Abstract
Mammalian cells are surrounded by neighboring cells and extracellular matrix (ECM), which provide cells with structural support and mechanical cues that influence diverse biological processes1. The Hippo pathway effectors YAP and TAZ are regulated by mechanical cues and mediate cellular responses to ECM stiffness2,3. Here we identified Ras-related GTPase RAP2 as a critical intracellular signal transducer that relays ECM rigidity signals to control mechano-sensitive cellular activities through YAP/TAZ. RAP2 is activated by low ECM stiffness, and RAP2 deletion blocks YAP/TAZ regulation by stiffness signals and promotes aberrant cell growth. Mechanistically, matrix stiffness acts through phospholipase Cγ1 (PLCγ1) to influence levels of phosphatidylinositol 4,5-bisphosphate (PIP2) and its product phosphatidic acid (PA), which activates RAP2 through PDZGEF1/2. At low stiffness, active RAP2 binds to and stimulates mitogen-activated protein kinase kinase kinase kinase 4/6/7 (MAP4K4/6/7) and Rho GTPase activating protein 29 (ARHGAP29), resulting in LATS1/2 activation and YAP/TAZ inhibition. RAP2 and YAP/TAZ play pivotal roles in mechano-regulated transcription, as YAP/TAZ deletion abolishes the ECM stiffness-responsive transcriptome. Our findings reveal RAP2 as a molecular switch in mechanotransduction, thereby defining a mechanosignaling pathway from ECM stiffness to the nucleus.
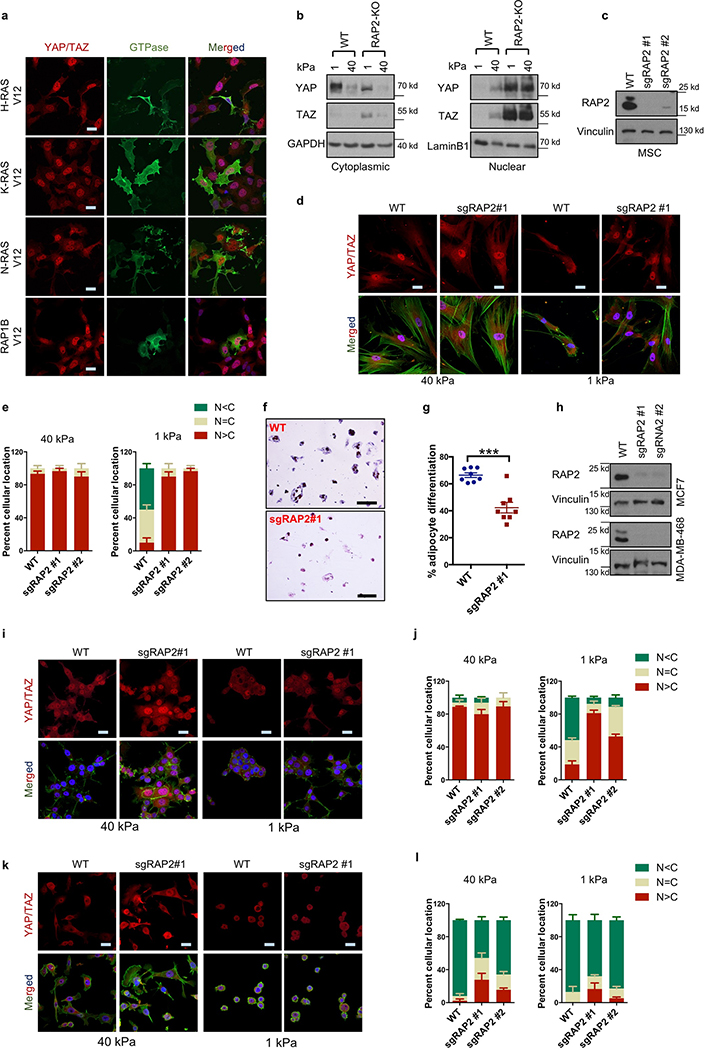
YAP/TAZ function as essential effectors of mechanotransduction to regulate cell proliferation and differentiation3–7. When cells are shifted from stiff to soft matrices, YAP/TAZ translocate from the nucleus to the cytoplasm, and are thus inactivated. However, the signaling mechanism from ECM stiffness to the Hippo pathway is unclear. Because small GTPases function as molecular switches in many biological processes8, we screened for small GTPases that affect YAP/TAZ localization in cells seeded on soft (1 kPa) or stiff (40 kPa) matrices (Supplemental information). RAP2A was identified since its overexpression induced cytoplasmic translocation of YAP/TAZ even on a stiff matrix (Fig. 1a). No other GTPases, including the closely related RAP1 and RAS, showed similar activity (Extended Data Fig. 1a).
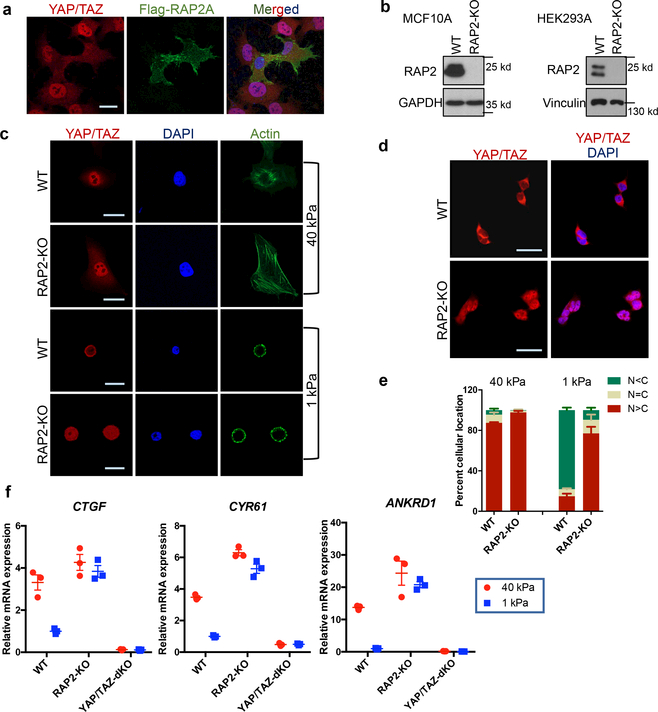
Figure 1|. RAP2 mediates YAP/TAZ regulation by ECM stiffness.
a. Overexpression of Flag-RAP2A induces YAP/TAZ cytoplasmic translocation in HEK293A cells on a stiff (40 kPa) matrix. Merged, combined signals from YAP/TAZ (red), Flag (green), and DAPI (blue). b. Immunoblot showing RAP2A/B/C deletion (RAP2-KO) in MCF10A and HEK293A cells. c. Immunofluorescence showing that RAP2-KO MCF10A cells, unlike WT cells, retain nuclear YAP/TAZ at low stiffness (1 kPa). The experiments in Panel b,c were repeated independently twice with similar results. d. RAP2A/B/C deletion in HEK293A cells blocks YAP/TAZ cytoplasmic localization by low stiffness. e. Quantification of YAP/TAZ localization, presented as mean+SEM, in HEK293A cells. N<C, less YAP/TAZ in nucleus than in cytoplasm. N=C, similar levels of YAP/TAZ in cytoplasm and nucleus. N>C, more YAP/TAZ in nucleus than in cytoplasm. f. RAP2 is required for regulation of YAP/TAZ target genes CTGF, CYR61, and ANKRD1 by stiffness in HEK293A cells. Data are presented as mean±SEM. For Panel e,f, n=3 biologically independent samples. Scale bar, 25 μm.
At high stiffness, both wild-type (WT) and RAP2A/B/C-triple knockout (RAP2-KO) MCF10A cells showed nuclear localization of YAP/TAZ (Fig. 1b,c). At low stiffness, WT cells exhibited mainly cytoplasmic YAP/TAZ, whereas RAP2-KO MCF10A cells retained YAP/TAZ in the nucleus (Fig. 1c). RAP2 deletion in HEK293A cells also suppressed low stiffness-induced YAP/TAZ cytoplasmic translocation (Fig. 1d,e, Extended Data Fig. 1b). YAP/TAZ target genes CTGF, CYR61, and ANKRD1 were repressed by low stiffness in WT cells, but not in the RAP2-KO cells (Fig. 1f). Similar results were observed in human mesenchymal stem cells (Extended Data Fig. 1c-e), in which RAP2 deletion suppressed their differentiation into adipocytes (Extended Data Fig. 1f,g). In the luminal breast cancer MCF7 cells, ECM stiffness modulated YAP/TAZ localization in a RAP2-dependent manner, whereas the basal type MDA-MB-468 showed constitutively cytoplasmic YAP/TAZ localization regardless of stiffness (Extended Fig. 1h-l). TWIST and β-catenin were reported to show nuclear-cytoplasmic shuttling in response to physical cues9,10. TWIST, but not β-catenin, displayed nuclear-cytoplasmic translocation in response to ECM stiffness (Extended Data Fig. 2a). However, RAP2 deletion had no obvious impact on TWIST localization.
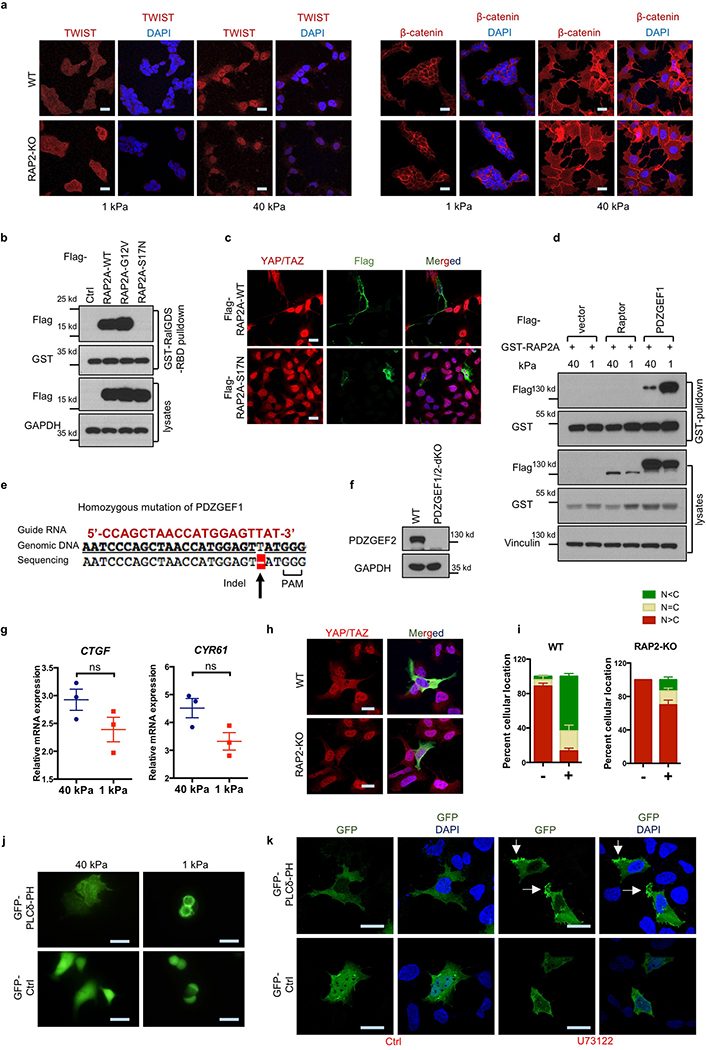
Activity of small GTPases is switched on and off by GTP- and GDP-binding, respectively. A RalGDS-RBD pulldown assay showed that low stiffness promotes RAP2 GTP-binding (Extended Data Fig. 2b, Fig. 2a). Unlike WT RAP2A, the GTP-binding-deficient mutant RAP2A-S17N did not induce cytoplasmic translocation of YAP/TAZ (Extended Data Fig. 2b,c). RAP2 interaction with its activators PDZGEF1/211-13 was enhanced by low stiffness (Extended Data Fig. 2d). We generated PDZGEF1/2-dKO cells (Extended Data Fig. 2e,f) and discovered that they were defective in YAP/TAZ cytoplasmic translocation (Fig. 2b,c) and target gene repression (Extended Data Fig. 2g) in response to low stiffness. PDZGEF1/2 deletion blunted RAP2 activation by low stiffness (Fig. 2d), while PDZGEF1 overexpression induced YAP/TAZ cytoplasmic translocation in WT but not RAP2-KO cells (Extended Data Fig. 2h,i).
