Abstract
Pancreatic ductal adenocarcinoma (PDA) is characterized by epithelial mutations in KRAS and prominent tumor-associated inflammation, including macrophage infiltration. But knowledge of early interactions between neoplastic epithelium and macrophages in PDA carcinogenesis is limited. Using a pancreatic organoid model, we found that the expression of mutant KRAS in organoids increased i) ductal to acinar gene expression ratios, ii) epithelial cells proliferation, and iii) colony formation capacity in vitro, and endowed pancreatic cells with the ability to generate neoplastic tumors in vivo. KRAS mutations induced a pro-tumorigenic phenotype in macrophages. Altered macrophages decreased epithelial Pigment Epithelial Derived Factor (PEDF) expression and induced a cancerous phenotype. We validated our findings using annotated patient samples from The Cancer Genome Atlas (TCGA) as well as in our human PDA specimens. Epithelium-macrophage cross talk occurs early in pancreatic carcinogenesis where KRAS directly induces cancer-related phenotypes in epithelium, and also promotes a pro-tumorigenic phenotype in macrophages, in turn augmenting neoplastic growth.
Keywords: Pancreas cancer, KRAS, macrophage, PEDF
INTRODUCTION
Several epithelial cancers1 are associated with inflammation, especially pancreatic ductal adenocarcinoma (PDA)2, 3. PDA remains the most fatal cancer and by 2030 is projected to rank among the top three leading causes of cancer-related death 4. Therefore, improvement in our understanding of the molecular mechanisms causing PDA is highly needed to address our unmet medical need of achieving more effective treatments.
PDA is characterized by ubiquitous mutation of KRAS in epithelium and a prominent tumor-associated inflammation in the tumor microenvironment. The latter is important as inflammation and immune cell infiltrates are linked both to predisposition to cancer formation5 as well as to prognosis6 in PDA. Among immune cells in the tumor microenvironment, macrophages in particular, have been shown to contribute to disease progression and to responses to chemotherapy7–11. Nonetheless, macrophages can undergo phenotypic alterations (plasticity) during the neoplastic process, by losing their anti-tumorigenic/cytotoxic capacities and gaining pro-tumorigenic characteristics12–14. This plasticity is influenced by signals from surrounding tissue15, including cancerous epithelium16. However, the temporal interplay between transforming epithelial cells and surrounding macrophages in PDA remains peculiar. We hypothesize that epithelial KRAS mutations, indispensable oncogenic events in PDA, induce pro-tumorigenic phenotypes in macrophages, providing a permissive microenvironment to further promote epithelial cancerization. Here we test our hypothesis using organoid models to study early events in PDA carcinogenesis. We verify the relevance of our findings in human PDAs using a gene expression database and archived tissue.
Material and Method
Pancreas Organoid Development
First, pancreatic ducts were isolated with collagenase-based media from the pancreases of 10 week old B6 mice, and deposited in liquid Matrigel. We seeded the mixture in plates and added organoid culture medium (see supplementary table 1), as was previously described17. The first passage was done 21 days later, and was repeated every 10 days.
Transfection and sorting
The lentiviral expression clone eGFP-KRAS G12D (R700-M05-658) was obtained through the RAS Program at the Frederick National Laboratory for Cancer Research. Lentiviral particles were prepared as previously described18. Briefly, HEK-293T cells were grown to 40% confluence in the absence of antibiotics. Then the lentiviral, psPAX2, and VSVG plasmids were mixed in a 3:3:1 ratio in Optimem and Fugene for 15 minutes. The mixture was then added to HEK-293T cells in a humidified atmosphere supplemented with 5.1% CO2 at 37°C. After 48 hours, the supernatant was collected and centrifuged and then passed through a 0.45 um filter.
Organoids were mechanically disrupted by pipetting and then centrifuged. The pellet was suspended in the viral medium supplemented with 8 ug/mL polybrene overnight in a humidified atmosphere supplemented with 5.1% CO2 at 37°C. The following day, organoid suspensions were harvested and washed three times. Organoids were seeded on organoid culture medium supplemented with 2 ug/ml blastacydin (gibco by life technologies). After 7 days, organoids were visualized for GFP expression (Fig. 1). The transfection efficiency was about 50%, with approximately half of organoids containing GFP+ cells. To further purify for the KRAS carrying organoids, GFP sorting was performed. Organoids were dissociated using the TrypLE Express (gibco by life technologies #2016-03) dissociating buffer. The dissociated organoids were pelleted by centrifuging for 5 minutes. Dissociated cells were resuspended at 1 × 107 cells/ml in 1 ml sorting medium (DMEM with 2% FBS). GFP positive cells were sorted by a MoFlo Astrios sorter and were reseeded on the plates in organoid media. Purified KRAS carrying organoids were grown for 7 days. We confirmed by light microscopy that 90% of organoids contained GFP+ cells. Organoids were passaged 10 days after sorting; these organoids were used for the histology and functional experiments described below. Control organoids were treated only with empty lentivirus (not carrying KRAS vector) and are referred as wild type (WT) throughout the study.
Figure 1.
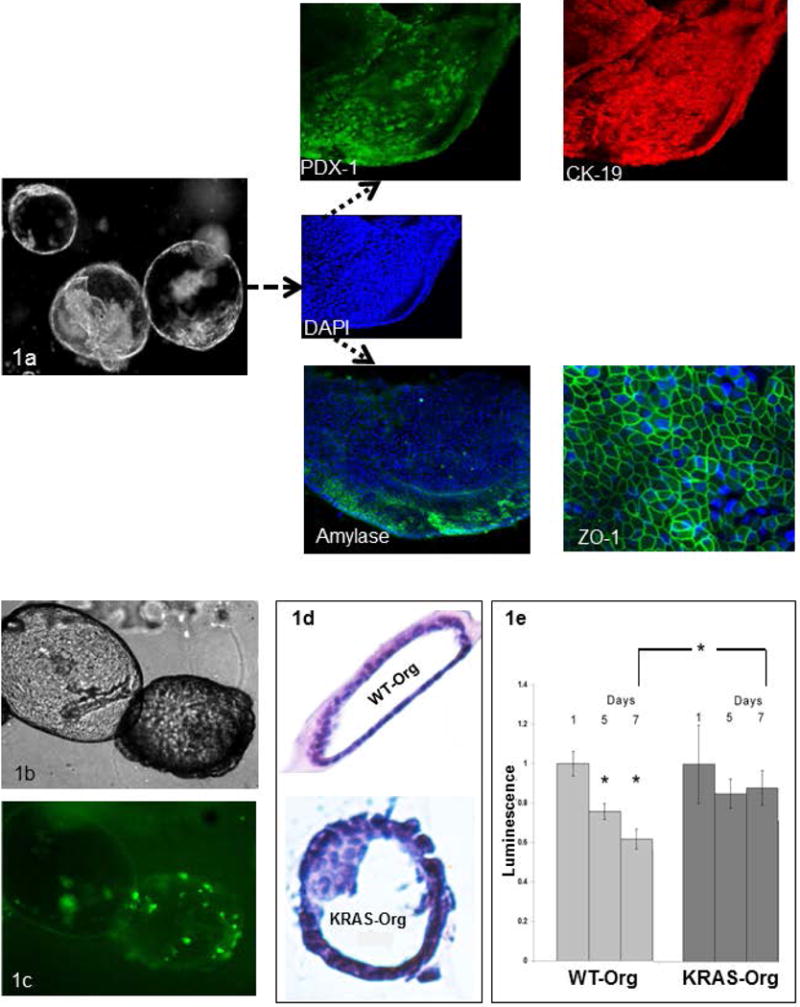
Pancreas organoids retain the characteristics of the tissue of their origin, and show neoplastic changes and increased survival upon transfection with KRAS mutation; (a) Morphology of day 21 organoids on the left; higher magnification image of one organoid; inset, stained with DAPI (blue), PDX1/green, CK19/red, acinar marker (Amylase/green) and tight junction protein (ZO-1/green); (b) Morphology of the transfected pancreatic organoids with human KRASG12D mutation-carrying vector 10 days after transfection (c) tagged with GFP; (d) H&E staining of a wild-type (WT) organoid compared to mutated KRAS carrying organoid with enlarged, disorganized, crowded and hyperchromatic nuclei along with multilayering of cells, neoplastic changes typically preceding frank invasive carcinoma (ie, features of pancreatic intraepithelial neoplasia).; (e) Cell viability in WT and mutated KRAS carrying organoids at day 5 and 7 days compared to day 1, bar graphs show means ± SE of n=3 experiments, asterisk indicates p<0.05.
