguyen T, Schmandke A, Di Giovanni S, 2010. HDAC inhibition promotes neuronal outgrowth and counteracts growth cone collapse through CBP/p300 and P/CAF-dependent p53 acetylation. Cell Death Differ 17, 1392–1408. [DOI] [PubMed] [Google Scholar]
- Graff J, Tsai LH, 2013. Histone acetylation: molecular mnemonics on the chromatin. Nat. Rev. Neurosci 14, 97–111. [DOI] [PubMed] [Google Scholar]
- Graff J, Joseph NF, Horn ME, Samiei A, Meng J, Seo J, Rei D, Bero AW, Phan TX, Wagner F, et al. , 2014. Epigenetic priming of memory updating during reconsolidation to attenuate remote fear memories. Cell 156, 261–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer PL, Greenberg ME, 2008. From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron 59, 846–860. [DOI] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, et al. , 2009. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature 459, 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Ma DK, Mo H, Ball MP, Jang MH, Bonaguidi MA, Balazer JA, Eaves HL, Xie B, Ford E, et al. , 2011a. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat. Neurosci 14, 1345–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H, 2011b. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145, 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Shin JH, Shin J, Li H, Xie B, Zhong C, Hu S, Le T, Fan G, et al. , 2014. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci 17, 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider R, Hennion M, Vidal RO, Shomroni O, Rahman RU, Rajput A, Centeno TP, van Bebber F, Capece V, Garcia Vizcaino JC, et al. , 2016. DNA methylation changes in plasticity genes accompany the formation and maintenance of memory. Nat. Neurosci 19, 102–110. [DOI] [PubMed] [Google Scholar]
- Hemstedt TJ, Lattal KM, Wood MA, 2017. Reconsolidation and extinction: using epigenetic signatures to challenge conventional wisdom. Neurobiol. Learn Mem 142, 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y, Ihara K, Masuda T, Yamamoto T, Iwata I, Takahashi A, Awata H, Nakamura N, Takakura M, Suzuki Y, et al. , 2016. Shifting transcriptional machinery is required for long-term memory maintenance and modification in Drosophila mushroom bodies. Nat. Commun 7, 13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson AN, Deng JV, Cohen S, West AE, 2012. Phosphorylation of MeCP2 at Ser421 contributes to chronic antidepressant action. J. Neurosci 32, 14355–14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S, Smith DM, Obrietan K, Donahue R, Wade C, Storm DR, 1998. Stimulation of cAMP response element (CRE)-mediated transcription during contextual learning. Nat. Neurosci 1, 595–601. [DOI] [PubMed] [Google Scholar]
- Iourgenko V, Zhang W, Mickanin C, Daly I, Jiang C, Hexham JM, Orth AP, Miraglia L, Meltzer J, Garza D, et al. , 2003. Identification of a family of cAMP response element-binding protein coactivators by genome-scale functional analysis in mammalian cells. Proc. Natl. Acad. Sci. U. S. A 100, 12147–12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarome TJ, Lubin FD, 2014. Epigenetic mechanisms of memory formation and reconsolidation. Neurobiol. Learn Mem 115, 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing X, Sui WH, Wang S, Xu XF, Yuan RR, Chen XR, Ma HX, Zhu YX, Sun JK, Yi F, et al. , 2017. HDAC7 ubiquitination by the E3 ligase CBX4 is involved in contextual fear conditioning memory formation. J. Neurosci 37, 3848–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josselyn SA, Kida S, Silva AJ, 2004. Inducible repression of CREB function disrupts amygdala-dependent memory. Neurobiol. Learn Mem 82, 159–163. [DOI] [PubMed] [Google Scholar]
- Kaas GA, Zhong C, Eason DE, Ross DL, Vachhani RV, Ming GL, King JR, Song H, Sweatt JD, 2013. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron 79, 1086–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang W, Hebert JM, 2015. FGF signaling is necessary for neurogenesis in young mice and sufficient to reverse its decline in old mice. J. Neurosci 35, 10217–10223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida S, Serita T, 2014. Functional roles of CREB as a positive regulator in the formation and enhancement of memory. Brain Res. Bull 105, 17–24. [DOI] [PubMed] [Google Scholar]