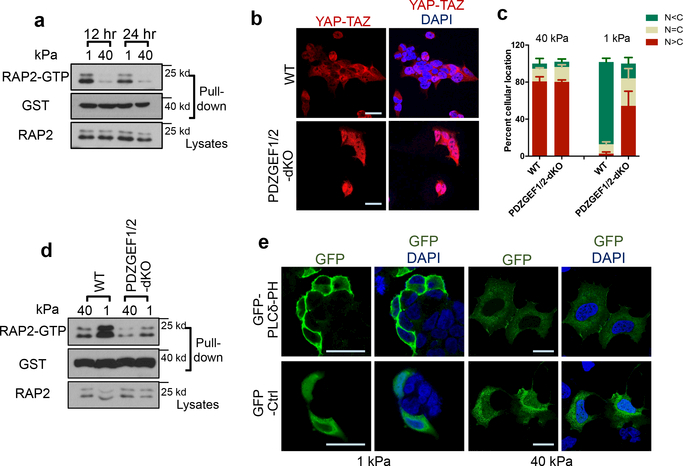
Figure 2|. ECM stiffness acts via PDZGEF1/2 to regulate RAP2.
a. RAP2 is activated by low stiffness. Pulldown of GTP-bound RAP2 from cells at 1 kPa and 40 kPa using GST-RalGDS-RBD. b. PDZGEF1/2 deletion compromises YAP/TAZ translocation at 1 kPa. c. Quantification of YAP/TAZ localization, presented as mean+SEM, in Panel b. n=3 (40 kPa) or 4 (1 kPa) biologically independent samples. d. PDZGEF1/2 mediate RAP2 regulation by stiffness. Experiments were similar to Panel a. e. High stiffness reduces PIP2 enrichment at the plasma membrane. A GFP-tagged PIP2 reporter PLCδ-PH domain was transfected into cells and detected with anti-GFP antibodies. The experiments in Panel a,d,e were repeated independently twice with similar results. Scale bar, 25 μm.
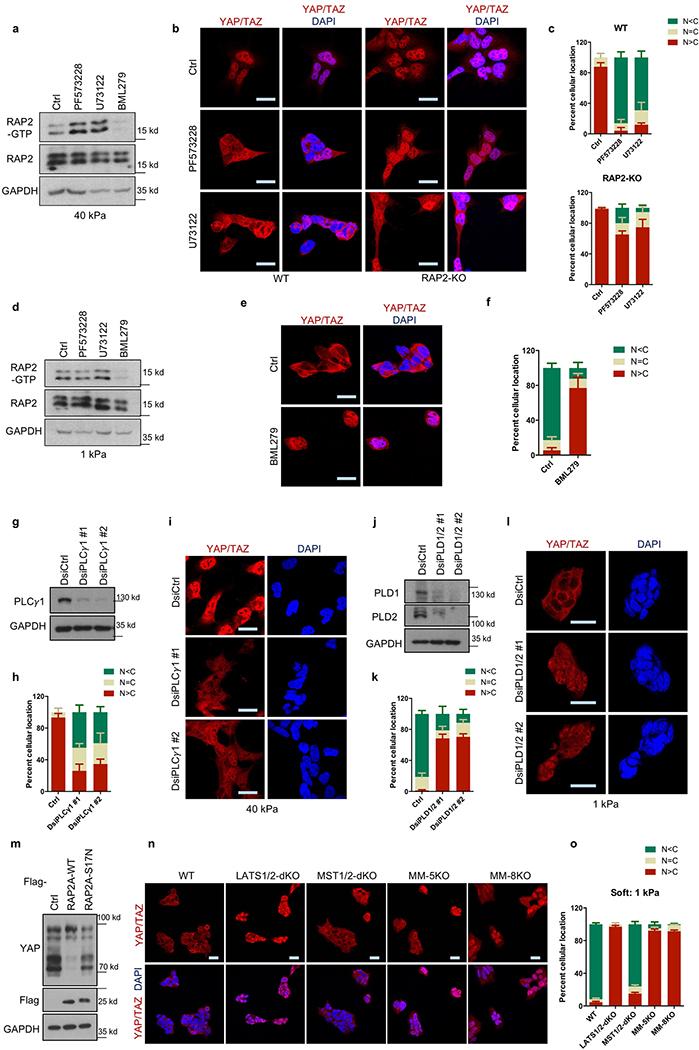
PIP2 activates RAP2 through PDZGEF1/2 at the plasma membrane after PIP2 is converted to PA by phospholipase D1/2 (PLD1/2)13. Using a GFP-tagged PIP2 reporter, we observed that PIP2 was enriched at the plasma membrane at low stiffness (Fig. 2e, Extended Data Fig. 2j). Focal adhesions decrease PIP2 through activating PLC𝛾114,15. Inhibition of PLC𝛾1 by U73122 induced PIP2 accumulation (Extended Data Fig. 2k). We hypothesized that ECM stiffness regulates RAP2 and YAP/TAZ through modulating focal adhesion and local PIP2 abundance at plasma membrane. Focal adhesion kinase (FAK) inhibitor PF573228 or PLC𝛾1 inhibitor U73122 increased RAP2 GTP-binding and YAP/TAZ cytoplasmic translocation (Extended Data Fig. 3a-c). In contrast, PLD1/2 inhibitor BML279 reduced RAP2 GTP-binding and accumulated YAP/TAZ in the nucleus (Extended Data Fig. 3d-f). The effects of PIP2 on YAP/TAZ localization were confirmed by PLC𝛾1- and PLD1/2-knockdown experiments (Extended Data Fig. 3g-l).
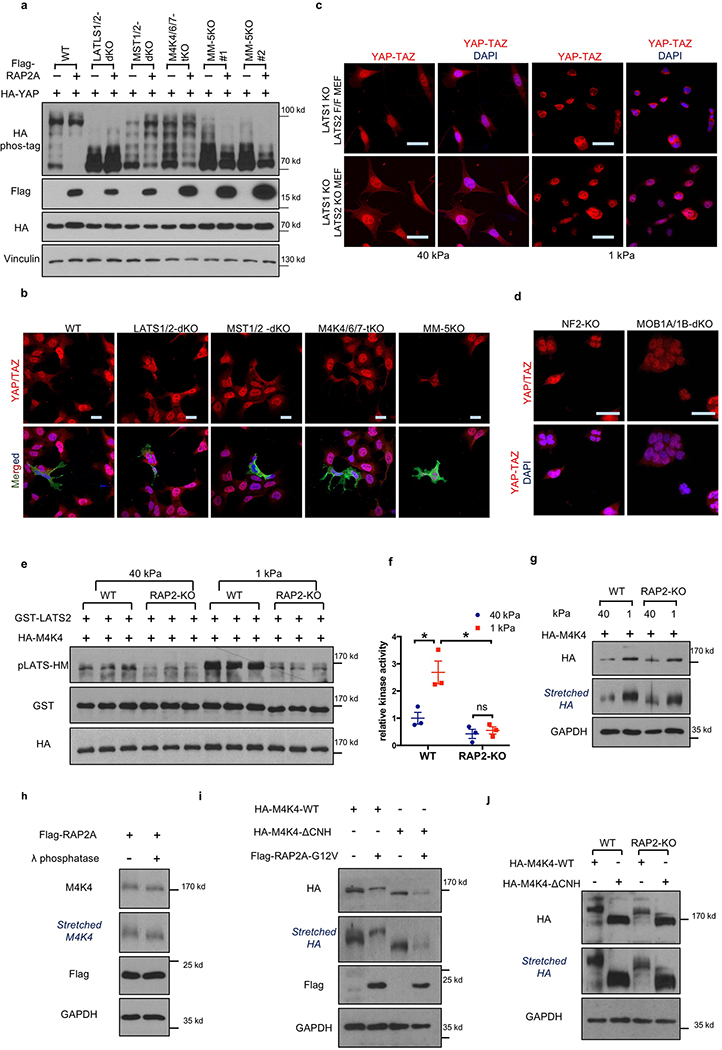
The nuclear-cytoplasmic shuttling of YAP/TAZ is generally controlled by LATS1/2-dependent phosphorylation2. However, the role of Hippo kinase cascade in YAP/TAZ regulation by mechanotransduction is not well-defined4–7. We found that low ECM stiffness induced phosphorylation of LATS1/2 and YAP/TAZ in WT cells, and this was significantly blunted in RAP2-KO cells (Fig. 3a). Furthermore, RAP2 induced YAP phosphorylation in a GTP-binding-dependent manner (Extended Data Fig. 3m). We proposed that RAP2 controls YAP/TAZ localization via the Hippo pathway. Consistent with this notion, deletion of LATS1/2 or combined deletion of MST1/2 and MAP4Ks abolished the YAP/TAZ regulation by ECM rigidity (Extended Data Fig. 3n,o). Hippo pathway core components were similarly required for RAP2 to induce YAP/TAZ phosphorylation and cytoplasmic translocation (Extended Data Fig. 4a,b). The role of LATS in this regulation was further confirmed in LATS1/2-dKO mouse embryonic fibroblasts, as well as NF2-KO or MOB1A/1B-dKO HEK293A cells (Extended Data Fig. 4c,d)16,17.
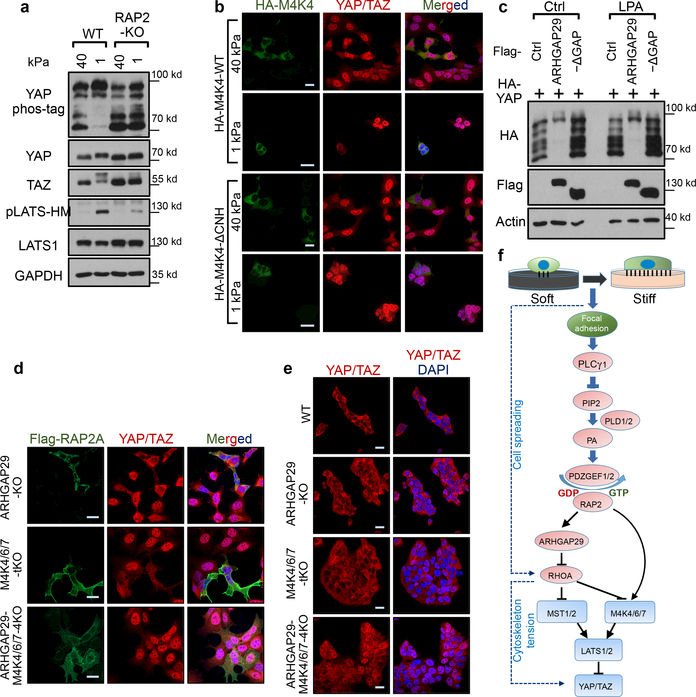
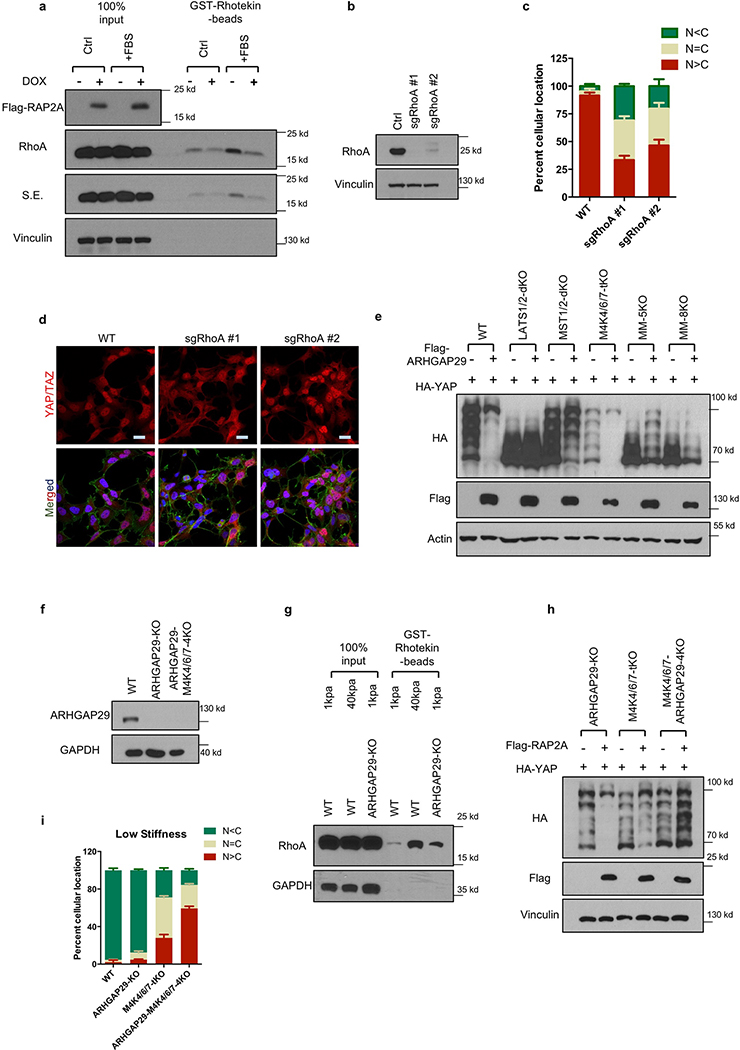
Figure 3.|. RAP2 inhibits YAP/TAZ through MAP4K4/6/7 and ARHGAP29.
a. RAP2 is important for stiffness-regulated phosphorylation of LATS1/2 and YAP/TAZ. Phos-tag gel detects YAP phosphorylation by mobility shift. pLATS-HM detects LATS phosphorylation in the hydrophobic motif. b. Expression of MAP4K4-WT, but not the Citron domain-deleted mutant (ΔCNH), rescued YAP/TAZ localization in MM-8KO (MST1/2-MAP4K1/2/3/4/6/7–8KO) cells at low stiffness. Merged, combined signals from HA-MAP4K4 (green), YAP/TAZ (red), and DAPI (blue). c. Overexpression of ARHGAP29 WT, but not the GAP domain-deleted mutant (-ΔGAP), induced YAP phosphorylation. LPA treatment, which activates RhoA, is indicated. d. Knockout of ARHGAP29 and MAP4K4/6/7 blocked RAP2A-induced YAP/TAZ cytoplasmic localization. Merged, combined signals from Flag-RAP2A (green), YAP/TAZ (red), and DAPI (blue). e. Co-deletion of ARHGAP29 and MAP4K4/6/7 blocked YAP/TAZ cytoplasmic translocation at 1 kPa. For Panel a-e, the experiments were repeated twice independently with similar results. f. A model showing a signaling axis by which stiffness regulates YAP/TAZ. The dotted lines indicate an actin/cytoskeleton-mediated pathway reported previously (ref 7). Scale bar in, 25 μm.
