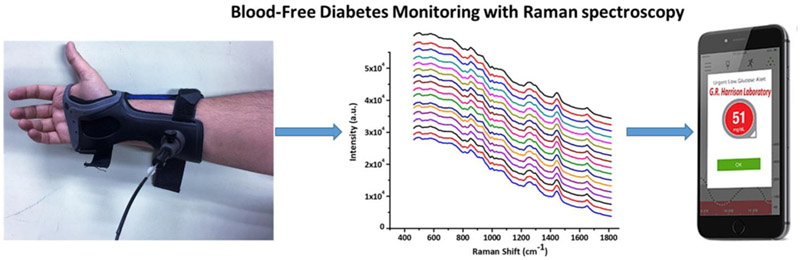
Abstract
Optical monitoring of blood glucose levels for non-invasive diagnosis is a growing area of research. Recent efforts in this direction have been inclined towards reducing the requirement of calibration framework. Here we are presenting a systematic investigation on the influence of variation in the ratio of calibration and validation points on the prospective predictive accuracy of spectral models. A fiberoptic probe coupled Raman system has been employed for transcutaneous measurements. Limit of agreement analysis between serum and partial least-square regression predicted spectroscopic glucose values has been performed for accurate comparison. Findings are suggestive of strong predictive accuracy of spectroscopic models without requiring substantive calibration measurements.
Keywords: Diabetes, Raman spectroscopy, glucose sensing, partial least square regression
Introduction
Diabetes mellitus (DM) caused by impairment in the glucose metabolism is a worldwide epidemic, it is estimated that by the year of 2035 there will be approximately 592 million people affected by this disease [1,2]. DM is caused by a defect in insulin secretion and results in a change in the blood glucose levels. Therefore, DM is diagnosed or monitored by measuring levels of glucose in the blood. It is important to frequently monitor blood glucose levels and maintain it within the prescribed range to avoid secondary complications including strokes, heart attacks, blindness and coma [3,4]. In addition, DM patients with an established diagnosis of insulin-dependent diabetes (all type-I, and many type-II) require frequent glucose measurements for monitoring and adjustment of insulin doses. Currently, DM patients use the hand-held glucose-monitoring device to monitor the BG levels. The device requires small blood sample (<1 μL) obtained by a ‘finger-pricking’ followed by electrochemical sensing using a portable ‘glucometer’. Unfortunately, the procedure is inconvenient and painful; thus, resulting in poor patient compliance [5,6]. Other clinical tests require either blood or interstitial fluids for measurement of glucose levels. In this regard, it is important to develop a device that can enable non-invasive monitoring of blood-glucose levels.
Significant research has been undertaken in the quest of identifying a novel diagnostic tool for managing DM [7–9]. This research can be broadly categorized into electrochemical and optical sensing approaches. Electrochemical sensing of blood glucose levels utilizes enzymatic or non-enzymatic methods. Optical methods can be further subdivided into fluorophores (fluorescein) and non-fluorophores based approaches [10]. Recent advancement in the instrumentation and analysis methods has provided a great momentum to non-fluorophores based approaches such as near-infrared (NIR) absorption spectroscopy, optical coherence tomography (OCT), photoacoustic spectroscopy and polarization spectroscopy for glucose sensing [11,12,7,13]. Our laboratory has pioneered the applications of NIR Raman spectroscopy (RS) for measuring blood glucose, urea, lactic acid, and cholesterol levels [14–19]. RS is based on inelastic scattering of light, where changes in the molecular polarizability lead to a shift in the wavelengths associated with specific chemical bonds and this, in turn, provides a molecular fingerprint with both qualitative and quantitative information. Other benefits of RS include non-destructive and label-free nature, no sample preparation requirement, minimum water interference and real-time evaluation. Overall methodology of Raman spectroscopic glucose sensing involves transcutaneous spectral measurement followed by feature extraction using multivariate chemo metric modeling approaches such as partial least square regression (PLSR), principal component regression (PCR) or support vector regression (SVR) followed by cross-validation. Typically, a calibration model is developed by tying up the blood glucose values with corresponding Raman spectra. Leave-one-out