Organoid H&E
Agarose organoid cell blocks were made using 1% agarose (cell culture grade) dissolved in isosmotic PBS. Once the temperature was lowered to 50°C, molten agarose was carefully mixed with WT and KRAS mutant organoids to obtain an agarose-organoid-suspension. The suspension was quickly used to overlay 2% compact agarose contained in Eppendorf tubes and allowed to solidify at 4°C. Solidified agarose cell blocks were extracted from the Eppendorf tubes by tapping the tubes upside down on a flat surface. All agarose cell blocks were sliced in half, placed in a tissue cassette, and fixed with 10% formalin. After overnight fixing, agarose cell blocks were processed for paraffin embedding and transversely cut into 5 μm and 6 μm thick sections, respectively. Multiple sections were stained with hematoxylin and eosin (H&E) stain.
Cell Culture and Co-culture Experiments
RAW 264.7 (ATCC® TIB71™) were cultured with Dulbecco’s Modified Eagle’s Medium (Catalog No. 302002) with addition of fetal bovine serum to a final concentration of 10%. Corning Permeable Plates (Product # 3450) were used for co-culture experiments. Macrophage RAW 264.7 cells when grown to 80% confluence were seeded to the insert well in equal numbers, and the organoid cells were grown in the organoid media on the plate well within the Matrigel. Matrigel was placed in the plate so as to avoid any physical contact between Matrigel and the insert. Macrophage RAW 264.7 cells were grown in a supplemented basic media and incubated as the epithelial organoid cells were. To study dynamic changes in the expression of M1 and M2 markers in the macrophages, RAW 264.7 cells in the insert were collected for protein as well as mRNA expression analysis after 48 hours of co-culture. Organoids in the matrigel in the plates were analyzed for pancreatic neoplastic changes in the epithelium in response to macrophage co-culture as described below. Alterations in ductal/acinar expression were studied in the pancreas organoids using RT-qPCR of acinar-ductal markers (RT-PCR section).
Mouse neoplastic cells (PanIN)19 (a kind gift of Dr. Paul Grippo, UIC, Chicago) were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) (Sigma, Saint Louis, MO) and 1% penicillin – streptomycin (Life Technologies, Grand Island, NY) in a water jacketed incubator at 37 °C with 5.1% CO2. PanIN cells (0.1 × 106) were mono- and co-cultured in 6 – well plates. RAW 264.7 cells (0.4 × 106 per well) were co-cultured in Corning Transwell® Permeable Supports (3450, Corning, Manassas, VA). All cells were seeded and allowed to adhere for 24 hrs. The next day medium was replaced for all cells and treated with either vehicle or 20 μM erlotinib (Selleckchem, Houston, TX) for 24 hrs. For immunoblotting, all cells were washed with Hank’s Balanced Salt Solution (Corning, Manassas, VA) and harvested in RIPA buffer containing 1 mM Na3VO4 (New England Biolabs, Ipswich, MA), 5 mM NaF (New England Biolabs, Ipswich, MA) and 1:200 dilution of protease inhibitor cocktail set III (Sigma, Saint Louis, MO) (Western blot section).
Primary macrophages
Spleen from 6–8 weeks old B6 mice were collected and placed in 2% FBS/PBS solution. Spleen was squeezed, passed through a 40um filter to 50ml tube, and centrifuged at 1200 rpm for 10 minutes at 4 °C. After supernatant removal, prewarmed RBC lysis buffer was added to the pellet, mixed well, and kept at room temperature for 5 min. PBS was added to the conical tube, and the solution underwent another centrifugation step. Supernatant was removed and and cells were resuspended with PBS. Cells were counted, underwent repeat centrifugation and were resuspended, this time in condition medium (CM). (RPMI containing 10% of FBS and 20% of L929 conditioned media). Cells were cultured in 6-well tissue culture plates (5× 106 cells/well) in CM at 37oC and 5% CO2 in an incubator. After 3 days of culture, non-adherent cells were removed as the entire medium was replaced on day 3 to enrich for adherent cells. This was repeated again on day 6 and adherent cells were harvested on day 7. On day 8 the cells were treated with CM (control), and organoids medium vs. supernatants from WT and KRAS organoids for 24 and 48 hours. Cells were collected for RNA isolation.
Western blots
Immunoblotting in macrophages was done for Arg-1 and iNOS to differentiate pro-tumorigenic (M2) from cytotoxic (anti-tumorigenic, M1) macrophages. For immunoblotting in organoids, the medium was removed and organoids were washed with PBS several times while in Matrigel. Corning Cell Recovery Solution was added to Matrigel, and samples were kept on ice for an hour until the Matrigel dissolved. Organoids were isolated by centrifugation from the dissolved Matrigel. Tris-triton buffer (Bio-world CAT#4202031) with protease inhibitor and sonication were used to liquefy the organoid pellets. Protein concentration was measured and 10 ug total protein plus 20 ul of laemmli sample buffer (Bio-RAD) was prepared. Similarly, protein quantification was performed for PanIN cells. Samples were loaded onto SDS gels and transferred to nitrocellulose blotting Membrane (GE Healthcare life Sciences CAT#10600004). The membranes were incubated overnight at 4 °C with primary antibodies, washed, and further incubated with corresponding secondary antibodies, and developed using ECL solutions. Antibodies included Arginase-1 (Cell Signaling #9819), iNOS (BD Transduction Laboratories# 610332), PEDF (H125; SC-25594), p-ERK (Cell Signaling #9101), and phosphorylated EGFR (Cell Signaling #3777), total EGFR (Cell Signaling #4267), GAPDH (Cell Signaling #5174) and rabbit secondary (Cell Signaling #7074).
RT-PCR and qPCR analysis
RNA was extracted using the RNeasy Mini RNA Extraction Kit (Qiagen). cDNA was prepared using the high capacity cDNA reverse transcription kit from the manufacturer (Applied Biosystems, Foster City, CA). Real time PCR was done on an Applied Biosystems 7900HT Fast apparatus using primers (IDT, Coralville, IA) and Fast Sybr green (Applied Biosystems). The quantitative analysis was calculated from the ΔΔCt values normalized against β-actin which was used as a housekeeping gene.
Transcription of macrophage markers including Arg-1, (associated with a pro-tumorigenic/M2 state) as well as iNOS and IL-6 (for the cytotoxic/M1 phenotype) were determined and normalized to β-actin. We measured changes in macrophage gene expression after 24 to 48 hours of co-culturing with organoids (controls were matrigel and medium alone). Fold changes were calculated for co-culturing with KRASWT-Org and KRASG12D-Org, separately. The M2 to M1 ratio was calculated as the average fold expression change of differentially altered genes of the M2 gene (Arg-1) over the average fold expression changes of the M1 gene (IL-6).
Macrophage effects on gene expression profiles of organoids for acinar (Amylase), and ductal (CFTR) markers were measured. For this purpose we compared changes in expression levels of the acinar and ductal markers in organoids after co-culturing with RAW 264.7 macrophages, over controls (parallel organoid culture from the same batch with no macrophage incubation). The Ductal-Acinar (D-A) ratio was calculated as the average fold changes in gene expression of CFTR over average fold expression changes of Amylase. Primer sequences are listed in Supplementary Table 1.
Organoid Immunofluorescence
Organoid immunofluorescence staining was done on 8 well chamber slides (Nunc, Rochester, NY). Organoid pellets were isolated from the Matrigel using Corning Cell Recovery Solution as described above. The organoids were fixed with warm 4% PFA on chamber slides for one hour at RT and then permeabilized with 1% Triton-X100. The samples were blocked with a solution of 3% goat serum/1% BSA/0.2% Triton X-100 for 1 hour. Primary staining for the organoids was done overnight in a humidified chamber. This step was followed by incubation with the appropriate secondary antibodies conjugated to Alexa Flour 488. These were washed and further stained for DAPI and mounted using Flouromount aqueous mounting medium (Sigma Aldrich). Organoids were imaged using a Zeiss LSM 700 confocal microscope (Zeiss, Oberkochen, Germany). The following antibodies were used: Anti-PDX-1 (B-11) (sc-390792), Anti-Cytokeratin 19 (CK19) (ab#15463), Anti-Amylase (C-20) (sc-12821), Anti-ZO-1 (Invitrogen™ Catalog #:61-7300).