- Kida S, Josselyn SA, Pena de Ortiz S, Kogan JH, Chevere I, Masushige S, Silva AJ, 2002. CREB required for the stability of new and reactivated fear memories. Nat. Neurosci 5, 348–355. [DOI] [PubMed] [Google Scholar]
- Kim MS, Akhtar MW, Adachi M, Mahgoub M, Bassel-Duby R, Kavalali ET, Olson EN, Monteggia LM, 2012. An essential role for histone deacetylase 4 in synaptic plasticity and memory formation. J. Neurosci 32, 10879–10886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizilbash AH, Vanderploeg RD, Curtiss G, 2002. The effects of depression and anxiety on memory performance. Arch. Clin. Neuropsychol 17, 57–67. [PubMed] [Google Scholar]
- Klann E, Dever TE, 2004. Biochemical mechanisms for translational regulation in synaptic plasticity. Nat. Rev. Neurosci 5, 931–942. [DOI] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M, 2004. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron 42, 961–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs KA, Steullet P, Steinmann M, Do KQ, Magistretti PJ, Halfon O, Cardinaux JR, 2007. TORC1 is a calcium- and cAMP-sensitive coincidence detector involved in hippocampal long-term synaptic plasticity. Proc. Natl. Acad. Sci. U. S. A 104, 4700–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lattal KM, Wood MA, 2013. Epigenetics and persistent memory: implications for reconsolidation and silent extinction beyond the zero. Nat. Neurosci 16, 124–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. , 2007. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176. [DOI] [PubMed] [Google Scholar]
- Levenson JM, O’Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD, 2004. Regulation of histone acetylation during memory formation in the hippocampus. J. Biol. Chem 279, 40545–40559. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, Malone LM, Sweatt JD, 2006. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J. Biol. Chem 281, 15763–15773. [DOI] [PubMed] [Google Scholar]
- Li S, Zhang C, Takemori H, Zhou Y, Xiong ZQ, 2009. TORC1 regulates activity-dependent CREB-target gene transcription and dendritic growth of developing cortical neurons. J. Neurosci 29, 2334–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhong X, Chau KF, Williams EC, Chang Q, 2011. Loss of activity-induced phosphorylation of MeCP2 enhances synaptogenesis, LTP and spatial memory. Nat. Neurosci 14, 1001–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wang Y, Gao T, Pan Z, Cheng H, Yang Q, Cheng Z, Guo A, Ren J, Xue Y, 2014. CPLM: a database of protein lysine modifications. Nucleic Acids Res 42, D531–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Atalaya JP, Barco A, 2014. Can changes in histone acetylation contribute to memory formation? Trends Genet 30, 529–539. [DOI] [PubMed] [Google Scholar]
- Lubin FD, Sweatt JD, 2007. The IkappaB kinase regulates chromatin structure during reconsolidation of conditioned fear memories. Neuron 55, 942–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD, 2008. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J. Neurosci 28, 10576–10586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H, 2009. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 323, 1074–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox SA, Watts CS, Schafe GE, 2013. p300/CBP histone acetyltransferase activity is required for newly acquired and reactivated fear memories in the lateral amygdala. Learn. Mem 20, 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, 2004. Pathways towards and away from Alzheimer’s disease. Nature 430, 631–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice T, Duclot F, Meunier J, Naert G, Givalois L, Meffre J, Celerier A, Jacquet C, Copois V, Mechti N, et al. , 2008. Altered memory capacities and response to stress in p300/CBP-associated factor (PCAF) histone acetylase knockout mice. Neuropsychopharmacology 33, 1584–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M, Siegelbaum SA, Kandel ER, 2012. Synapses and memory storage. Cold Spring Harb. Perspect. Biol 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuown SC, Barrett RM, Matheos DP, Post RJ, Rogge GA, Alenghat T, Mullican SE, Jones S, Rusche JR, Lazar MA, et al. , 2011. HDAC3 is a critical negative regulator of long-term memory formation. J. Neurosci 31, 764–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N, 2012. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151, 1417–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendioroz M, Celarain N, Altuna M, Sanchez-Ruiz de Gordoa J, Zelaya MV, Roldan M, Rubio I, Larumbe R, Erro ME, Mendez I, et al. , 2016. CRTC1 gene is differentially methylated in the human hippocampus in Alzheimer’s disease. Alzheimers Res. Ther 8, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mews P, Donahue G, Drake AM, Luczak V, Abel T, Berger SL, 2017. Acetyl-CoA synthetase regulates histone acetylation and hippocampal memory. Nature 546, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan EM, Breuillaud L, Seredenina T, Magistretti PJ, Halfon O, Luthi-Carter R, Cardinaux JR, 2016a. Involvement of the agmatinergic system in the depressive-like phenotype of the Crtc1 knockout mouse model of depression. Transl. Psychiatry 6, e852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan EM, Halfon O, Magistretti PJ, Cardinaux JR, 2016b. The HDAC inhibitor SAHA improves depressive-like behavior of CRTC1-deficient mice: possible relevance for treatment-resistant depression. Neuropharmacology 107, 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD, 2007. Covalent modification of DNA regulates memory formation. Neuron 53, 857–869. [DOI] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, Rivera IM, Rubio MD, Rumbaugh G, Sweatt JD, 2010. Cortical DNA methylation maintains remote memory. Nat. Neurosci 13, 664–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K, Dempster E, Mill J, Giese KP, 2012. Long-lasting regulation of hippocampal Bdnf gene transcription after contextual fear conditioning. Genes Brain Behav 11, 651–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti P, Levenson JM, Battaglia F, Atkinson R, Teague R, Antalffy B, Armstrong D, Arancio O, Sweatt JD, Zoghbi HY, 2006. Learning and memory and synaptic plasticity are impaired in a mouse model of Rett syndrome. J. Neurosci 26, 319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris MJ, Adachi M, Na ES, Monteggia LM, 2014. Selective role for DNMT3a in learning and memory. Neurobiol. Learn Mem 115, 30–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nader K, Schafe GE, Le Doux JE, 2000. Fear memories require protein synthesis in the amygdala for reconsolidation after retrieval. Nature 406, 722–726. [DOI] [PubMed] [Google Scholar]
- Nonaka M, Kim R, Fukushima H, Sasaki K, Suzuki K, Okamura M, Ishii Y, Kawashima T, Kamijo S, Takemoto-Kimura S, et al. , 2014. Region-specific activation of CRTC1-CREB signaling mediates long-term fear memory. Neuron 84, 92–106. [DOI] [PubMed] [Google Scholar]
- Nott A, Cheng J, Gao F, Lin YT, Gjoneska E, Ko T, Minhas P, Zamudio AV, Meng J, Zhang F, et al. , 2016. Histone deacetylase 3 associates with MeCP2 to regulate FOXO and social behavior. Nat. Neurosci 19, 1497–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira AM, Wood MA, McDonough CB, Abel T, 2007. Transgenic mice expressing an inhibitory truncated form of p300 exhibit long-term memory deficits. Learn. Mem 14, 564–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira AM, Estevez MA, Hawk JD, Grimes S, Brindle PK, Abel T, 2011. Subregion-specific p300 conditional knock-out mice exhibit long-term memory impairments. Learn. Mem 18, 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira AM, Hemstedt TJ, Bading H, 2012. Rescue of aging-associated decline in Dnmt3a2 expression restores cognitive abilities. Nat. Neurosci 15, 1111–1113. [DOI] [PubMed] [Google Scholar]