or K-fold cross-validation methods are employed to establish the accuracy of spectral models against the reference values. Our long-term efforts in realizing the goal of non-invasive Raman glucose sensing have been evaluated by clamping studies in animals and oral glucose tolerance test (OGTT) in humans [17,20,21]. Recently Shao et al. has shown that by focusing laser directly in to the blood stream of mouse models, problem of strong background can be avoided to some extent [22]. Shih et al. has demonstrated feasibility of non-invasive transcutaneous measurement of blood glucose using dogs [20]. Scholtes-Timmerman et al. reported a human trial involving 186 subjects using a system with fairly large spot size (8 mm). PLS modeling coupled with cross validation yielded encouraging results [23]. Weber et al. developed a tabletop confocal Raman system to collect signals from interstitial fluid and tested this set-up on a group of 35 patients[24]. Even though results of these studies have provided substantial evidence in support of RS for monitoring glucose level one of the major hindrances in successful translation of spectroscopic glucose sensors for routine clinical usage is the unavailability of appropriate exit criterion for analyzing the unknown samples [21]. Efforts have been made towards developing a universal calibration model. However, it is limited by non-linearity of glucose concentration, site, and inter-personal-variance in terms of skin color and thickness, basal metabolic rates and hydration status [25]. To overcome these issues, ‘patient-specific’ calibration strategies with minimum blood reference values are preferred. Our recent efforts have been inclined towards minimizing the requirement of blood reference values for calibration [26]. Here we are presenting a systematic investigation of the predictive accuracy dependence of spectroscopic models on the ratio of calibration and validation points. Three different calibration strategies using 50%, 30% and 18% of the total data as a reference have been employed and prediction accuracies of spectroscopic models are evaluated. In contrast to earlier studies where the accuracy of spectroscopic models was mostly accessed by ‘leave-one-out cross-validation’, here we have employed an ‘independent test prediction’ approach to obtain a realistic prospective read-out [17,21,20]. Also, an accurate reference method based on serum glucose has been used for better correlation. Findings of the study will be helpful in not only reducing the requirement of unnecessary finger-pricks and associated discomfort but also providing an appropriate validation to RS methods with respect to the clinical OGTT.
Materials and methods
Oral glucose tolerance testing
This pilot study was conducted at the Clinical Research Center of MU-Institute of Clinical and Translational Sciences, University of Missouri – Columbia. The study protocol was approved by Health Sciences Institutional Review Board (Protocol number: 2002948) at University of Missouri – Columbia. Written informed consent prior to participation was obtained from each of the participants of the study. A total of 23 healthy non-diabetic and non-pregnant volunteers aged 18 years or older (median age: 33 years) were chosen for the study. Of the 20 qualified volunteers, 45% were male and remaining 55% were female. Fasting blood glucose (FBG) values less than or equal to 125mg/dL were set as an inclusion criterion for the subjects in the study. The blood glucose levels of the subjects were checked prior to the study and those qualified were given a standard glucose drink, used in clinics for OGTT (75 g of glucose-rich beverage Dextrose, Azer Scientific Inc.). The spectral acquisition was initiated immediately after the drink and thereafter every 10 minutes for 160 minutes. Serum glucose measurements were performed concurrently every 10 minutes using YSI glucose analyzer. Finger prick measurements were also performed every 30 minutes using Accucheck™ blood glucose meters to corroborate YSI-measurements.