MAP4K4, TNIK (MAP4K7), and ARHGAP29 are RAP2 effectors18–20. Notably, ARHGAP29 is one of the RhoGAPs that are transcriptionally activated by YAP21,22. MAP4K4 kinase activity was stimulated by low stiffness in WT but not RAP2-KO cells (Extended Data Fig. 4e,f). Moreover, low stiffness induced MAP4K4 phosphorylation, as indicated by reduced mobility, in a RAP2-dependent manner (Extended Data Fig. 4g,h). Deletion of the RAP2-interacting citron domain19 in MAP4K4 abolished its regulation by RAP2 (Extended Fig. 4i,j), and the mutant also failed to rescue the YAP/TAZ translocation defect in MST1/2-MAP4Ks-8KO cells (Fig. 3b). Interestingly, a recent study showed that the Drosophila MAP4K4/6/7 homolog Msn regulates Yki in response to tension23. In addition, RAP2A overexpression led to RhoA inactivation (Extended Data Fig. 5a), a potent activator for YAP/TAZ7,16 (Extended Data Fig. 5b-d). This action of RAP2A on RhoA was mediated by ARHGAP29, because YAP phosphorylation induced by ARHGAP29 required its Rho-GAP domain and the Hippo kinase cascade (Fig. 3c, Extended Data Fig. 5e). ARHGAP29 deletion compromised RhoA inactivation caused by low stiffness (Extended Data Fig. 5f,g). Therefore, RAP2 acts through MAP4K4/6/7 and ARHGAP29 to inhibit YAP/TAZ. Consistent with this finding, MAP4K4/6/7-ARHGAP29–4KO cells were resistant to RAP2-induced cytoplasmic translocation and phosphorylation of YAP/TAZ (Fig. 3d, Extended Data Fig.5h), and displayed impaired responses to ECM stiffness (Fig. 3e, Extended Data Fig. 5i). Collectively, our data reveal a signaling axis linking matrix stiffness to YAP/TAZ regulation via focal adhesion→PLC𝛾1→PIP2→PA→PDZGEF1/2→RAP2→ARHGAP29/MAP4K→LATS1/2 (Fig. 3f), which works in parallel to the cell spreading/RhoA/cytoskeleton tension-mediated YAP/TAZ translocation mechanism proposed previously4,7.
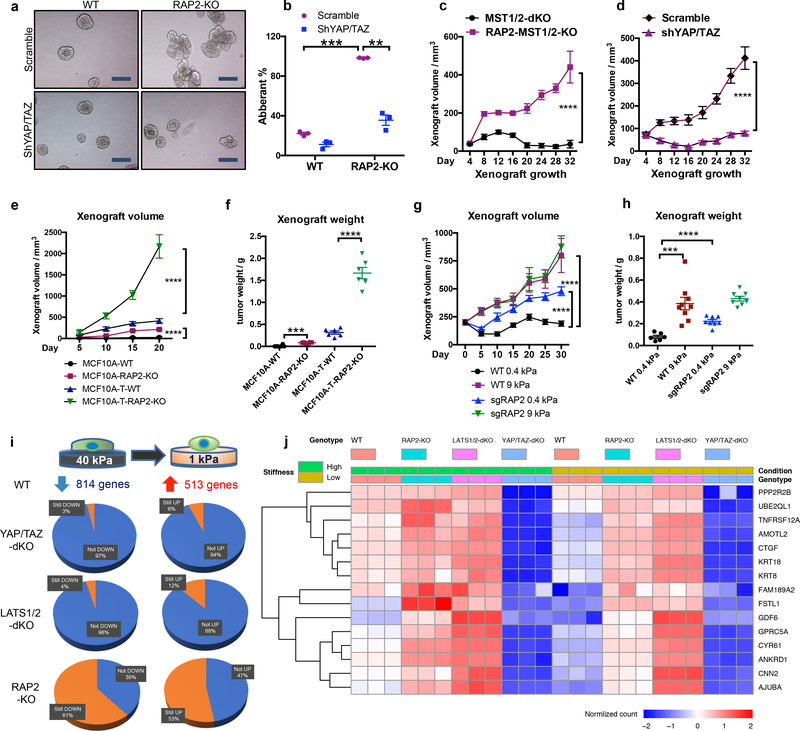
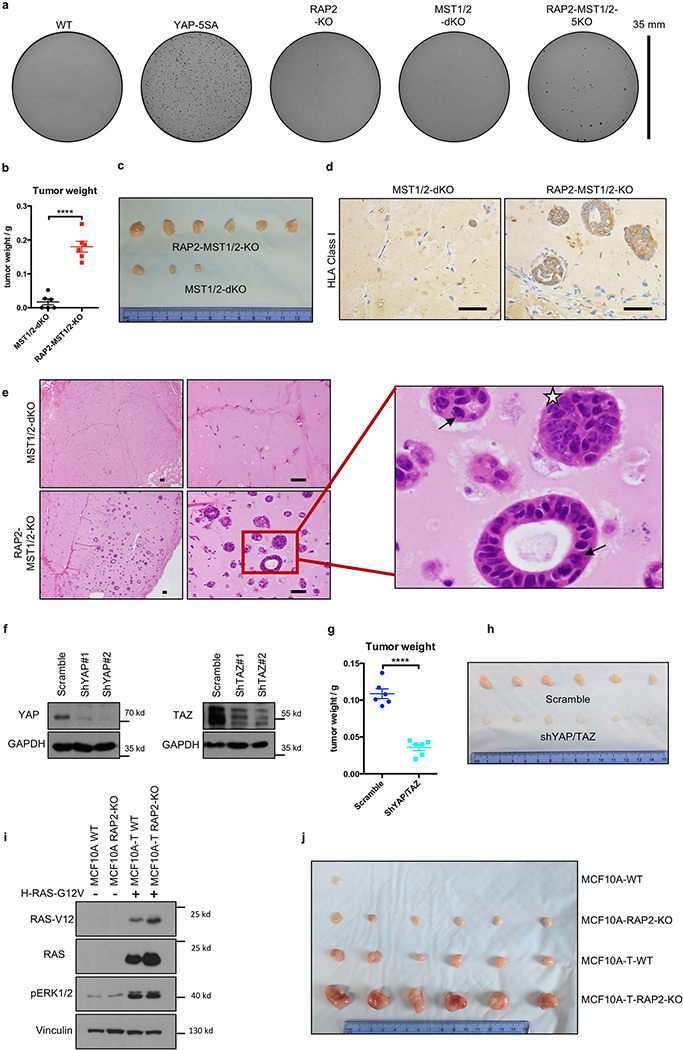
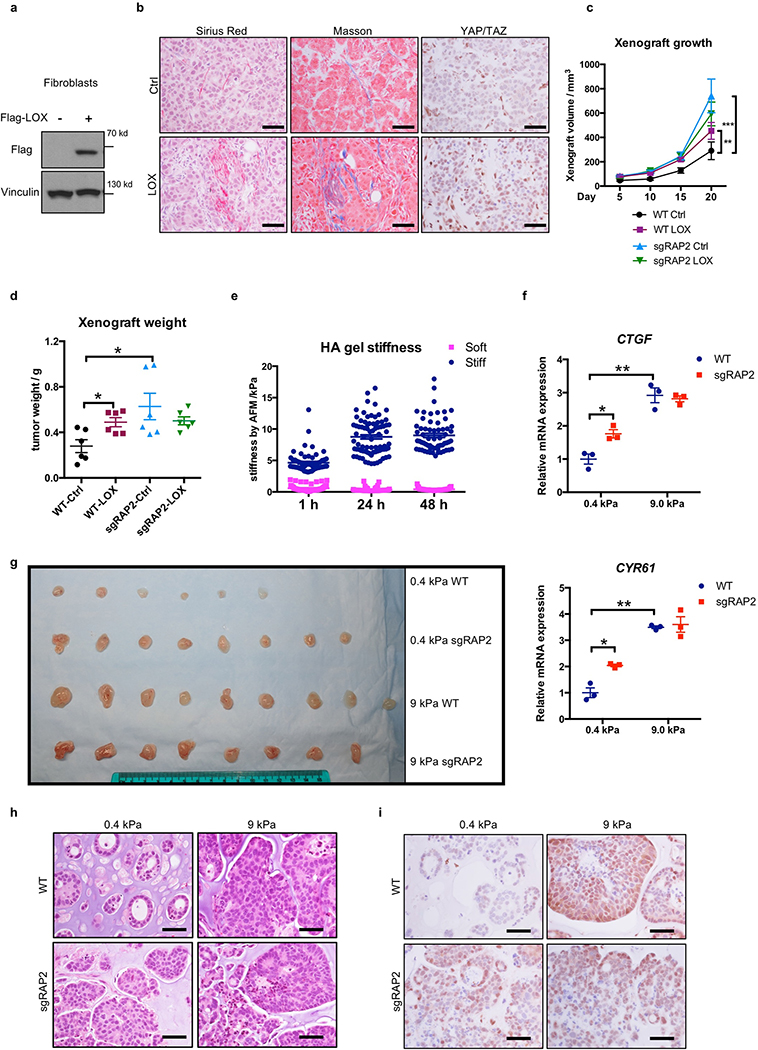
Figure 4.|. RAP2 suppresses cell transformation and regulates the ECM stiffness transcriptome through Hippo/YAP.
a. YAP/TAZ knockdown suppresses the aberrant acinus growth caused by RAP2A/B/C deletion in MCF10A cells. Scale bar, 100 μm. b. Quantification of aberrant acini from 3 hydrogels is shown as mean±SEM. Two-tailed t-test: ***, RAP2-KO vs. WT, p=0.00022; **, shYAP/TAZ vs. Scramble, p=0.0067. c. MST1/2-dKO and RAP2-MST1/2-KO MCF10A cells were s.c. injected into NOD/SCID mice. The tumor volume is shown as mean±SEM. ****, two-way ANOVA test, n=6 biologically independent xenografts, p<0.0001. d. RAP2-MST1/2-KO MCF10A cells with scramble or YAP/TAZ-targeting shRNAs were injected into NOD/SCID mice. The tumor volume is presented as mean±SEM. ****, two-way ANOVA test, n=6 biologically independent xenografts, p<0.0001. e. MCF10A and MCF10A-T cells with RAP2 deletion were injected into nude mice. The tumor volume is presented as mean+SEM. ****, two-way ANOVA test, n=6 biologically independent xenografts, p<0.0001. f. Tumor weights on Day 20 from Panel e are presented as mean±SEM. Two-tailed t-test: ***, n=6, p=0.0001; ****, n=6, p = 0.00007. g. MCF7 cells with lentivirus-mediated CRISPR deletion of RAP2 (sgRAP2) embedded in 200 μl 0.4 or 9.0 kPa HA gels were injected into nude mice. The xenograft volume was shown as mean±SEM. ****, two-way ANOVA test, n=6 (WT, 0.4 kPa), 8 (sgRAP2, 0.4 or 9.0 kPa), or 9 (WT, 9.0 kPa) biologically independent xenografts, p < 0.0001. h. Tumor weights on Day 31 from Panel g are presented as mean±SEM. Two-tailed t-test: ***, n=6 or 9, p = 0.0006; ****, n=6 or 8, p=0.00002. i. Expression of genes down- or up-regulated by low stiffness in WT cells was assessed by RNA-sequencing and compared with YAP/TAZ-dKO, LATS1/2-dKO, and RAP2-KO cells. j. A Heat-map of representative ECM-responsive genes regulated by RAP2 and the Hippo pathway.