- Owen BM, Bookout AL, Ding X, Lin VY, Atkin SD, Gautron L, Kliewer SA, Mangelsdorf DJ, 2013. FGF21 contributes to neuroendocrine control of female reproduction. Nat. Med 19, 1153–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra-Damas A, Valero J, Chen M, Espana J, Martin E, Ferrer I, Rodriguez-Alvarez J, Saura CA, 2014. Crtc1 activates a transcriptional program deregulated at early Alzheimer’s disease-related stages. J. Neurosci 34, 5776–5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra-Damas A, Chen M, Enriquez-Barreto L, Ortega L, Acosta S, Perna JC, Fullana MN, Aguilera J, Rodriguez-Alvarez J, Saura CA, 2017. CRTC1 function during memory encoding is disrupted in neurodegeneration. Biol. Psychiatry 81, 111–123. [DOI] [PubMed] [Google Scholar]
- Peixoto L, Abel T, 2013. The role of histone acetylation in memory formation and cognitive impairments. Neuropsychopharmacology 38, 62–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, Agis-Balboa RC, Cota P, Wittnam JL, Gogol-Doering A, Opitz L, et al. , 2010. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science 328, 753–756. [DOI] [PubMed] [Google Scholar]
- Pietrocola F, Galluzzi L, Bravo-San Pedro JM, Madeo F, Kroemer G, 2015. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab 21, 805–821. [DOI] [PubMed] [Google Scholar]
- Reddy PH, Beal MF, 2008. Amyloid beta, mitochondrial dysfunction and synaptic damage: implications for cognitive decline in aging and Alzheimer’s disease. Trends Mol. Med 14, 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison AJ, Nestler EJ, 2011. Transcriptional and epigenetic mechanisms of addiction. Nat. Rev. Neurosci 12, 623–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko A, Dawlaty MM, Seo J, Cheng AW, Meng J, Le T, Faull KF, Jaenisch R, Tsai LH, 2013. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron 79, 1109–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sando R 3rd, Gounko N, Pieraut S, Liao L, Yates J 3rd, Maximov A, 2012. HDAC4 governs a transcriptional program essential for synaptic plasticity and memory. Cell 151, 821–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saura CA, Cardinaux JR, 2017. Emerging roles of CREB-Regulated transcription coactivators in brain physiology and pathology. Trends Neurosci 40, 720–733. [DOI] [PubMed] [Google Scholar]
- Screaton RA, Conkright MD, Katoh Y, Best JL, Canettieri G, Jeffries S, Guzman E, Niessen S, Yates JR 3rd, Takemori H, et al. , 2004. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell 119, 61–74. [DOI] [PubMed] [Google Scholar]
- Sekeres MJ, Mercaldo V, Richards B, Sargin D, Mahadevan V, Woodin MA, Frankland PW, Josselyn SA, 2012. Increasing CRTC1 function in the dentate gyrus during memory formation or reactivation increases memory strength without compromising memory quality. J. Neurosci 32, 17857–17868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AJ, Kogan JH, Frankland PW, Kida S, 1998. CREB and memory. Annu. Rev. Neurosci 21, 127–148. [DOI] [PubMed] [Google Scholar]
- Sun H, Kennedy PJ, Nestler EJ, 2013. Epigenetics of the depressed brain: role of histone acetylation and methylation. Neuropsychopharmacology 38, 124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Josselyn SA, Frankland PW, Masushige S, Silva AJ, Kida S, 2004. Memory reconsolidation and extinction have distinct temporal and biochemical signatures. J. Neurosci 24, 4787–4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Fukushima H, Mukawa T, Toyoda H, Wu LJ, Zhao MG, Xu H, Shang Y, Endoh K, Iwamoto T, et al. , 2011. Upregulation of CREB-mediated transcription enhances both short- and long-term memory. J. Neurosci 31, 8786–8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Kasuya Y, Itoh Y, Ota Y, Zhan P, Asamitsu K, Nakagawa H, Okamoto T, Miyata N, 2013. Identification of highly selective and potent histone deacetylase 3 inhibitors using click chemistry-based combinatorial fragment assembly. PLoS One 8, e68669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, Irier H, Upadhyay AK, Gearing M, Levey AI, et al. , 2011. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci 14, 1607–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao QQ, Sun YM, Liu ZJ, Ni W, Yang P, Li HL, Lu SJ, Wu ZY, 2014. A variant within FGF1 is associated with Alzheimer’s disease in the Han Chinese population. Am. J. Med. Genet. B Neuropsychiatr. Genet 165B, 131–136. [DOI] [PubMed] [Google Scholar]
- Thorns V, Masliah E, 1999. Evidence for neuroprotective effects of acidic fibroblast growth factor in Alzheimer disease. J. Neuropathol. Exp. Neurol 58, 296–306. [DOI] [PubMed] [Google Scholar]