Raman set-up and spectral acquisition
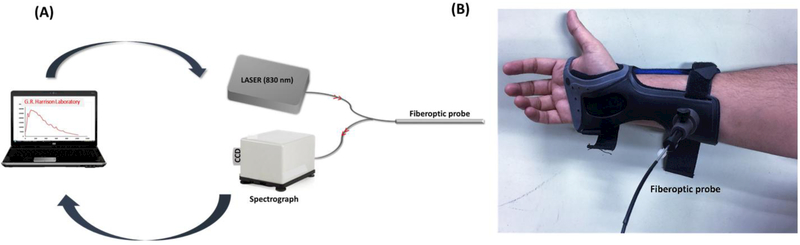
In contrast to our previous free-space glucose Raman instruments, a fiber-optic probe coupled glucose Raman unit was used in the present study, Fig. 1A [17,20]. A wrist support with a small hole to hold the probe at the same tissue spot over the course of the experiment was fabricated (Fig. 1B). This wrist support minimizes unwanted light interference and allows Raman measurements under the ambient room light. A diode laser (Process Instruments) of 830 nm is used as an excitation source. Excitation light is launched into the central fiber of the probe. Fiber background signals are removed by incorporating a short pass filter at the other end of the fiber. The filtered excitation beam is delivered into the tissue through a sapphire ball lens and back scattered signal is collected by the same lens. The Rayleigh scattered light is eliminated by a long pass filter in front of the six collection fibers surrounding one central excitation fiber, and the filtered Raman signal is delivered to the imaging spectrograph. Collection fibers are aligned as a line at the spectrograph entrance and the signal is dispersed by an imaging spectrograph (LS785, Princeton Instruments). Dispersed spectrum is detected by a back-illuminated deep-depletion CCD (Spec-10:400BR-XTE, Princeton Instruments). The Raman instrument for glucose monitoring is built inside of a portable cart (84cm × 48cm × 100cm) for easy transfer between laboratory and clinical research center. Spectral preprocessing steps included standard normal variate (SNV) correction to remove scaling differences from the spectra followed by subtraction of 3rd order polynomial function for objective baseline correction.
Fig. 1.
(A) Schematic of portable Raman system employed in the study and (B) Fiber optic probe-holding adapter
Data analysis
Partial least square regression (PLSR) and Clarke error grid analysis using MATLAB based in-house codes was employed. Blood glucose values obtained by serum measurements and corresponding Raman spectra at 17-time points during the course of OGTT of 20 patients was recorded. Data from 2 patients weren’t included due to poor spectral quality. Three different calibration models using 50, 30 and 18% corresponding roughly to 9/17, 5/17 and 3/17 spectra of individual OGTT were developed. Rest of 50, 70 and 82% data were used as independent test data. Appropriate ranks of PLS calibration ranging from 3 to 5 were used in accordance with the general assumption of having samples numbers at least 3 times to the rank of PLS calibration [27].
Results and Discussion
In the present study, a calibration model is developed by tying up serum glucose values with corresponding Raman spectra. Typical transcutaneous Raman spectra acquired from one of the subjects during the course of OGTT is shown in Fig. 2A. Corroborating with the spectral profiles noted in our previous studies, major spectral features are suggestive of collagen, lipids and structural proteins [26,28]. Strongest peak at 1445 cm−1 is assigned to CH2 stretching, other features at 859 cm−1, 938 cm−1 (collagen), 1004 cm−1 (phenylalanine), 1273 cm−1, 1302 cm−1 (amide III) and 1655 cm−1 (amide I) were also observed. Even though near infrared (NIR) excitation is used, as the spectra were obtained transcutaneous, large fluorescence envelope is observed, Fig. 2A. Expectedly, small Raman bands related to glucose are overwhelmed by the background and thus necessitating requirement of multivariate analysis to identify these variations with respect to time and serum glucose concentration. Mean OGTT curve along with standard deviation with every 10-minute read-out is shown in Fig. 2B. OGTT is a standard diabetes screening procedure, where a subject is given a glucose-rich drink to induce a substantive rise in the blood glucose level. Blood samples are then withdrawn using an intravenous (IV) line at specific time intervals (10 mins) to investigate the rate of clearance of glucose from the blood and therefore to infer the effectiveness of the subject’s insulin-based glucose regulation mechanism. Standard bell-shaped OGTT curve of subjects shown in Fig. 2B suggests healthy nature of the participants in the study.
Fig. 2.