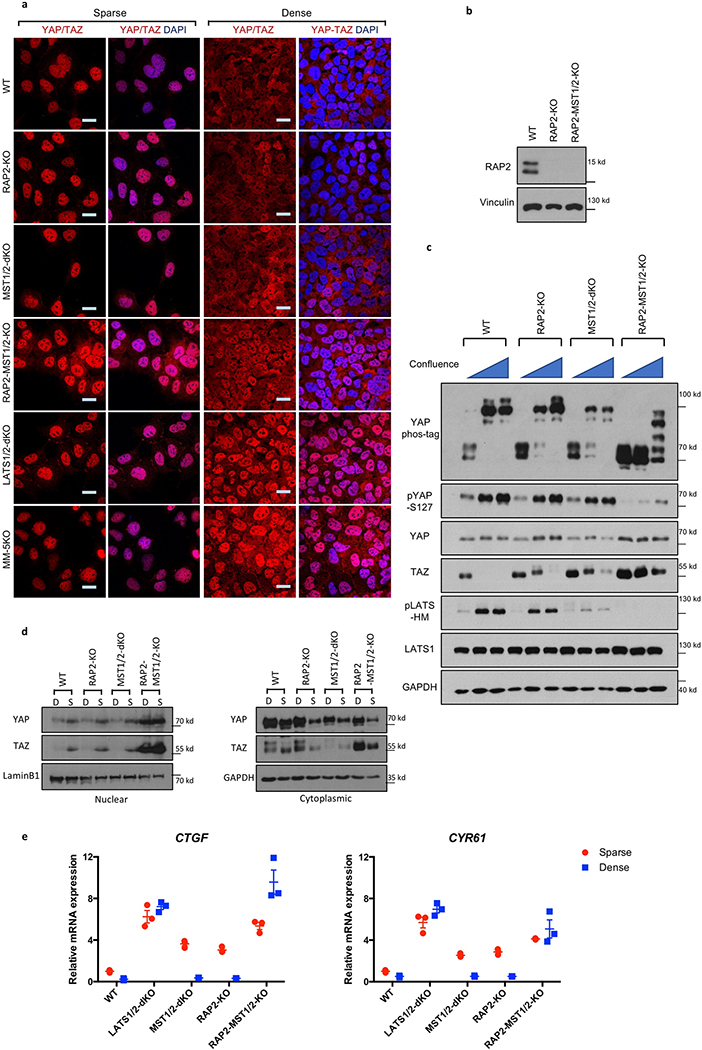
RAP2 is activated by cell-cell contact12, which also presents mechanical cue to cells and inhibits YAP/TAZ2,17. RAP2 deletion moderately increased nuclear YAP/TAZ at high confluence (Extended Data Fig. 6a,b). Combined deletion of RAP2 and MST1/2 resulted in stronger YAP/TAZ nuclear accumulation and gene transactivation (Extended Data Fig. 6a-e). Deletion of both RAP2 and MST1/2 is required to blunt LATS phosphorylation and YAP/TAZ inactivation, suggesting that confluency signaling is rather complex and additional routes, such as cellular junctions, contribute to LATS1/2 activation.
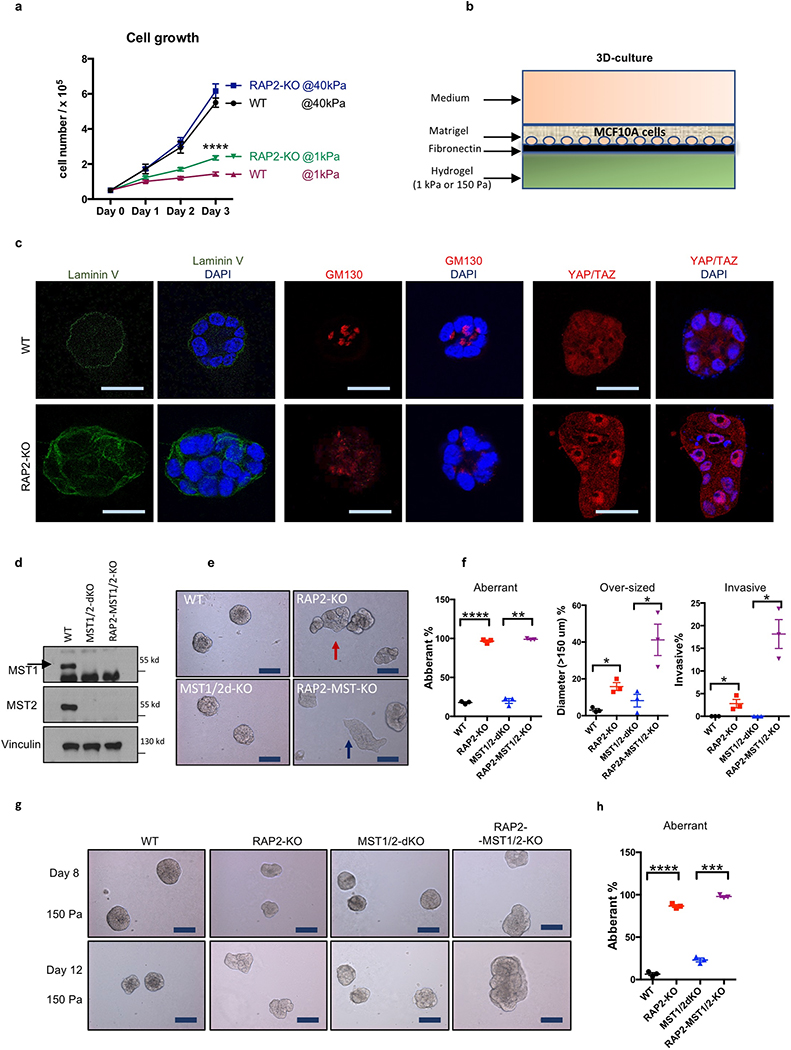
RAP2 deletion selectively enhanced cell growth only at low stiffness (Extended Data Fig. 7a). To assess RAP2’s role in tumorigenesis, we performed three assays: acinus formation, anchorage-independent growth, and xenotransplantation. First, we applied a 3D-culture system with low stiffness hydrogels to assay MCF10A acinus formation9 (Extended Data Fig. 7b), the aberrancy of which represents irregular cell growth and malignant transformation9,24. WT cells formed normal acini while RAP2-KO cells generated multi-acinar structures (Fig. 4a,b, Extended Data Fig. 7c), and YAP/TAZ knockdown significantly reduced aberrant acini. Since MST1/2 mediate some physical signals independent of RAP2 (Extended Data Fig. 6), we generated RAP2A/B/C-MST1/2–5KO (RAP2-MST1/2-KO) MCF10A cells (Extended Data Fig. 7d). While MST1/2-dKO cells formed relatively normal acini, the RAP2-MST1/2-KO cells formed large acini with invasive behaviors (Extended Data Fig. 7e,f) even at 150 Pa, a stiffness mimicking normal breast tissue (Extended Data Fig. 7g,h). Second, colony formation assay in soft agar showed that RAP2-MST1/2-KO cells conferred anchorage-independent growth (Extended Data Fig. 8a). Third, RAP2-MST1/2-KO cells showed significant xenograft growth in immune-deficient mice while MST1/2-dKO cells failed to sustain growth (Fig. 4c, Extended Data Fig. 8b,c). RAP2-MST1/2-KO xenografts contained abundant MCF10A cells recapitulating acinus and duct formation of breast tissue, while MST1/2-dKO xenografts consisted of mainly host cells with a small number of MCF10A cells (Extended Data Fig. 8d,e). Moreover, the RAP2-MST1/2-KO cells displayed architectural and cytological atypia with signs of malignancy (Extended Data Fig. 8e). Consistently, YAP/TAZ knockdown suppressed xenograft growth (Fig. 4d, Extended Data Fig. 8f-h). The RAP2’s function in stiffness-regulated growth was confirmed in a xenograft model using H-RAS-V12 expressing MCF10A cells25,26 (Fig. 4e,f, Extended Data Fig. 8i,j). We used LOX-overexpressing fibroblasts or semi-synthetic hyaluronan-derived hydrogels (soft: 0.40 ± 0.03 kPa; stiff: 8.98 ± 0.33 kPa)27,28 to assess the effect of stiffness on cell growth in xenograft models (Extended Data Fig. 9). Under low stiffness or with control fibroblasts, RAP2-KO cells grew significantly larger xenografts than WT cells whereas the growth advantage of RAP2-KO cells diminished at high stiffness or in presence of LOX-overexpressing fibroblasts (Fig. 4g,h, Extended Data Fig. 9).
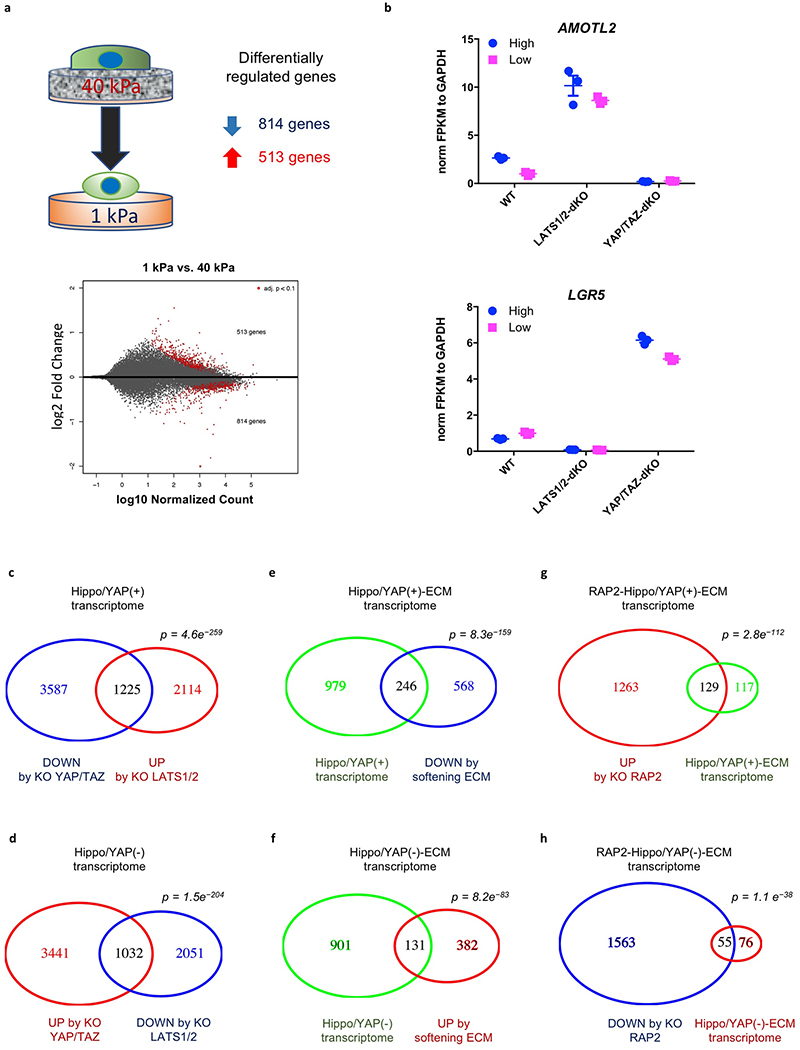
YAP/TAZ are transcriptional co-activators that generate functional output through gene transcription2. RNA-seq was performed with RAP2-KO, LATS1/2-dKO, and YAP/TAZ-dKO HEK293A cells to investigate their transcriptional responses to ECM stiffness (Fig. 4i, Extended Data Fig. 10a). 814 genes were down-regulated and 513 genes were up-regulated by low stiffness in WT cells. These genes are involved in metabolic processes, such as RNA and macromolecule biosynthesis, and morphogenesis (Supplemental information). YAP/TAZ are ‘gate-keepers’ that are responsible for virtually all the stiffness-responsive genes, as deletion of YAP/TAZ or LATS1/2 abolished most of the up- or down-regulations of these genes (Fig. 4i), such as AMOTL2 and LGR5 (Extended Data Fig. 10b). Consistent with its role in mechano-signaling, RAP2 deletion completely abolished the changes in expression of 40–50% (not including those partially blunted) of stiffness-responsive genes (Fig. 4i).
To assess the interplay of RAP2 and Hippo pathway components in stiffness-dependent gene regulation, we enriched YAP/TAZ-activating genes that were down-regulated by YAP/TAZ knockout at high stiffness and upregulated by LATS1/2 knockout at low stiffness, and YAP/TAZ-repressing genes that were conversely regulated (Extended Data Fig.10c,d). These genes defined a ECM-Hippo transcriptome that comprised nearly a third of the genes affected by ECM stiffness (Extended Data Fig. 10e,f). RAP2 deletion completely abolished the responses to ECM in about 50% of ECM-Hippo transcriptome genes (Extended Data Fig. 10g,h), not to mention that RAP2 deletion also partially compromised many genes. The RAP2-regulated ECM-Hippo transcriptome revealed that RAP2 controls genes involved in cell growth and adhesion (e.g. CTGF, CYR61), as well as morphogenesis and development (e.g. KRT8, KRT18, GDF6), through the Hippo pathway to respond to ECM stiffness (Fig. 4j, Supplemental information).