- Thorns V, Licastro F, Masliah E, 2001. Locally reduced levels of acidic FGF lead to decreased expression of 28-kda calbindin and contribute to the selective vulnerability of the neurons in the entorhinal cortex in Alzheimer’s disease. Neuropathology 21, 203–211. [DOI] [PubMed] [Google Scholar]
- Turner CA, Watson SJ, Akil H, 2012. The fibroblast growth factor family: neuromodulation of affective behavior. Neuron 76, 160–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler WJ, Alonso M, Bramham CR, Pozzo-Miller LD, 2002. From acquisition to consolidation: on the role of brain-derived neurotrophic factor signaling in hippocampal-dependent learning. Learn. Mem 9, 224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S, Shumyatsky GP, 2017. Synaptically localized transcriptional regulators in memory formation. Neuroscience 370, 4–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S, Hara K, Kobayashi A, Otsuki K, Yamagata H, Hobara T, Suzuki T, Miyata N, Watanabe Y, 2011. Epigenetic status of Gdnf in the ventral striatum determines susceptibility and adaptation to daily stressful events. Neuron 69, 359–372. [DOI] [PubMed] [Google Scholar]
- Uchida S, Teubner BJ, Hevi C, Hara K, Kobayashi A, Dave RM, Shintaku T, Jaikhan P, Yamagata H, Suzuki T, et al. , 2017a. CRTC1 nuclear translocation following learning modulates memory strength via exchange of chromatin remodeling complexes on the fgf1 gene. Cell Rep 18, 352–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S, Yamagata H, Seki T, Watanabe Y, 2017b. Epigenetic mechanisms of major depression: targeting neuronal plasticity. Psychiatry Clin. Neurosci 10.1111/pcn.12621. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Villain H, Florian C, Roullet P, 2016. HDAC inhibition promotes both initial consolidation and reconsolidation of spatial memory in mice. Sci. Rep 6, 27015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Coelho CM, Li X, Marek R, Yan S, Anderson S, Meyers D, Mukherjee C, Sbardella G, Castellano S, et al. , 2012. p300/CBP-associated factor selectively regulates the extinction of conditioned fear. J. Neurosci 32, 11930–11941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB, 2009. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AE, Greenberg ME, 2011. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb. Perspect. Biol 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MA, Kaplan MP, Park A, Blanchard EJ, Oliveira AM, Lombardi TL, Abel T, 2005. Transgenic mice expressing a truncated form of CREB-binding protein (CBP) exhibit deficits in hippocampal synaptic plasticity and memory storage. Learn. Mem 12, 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood MA, Attner MA, Oliveira AM, Brindle PK, Abel T, 2006. A transcription factor-binding domain of the coactivator CBP is essential for long-term memory and the expression of specific target genes. Learn. Mem 13, 609–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu QF, Yang L, Li S, Wang Q, Yuan XB, Gao X, Bao L, Zhang X, 2012. Fibroblast growth factor 13 is a microtubule-stabilizing protein regulating neuronal polarization and migration. Cell 149, 1549–1564. [DOI] [PubMed] [Google Scholar]
- Xu W, Kasper LH, Lerach S, Jeevan T, Brindle PK, 2007. Individual CREB-target genes dictate usage of distinct cAMP-responsive coactivation mechanisms. EMBO J. 26, 2890–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata H, Chen Y, Akatsu H, Kamino K, Ito J, Yokoyama S, Yamamoto T, Kosaka K, Miki T, Kondo I, 2004. Promoter polymorphism in fibroblast growth factor 1 gene increases risk of definite Alzheimer’s disease. Biochem. Biophys. Res. Commun 321, 320–323. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Wu H, Li S, Chen Q, Cheng XW, Zheng J, Takemori H, Xiong ZQ, 2006a. Requirement of TORC1 for late-phase long-term potentiation in the hippocampus. PLoS One 1, e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L, Chen WG, Lin Y, Savner E, Griffith EC, et al. , 2006b. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron 52, 255–269. [DOI] [PMC free article] [PubMed] [Google Scholar]