(A) Typical transcutaneous Raman spectrum acquired over the course of OGTT. Spectrum at time points 0, 100 and 160 minutes are marked as T0, T100 and T160, respectively. (B) Mean OGTT curve along with standard deviation obtained from all the participants of the study
Choosing an appropriate ‘local calibration model’ is a prerequisite for employing regression analysis in spectroscopic quantitative measurements. Raman spectra were acquired at 17-time points (0 to 160 minute) on 20 patients during OGTT. Random sampling approach was employed and in the first step, 50% of the total data was used for developing a calibration model. Rest of data were used as ‘independent test data’. Clarke error grid analysis (EGA), approved by food and drug administration (FDA) is one of the widely utilized methods for evaluating clinical accuracy of new method/devices in estimating blood glucose with respect to a reference method [29,30]. The Clarke error grid plots with independent test data predictions are shown in Fig. 3. As per the FDA, guidelines zone, A and B in the plots correspond to the area with ± 20% error rate and are clinically acceptable. Rest of the areas C, D, E are clinically irrelevant and considered to be potentially dangerous. As shown in Fig. 3A, most of the ‘independent test data’ fall into the clinically acceptable zone A (71%) and zone B (26%) region of the error grid. Further, we reduced the number of calibration point to 30% and 18% of the total data using random sampling approach. As can be seen from Fig. 3B and C, a total of 97 and 96% of the test data for these two calibrations strategies, respectively are still in the clinically relevant zones of the Clarke error grid. Overall findings of EGA are suggestive of high accuracy of spectroscopic models for predicting analogues glucose values. As shown in Table 1, clinically valid predictions obtained with three calibration strategies further supports applicability of transcutaneous spectroscopic measurements for glucose monitoring.
Fig. 3.
Clarke error grid plots showing independent test prediction accuracy of spectroscopic models with different number of calibration points. (A) 50% (B) 30% (C) 18%
Table 1.
Summary of predictive accuracy of spectroscopic models with different calibration strategies
| Spectral Model | Independent predictions in A region of the Clarke error grid |
Independent predictions in B region of the Clarke error grid |
Correlation coefficient (R values) |
Bias among reference and predicted glucose values (mg/dl) |
|---|---|---|---|---|
| 50% calibration + 50 % validation |
71.05% | 26.31% | 69.36% | −1.02 |
| 30% calibration + 70 % validation |
70.23% | 27.90% | 65.61% | 1.23 |
| 18% calibration + 82 % validation |
60.65% | 36.06% | 53.33% | −4.47 |
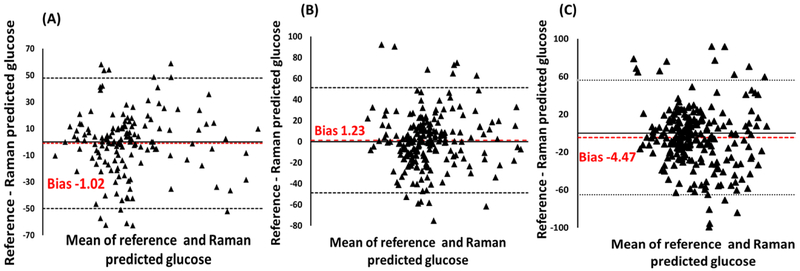
The measurement of a variable with two different methods will always have some degree of error as neither can provide unequivocally same value. An accurate estimation of the degree of agreement can provide valuable information about the efficiency of both methods. The correlation coefficient (r) and coefficient of determination (r2) are two widely utilized parameters to access the linear relationship between two measurements [31]. As shown in Table 1, an average correlation value of ~63% was observed. However, this value is not reliable as it measures only the strength of relation or percentage common variance between two variables, not the agreement between them. Limit of agreement (LOA) is quantified by defining mean and standard deviation(s) of the difference between two methods. The difference between the measurements (A-B) is plotted against the mean of difference (A+B)/2. Two different methods are considered to be in good agreement if ~95% data points are in the ± 2 standard deviations (1.96) of the mean difference. Bland-Altman (B&A) plots are generally employed to perform ‘the analysis of difference’ between two quantitative measurements [32]. The analysis of difference plots using YSI-analyzer values as a reference method against Raman spectroscopic predictions are shown in Fig. 4. Parameter ‘bias’ is an indicator of the average difference in the predicted and reference glucose values. As shown in Table 1, spectral models with 50, 30 and 18% calibration points have ‘bias’ of −1.02, 1.23 and −4.47 mg/dl, respectively. Most of the points (>95%) are in the ‘range of agreement’ suggesting analogous blood glucose values predicted by Raman measurements. Overall, the results obtained are an indicator of the high accuracy of spectroscopic models in predicting independent test samples even with minimum number of calibration points.