This study has uncovered RAP2 as an intracellular mechanotransducer, which relays extracellular mechanical signals to transcriptional regulation through the Hippo pathway. ECM stiffness acts through RAP2 and its downstream Hippo kinase cascade to modulate a YAP/TAZ-mediated mechano-responsive transcriptome. The identification of this new signaling axis provides mechanistic insights into how cellular machinery is driven by mechanical stimuli.
Methods
Cell culture
HEK293A were maintained in DMEM containing 10% fetal bovine serum. MCF10A cells were maintained as previously described24. The pre-malignant derivative of MCF10A cells, MCF10A-T, were generated by infecting MCF10A cells with a lentiviral vector that expressing a constitutively active H-Ras mutant (G12V) as previously described25,26. Adipocyte-derived mesenchymal stem cells (MSCs) were cultured in HMSC growth medium from Cell Applications Inc, and were differentiated into adipocytes and stained according to a modified protocol29. MCF7 and MDA-MB-468 were maintained in DMEM/F12 with 10% FBS. Insulin (0.01 mg/ml) was used for maintaining MCF7 cells.
Plasmids
Flag-pLJM1-RAP2A and pRK5-HA-GST-RAP2A plasmids were kindly provided by Dr. David M. Sabatini (Addgene #19311 & 14952). PDZGEF1 and ARHGAP29 coding sequences were subcloned from cDNA clones BC117321 and BC093741 (Transomics Technology), respectively.
GFP-C1-PLCdelta-PH was a gift from Dr. Tobias Meyer (Addgene #21179).
CRISPR/Cas9 system was used to delete genes in HEK293A, MCF10A, MSC, MCF7 and MDA-MB-468 cells. The plasmids px459 v2 and lentiCRISPR v2 were provided by Dr. Feng Zhang (Addgene #62988 & #52961).
The sgRNA sequences targeting individual genes were as following:
RAP2A #1: GATGCGCGAGTACAAAGTGG RAP2A #2: GTATTTCTCGATGAAGGTGC RAP2B #1: CATGAGAGAGTACAAAGTGG RAP2B #2: GGAGCCCGTCACGAACTGCA RAP2C #1: GGTGAAGGTGAGACTCATGA RAP2C #2: AGTGACAAACTGCACAGTAA PDZGEF1 (RAPGEF2) #1: CCCATAACTCCATGGTTAGC PDZGEF1 (RAPGEF2) #2: CCAGCTAACCATGGAGTTAT PDZGEF2 (RAPGEF6) #1: TCAACGCCTGCCTAGCGCCA PDZGEF2 (RAPGEF6) #2: GCCACCCGAGCGGACTCCCG ARHGAP29 #1: CTCTACTTACATATTTCCAA ARHGAP29 #2: AGTTATTCATATACGTCTAG
RAP2-GTP and RhoA-GTP binding assay
GST-RalGDS-RBD-expressing BL21 strain was kindly provided by Dr. Rick Firtel, and GST-Rhotekin-RBD-expressing BL21 strain was a gift from Dr. Joan Heller Brown at the University of California, San Diego. The recombinant proteins were purified and stored as bound to glutathione-agarose beads. The binding of RAP2-GTP and RHOA-GTP from cell lysates to RalGDS-RBD-agarose and Rhotekin-RBD-agarose beads, respectively, was performed in a buffer containing 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 25 mM MgCl2, 10% glycerol, 0.5% NP-40 substitute, 1 mM DTT.
Cell culture with two-dimensional or three-dimensional polyacrylamide-based hydrogels
Two-dimensional culture on hydrogels of high (40.40 ± 2.39 kPa) or low (1.00 ± 0.31 kPa) stiffness was described elsewhere30. 10 μg/ml human placenta fibronectin or 25–50 ug/ml rat tail Collagen I were used to coat the Sulfo-Sanpah-activated hydrogels according to the preferences of the cell lines. Three-dimensional culture followed a protocol with the modification that the Matrigel base was replaced with fibronectin-coated hydrogels9,24.
Staining and microscopy
For immunofluorescence, cells were fixed in 4% formaldehyde/PBS for 10 min and then were treated with 0.1% Triton X-100 or Saponin (only for staining PIP2 GFP reporter) for 15 min. After blocking, the cells were stained with corresponding antibodies. Most images were captured with a Nikon Eclipse Ti confocal microscope and then were exported from NIS elements imaging software. Images in Fig. 2d, Ext Data Fig. 4c,d were taken with Olympus FV1000 confocal microscope, and Image J was used to merge the signals from channels. For immunohistochemistry, xenografts were subject to heat-induced antigen retrieval using 10 mM sodium citrate buffer followed by 3% H2O2 for 30 min to quench endogenous peroxidase activity. Sections were incubated overnight at 4°C with YAP/TAZ antibody and detected using Vectastain elite ABC kit and DAB Peroxidase Substrate kit (Vector Laboratories) according to the manufacturer’s protocol.
Preparation of semi-synthetic hyaluronan-derived hydrogels
Under aseptic conditions, Glycosil (ESI Bio, cat # GS222), Gelin-S, (ESI Bio, cat # GS231), and Extralink (ESI Bio, cat # GS3006) were dissolved in degassed water (ESI Bio, cat # GS240) according to the manufacturer’s directions. To make soft hydrogels, stock concentrations of 10 mg/mL Glycosil, 10 mg/mL Gelin-S, and 5 mg/mL Extralink were made per manufacturer’s directions. To make stiff hydrogels, concentrated stocks of Glycosil and Extralink were prepared by solubilization in reduced volumes to make 2x Glycosil and 5x Extralink. Prior to use, aliquots were taken from each vial to make solutions at 1:5 ratios of Extralink: (Glycosil + Gelin-S) and 5x Extralink: (2x Glycosil + Gelin-S) for soft and stiff hydrogels respectively. For all conditions, the amount of Gelin-S was kept constant to ensure the same number of gelatin-based cell binding sites.
In order to perform atomic force microscopy, the Glycosil and Gelin-S components were mixed thoroughly and then the Extralink was added to initiate gelation. Fifty μL of the mixture was added drop-wise to DCMS-treated glass slides and a methacrylated coverslip was placed on top. Each sample was prepared in triplicate. The slides were then incubated at 37°C until complete gelation. For the samples tested, this occurred within 30–40 minutes of incubation at 37°C. Upon gelation, hydrogel stiffness was measured by AFM. Hydrogels were then placed in PBS containing 1% antibiotic/antimycotic at 37°C and the stiffness was measured at 1, 24, and 48 hours after mixing.
AFM measurement procedure
AFM was performed to measure hydrogel stiffness as previously described31. Briefly, indentations were performed using a pyrex-nitride probe with a pyramid tip (spring constant ~0.04 N/m, 35º half-angle opening, NanoAndMore USA Corporation, cat # PNP-TR) connected to a MFP-3D Bio Atomic Force Microscope (Oxford Instruments) mounted on a Ti-U fluorescent inverted microscope (Nikon Instruments). Probes were calibrated using the Igor 6.34A software (WaveMetrics). Samples were then loaded on the AFM, submersed in PBS, and indented at a velocity of 2 μm/s with a trigger force of 2 nN. ~20 force measurements were performed over a 90 μm x 90 μm region per gel. Measurements were made each day for three separate gels per condition. Elastic modulus was calculated based on a Hertz-based fit using a built-in code written in the Igor 6.34A software.
Animal studies.
8–9 weeks old female NOD/SCID mice were purchased from Jackson Lab, and 8–9 weeks old female nude mice were provided by the animal care program at University of California, San Diego. The mice were hosted in a special pathogen-free room under standard 12:12-h light/dark cycle, fed with standard rodent chow and water ad libitum, randomized before experiments. The sample size choice was not pre-determined for each experiment. When comparing RAP2-MST1/2-KO MCF10A cell and MST1/2-dKO MCF10A cells, a total of 5 × 106 MCF10A cells in 50% high concentration Matrigel (BD Bioscience) dissolved in PBS were subcutaneously inoculated in a NOD/SCID mouse. When comparing RAP2-KO MCF10A and MCF10A-T cells with WT MCF10A and MCF10A-T cells, 5 × 106 cells in 50% Matrigel were injected into nude mice subcutaneously. When comparing RAP2-KO with WT MCF7 cells, 2 × 106 MCF7 cells and 4 × 105 LOX-expressing or control NIH3T3 cells in 50% Matrigel/PBS were co-injected subcutaneously into nude mice. For the semi-synthetic hyaluronan-derived hydrogels, 2 × 106 MCF7 cells in 20 μl PBS suspension were embedded into 200 μl of soft and stiff formulations as described in the preparation of hydrogels. After brief gelation (5 min) at room temperature, the cell-laden hydrogels were subcutaneously injected into nude mice. The investigators are blinded to group allocations during data collection and analyses. All the procedures followed the NIH guidelines for the care and use of laboratory animals and the IACUC at the University of California, San Diego approved the experiments. For subcutaneous tumor growth, the maximum single tumor cannot exceed 2 cm in diameter in mice according to the guidelines provided by the animal care program at University of California, San Diego, and no experiments in this study generated tumor burden over this limit.
RNA interference
Lentiviral vectors pLKO.1-hygromycin (Addgene #24150) and pLKO.1-Blasticidin (Addgene #26655) were used to clone the following sense sequences to knock down human YAP or TAZ:
Negative control: CCTAAGGTTAAGTCGCCCTCG (cloned into pLKO.1-hygromycin and pLKO.1-blasticidin); YAP#1: GCCACCAAGCTAGATAAAGAA (pLKO.1-hygromycin); YAP#2: GACATCTTCTGGTCAGAGATA (pLKO.1-hygromycin); TAZ#1: GCGTTCTTGTGACAGATTATA (pLKO.1-blasticidin); TAZ#2: GCTCATGAGTATGCCCAATGC (pLKO.1-blasticidin). The resulting plasmids were used to package lentiviruses and the target cells were infected with an MOI of 0.5.
Duplex siRNAs targeting PLC𝛾1 and PLD1/2 were purchased from Integrated DNA Technologies, Inc. and transfected into cells with RNAiMAX (ThermoFisher Scientific). The sequences are as following
PLC𝛾1 #1 sense strand 5’-rGrArCrCrUrCrArUrCrArGrCrUrArCrUrArUrGrArGrArAAC-3’; anti-sense strand 5’-rGrUrUrUrCrUrCrArUrArGrUrArGrCrUrGrArUrGrArGrGrUrCrArA-3’. PLC𝛾1 #2 sense strand 5’-rGrGrCrArArGrArArGrUrUrCrCrUrUrCrArGrUrArCrArATC-3’; anti-sense strand 5’-rGrArUrUrGrUrArCrUrGrArArGrGrArArCrUrUrCrUrUrGrCrCrUrU-3’. PLD1 #1 sense strand 5’-rGrUrGrGrArUrArArArUrUrArCrArUrArUrCrArUrUrCrUGT-3’; antisense strand 5’- rArCrArGrArArUrGrArUrArUrGrUrArArUrUrUrArUrCrCrArCrUrG-3’. PLD1 #2 sense strand 5’-rArCrUrGrGrArArGrArUrUrArCrUrUrGrArCrArArArGrATA-3’; anti-sense strand 5’-rUrArUrCrUrUrUrGrUrCrArArGrUrArArUrCrUrUrCrCrArGrUrUrG-3’. PLD2 #1 sense strand 5’-rArArCrCrArArGrArArGrArArArUrArCrCrGrUrCrArUrUTT-3’; anti-sense strand 5’-rArArArArUrGrArCrGrGrUrArUrUrUrCrUrUrCrUrUrGrGrUrUrGrU-3’. PLD2 #2 sense strand 5’-rCrUrCrUrArCrArUrUrGrArGrArArUrCrArGrUrUrCrUrUCA-3’; anti-sense strand 5’-rUrGrArArGrArArCrUrGrArUrUrCrUrCrArArUrGrUrArGrArGrGrA-3’.
Quantitative real-time PCR
Total RNAs were extracted with a kit from Qiagen. Reverse transcription was performed with iScript from Bio-Rad. The real-time PCR was performed with the Applied Biosystems 7300 with primers targeting CTGF, CYR61, and ANKRD1: CTGF-Forward: 5’-CCAATGACAACGCCTCCTG-3’, CTGF-Reverse: 5’-TGGTGCAGCCAGAAAGCTC-3’; CYR61-Forward: 5’-AGCCTCGCATCCTATACAACC-3’, CYR61-Reverse: 5’-TTCTTTCACAAGGCGGCACTC-3’; ANKRD1-Forward: 5’-GTGTAGCACCAGATCCATCG-3’, ANKRD1-Reverse: 5’-CGGTGAGACTGAACCGCTAT-3’. The gene expression was normalized to GAPDH: Forward: 5’-TGCACCACCAACTGCTTAGC-3’; Reverse: 5’- GGCATGGACTGTGGTCATGAG-3’.