Xenograft model
WT and KRAS organoids that were in culture for 20 days were used for xenografting. Approximately 50,000 cells mixed in ice cold liquid Matrigel in a total of 200 microliter were subcutaneously injected into the flanks of 8-week-old female immunocompromised NOD/SCID/IL-2Ry null (NSG) mice following a protocol approved by the Institutional Animal Care and Use Committee of Rush University. Both flanks of mice were injected. Routine animal care was provided. Tumor growths were monitored. Animals were sacrificed after 10-12 weeks and Xenograft lesions were isolated. The tumors were fixed in paraffin. Paraffin-embedded tissue was used for hematoxylin and eosin (H&E) staining. Pictures were taken with an Olympus BX46 microscope, and were reviewed by a pathologist specialized in gastrointestinal diseases (B.B.S), who was blind to treatment groups.
Colony Formation assay
WT (Wild type) and mutant KRAS organoids (2×104-30×104 cells) were harvested and plated in 3.0 ml of top-layer medium consisting of supplemented DMEM/F12 and 0.4% UltraPure Agarose (Invitrogen, Grand Island, NY). The contents were then poured on top of a 2 ml bottom layer consisting of 0.6% agarose in 6-well tissue culture plates. Organoid medium was replaced weekly. After three weeks of incubation, the colonies were stained overnight with Nitro Blue Tetrazolium (Sigma, St. Louis, MO) and imaged next day using a VersaDoc imager (Bio-Rad, Hercules, CA). Each experiment was repeated 3 times from separate batches of organoids, and each time three parallel plates for each condition were run and counted.
Cell Viability Assay
Organoids were seeded at equal number of cells per well in Matrigel in opaque-walled 96 well plates in media without growth factors. The number of viable cells in culture based on their metabolic activity by ATP quantification was determined after optimization of the CellTiter-Glo® Luminescent Cell Viability Assay for our organoid model. In brief, appropriate amounts of CellTiter-Glo reagent (Promega, Madison, WI) from the Cell-Titer-Glo luminescent cell viability assay (Promega) were added to each well per the manufacturer’s protocol. After 10 min incubation at room temperature, luminescence was measured using a SpectraMax M5 fluorescence plate reader (Molecular Devices, Sunnydale, CA). Background luminescence from a well containing Matrigel only was subtracted from each subsequent well containing organoid samples. Measurements for days 1, 5,and 7 for each organoid group were normalized to day 1 of each respective group. The dynamic survival capacity of organoids during days 5, and 7 was measured, normalized to day 1 for each experiment, and compared between WT and mutant KRAS organoids.
Human specimens and staining
Human pancreatic cancer paraffin embedded tissue blocks (n=29) were obtained from the Rush University Medical Center pathology department in a de-identified fashion under an institutional IRB (#16051106). These blocks contained pancreatic samples from consecutive patients with pathology-proven pancreatic adenocarcinoma. Five um thick sections were cut from these blocks and used for Immunohistochemistry (IHC) and Immunofluorescence (IF). Briefly, for IHC, slides were baked at 60°C for 30 minutes and then placed in xylene for de-paraffinization. Slides were then rehydrated in serially graded alcohol. Endogenous peroxidase was inhibited with 0.3% H2O2, blocked with 3% Goat serum at room temperature for an hour, and then incubated overnight with the anti-PEDF antibody (Abcam/ab180711) diluted (1:200) in DAKO antibody diluent (Catalog # S0809). The sections were then incubated in horse-radish-peroxidase conjugated secondary antibody for 1 hour. Color was developed with the DAB kit (SK-100) using the manufacturer’s protocol. The slides were then counterstained with Hematoxylin and bluing solution for light field microscopy. For IF, antigen retrieval was done in Citrate buffer (DAKO target retrieval solution) using a steamer for 15 minutes. After blocking in 3% Donkey serum for 20 minutes, slides were incubated overnight with primary antibody [CK 19, Abcam/ab15463 – 1:100; CD 163, NCL-L-CD163/clone 10D6 – 1:200; CD 68; Abcam/ab955 – 1:200], followed by incubation with secondary fluorochrome tagged antibodies (Invitrogen, Alexa fluor Donkey Anti-Rabbit and Donkey anti mouse) for 1 hour (1:250). Sections were then stained with DAPI and mounted using mounting medium (DAPI Vectashield mounting, Catalog # H-1200).
Imaging and Quantification
Sequential sections (5 um) were cut from the human PDA samples (n=29) and used for 2 sets of staining experiments. One was co-stained with CK 19 and CD 68 (to assess for total macrophage infiltration within the tumor) and the other co-stained with CK 19 and CD 163 (to assess for the proportion of M2 subtype macrophage infiltration).
Cross sectional images for each sample were taken in such a way that each area on one slide (co-stained with CK 19 and CD 68) was approximately correlated to the same area on the next subsequent cut section (co-stained with CK 19 and CD 163). These images were quantified and hence allowed for an approximate evaluation of the correlation between the amount of pan-macrophage staining and M2 macrophage subtype staining within the same area of each tissue sample.
All microscope images were taken using Carl Zeiss confocal microscope (LSM 700). At least 4-5 (10× magnification) images were taken from different fields of each slide as mentioned above to represent the tumor. Images were quantified as a percent of the total area stained per high power filed (10×) and ImageJ software was used for quantification.
Study approval
All experiments involving the use of mice and human specimens were done under protocols approved by either the Institutional Animal Care and Use Committee or the IRB at Rush University of Medical Science.
Statistical Analysis
SPSS version 23 (SPSS, Inc., Chicago, IL, USA) was used for analyses. Numeric results (i.e., qPCR data, MTT proliferation assay, colony formation assay) are presented as mean +/− S.E., and were compared using two-tailed ANOVA tests. P<0.05 is considered significant and is noted by the asterisks in the figures as appropriate. GraphPad Prism was used to generate figures. Gene expression of 186 Human Pancreas Adenocarcinomas from The Cancer Genome Atlas (TCGA) was queried through cBioportal.org.
RESULTS
Pancreas organoids retain the characteristics of the tissue of their origin
We developed and propagated pancreatic organoids from untransformed wild-type murine pancreatic duct cells (Figure 1). After 21 days, spherical structures formed, with a layer of ductal epithelial cells in the periphery giving rise to budding spheres and new organoids, a process observed after each passage. As expected, and in line with their ductal origin, organoids had high expression of the ductal protein CK19. Consistent with their progenitor properties as stem-like cells, organoids expressed PDX1, and also contained cells from the acinar (+ for amylase) lineage albeit in lower proportions (Figure 1). Thus, pancreatic organoids recapitulate the physiological state of the tissue of origin, and consist of heterogeneous populations of epithelial cells17, 20, which allows in vitro assessment of acinar to ductal differentiation, an early event in pancreatic neoplastic transformation21.
KRAS mutation causes neoplastic structural and phenotypic changes in pancreatic organoids
Mutations in KRAS are early and ubiquitous events in PDA22. We infected pancreatic organoids with a GFP-tagged human KRASG12D mutation-carrying vector, the most common mutation present in human PDA23. Compared to wild-type (WT) organoids (KRASWT-org), KRASG12D-organoids, showed cell-crowding associated with disorganization of normal structures after 10 days (Figure 1b-c). On histology, KRASG12D-org showed multilayered epithelial cells in the periphery, disorganized hyperchromatic cells, and overlapping nuclei (Figure 1d). These are all features of early pancreatic intraepithelial neoplasms (PanIN)24, similar to recently reported PanIN driven organoids from a KRAS mouse model20. Cell viability, indicative of survival capacity, was significantly reduced in WT organoids over 5-7 days, while the viability of KRASG12D-org was not. Viability of KRASG12D-org at day 7 remained significantly higher than that in WT organoids (Figure 1e). The effect of KRAS mutation on cellular growth was measured by colony formation assay. KRASG12D-org formed more colonies than KRASWT-org, indicating these cells can grow anchorage-independently (Figure 2a). The increased survival and proliferation in oncogenic KRAS-carrying organoids was associated with augmented mitogenic signaling and phosphorylated ERK, a KRAS downstream effector that is critical for pancreatic carcinogenesis25 (Figure 2c). Subcutaneous injection of KRASG12D-org to immunocompromised mice formed PDA-like neoplastic lesions, while KRASWT-org formed normal-appearing ductal structures and no neoplastic lesions (3/4 vs. 0/6 respectively, Fisher’s exact P<0.05; Figure 2d-f).
Figure 2.