Fig. 4.
Bland Altman plot demonstrating degree of agreement between reference and Raman predicted values with different number of calibration points. (A) 50% (B) 30% (C) 18%. (LOA-limit of agreement)
As glucose constitutes less than 0.1% of human tissue by weight, spectral differences induced by concentration changes are mitigated by skin variations during transcutaneous measurements [3,25]. Therefore, use of multivariate algorithms to identify the subtle changes in a temporal manner becomes a necessity. Regression methods coupled with different cross-validation strategies have been employed largely to link the spectral changes with glucose concentration. However, even though cross-validation is widely used its validity for the larger data set and prospective predictions is debatable [33]. As the probability of having an equivalent pair of spectrum and reference blood glucose values increases in larger data set, the root mean square error calculation can turn overly optimistic. Therefore, partitioning of samples into independent calibration and validation sets is a preferred approach. Although it hasn’t been studied in great detail with respect to glucose sensing, available reports suggest it to be a heuristic task [33]. Random sampling approach for selecting a representative sub set of calibration and validation samples from the pool of data is commonly employed because of its simplicity and the fact that group of data randomly extracted from a larger set follows the statistical distribution of the entire set [34]. The main objective of spectroscopic glucose sensing methods is to replace/reduce the finger pricks or blood withdrawals required for glucose monitoring. However, all the spectroscopic devices require a calibration framework to predict unknown concentrations. Minimizing the number of calibration steps needed for continuous monitoring using non-invasive devices can help in comprehending the ultimate goal. In the present study, we have evaluated prediction accuracy of Raman spectroscopic sensors with different number of calibration points during OGTT. Spectra were randomly divided into three different calibration sets with 50, 30 and 18% of the total data set. In all three cases, ~97% of the independent test data falls into the clinically acceptable area of the Clarke error grid. The rise in number of predictions in zone B of the error grid with reducing calibration points is expected due to non-linear nature of spectroscopic signatures and decreasing variance cover. Further, the fact that random sampling approach employed in the present study doesn’t always guarantee the inclusion of borderline samples in calibration models, could be another factor. The bias between the predicted and reference value is a good indicator of the predictive power of calibration models. As per the ISO15197 guidelines a difference of 15 mg/dl (for values <100 mg/dl) or 15% (for values ≥100 mg/dl) is considered to be acceptable accuracy of new devices [35]. As shown in Bland Altman plots and Table 1, the bias between predicted and reference blood glucose values was minimal for spectral model with highest number of calibration points (50%) followed by 30 and 18%. These values were in the acceptable range, suggesting small number of calibration points can also lead to a good predictive accuracy, provided accurate blood referencing methods are used.
Overall findings of the present study further provided evidence in support of the prospective application of Raman spectroscopic methods for non-invasive glucose monitoring. In contrast to earlier studies, by utilizing an accurate reference method we have successfully demonstrated the strong predictive accuracy of spectroscopic models with minimum calibration information. Further efforts are underway to examine the predictive accuracy in DM patients. Our future studies will also focus on improving the accuracy by replacing random sampling methods with new algorithms for more effective partitioning of calibration and validation sets to improve the predictive ability and robustness of the resulting model.
Acknowledgments
This work is supported by NIH P41-EB015871-30 and Samsung Advanced Institute of Technology (Seoul, South Korea). PTCS acknowledge support from U01-NS090438-03, R21-NS091982-01, R01-HL121386-03, the Singapore-MIT Alliance 2 (Cambridge, MA, USA), the Biosym IRG of Singapore-MIT Alliance Research and Technology Center (Cambridge, MA, USA), and Hamamatsu Corporation (Hamamatsu City, Japan). AU thanks Professor Elizabeth J. Parks (Department of Nutrition and Exercise Physiology, and Division of Gastroenterology and Hepatology, School of Medicine, University of Missouri- Columbia) for providing the YSI analyzer and Nhan T Le (Department of Nutrition and Exercise Physiology) for helping us with calibration and use of the instrument. Intramural Funding for this work was provided by Office of Medical Research, School of Medicine, University of Missouri- Columbia.