RNA sequencing and bioinformatics analysis
Total RNAs were extracted by TRIzol (Thermo Fisher Scientific) from HEK293A cells seeded on high and low stiffness fibronectin-coated hydrogels. Three replicates for each sample were generated and analyzed. The resulting RNA was then used to prepare libraries using Illumina TruSeq Stranded mRNA Library Prep Kit Set A (Illumina, RS- 122–2101) or Set B (Illumina, RS-122–2102). The libraries were sequenced using Illumina HiSeq 4000 (single end 50bp). Reads were aligned to the hg19 reference genome using STAR32. Only uniquely mapped reads were kept for further analysis. Number of reads for each gene were counted using htseq-count33 according to Gencode human annotation release 24. DeSeq234 was used to identify differential expressed genes with default parameters. Genes with adjusted p-value < 0.1 were considered as differentially expressed. GO and KEGG enrichment analysis of differential expressed genes was performed using DAVID35.
Statistical analysis
Microsoft Excel was used for t-tests, and Graphpad Prism v6 was used for two-way ANOVA tests. When p value is smaller than 0.0001, Graphpad Prism v6 does not provide a precise p value and instead only shows a range of p < 0.0001.
Data availability
The source data for the graph representations can be found in the online version of the paper. For uncropped images of Western blot data, see Supplementary Figure 1. The RNA sequencing data is available in GEO DataSets with the accession number GSE98547. All other data that support the findings of this study are available upon request from the corresponding author.
Extended Data
Extended Data Figure 1. RAP2 is involved in YAP/TAZ regulation by matrix stiffness.
a.YAP/TAZ localization is not significantly affected by H-, K-, N-RAS, or RAP1B overexpression. HEK293A cells were cultured on high stiffness hydrogels. Overexpression of HA-tagged H-RAS, K-RAS, N-RAS, and Flag-tagged RAP1B are indicated. Merged, combined signals of YAP/TAZ (red), transfected small GTPases (green), and DAPI (blue, staining for DNA). Scale bar, 25 μm. The images are representative of two independent experiments with similar results.
b. Subcellular fractionation of WT and RAP2A/B/C-triple knock (RAP2-KO) HEK293A cells at low or high stiffness. The images are representative of two independent experiments with similar results.
c. CRISPR-mediated RAP2A/B/C deletion in adipocyte-derived mesenchymal stem cells (MSCs) by lentiviral transduction. Single guide RNAs targeting RAP2A, RAP2B, and RAP2C (sgRAP2) were individually cloned into lentiCRISPR v2 (Addgene #52961) plasmids. sgRAP2 #1 and #2 were two sets of three single guide RNAs targeting RAP2A, RAP2B, and RAP2C with unique sequences for each RAP2 isoform. Adipocyte-derived MSCs were infected with the sgRAP2 lentiviruses with a MOI of 10 and selected by puromycin. After puromycin selection, the pooled cells were examined for the RAP2 protein expression. The images are representative of two independent experiments with similar results.
d. RAP2 is required for the low stiffness-induced cytoplasmic localization of YAP/TAZ in MSCs. The MSCs with CRISPR-mediated RAP2A/B/C deletion were seeded on 40 kPa and 1 kPa collagen-coated hydrogels, cultured for 24 hours, and then stained for YAP/TAZ. The results are representative of 3 biologically independent samples showing similar results. Merged, combined signals of YAP/TAZ (red), F-actin (green), and DAPI (blue). Scale bar, 25 μm.
e. The YAP/TAZ localization distribution is presented as mean+SEM for cells with more nuclear (N>C), more cytoplasmic (N<C), or even (N=C) distribution of YAP/TAZ. n=3 biologically independent samples. Scale bar, 25 μm.
f. RAP2 plays an important role in adipocyte differentiation at low stiffness. Representative images of Oil-Red O staining of adipocyte-derived MSCs that were treated with adipocyte differentiation medium for 15 days. The MSCs were grown on 1 kPa hydrogels. Scale bar, 200 μm.
g. Quantification of Oil-Red O-positive cells. The results are presented as mean±SEM. ***, two-tailed t-test, n=8 biological independent samples, p = 0.00026.
h. CRISPR-mediated RAP2A/B/C deletion in MCF7 and MDA-MB-468 by lentiviral transduction. The experiments were similarly performed as for RAP2A/B/C deletion in MSCs (Panel c). The images are representative of two independent experiments with similar results.
i. RAP2 deletion promotes YAP/TAZ nuclear localization in MCF7 at low stiffness. The images are from 3 independent experiments showing similar results. Merged, combined signals of YAP/TAZ (red), F-actin (green), and DAPI (blue). Scale bar, 25 μm.
j. Quantification of YAP/TAZ localization in MCF7 cells. The YAP/TAZ localization distribution is presented as mean+SEM. n=3 biologically independent samples.
k. YAP/TAZ in MDA-MB-468 cells are not significantly regulated by matrix stiffness. The images are from 3 independent experiments showing similar results. Merged, combined signals of YAP/TAZ (red), F-actin (green), and DAPI (blue). Scale bar, 25 μm.
l. Quantification of YAP/TAZ localization in MDA-MB-468 cells. The YAP/TAZ localization distribution is presented as mean+SEM. n=3 biologically independent samples.
Extended Data Figure 2. RAP2 is activated at low matrix stiffness involving PDZGEF1/2.
a. RAP2 has no effect on localization of TWIST and β-catenin at different matrix rigidities. Immunofluorescence staining of TWIST and β-catenin in WT and RAP2-KO HEK293A cells grown on soft (1 kPa) and stiff (40 kPa) fibronectin-coated hydrogels. Low stiffness induced cytoplasmic localization of TWIST similarly in WT and RAP2-KO cells. Stiffness had no significant effect on β-catenin localization in HEK293A cells. Scale bar, 25 μm. The images are representative of 3 independent experiments with similar results.
b. GST-RalGDS-RBD specifically binds to the active RAP2A in the pulldown assay. HEK293A cells were transfected with plasmids expressing RAP2A WT, G12V mutant (constitutively GTP-binding), or S17N mutant (GTP-binding deficient) and then seeded onto soft hydrogels. The cells were lysed 24 hours after seeding and the lysates were incubated with glutathione agarose beads that were pre-loaded with GST-RalGDS-RBD. The beads were washed and subjected to Western blot analyses with the indicated antibodies. The images are representative of 2 independent experiments with similar results.
c. GTP-binding/activity of RAP2A is required to induce YAP/TAZ cytoplasmic translocation. Merged, combined signals from YAP/TAZ (red), Flag (green), and DAPI (blue). Scale bar, 25 μm. The images are representative of 2 independent experiments with similar results.
d. ECM stiffness regulates the interaction between RAP2A and PDZGEF1. GST-RAP2A plasmids were co-transfected with Flag-Raptor (a negative control) or Flag-PDZGEF1 into HEK293A cells. The cells were thereafter seeded on stiff and soft hydrogels. 24 hours after seeding, the cells were lysed and the lysates were incubated with glutathione agarose beads for 6 hours. Then the beads were washed and subjected to Western blot analyses. The images are representative of 2 independent experiments with similar results.
e. Sanger DNA sequencing confirmed the homozygous deletion of a ‘T’ nucleotide in PDZGEF1 genomic DNAs in the PDZGEF1/2-dKO HEK293A cells.
f. Western blot showing the absence of PDZGEF2 expression in PDZGEF1/2-dKO HEK293A cells. The images are representative of 2 independent experiments with similar results.
g. The repression on YAP/TAZ target genes CTGF and CYR61 by low stiffness was compromised in PDZGEF1/2-dKO cells. Expression of CTGF and CYR61 in PDZGEF1/2-dKO HEK293A cells on soft and stiff matrices was measured by quantitative Real-time PCR. ns, not significant, two-tailed t-test, n=3 biological independent samples. Data are represented as mean±SEM.
h. RAP2 is required for PDZGEF1 to induce cytoplasmic localization of YAP/TAZ. Immunofluorescence showing YAP/TAZ localization after ectopic expression of a Flag-tagged PDZGEF1 in WT and RAP2-KO cells at high stiffness. Merged, combined signals from Flag-PDZGEF1 (green), YAP/TAZ (red), and DAPI (blue). Scale bar, 25 μm.
i. Quantification of the results in Panel h. ‘+’ denotes the Flag-PDZGEF1-transfected cells and ‘−’ denotes cells that were not transfected. Data are represented as mean+SEM. n=3 biologically independent samples.
j. Stiffness influences cellular PIP2. The PIP2 reporter GFP-PLCδ-PH domain, which binds to PIP2, was imaged with a Nikon inverted microscope in cells at low or high stiffness. Cells grown at high stiffness display diffused PIP2 localizations while cells at low stiffness show enrichment of PIP2 at the plasma membrane. The image represents 2 independent experiments with similar results. Scale bar, 25 μm.
k. Inhibition of PLC alters cellular PIP2 distribution. Immunofluorescence of cells treated with 5 μM PLC inhibitor U73122 at high stiffness. GFP was detected with anti-GFP immunofluorescence. PIP2 enrichment, indicated by arrows, was observed. Scale bar, 25 μm. The image represents 2 independent experiments with similar results.
Extended Data Figure 3. FAK, PLC𝛾1, and PLD1/2 are involved in RAP2 activation and Hippo pathway activation in response to stiffness.
a. FAK inhibitor PF573228 and PLC inhibitor U73122 promote RAP2-GTP loading at high stiffness. The images represent two biologically independent experiments with similar results.
b. RAP2 functions downstream of FAK and PLC to regulate YAP/TAZ localization. YAP/TAZ localization was imaged in cells cultured at high stiffness and treated with 10 μM FAK inhibitor PF573228 or 5 μM PLC inhibitor U73122. Scale bar, 25 μm. The image represents 3 (WT treated with PF573228 or U73122; RAP2-KO ctrl) or 4 (WT ctrl; RAP2-KO treated with PF573228 or U73122) biologically independent samples with similar results.
c. The localization distribution in Panel b is presented as mean+SEM. n=3 (WT treated with PF573228 or U73122; RAP2-KO ctrl) or 4 (WT ctrl; RAP2-KO treated with PF573228 or U73122) biologically independent samples.
d. PLD1/2 inhibitor BML279 suppresses RAP2-GTP binding at low stiffness. The images represent two independent experiments with similar results.
e. Inhibition of PLD increases nuclear YAP/TAZ in cells at low stiffness. Cells growing at low stiffness were treated with 5 μM PLD inhibitor BML279. Scale bar, 25 μm. The image represents 7 (ctrl) or 5 (BM279) biologically independent samples with similar results.
f. The localization distribution in Panel e is presented as mean+SEM. n=7 (ctrl) or 5 (BML279) biologically independent samples.
g. Western blot showing PLC𝛾1 knockdown by Duplex siRNAs (DsiRNAs). Two independent siRNAs were used. The image represents 2 independent experiments with similar results.
h. PLC𝛾1 knockdown decreases nuclear YAP/TAZ at high stiffness. Quantification of YAP/TAZ localization in PLC𝛾1 knockdown and control cells growing on 40 kPa hydrogels. The YAP/TAZ localization distribution is presented as mean+SEM. n=3 biologically independent samples.
i. Representative images of YAP/TAZ localization in cells with PLC𝛾1 knockdown. Scale bar, 25 μm. The image represents 3 biologically independent experiments with similar results.
j. Western blot showing PLD1/2 knockdown. Two independent DsiRNAs were used. The image represents 2 independent experiments with similar results.
k. PLD1/2 knockdown increases nuclear YAP/TAZ at low stiffness. Quantification of YAP/TAZ localization in PLD1/2 knockdown and control cells growing on 1 kPa hydrogels. The YAP/TAZ localization distribution is presented as mean+SEM. The results are from 3 biologically independent samples.
l. Representative images of YAP/TAZ localization in cells with PLD1/2 knockdown. Scale bar, 25 μm. The image represents 3 biologically independent experiments with similar results.
m. WT, but not the GTP binding-deficient (S17N), RAP2A induces YAP phosphorylation. YAP phosphorylation was analyzed by phos-tag SDS-PAGE in HEK293A cells stably expressing WT or the S17N mutant RAP2A at high stiffness. The image represents 2 independent experiments with similar results.
n. Hippo pathway components are involved in YAP/TAZ regulation by stiffness. HEK293A cell lines with deletion of Hippo components were cultured at low stiffness (1 kPa). Deletion of Hippo components includes LATS1/2, MST1/2, MM-5KO (MST1/2-MAP4K4/6/7–5KO), and MM-8KO (MST1/2-MAP4K1/2/3/4/6/7–8KO). The image represents 3 biologically independent experiments with similar results. Scale bar, 25 μm.
o. Quantification of immunofluorescence of the samples in Panel n. The YAP/TAZ localization distribution is presented as mean+SEM. n=3 biologically independent samples.