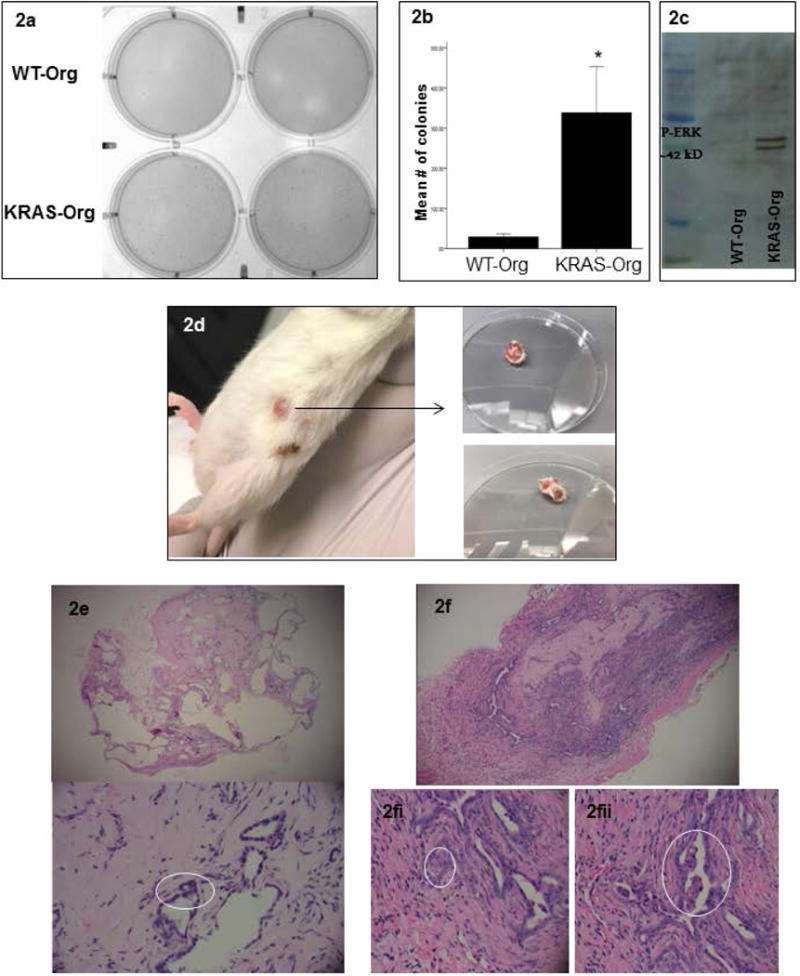
KRAS mutations in pancreas organoids induce cancer-related phenotypes in vitro and in vivo; (a) representative of a colony formation assay comparing WT and mutated KRAS carrying organoids; (b) quantification of colonies formed by WT and mutated KRAS carrying organoids, bar graphs show means ± SE of n=3 experiments, asterisk indicates p<0.05; (c) representative of a p-ERK western blot in the WT vs. mutated KRAS carrying organoids’ lysates; (d) KRAS carrying organoids formed tumors in NSG mice 10 weeks after subcutaneous injection; right, tumors after resection; (e) histology of the tissue formed 12 weeks after WT organoids injection: Top panel is a 4× view showing benign pancreatic ductal structures with variable degrees of dilatation. No internal papillary formations are seen. Bottom panel is a 10× view of benign ductal structures. Nuclei are small and bland (circled). (f) 4× view of a tumor formed from KRAS carrying organoids in NGS mice after 10 weeks showing back-to-back gland formation, densely cellular ductal structures with infiltrative edges representing an early invasive phenotype; (2.f.i) 10× view of an area of cells with large nuclei that are not uniformly bland, showing noticeable nucleoli; (2.f.ii) 10× view of luminal papillary structures, reminiscent of pancreatic intraepithelial neoplasia lesions.
KRAS mutations in epithelial organoids affect macrophage phenotype, and altered macrophages promote cancerous behavior in epithelial organoids
We hypothesized that early neoplastic events in pancreatic epithelium alter macrophage phenotype in their microenvironment, which can facilitate PDA progression13, 26. We modeled interactions between epithelium and macrophage compartments in vitro, and established co-cultures of organoids and macrophages. Wild-type or mutant pancreatic organoids were incubated with murine macrophages (RAW 264.7). Changes in the expression of Arg-1 (marker of pro-tumorigenic states/M2), iNOS and IL-6 (markers of cytotoxic/anti-tumor phenotypes, M1) were analyzed during 48h and normalized to the housekeeping gene β-actin. Co-culturing with KRASG12D-org shifted the macrophages towards M2, with an increase in M2/M1 ratios (Figure 3a). Similar pattern was observed when primary macrophages were incubated with supernatant of KRASG12D-org vs. KRASWT-org, where KRASG12D-org resulted in a significantly higher expression of Arg-1 (Figure 3b). The effect of KRAS carrying organoids on macrophages was verified at the protein level, where the RAW 264.7 expression of Arginase-1 increased after incubation with KRASG12D-org, while iNOS was unchanged (Figure 3c). Thus, epithelial KRAS mutations switch macrophages towards a pro-tumorigenic phenotype, which may favor neoplastic transformation, and which is what we studied next.
Figure 3.
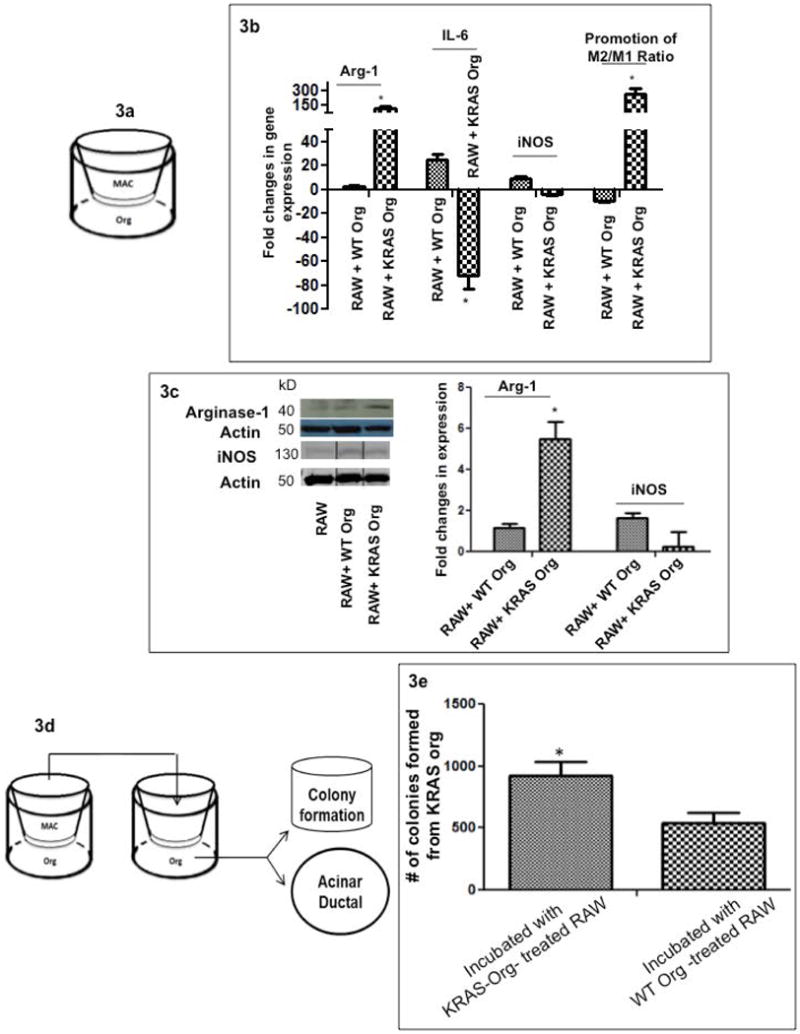
Epithelial-Macrophage interactions; (a) Pancreas epithelial organoids and mouse macrophages (RAW 264.7) were co-cultured; Fold changes in gene expression of Arg-1, iNOS and IL-6, normalized to the housekeeping gene (β-actin) expression in RAW 264.7 over 48 hours co-culturing with WT and mutated KRAS carrying organoids, compared to incubation with control (only Matrigel and medium). Macrophage incubation with KRAS carrying organoids significantly increased M2/M1 ratios with time, bar graphs show means ± SE of n=3 experiments; (b) Fold changes in gene expression of Arg-1, iNOS and IL-6, normalized to the housekeeping gene (β-actin) expression in primary macrophages over 48 hours incubated with supernatant of WT vs. mutated KRAS carrying organoids. Macrophage incubation with KRAS carrying organoids significantly increased Arg-1 (M2 marker) by time, bar graphs show means ± SE of n=3 experiments (c) RAW 264.7 expression of Arg-1 and iNOS proteins after 48 hours incubation with control (Matrigel and organoid medium), WT and KRAS carrying organoids; representative experiment is shown; (d) schematic of the in vitro model to assess the functional effect of phenotypic alterations in macrophages on the epithelial organoids. Equal numbers of RAW 264.7 cells were first incubated for 48 hours with WT and KRAS carrying organoids; next the pre-treated macrophages were transferred to a new co-culture system with epithelial organoids to assess acinar-ductal markers (Supplemental Figure 3) and colony formation capacity (Figure 3e); (e) comparison of number of colonies formed in KRASG12D-org after incubation with macrophages pre-treated with WT and KRAS carrying organoids, bar graphs show means ± SE of n=3 experiments.