Footnotes
Conflict of interest
Authors declare no conflict of interest.
References
- 1.American Diabetes A Economic costs of diabetes in the U.S. in 2012. Diabetes care. 2013. April;36(4): 1033–46. PubMed PMID: . Pubmed Central PMCID: 3609540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes care. 2004. May;27(5): 1047–53. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 3.Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001. December 13;414(6865):813–20. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 4.Zimmet P, Alberti KG, Shaw J. Global and societal implications of the diabetes epidemic. Nature. 2001. December 13;414(6865):782–7. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 5.American Diabetes A. Standards of medical care in diabetes--2009. Diabetes care. 2009. January;32 Suppl 1:S13–61. PubMed PMID: . Pubmed Central PMCID: 2613589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olansky L, Kennedy L. Finger-Stick Glucose Monitoring: Issues of accuracy and specificity. Diabetes care. 2010;33(4):948–9. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khalil OS. Spectroscopic and clinical aspects of noninvasive glucose measurements. Clin Chem. 1999. February;45(2):165–77. PubMed PMID: . [PubMed] [Google Scholar]
- 8.Bruen D, Delaney C, Florea L, Diamond D. Glucose Sensing for Diabetes Monitoring: Recent Developments. Sensors. 2017. August 12; 17(8). PubMed PMID: . Pubmed Central PMCID: 5579887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C, Zhao XL, Li ZH, Zhu ZG, Qian SH, Flewitt AJ. Current and Emerging Technology for Continuous Glucose Monitoring. Sensors. 2017. January 19;17(1). PubMed PMID: . Pubmed Central PMCID: 5298755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pickup JC, Hussain F, Evans ND, Rolinski OJ, Birch DJS. Fluorescence-based glucose sensors. Biosens Bioelectron. 2005 2005/June/15/;20(12):2555–65. [DOI] [PubMed] [Google Scholar]
- 11.Vashist SK. Non-invasive glucose monitoring technology in diabetes management: a review. Anal Chim Acta. 2012. October 31;750:16–27. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 12.Wang HC, Lee AR. Recent developments in blood glucose sensors. J Food Drug Anal. 2015. June;23(2): 191–200. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khalil OS. Non-invasive glucose measurement technologies: an update from 1999 to the dawn of the new millennium. Diabetes Technol Ther. 2004. October;6(5):660–97. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 14.Kong CR, Barman I, Dingari NC, Kang JW, Galindo L, Dasari RR, et al. A novel non-imaging optics based Raman spectroscopy device for transdermal blood analyte measurement. AIP advances. 2011. September;1(3):32175 PubMed PMID: . Pubmed Central PMCID: 3217291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berger AJ, Koo TW, Itzkan I, Feld MS. An enhanced algorithm for linear multivariate calibration. Anal Chem. 1998. February 01;70(3):623–7. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 16.Berger AJ, Koo TW, Itzkan I, Horowitz G, Feld MS. Multicomponent blood analysis by near-infrared Raman spectroscopy. Appl opt. 1999. May 01;38(13):2916–26. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 17.Enejder AM, Scecina TG, Oh J, Hunter M, Shih WC, Sasic S, et al. Raman spectroscopy for noninvasive glucose measurements. J Biomed Opt. 2005. May-Jun;10(3):031114 PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 18.Hanlon EB, Manoharan R, Koo TW, Shafer KE, Motz JT, Fitzmaurice M, et al. Prospects for in vivo Raman spectroscopy. Phys Med Biol. 2000. February;45(2):R1–59. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 19.Koo TW, Berger AJ, Itzkan I, Horowitz G, Feld MS. Reagentless blood analysis by near-infrared Raman spectroscopy. Diabetes Technol Ther. 1999. ; 1(2): 153–7. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 20.Shih WC, Bechtel KL, Rebec MV. Noninvasive glucose sensing by transcutaneous Raman spectroscopy. J Biomed Opt. 2015. May;20(5):051036 PubMed PMID: . Pubmed Central PMCID: 4330710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pandey R, Paidi SK, Valdez TA, Zhang C, Spegazzini N, Dasari RR, et al. Noninvasive Monitoring of Blood Glucose with Raman Spectroscopy. Acc Chem Res. 2017. February 21;50(2):264–72. PubMed PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shao J, Lin M, Li Y, Li X, Liu J, Liang J, et al. In vivo blood glucose quantification using Raman spectroscopy. Plos One. 2012;7(10):e48127 PubMed PMID: . Pubmed Central PMCID: 3485012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scholtes-Timmerman MJ, Bijlsma S, Fokkert MJ, Slingerland R, van Veen SJ. Raman spectroscopy as a promising tool for noninvasive point-of-care glucose monitoring. J Diabetes Sci Technol. 2014. September;8(5):974–9. PubMed PMID: . Pubmed Central PMCID: PMC4455378. Epub 2014/07/20. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lundsgaard-Nielsen SM, Pors A, Banke SO, Henriksen JE, Hepp DK, Weber A. Critical-depth Raman spectroscopy enables home-use non-invasive glucose monitoring. Plos One. 2018;13(5):e0197134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lipson J, Bernhardt J, Block U, Freeman WR, Hofmeister R, Hristakeva M, et al. Requirements for calibration in noninvasive glucose monitoring by Raman spectroscopy. J Diabetes Sci Technol. 2009. March 01;3(2):233–41. PubMed PMID: . Pubmed Central PMCID: 2771519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spegazzini N, Barman I, Dingari NC, Pandey R, Soares JS, Ozaki Y, et al. Spectroscopic approach for dynamic bioanalyte tracking with minimal concentration information. Sci Rep. 2014. November 12;4:7013 PubMed PMID: . Pubmed Central PMCID: 4894421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi D, Berger AJ. Chemical concentration measurement in blood serum and urine samples using liquid-core optical fiber Raman spectroscopy. Appl opt. 2007. April 01;46(10):1726–34. PubMed PMID: . Epub 2007/03/16. eng. [DOI] [PubMed] [Google Scholar]
- 28.Lui H, Zhao J, McLean D, Zeng H. Real-time Raman spectroscopy for in vivo skin cancer diagnosis. Cancer res. 2012. May 15;72(10):2491–500. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 29.Clarke WL, Cox D, Gonderfrederick LA, Carter W, Pohl SL. Evaluating Clinical Accuracy of Systems for Self-Monitoring of Blood-Glucose. Diabetes care. 1987. Sep-Oct;10(5):622–8. PubMed PMID: . English. [DOI] [PubMed] [Google Scholar]
- 30. Stockl D, Dewitte K, Fierens C, Thienpont LM. Evaluating clinical accuracy of systems for selfmonitoring of blood glucose by error grid analysis - Comment on constructing the “upper A-line”. Diabetes care. 2000. November;23(11):1711–2. PubMed PMID: . English. [DOI] [PubMed] [Google Scholar]
- 31.Giavarina D Understanding Bland Altman analysis. Biochem Med. 2015;25(2): 141–51. PubMed PMID: . Pubmed Central PMCID: 4470095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bland JM, Altman DG. Measuring agreement in method comparison studies. Stat methods Med res. 1999. June;8(2):135–60. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 33.Galvao RK, Araujo MC, Jose GE, Pontes MJ, Silva EC, Saldanha TC. A method for calibration and validation subset partitioning. Talanta. 2005. October 15;67(4):736–40. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 34.Daszykowski M, Walczak B, Massart DL. Representative subset selection. Anal Chim Acta. 2002 2002/September/10/;468(1):91–103. [Google Scholar]
- 35.Freckmann G, Schmid C, Baumstark A, Rutschmann M, Haug C, Heinemann L. Analytical Performance Requirements for Systems for Self-Monitoring of Blood Glucose With Focus on System Accuracy: Relevant Differences Among ISO 15197:2003, ISO 15197:2013, and Current FDA Recommendations. J Diabetes Sci Technol. 2015. July;9(4):885–94. PubMed PMID: . Pubmed Central PMCID: PMC4525642. Epub 2015/04/16. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]