Extended Data Figure 4. RAP2 activates MAP4K4 and induces YAP phosphorylation.
a. Hippo pathway components are required for RAP2A to induce YAP/TAZ phosphorylation. RAP2A is overexpressed in the indicated knockout HEK293A cells. The Western images represent 2 independent experiments with similar results. Two clones of MM-5KO cells were used for this experiment.
b. RAP2A acts through the Hippo pathway to induce YAP/TAZ cytoplasmic localization. Flag-RAP2A was transfected into HEK293A cell lines with deletion of different core Hippo pathway components as indicated. YAP/TAZ localization was analyzed by immunofluorescence. Merged, combined signals from Flag-RAP2A (green), YAP/TAZ (red), and DAPI (blue). Scale bar, 25 μm. The image represents 3 biologically independent experiments with similar results.
c. YAP/TAZ localization in LATS1−/− LATS2 Flox/Flox (LATS1-KO LATS2-F/F) and LATS1−/− LATS2−/− (LATS1/2-dKO) MEFs. Scale bar, 25 μm. The image represents 2 biologically independent experiments with similar results.
d. YAP/TAZ localization in NF2-KO and MOB1A/1B-dKO HEK293A cells. Scale bar, 25 μm. The image represents 2 biologically independent experiments with similar results.
e. Low matrix stiffness activates MAP4K4 in a RAP2-dependent manner. Plasmids expressing HA-tagged MAP4K4 were transfected into WT and RAP2-KO HEK293A cells. 24 hours after seeding on 40 kPa or 1 kPa hydrogels, HA-MAP4K4 proteins were immuno-precipitated and then used for in vitro kinase assay, in which recombinant full-length GST-tagged LATS2 protein was used as a substrate. The LATS2 phosphorylation by MAP4K4 was detected by an a phospho-specific antibody recognizing the phosphorylated hydrophobic motif of LATS1/2. Results were from triplicated experiments that were biologically independent.
f. Quantification of the kinase assay (in Panel e) shown as mean±SEM. The relative LATS2 phosphorylation was normalized by the protein levels of HA-MAP4K4 and defined as MAP4K4 kinase activity. *, n=3 biologically independent samples, two-tailed t-test, p = 0.027 (WT-1 kPa vs. RAP2 KO-1 kPa) or 0.037 (WT-1 kPa vs. WT-40 kPa); ns, not significant.
g. The lysates of WT and RAP2-KO HEK293A cells growing on stiff and soft hydrogels were analyzed by Western blot for MAP4K4 migration. ‘Stretched’ image was generated by vertically extending the same Western image directly above it in order to better visualize the MAP4K4 mobility shift. The image represents 2 independent experiments with similar results.
h. Phosphatase treatment increases MAP4K4 migration on SDS-PAGE. Lambda phosphatase was used to treat cell lysates of RAP2A-expressing HEK293A cells. Then the cell lysates were analyzed by Western blot. These results indicated that the altered migration of MAP4K4 was correlated with its phosphorylation. The image represents 2 independent experiments with similar results.
i. RAP2A promotes MAP4K4 phosphorylation (slower band migration) dependent on its citron domain. Flag-RAP2A-G12V plasmid was co-transfected with MAP4K4 WT or the citron domain deletion mutant (ΔCNH) into HEK293A cells. The image represents 2 independent experiments with similar results.
j. RAP2 is required for the reduced MAP4K4 mobility. WT and citron domain deletion mutant of MAP4K4 were transfected into WT and RAP2-KO HEK293A cells. The image represents 2 independent experiments with similar results.
Extended Data Figure 5. RAP2 inhibits RhoA GTPase through ARHGAP29.
a. RAP2A expression inhibits endogenous RhoA GTP-binding. HEK293A cells with doxycycline-inducible expression of RAP2A were established and the expression of Flag-RAP2A was induced by doxycycline. RhoA activity was determined by a GST-Rhotekin-RBD pulldown assay. S.E. denotes a short exposure of the RhoA Western blot. The image represents 2 independent experiments with similar results.
b. Western blot confirms CRISPR-mediated RhoA gene editing. HEK293A cells were transfected with CRISPR plasmids targeting RhoA and selected with puromycin for 3 days. Two different guide RNAs were used to generate two RhoA knockout pools (sgRhoA #1 and #2)16. The image represents 2 independent experiments with similar results.
c. CRISPR-mediated deletion of RhoA leads to increased cytoplasmic localization of YAP/TAZ in HEK293A at high stiffness. The localization distribution is presented as mean+SEM. n=6 biologically independent samples.
d. Representative immunofluorescence images for Panel c. Scale bar, 25 μm.
e. ARHGAP29 induces YAP phosphorylation in a Hippo pathway-dependent manner. MM-5KO, a HEK293A clone with deletion of MST1/2-MAP4K4/6/7 genes. MM-8KO, deletion of MST1/2-MAP4K1/2/3/4/6/7 genes. YAP phosphorylation was detected by phos-tag gels. The image represents two independent experiments with similar results.
f. Immunoblot showing deletion of ARHGAP29 in HEK293A WT and MAP4K4/6/7-tKO cells. The image represents 2 independent experiments with similar results.
g. Deletion of ARHGAP29 compromises Rho inhibition by low matrix stiffness. WT and ARHGAP29-KO HEK293A cells were cultured at indicated stiffness and then assayed for RhoA activity with GST-Rhotekin-RBD binding assay. The image represents two independent experiments with similar results.
h. Combined deletion of ARHGAP29 and MAP4K4/6/7 abolishes YAP phosphorylation induced by RAP2A. HA-YAP was co-transfected with vector or Flag-RAP2A into HEK293A cells cultured at high stiffness. HA-YAP phosphorylation was detected by phos-tag SDS PAGE. The image represents 2 independent experiments with similar results.
i. Combined deletion of ARHGAP29 and MAP4K4/6/7 blocks low stiffness-induced YAP cytoplasmic localization. Quantification of YAP/TAZ localization in HEK293A cells with deletion of ARHGAP29 and/or MAP4K4/6/7 at low stiffness in Fig. 3e. The YAP/TAZ localization distribution is presented as mean+SEM. n=3 biologically independent samples.
Extended Data Figure 6. RAP2 contributes to cell density-induced YAP/TAZ inactivation.
a. YAP/TAZ cytoplasmic translocation caused by cell contact involves RAP2 and the Hippo pathway. Scale bar, 25 μm. MM-5KO is a HEK293 cell line with deletion of MST1/2-MAP4K4/6/7 five genes. RAP2-MST-KO denotes the cell line with deletion of RAP2A/B/C-MST1/2 five genes. The image represents 3 biologically independent experiments with similar results.
b. Western blot showing absence of RAP2 proteins in RAP2-KO and RAP2-MST1/2-KO cells. Note that part of the results is shown in Fig. 1b and are from the same experiment. The image represents 2 independent experiments with similar results.
c. YAP/TAZ phosphorylation induced by cell contact requires RAP2 and MST1/2. The Western blot shows YAP phosphorylation of cells with low, medium, or high confluences. The image represents 2 independent experiments with similar results.
d. Deletions of RAP2A/B/C and MST1/2 interfere with YAP/TAZ regulation by cell density. Subcellular fractionations were performed for WT, RAP2-KO, MST1/2-dKO, RAP2-MST1/2-KO HEK293A cells cultured at low (S, sparse) or high density (D, dense). GAPDH and LaminB1 are markers for cytoplasmic and nuclear fraction, respectively. The image represents 2 independent experiments with similar results.
e. RAP2A/B/C and MST1/2 are required for YAP/TAZ target gene regulation by cell density. Quantitative RT-PCR was performed to determine the expression of YAP/TAZ target genes CTGF and CYR61 of the above cells at low or high confluence. Data are represented as mean±SEM. n=3 biologically independent samples.
Extended Data Figure 7. RAP2 prevents aberrant acinus growth of MCF10A cells on soft matrices.
a. RAP2 deletion selectively enhances HEK293A cell growth on soft matrix. WT and RAP2-KO HEK293A cells were seeded on stiff or soft matrices, and cell numbers were recorded as mean± SEM every day. ****, two-way ANOVA test, p < 0.0001, WT vs. RAP2-KO cells cultured at 1 kPa, n=3 biologically independent samples.
b. Diagram showing the model of 3-D culture of MCF10A cells.
c. RAP2 deletion causes abnormal acinus growth and cell polarity defects in MCF10A cells. Immunofluorescence staining of acini for cell polarity markers, Laminin V and GM130, and YAP/TAZ in WT and RAP2-KO MCF10A cells cultured for 6 days were shown. Scale bar, 25 μm. The images represent 3 independent experiments with similar results.
d. Western blot showing deletion of MST1/2 genes in WT and RAP2-KO cells. The arrow indicates the specific band for MST1. The image represents 2 independent experiments with similar results.
e. Representative images showing acinus formation of various genetically engineered MCF10A cells at 1 kPa. Red arrows indicate aberrant acini. Blue arrows indicate invasive cell morphology. Scale bar, 100 μm. The results are representative of two independent experiments with similar results.
f. Quantification of aberrant, oversized, and invasive acini in Panel e. The percentage of the cells is presented as mean±SEM. Two-tailed t-tests were used for statistical analyses of aberrant and oversized clones in n=3 biologically independent samples. Aberrant: ****, p=0.0000062; ***, p= 0.0012; Over-sized: *, p= 0.017 (for WT vs. RAP2-KO) or 0.046 (for MST1/2-dKO vs. RAP2-MST1/2-KO). For analysis of invasive clones, one-tailed t-test was used as no invasive clones were observed in either WT or MST1/2-dKO cells. *, p = 0.047 (for WT vs. RAP2-KO) or 0.029 (for MST1/2-dKO vs. RAP2-MST1/2-KO). n=3 biologically independent samples.
g. Representative images showing acinus formation of various genetically engineered MCF10A cells at 150 Pa. Scale bar, 100 μm. The image represents 2 biologically independent experiments with similar results.
h. Quantification of aberrant acini in Panel g. The results are from 3 biologically independent samples. Two-tailed t-tests were used for statistical analyses of aberrant clones in n=3 biologically independent samples. ****, p = 0.000045; ***, 0.00011.
Extended Data Figure 8. RAP2 deletion contributes to aberrant acinus growth and tumorigenesis of MCF10A cells in a YAP/TAZ-dependent manner.
a. Representative images showing soft-agar assays of MCF10A cells. Overexpression of the constitutively active YAP-5SA (mutation of all the five LATS1/2 phosphorylation serines to alanines in YAP) strongly promotes anchorage-independent growth. Combined deletion of RAP2 and MST1/2, but not either group alone, also causes anchorage-independent growth of MCF10A cells. The image represents 3 biologically independent experiments with similar results. Scale bar, 35 mm.
b. RAP2 inhibits tumorigenicity of MST1/2-dKO MCF10A cells. MST1/2-dKO and RAP2-MST1/2-KO MCF10A were injected into NOD/SCID mice for tumor growth. Tumor weight on Day 32 after injection is presented as mean±SEM. ****, n=6 biologically independent xenografts, p = 0.000025. A two-tailed t-test was performed.
c. An image showing the tumor size. Only 3 very small xenografts were recovered from the initial 6 s.c. injections for MST1/2-dKO cells. The image represents 6 biologically independent xenografts for each group that were initially made in the NOD/SCID mice.
d. Immunohistochemistry staining with an antibody recognizing human HLA Class I. The results showed that only the acinus structures in the xenografts were formed by MCF10A cells. The stroma cells negative for HLA Class I were derived from the host mice. The image represents 2 biologically independent experiments with similar results. Scale bar, 50 μm.
e. Hematoxylin & eosin staining of the xenografts from the MST1/2-dKO MCF10A cells revealed that largely hypocellular connective tissue was observed as stroma from the host animals. In contrast, RAP2-MST1/2-KO xenografts showed MCF10A cell-derived acinus and duct structures exhibiting nuclear polymorphisms, irregular nuclear contour, hyperchromasia, prominent nucleoli (noted by the star in the image), and pathological mitosis (noted by the arrows in the image). The image represents 2 biologically independent experiments with similar results. Scale bar, 50 μm.
f. Western blot showing knockdown of YAP or TAZ by lentiviral shRNAs in the RAP2-MST1/2-KO MCF10A cells. shYAP#2 and shTAZ#1 were used for the xenograft studies. The image represents 2 independent experiments with similar results.
g. Knockdown of YAP/TAZ inhibits tumor growth of RAP2-MST1/2-KO MCF10A cells. Tumor weight on Day 32 is presented as mean±SEM. ****, a two-tailed t-test was performed, n=6, p = 0.000010.
h. The image of the xenografts in NOD/SCID mice, in which 6 biologically independent xenografts were generated for each group.
i. Western blot showing the expression of H-Ras-G12V mutant in MCF10A-T cells, which were generated by expression of the oncogenic mutant H-RAS. H-RAS expression activates ERK whereas RAP2 deletion has no effect on ERK. The comparable phosphorylation levels of ERK1/2 in WT and RAP2-KO MCF10A-T cells indicate that the difference in xenograft growth was not due to difference in ERK1/2 activity. The image represents two independent experiments with similar results.
j. The image of the MCF10A and MCF10A-T xenografts in nude mice, in which 6 biologically independent xenografts were generated for each group and yielded similar results.