First we assessed macrophage effects on acinar (Amylase) and ductal (CFTR) expression and on ductal-Acinar (D-A) ratios, an indirect estimate for acinar-ductal differentiation21. We used CFTR as a ductal marker as its expression occurs only during differentiation of undifferentiated pancreatic epithelium to ductal cells27. Further, in our organoids, CFTR expression is not as abundant at baseline as the other ductal markers (i.e., CK19) (Supplementary Figure 1), making it a suitable marker for monitoring ductal differentiation. Incubation with RAW 264.7 increased the D-A ratio in pancreatic organoids (Supplementary Figure 2).
Next we examined whether the phenotypic alterations in macrophages caused by incubation with KRASG12D-org, could promote the expression of ductal markers in epithelial organoids. Macrophages were incubated with KRASG12D or WT-organoids. Treated macrophages were transferred to new wells where pancreatic organoids were seeded (Figure 3d). Macrophages pre-treated with KRASG12D-org increased D-A in WT-organoids, as compared to those pretreated with WT-organoids (Supplementary Figure 3). We then determined whether these effects resulted in the promotion of cancer-related outcomes in the organoids. Macrophages incubated with KRASG12D induced greater numbers and more prominent colonies in KRASG12D-org compared to MAC-WT-org (Figure 3e). Overall, macrophages exposed to KRAS-org shifted towards expressing pro-tumorigenic markers, and promoted a ductal fate and cancerous phenotype in the epithelium.
In order to determine, in pancreatic cancer patients, the association between ductal phenotypes with the herein studied macrophage’s markers, we turned to The Cancer Genome Atlas (TCGA)28. We observed that CK19 (a ductal marker) expression was inversely correlated with the expression of iNOS and IL6R, cytotoxic genes (Supplementary Figure 4a-b).
We then evaluated the association of tumor-associated macrophages with epithelial ductal invasion in 29 human PDA samples. First, slides were co-stained and quantified for CK 19 (ductal marker) and CD 163 staining (M2 macrophage subtype marker). Areas of higher expression for CK 19/higher ductal invasion showed a higher CD 163 expression/M2 macrophage infiltration as shown in representative images (Figure 4a). This staining was quantified and a significantly positive correlation between the two markers (correlation co-efficient, r=0.65) was found (Figure 4b).
Figure 4.
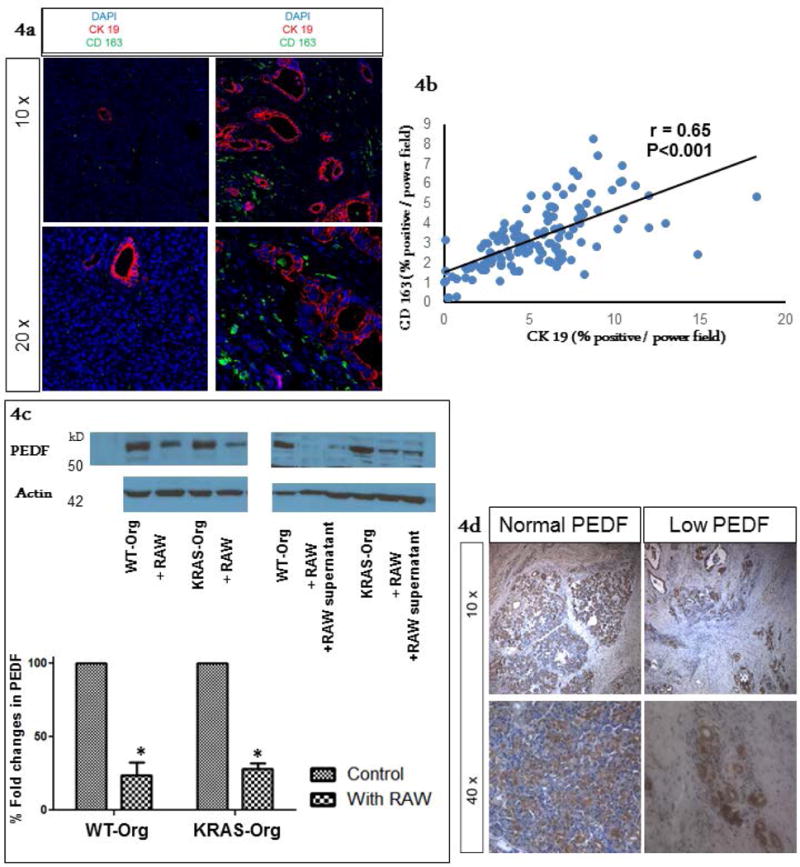
Ductal invasion association with pro-tumorigenic macrophage infiltration, and loss of epithelial PEDF. (a) Archived human PDA specimen (#29) were co-stained with CK19 (pancreatic ductal marker) and CD 163 (M2 macrophage subtype marker). Representative images show cancerous areas with an increase in CK 19 staining, associated with an increase in peri-tumoral M2 macrophage subtype infiltration in Right panels, while Left panels represent an area of low CK19 expression with a low M2 marker concentration. Low (10×) and high (20×) magnification views of similar density areas within the same sample are depicted in top and bottom rows, respectively. (b) Four to five random images from each specimen were quantified for both CK 19 and CD 163 staining and a correlation coefficient was calculated. As shown by the graph there was a significantly positive correlation between areas with CK 19 staining and CD 163 staining (r = 0.65), suggesting that ductal invasion is associated with pro-tumorigenic (M2 subtype) macrophage infiltration. (c) PEDF expression in organoids was not affected by KRAS mutations while it was downregulated after incubation with macrophages (left panel; a representative western blot), an effect likely exerted by a macrophage secretory factor (right panel; a representative western blot), bar graphs show means ± SE of n=3 experiments, asterisk indicates p<0.05. (d) Immunohistochemical staining for PEDF expression was done on the subsequent sections of the above samples. As shown in representative images, we observed normal expression of PEDF in peri-tumoral tissue while expression of PEDF was low within areas of more malignant ductal structures, where we had often observed high infiltration of M2 type macrophages.
To assess the approximate proportion of M2 subtypes to total macrophage infiltration in areas of ductal invasion, sequential sections from the same samples were co-stained with CD 68 (pan-macrophage marker) and CK 19. As expected we observed a significant positive correlation (correlation co-efficient, r= 0.8) between total macrophage infiltrations with areas of higher ductal expression (Supplementary Figure 5a). Next, we imaged and quantified CD 68 and CD 163 staining in similar fields from the two subsequent sections of each sample (Supplementary Figure 5b). CD 68 and CD 163 “index” was calculated by dividing CK 19 percent area staining with its corresponding CD 68 or 163 percent area staining within the same image. There was a significant positive correlation (correlation co-efficient, r= 0.71) between the CD 68 index and CD 163 index (Supplementary Figure 5c). These data show that as CK 19 expression within an area of the tumor increases so does the macrophage infiltration, a higher proportion of which are consistent with the M2 subtypes. This further confirms a feed-forward interaction between transformed ductal epithelium and macrophages in human PDA10, 29.
Macrophages reduce epithelial Pigment Epithelial Derived Factor (PEDF)
Decreased PEDF expression has been previously associated with enhanced macrophage infiltration30 and PDA progression19. We have observed enhanced growth in our pancreatic organoids due to KRAS mutations, which was exacerbated by the presence of macrophages. Here we assessed modulation of PEDF expression during early neoplastic transformation, in response to KRAS mutations and/or macrophages.
KRAS mutations did not decrease PEDF expression, but macrophages did downregulate epithelial PEDF expression, an effect likely exerted by a macrophage secretory factor (Figure 4c). Furthermore, PEDF tissue staining in our archived human PDA samples revealed that PEDF expression is lowest in areas of increased ductal expression, corresponding to enhanced tumor-associated macrophages (Figure 4d). Analysis of TCGA confirmed an inverse association of ductal (CK19) and PEDF expression in PDA (Supplementary Figure 6).
Macrophages reduce epithelial PEDF via EGF/EGFR pathway
We further explored possible mechanisms that underlie macrophage-induced epithelial PEDF reduction. Tumor-associated macrophages can secrete epidermal growth factor (EGF), which activates epithelial EGFR pathway, and exerts pro-tumorigenic effects on the neoplastic epithelium 31, 32. We first screened EGF and EGFR expression in our co-cultured macrophages and pancreas organoids (WT) by quantitative PCR. EGF was highly expressed in macrophages, but not in epithelial organoids, whereas EGFR was highly expressed in epithelial organoids (Figure 5a), suggesting possible involvement of macrophage EGF-epithelial EGFR axis in delivering macrophage effects’ on the epithelium in our model. This pattern was not present for VEGF/VEGFR, another growth factor involved in pro-tumorigenic effects of macrophages33; No significant changes were observed in the expression of VEGF and VEGFR between macrophages and organoid epithelium (Figure 5a). Next, we directly tested whether macrophages promote epithelial PEDF downregulation through the EGF/EGFR axis, using pancreas neoplastic epithelial (PanIN) cells. We first verified down-regulation of the expression of PEDF in the co-culture of PanIN cells with macrophages (Figure 5b). EGFR inhibitor (erlotinib) was used to block EGFR in epithelial cells. The effect of erlotinib on EGF/EGFR signaling within pancreas neoplastic epithelial cells (PanIN cells) was verified by reduced expression of EGFR phosphorylation (Figure 5c). Erlotinib was then added to the co-culture of macrophages with PanIN cells, and the influence of EGFR inhibition on PEDF expression was measured. Significant recovery of epithelial PEDF expression was observed upon EGFR inhibition (Figure 5c).
Figure 5.
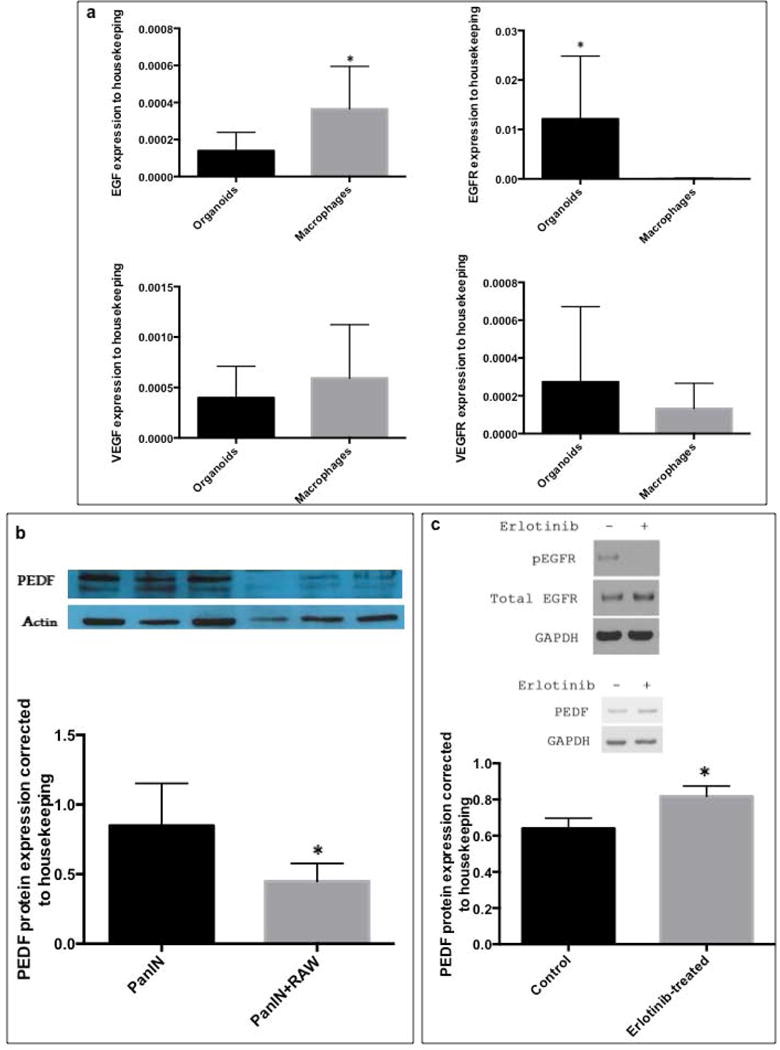
Macrophage induced loss of epithelial PEDF ; (a) Organoids and RAW 264.7 macrophages were collected after 48 hours of co-culture; RNA was isolated, and expression of EGF, EGFR, VEGFC and VEGFR3 genes were measured by quantitative PCR, normalized to expression of a housekeeping gene (β-actin) and compared between organoid and macrophages; bar graphs show means ± SE of n=6 experiments, asterisk indicates p<0.05 (b) Representative graph of the expression of PEDF and Actin proteins in PanIN cells after 48 hours incubation with RAW 264.7, PEDF expression was normalized to Actin and compared between mono- and co-cultured PanIN cells with macrophages; bar graphs show means ± SE of n=3 experiments, asterisk indicates p<0.05 (c) Upper panel shows effect of Erlotinib on phosphorylated EGFR to total EGFR ratios in PanIN cells after 24 hours: Erlotinib at 20 μM reduced EGFR phosphorylation; Lower panel is a representative immunoblot of mono and co-cultured PanIN cells with RAW 264.7 macrophages in the absence and presence of 20 μM erlotinib. PEDF expression increased more in Erlotinib treated cells than in vehicle/DMSO treated cells; the bar graphs show means ± SE of n=6 experiments, asterisk indicates p<0.05.
DISCUSSION
Using an organoid culture, we showed that the most common KRAS mutation in human PDA results in epithelial neoplastic transformation. Successful implementation of organoid-macrophage co-culture enabled us to study dynamic interactions between transformed epithelium and macrophages. Bidirectional signaling between epithelium and macrophages was noted, where epithelial KRAS promoted pro-tumorigenic expression patterns in macrophages that in turn augmented cancerous phenotypes in the epithelium.
PDA almost invariably harbors mutations in the KRAS gene, among which KRASG12D is the most common. Introduction of the KRASG12D mutation in our pancreas organoids induced neoplastic changes as evidenced by histologic transformation and increased proliferation and survival rates of the organoids, as well as their capacity to from PDA-like neoplasms in vivo. These results are consistent with prior animal data that highlighted the critical role of oncogenic KRAS in PDA formation34; induction of pancreas-specific KRASG12D in mice promoted epithelial neoplastic transformation similar to the observation in our organoid model.
Epithelial KRAS activation may also contribute to the inflammatory stroma by upregulating signaling between epithelium and microenvironment34, 35. Previous studies indicated that KRAS-dependent production of chemoattractants and adhesion molecules could be involved in macrophage recruitment in PDA36–38. However effects of epithelial KRAS on the macrophage’s phenotype have not been adequately studied. We found that epithelial KRAS mutations switch macrophages towards a pro-tumorigenic phenotype, assessed by macrophage’s gene expression pattern as well as their capacity to promote colony formation and ductal to acinar expression ratio in the organoids. Association of transforming ductal epithelium with a macrophage shift from cytotoxic to pro-tumorigenic phenotype underlines an active interaction between cancerous epithelium and macrophages in disease progression26. This is clinically relevant given our observation that expression of a ductal marker (CK19) was inversely correlated with cytotoxic genes in human PDAs from TCGA database and positively correlated with infiltrating tumor-associated macrophages in our archived human PDA tissues. iNOS and IL6 associate with cytotoxic capacity of macrophages. However their overall role in tumorigenesis could be cell, environment, time and concentration dependent39–42. For example, while epithelial source of iNOS could help tumor progression43, iNOS from stromal cells (i.e., macrophages), especially at high levels, is mostly cytotoxic against tumor cells44, 45. Similar pattern is found in pancreas cancer where increased stromal iNOS exerts anti-tumor activity46.