Extended Data Figure 9. RAP2 deletion selectively promotes MCF7 malignancy at low stiffness in vivo.
a. Western blot showing the LOX overexpression in NIH3T3 fibroblasts. Ectopic LOX expression is known to promote cross-linking of ECM proteins and thus increase matrix stiffness28. The image represents 2 independent experiments with similar results.
b. RAP2 is involved in the xenograft growth enhancement caused by LOX-overexpressing fibroblasts. Xenografts were generated with co-injection of 0.4 × 106 fibroblasts (control and Flag-LOX-overexpressing) and 2.0 × 106 MCF7 cells. The xenografts were harvested on Day 6 and then were examined for collagen deposition and crosslinking and YAP/TAZ localization. LOX overexpression led to a woven-like structure of collagens in the xenografts based on the Sirius Red staining (red color for collagens) and Masson staining (blue color for collagens), and increased nuclear YAP/TAZ proteins. The image represents 3 biologically independent experiments with similar results. Scale bar, 50 μm.
c. LOX-induced tumor growth requires RAP2. Xenografts were generated with co-injection of 0.4 × 106 NIH3T3 and 2.0 × 106 MCF7 cells. The growth of the xenografts with different combinations of NIH3T3 and MCF7 cells was measured and shown as mean±SEM. RAP2A/B/C deletion promoted MCF7 tumor growth as well as masked the enhancement effects by the co-injected LOX expressing fibroblasts. **, two-way ANOVA test, (WT MCF7 + WT NIH3T3) cells vs (WT MCF7 +LOX-overexpressing NIH3T3) cells, n=6 biologically independent samples, p < 0.0027. ***, two-way ANOVA test, (sgRAP2 MCF7 + WT NIH3T3) cells vs (WT MCF7 + WT NIH3T3) cells, n=6 biologically independent samples, p = 0.002.
d. The tumor weights were shown as mean±SEM. *, two-tailed t-test, n=6 biologically independent xenografts, p=0.014 for (WT MCF7 + WT NIH3T3) cells vs (WT MCF7 +LOX-overexpressing NIH3T3) cells, or 0.029 (sgRAP2 MCF7 + WT NIH3T3) cells vs (WT MCF7 + WT NIH3T3) cells.
e. The elasticity/stiffness of the ‘soft’ and the ‘stiff’ semi-synthetic hyaluronan (HA)-derived gels was measured by atomic force microscopy. The results are presented as mean±SEM. The measurements were made for >55 times for each stiffness at each different time.
f. RAP2 inhibits expression of YAP/TAZ target gene CTGF and CYR61 at low stiffness, but not at high stiffness. Quantitative real-time PCR analyses of YAP/TAZ target genes CTGF and CYR61 in WT and sgRAP2 MCF7 cells (as in Extended Data Fig. 1g) cultured in vitro for 48 hours in the soft or the stiff HA gels. The relative mRNA levels are presented as mean±SEM. n=3 biologically independent samples. Two-tailed t-test was used for analyses. For CTGF, *, sgRAP2 vs. WT at 0.4 kPa, p=0.020; **, 0.4 kPa vs. 9.0 kPa for WT, p=0.0032. For CYR61, *, sgRAP2 vs. WT, p=0.025; **, 0.4 kPa vs. 9.0 kPa for WT, p=0.0033.
g. The image of the xenograft tumors. For the WT cells at 0.4 kPa, 8 independent xenografts were initially generated in nude mice. However, due to the animal deaths, only 6 xenografts were recovered. For WT cells at 9 kPa, 9 independent xenografts were generated. For sgRAP2 cells at both 0.4 kPa and 9 kPa, 8 independent xenografts were generated.
h. RAP2 deletion preferentially promotes MCF7 malignancy at low stiffness. The HE staining of the MCF7 xenografts. WT cells embedded in 0.4 kPa matrix produced mostly tubular and some cribriform structures while those embedded in 9 kPa matrix produced mostly solid nests, as well as more marked cellular pleomorphism and nuclear atypia. sgRAP2 cells already showed more malignant architecture and morphology at 0.4 ka, while at high stiffness (9 kPa) sgRAP2 and WT cells exhibited similar morphology. The image represents 3 biologically independent experiments with similar results. Scale bar, 50 μm.
i. Immunohistochemistry for YAP/TAZ in the xenografts. RAP2 deletion increased nuclear YAP/TAZ. The image represents 3 biologically independent experiments with similar results. Scale bar, 50 μm.
Extended Data Figure 10. RAP2 mediates regulation of ECM stiffness transcriptome by Hippo pathway.
a. MA-plot of WT cells in low stiffness vs. WT in high stiffness. Three biologically independent samples were assayed for each condition. Differentially expressed genes (adjusted p-value <0.1) are colored in red. P values for deferential expression were derived using Wald test and corrected using the procedure of Benjamini and Hochberg with default functions in DESeq2.
b. Dot plot showing expression level of AMOTL2 and LGR5. AMOTL2 and LGR5 are YAP/TAZ-dependent and stiffness-regulated genes. High and low denote high stiffness (40 kPa) and low stiffness (1 kPa). Data are presented as mean±SEM. n=3 biologically independent samples.
c. Venn diagram comparing down-regulated genes by YAP/TAZ knockout at high stiffness and up-regulated genes by LATS1/2 knockout at low stiffness. P-value: hypergenometric test. The result represents analyses of 3 biological replicates for each condition.
d. Venn diagram comparing up-regulated genes by YAP/TAZ knockout at high stiffness and down-regulated genes by LATS1/2 knockout at low stiffness. P-value: hypergeometric test. The result represents analyses of 3 biological replicates for each condition.
e. Venn diagram comparing overlapped genes in Panel c and down-regulated genes by low stiffness. P-value: hypergenometric test. The result represents analyses of 3 biological replicates for each condition.
f. Venn diagram comparing overlapped genes in Panel d and up-regulated genes by low stiffness. P-value: hypergeometric test. The result represents analyses of 3 biological replicates for each condition.
g. Venn diagram comparing overlapped genes in Panel e and up-regulated genes by RAP2 knockout at low stiffness. P-value: hypergenometric test. The result represents analyses of 3 biological replicates for each condition.
h. Venn diagram comparing overlapped genes in Panel f and down-regulated genes by RAP2 knockout at low stiffness. P-value: hypergeometric test. The result represents analyses of 3 biological replicates for each condition.
Supplementary Material
Acknowledgements
K.C.L., A.W.H., and S.W.P. are supported by the T32 GM007752 training grant, A.K. is by T32AR060712, and J.K.P by F32HL126406. K.L.G. is supported by grants from NIH (CA196878, CA217642, GM51586, DEO15964) as is A.J.E. (R21CA217735, R01CA206880).
A. J. E is also supported by NSF grant 1463689, and A.K. is supported by NSF graduate research fellowship program and ARCS/Roche Foundation Scholar Award in Life Science.
Footnotes
Competing interests
Dr. Kun-Liang Guan is a co-founder of Vivace Therapeutics.
Reference:
- 1.Humphrey JD, Dufresne ER & Schwartz MA Mechanotransduction and extracellular matrix homeostasis. Nat Rev Mol Cell Biol 15, 802–812, doi: 10.1038/nrm3896 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meng Z, Moroishi T & Guan KL Mechanisms of Hippo pathway regulation. Genes Dev 30, 1–17, doi: 10.1101/gad.274027.115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halder G, Dupont S & Piccolo S Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat Rev Mol Cell Biol 13, 591–600, doi: 10.1038/nrm3416 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Aragona M et al. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 154, 1047–1059, doi: 10.1016/j.cell.2013.07.042 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Codelia VA, Sun G & Irvine KD Regulation of YAP by mechanical strain through Jnk and Hippo signaling. Curr Biol 24, 2012–2017, doi: 10.1016/j.cub.2014.07.034 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wada K, Itoga K, Okano T, Yonemura S & Sasaki H Hippo pathway regulation by cell morphology and stress fibers. Development 138, 3907–3914, doi: 10.1242/dev.070987 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Dupont S et al. Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183, doi: 10.1038/nature10137 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Cherfils J & Zeghouf M Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev 93, 269–309, doi: 10.1152/physrev.00003.2012 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Wei SC et al. Matrix stiffness drives epithelial-mesenchymal transition and tumour metastasis through a TWIST1-G3BP2 mechanotransduction pathway. Nat Cell Biol 17, 678–688, doi: 10.1038/ncb3157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benham-Pyle BW, Pruitt BL & Nelson WJ Cell adhesion. Mechanical strain induces E-cadherin-dependent Yap1 and beta-catenin activation to drive cell cycle entry. Science 348, 1024–1027, doi: 10.1126/science.aaa4559 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Rooij J et al. PDZ-GEF1, a guanine nucleotide exchange factor specific for Rap1 and Rap2. J Biol Chem 274, 38125–38130 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Monteiro AC et al. Trans-dimerization of JAM-A regulates Rap2 and is mediated by a domain that is distinct from the cis-dimerization interface. Mol Biol Cell 25, 1574–1585, doi: 10.1091/mbc.E14-01-0018 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gloerich M et al. Rap2A links intestinal cell polarity to brush border formation. Nat Cell Biol 14, 793–801, doi: 10.1038/ncb2537 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Carloni V, Romanelli RG, Pinzani M, Laffi G & Gentilini P Focal adhesion kinase and phospholipase C gamma involvement in adhesion and migration of human hepatic stellate cells. Gastroenterology 112, 522–531 (1997). [DOI] [PubMed] [Google Scholar]
- 15.Zhang X et al. Focal adhesion kinase promotes phospholipase C-gamma1 activity. Proc Natl Acad Sci U S A 96, 9021–9026 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plouffe SW et al. Characterization of Hippo Pathway Components by Gene Inactivation. Mol Cell 64, 993–1008, doi: 10.1016/j.molcel.2016.10.034 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng Z et al. MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat Commun 6, 8357, doi: 10.1038/ncomms9357 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Myagmar BE et al. PARG1, a protein-tyrosine phosphatase-associated RhoGAP, as a putative Rap2 effector. Biochem Biophys Res Commun 329, 1046–1052, doi: 10.1016/j.bbrc.2005.02.069 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Machida N et al. Mitogen-activated protein kinase kinase kinase kinase 4 as a putative effector of Rap2 to activate the c-Jun N-terminal kinase. J Biol Chem 279, 15711–15714, doi: 10.1074/jbc.C300542200 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Taira K et al. The Traf2- and Nck-interacting kinase as a putative effector of Rap2 to regulate actin cytoskeleton. J Biol Chem 279, 49488–49496, doi: 10.1074/jbc.M406370200 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Qiao Y et al. YAP Regulates Actin Dynamics through ARHGAP29 and Promotes Metastasis. Cell Rep 19, 1495–1502, doi: 10.1016/j.celrep.2017.04.075 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Porazinski S et al. YAP is essential for tissue tension to ensure vertebrate 3D body shape. Nature 521, 217–221, doi: 10.1038/nature14215 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Q et al. Ingestion of Food Particles Regulates the Mechanosensing Misshapen-Yorkie Pathway in Drosophila Intestinal Growth. Dev Cell 45, 433–449 e436, doi: 10.1016/j.devcel.2018.04.014 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Debnath J, Muthuswamy SK & Brugge JS Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30, 256–268 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Yoh KE et al. Repression of p63 and induction of EMT by mutant Ras in mammary epithelial cells. Proc Natl Acad Sci U S A 113, E6107–E6116, doi: 10.1073/pnas.1613417113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dawson PJ, Wolman SR, Tait L, Heppner GH & Miller FR MCF10AT: a model for the evolution of cancer from proliferative breast disease. Am J Pathol 148, 313–319 (1996). [PMC free article] [PubMed] [Google Scholar]
- 27.Serban MA, Scott A & Prestwich GD Use of hyaluronan-derived hydrogels for three-dimensional cell culture and tumor xenografts. Curr Protoc Cell Biol Chapter 10, Unit 10 14, doi: 10.1002/0471143030.cb1014s40 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levental KR et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 139, 891–906, doi: 10.1016/j.cell.2009.10.027 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen JH et al. Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat Mater 13, 979–987, doi: 10.1038/nmat4051 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tse JR & Engler AJ Preparation of hydrogel substrates with tunable mechanical properties. Curr Protoc Cell Biol Chapter 10, Unit 10 16, doi: 10.1002/0471143030.cb1016s47 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Kaushik G, Fuhrmann A, Cammarato A & Engler AJ In situ mechanical analysis of myofibrillar perturbation and aging on soft, bilayered Drosophila myocardium. Biophys J 101, 2629–2637, doi: 10.1016/j.bpj.2011.10.042 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, doi: 10.1093/bioinformatics/bts635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anders S, Pyl PT & Huber W HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169, doi: 10.1093/bioinformatics/btu638 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550, doi: 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dennis G Jr. et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4, P3 (2003). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The source data for the graph representations can be found in the online version of the paper. For uncropped images of Western blot data, see Supplementary Figure 1. The RNA sequencing data is available in GEO DataSets with the accession number GSE98547. All other data that support the findings of this study are available upon request from the corresponding author.