Paracrine signaling that drives the effects of transforming epithelium on the macrophages needs to be further studied. Recently, in a KRAS model of PDA, tumor-associated macrophages increased with ductal metaplasia and neoplastic transformation, effects that appeared to depend on IL-13 released from cancer cells47. On the other hand, several pathways have been proposed to mediate the effects of tumor-associated macrophages on cancerous epithelium10, 13, 29. Here we found that expression of PEDF, a tumor suppressor in PDA, decreases in the transforming epithelium not due to KRAS mutation but upon exposure to macrophages. We also observed an inverse association of epithelial PEDF with ductal expression and macrophage infiltration in human PDAs. This is consistent with our previous report, where the absence of PEDF signaling in the KRASG12D model of PDA was associated with an enhanced macrophage infiltration into the tumor tissue19. Reduction in endogenous PEDF levels by the microenvironment was previously reported in prostate cancer30. Mechanistically, we found EGF/EGFR signaling as the pathway via which macrophages influence PEDF levels in pancreas neoplastic epithelial cells. No direct link between EGF/EGFR axis and PEDF has been reported previously. It is possible that EGFR directly or indirectly, via its several downstream effects, influences PEDF expression. EGFR activation can induce matrix metalloproteinases (MMPs)48, 49 that are associated with pancreas tumor invasion50. Previous studies showed MMPs can downregulate PEDF in pancreas51. VEGF upregulation, which could also occur in response to EGFR activation, has been shown to induce PEDF degradation as well51. The link between macrophage induced EGFR activation in the epithelium and downregulation of PEDF in pancreas cancer needs further investigations, and may explain the previously reported association of increased macrophage infiltration with decreased PEDF levels in more invasive human PDAs19. Altogether, these findings suggest that PEDF downregulation in epithelium may occur in response to the microenvironment. Reduction in epithelial PEDF can further facilitate PDA progression through several already established anti-tumorigenic mechanisms (i.e., pro-apoptotic, anti-proliferative, anti-angiogenic effects) 52, 53. Future gain-of-function experiments could establish PEDF as a potential target to offset pro-tumorigenic effects of macrophages on the epithelium.
Our study has limitations. We used RAW 264.7 cells, which has been widely utilized to study macrophage plasticity in vitro10. However we verified similar phenotypic changes in response to mutant KRAS carrying organoids using primary macrophages. We used changes in proportion of ductal/acinar gene expression only as an estimate of acinar to ductal differentiation. In this proof-of-concept study we showed epithelium-macrophage interactions during PDA development. Targeting macrophages in PDA requires a better understanding of their plastic role during tumor progression54. Identification of signaling underlying the bidirectional epithelium-macrophage cross-talk could reveal novel targets for PDA therapy.
Summary
Pancreas epithelial KRAS mutation can induce cancer-related phenotypes in vitro, and tumor formation in vivo. KRAS also induces a pro-tumorigenic phenotype in macrophages, further promoting cancerous behavior in epithelium. Epithelial PEDF expression, possibly via the EGFR pathway, decreases in response to macrophages. This is relevant in human PDA where a significant association between tumor-associated macrophages, ductal markers, and loss of PEDF is observed.
Supplementary Material
Supplementary Figure: 5: Association of ductal invasion with higher proportion of pro-tumorigenic (M2 subtype) macrophage infiltration. (a) Subsequent sections of the 29 human PDA specimens were co-stained with CK 19 and CD 68 (pan-macrophage marker); the image quantification revealed a significant positive correlation between the two markers (r=0.8, p<0.01). (b) Representative 20× magnification images of slides co-stained with CK 19/CD 68 (top panel) with corresponding areas on progressive sections from the same sample co-stained with CK19/CD 163 (bottom panel) showing that as CK 19 expression increases (left to right) so does macrophage staining for CD68 (pan macrophage, top panel), most of which are CD 163 (M2 subtype macrophage, bottom panel). (c) This was quantified by calculating the CD 68 index and CD 163 index in each imaged field (see the results text) followed by calculating the correlation between the two indices; A significant positive correlation between the CD 68 index and CD 163 index (r=0.71) was observed, suggesting that ductal invasion is associated with a higher proportion of M2 macrophage infiltration.
Supplementary Figure 6: Correlation between gene expressions of CK19 (KRT19) with PEDF (SERPINF1) in 186 human pancreas adenocarcinomas. Data and graph were queried from The Cancer Genome Atlas (TCGA).
Supplementary Table 1. Reagents used for organoid culture in this study.
Supplementary Table 2. Primers used for gene expression analysis in this study.
Supplementary Figure 1: Comparison of CK19 and CFTR expressions in WT and mutant KRAS carrying organoids, bar graphs show means ± SE of n=3 experiments, asterisk indicates p<0.05.
Supplementary Figure 2: The fold change in gene expression of Amylase (acinar marker) and CFTR (Ductal marker) in WT and mutant KRAS carrying organoids after 48 h incubation with RAW 264.7 macrophages, compared to control (no macrophage incubation). D-A ratios increased in both pancreatic organoids after macrophage incubation, bar graphs show means ± SE of n=3 experiments, asterisk indicates p<0.05.
Supplementary Figure 3: Relative gene expression of Amylase and CFTR and Ductal-Acinar (D-A) ratios in WT organoids after 48 hours incubation with RAW 264.7 which was pre-treated with KRAS carrying organoids relative to RAW 264.7 pre-treated with WT organoids, bar graphs show means ± SE of n=3 experiments.
Supplementary Figure 4: Correlation between gene expression of CK19 (KRT19) with (a) iNOS (NOS1) and (b) IL6R as part of IL6 signaling pathways, in 186 human pancreas adenocarcinomas. Data and graph was queried from The Cancer Genome Atlas (TCGA).
Acknowledgments
Grant Support: Faraz Bishehsari is supported by Rush Translational Sciences Consortium/Swim Across America; Abde Abukhdeir receives support from The Lynn Sage Foundation. Ali Keshavarzian is supported by NIH-NIAAA R01AA023417
Abbreviations
- PDA
Pancreatic ductal adenocarcinoma
- KRAS
Kirsten rat sarcoma virus
- PDX1
Pancreatic And Duodenal Homeobox 1
- CK19
Cytokeratin 19
- iNOS
inducible nitric oxide synthase
- IL-6
Interleukin 6
- CFTR
Cystic fibrosis transmembrane conductance regulator
- TCGA
The Cancer Genome Atlas
- CD163
Cluster of Differentiation 163
- PEDF
epithelial Pigment Epithelial Derived Factor
Footnotes
Disclosures None of the authors have any potential conflicts (financial, professional, or personal) related to the manuscript to disclose.
Author Contributions:
Faraz Bishehsari conceived and designed the experiments; Mahboobeh Mahdavinia, Abde Abukhdeir and Ali Keshavarzian helped in the study design and supervision; Faraz Bishehsari, Lijuan Zhang, Usman Barlass, Nailliw Preite, Sanja Turturro, Matthew S. Najor, Janet P Zayas and Abde Abukhdeir performed the experiments and acquired the data; Brandon B. Shetuni reviewed histopathology slides; Faraz Bishehsari analyzed the data and wrote the manuscript; Ali Keshavarzian, and Mahboobeh Mahdavinia edited the manuscript for intellectual content; all the authors approved the final draft.
Novelty and Impact: We used organoid models to determine the role of epithelial KRAS mutation, and its interplay with macrophages in pancreatic neoplastic transformation. We found cooperativity between transforming epithelium and macrophages in pancreatic carcinogenesis. A better understanding of the bidirectional epithelium-macrophage cross-talk could reveal novel targets for PDA therapy.
References
- 1.Moss SF, Blaser MJ. Mechanisms of disease: Inflammation and the origins of cancer. Nat Clin Pract Oncol. 2005;2:90–7. doi: 10.1038/ncponc0081. quiz 1 p following 113. [DOI] [PubMed] [Google Scholar]
- 2.Evans A, Costello E. The role of inflammatory cells in fostering pancreatic cancer cell growth and invasion. Front Physiol. 2012;3:270. doi: 10.3389/fphys.2012.00270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farrow B, Evers BM. Inflammation and the development of pancreatic cancer. Surg Oncol. 2002;10:153–69. doi: 10.1016/s0960-7404(02)00015-4. [DOI] [PubMed] [Google Scholar]
- 4.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer research. 2014;74:2913–21. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 5.Pinho AV, Chantrill L, Rooman I. Chronic pancreatitis: a path to pancreatic cancer. Cancer Lett. 2014;345:203–9. doi: 10.1016/j.canlet.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 6.Chang JH, Jiang Y, Pillarisetty VG. Role of immune cells in pancreatic cancer from bench to clinical application: An updated review. Medicine (Baltimore) 2016;95:e5541. doi: 10.1097/MD.0000000000005541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mielgo A, Schmid MC. Impact of tumour associated macrophages in pancreatic cancer. BMB Rep. 2013;46:131–8. doi: 10.5483/BMBRep.2013.46.3.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamasaki A, Kameda C, Xu R, Tanaka H, Tasaka T, Chikazawa N, Suzuki H, Morisaki T, Kubo M, Onishi H, Tanaka M, Katano M. Nuclear factor kappaB-activated monocytes contribute to pancreatic cancer progression through the production of Shh. Cancer Immunol Immunother. 2010;59:675–86. doi: 10.1007/s00262-009-0783-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bayne LJ, Beatty GL, Jhala N, Clark CE, Rhim AD, Stanger BZ, Vonderheide RH. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer Cell. 2012;21:822–35. doi: 10.1016/j.ccr.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liou GY, Doppler H, Necela B, Krishna M, Crawford HC, Raimondo M, Storz P. Macrophage-secreted cytokines drive pancreatic acinar-to-ductal metaplasia through NF-kappaB and MMPs. J Cell Biol. 2013;202:563–77. doi: 10.1083/jcb.201301001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Funamizu N, Hu C, Lacy C, Schetter A, Zhang G, He P, Gaedcke J, Ghadimi MB, Ried T, Yfantis HG, Lee DH, Subleski J, et al. Macrophage migration inhibitory factor induces epithelial to mesenchymal transition, enhances tumor aggressiveness and predicts clinical outcome in resected pancreatic ductal adenocarcinoma. International journal of cancer. 2013;132:785–94. doi: 10.1002/ijc.27736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long KB, Gladney WL, Tooker GM, Graham K, Fraietta JA, Beatty GL. IFNgamma and CCL2 Cooperate to Redirect Tumor-Infiltrating Monocytes to Degrade Fibrosis and Enhance Chemotherapy Efficacy in Pancreatic Carcinoma. Cancer Discov. 2016;6:400–13. doi: 10.1158/2159-8290.CD-15-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Habtezion A, Edderkaoui M, Pandol SJ. Macrophages and pancreatic ductal adenocarcinoma. Cancer Lett. 2016;381:211–6. doi: 10.1016/j.canlet.2015.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tremble LF, Forde PF, Soden DM. Clinical evaluation of macrophages in cancer: role in treatment, modulation and challenges. Cancer Immunol Immunother. 2017 doi: 10.1007/s00262-017-2065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagemann T, Biswas SK, Lawrence T, Sica A, Lewis CE. Regulation of macrophage function in tumors: the multifaceted role of NF-kappaB. Blood. 2009;113:3139–46. doi: 10.1182/blood-2008-12-172825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sica A, Saccani A, Bottazzi B, Bernasconi S, Allavena P, Gaetano B, Fei F, LaRosa G, Scotton C, Balkwill F, Mantovani A. Defective expression of the monocyte chemotactic protein-1 receptor CCR2 in macrophages associated with human ovarian carcinoma. Journal of immunology. 2000;164:733–8. doi: 10.4049/jimmunol.164.2.733. [DOI] [PubMed] [Google Scholar]
- 17.Huch M, Bonfanti P, Boj SF, Sato T, Loomans CJ, van de Wetering M, Sojoodi M, Li VS, Schuijers J, Gracanin A, Ringnalda F, Begthel H, et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 2013;32:2708–21. doi: 10.1038/emboj.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Shidfar A, Ivancic D, Ranjan M, Liu L, Choi MR, Parimi V, Gursel DB, Sullivan ME, Najor MS, Abukhdeir AM, Scholtens D, et al. Overexpression of lipid metabolism genes and PBX1 in the contralateral breasts of women with estrogen receptor-negative breast cancer. International journal of cancer. 2017;140:2484–97. doi: 10.1002/ijc.30680. [DOI] [PubMed] [Google Scholar]
- 19.Principe DR, DeCant B, Diaz AM, Mangan RJ, Hwang R, Lowy A, Shetuni BB, Sreekumar BK, Chung C, Bentrem DJ, Munshi HG, Jung B, et al. PEDF inhibits pancreatic tumorigenesis by attenuating the fibroinflammatory reaction. Oncotarget. 2016 doi: 10.18632/oncotarget.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, Jager M, Ponz-Sarvise M, Tiriac H, Spector MS, Gracanin A, Oni T, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–38. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Storz P. Acinar cell plasticity and development of pancreatic ductal adenocarcinoma. Nat Rev Gastroenterol Hepatol. 2017;14:296–304. doi: 10.1038/nrgastro.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maitra A, Hruban RH. Pancreatic cancer. Annu Rev Pathol. 2008;3:157–88. doi: 10.1146/annurev.pathmechdis.3.121806.154305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cicenas J, Kvederaviciute K, Meskinyte I, Meskinyte-Kausiliene E, Skeberdyte A, Cicenas J. KRAS, TP53, CDKN2A, SMAD4, BRCA1, and BRCA2 Mutations in Pancreatic Cancer. Cancers (Basel) 2017:9. doi: 10.3390/cancers9050042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reichert M, Rustgi AK. Pancreatic ductal cells in development, regeneration, and neoplasia. The Journal of clinical investigation. 2011;121:4572–8. doi: 10.1172/JCI57131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collisson EA, Trejo CL, Silva JM, Gu S, Korkola JE, Heiser LM, Charles RP, Rabinovich BA, Hann B, Dankort D, Spellman PT, Phillips WA, et al. A central role for RAF–>MEK–>ERK signaling in the genesis of pancreatic ductal adenocarcinoma. Cancer Discov. 2012;2:685–93. doi: 10.1158/2159-8290.CD-11-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng F, Li C, Li W, Gao Z, Guo K, Song S. Interaction between pancreatic cancer cells and tumor-associated macrophages promotes the invasion of pancreatic cancer cells and the differentiation and migration of macrophages. IUBMB Life. 2014;66:835–46. doi: 10.1002/iub.1336. [DOI] [PubMed] [Google Scholar]
- 27.Kadison AS, Maldonado TS, Crisera CA, Longaker MT, Gittes GK. In vitro validation of duct differentiation in developing embryonic mouse pancreas. J Surg Res. 2000;90:126–30. doi: 10.1006/jsre.2000.5876. [DOI] [PubMed] [Google Scholar]
- 28.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salmiheimo A, Mustonen H, Vainionpaa S, Shen Z, Kemppainen E, Puolakkainen P, Seppanen H. Tumour-associated macrophages activate migration and STAT3 in pancreatic ductal adenocarcinoma cells in co-cultures. Pancreatology. 2017;17:635–41. doi: 10.1016/j.pan.2017.04.013. [DOI] [PubMed] [Google Scholar]
- 30.Halin S, Rudolfsson SH, Doll JA, Crawford SE, Wikstrom P, Bergh A. Pigment epithelium-derived factor stimulates tumor macrophage recruitment and is downregulated by the prostate tumor microenvironment. Neoplasia. 2010;12:336–45. doi: 10.1593/neo.92046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carroll MJ, Kapur A, Felder M, Patankar MS, Kreeger PK. M2 macrophages induce ovarian cancer cell proliferation via a heparin binding epidermal growth factor/matrix metalloproteinase 9 intercellular feedback loop. Oncotarget. 2016;7:86608–20. doi: 10.18632/oncotarget.13474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams CB, Yeh ES, Soloff AC. Tumor-associated macrophages: unwitting accomplices in breast cancer malignancy. NPJ breast cancer. 2016:2. doi: 10.1038/npjbcancer.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin M, Li X, Tan S, Zhou HJ, Ji W, Bellone S, Xu X, Zhang H, Santin AD, Lou G, Min W. Tumor-associated macrophages drive spheroid formation during early transcoelomic metastasis of ovarian cancer. The Journal of clinical investigation. 2016;126:4157–73. doi: 10.1172/JCI87252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Collins MA, Bednar F, Zhang Y, Brisset JC, Galban S, Galban CJ, Rakshit S, Flannagan KS, Adsay NV, Pasca di Magliano M. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. The Journal of clinical investigation. 2012;122:639–53. doi: 10.1172/JCI59227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ancrile BB, O’Hayer KM, Counter CM. Oncogenic ras-induced expression of cytokines: a new target of anti-cancer therapeutics. Mol Interv. 2008;8:22–7. doi: 10.1124/mi.8.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liou GY, Doppler H, Necela B, Edenfield B, Zhang L, Dawson DW, Storz P. Mutant KRAS-induced expression of ICAM-1 in pancreatic acinar cells causes attraction of macrophages to expedite the formation of precancerous lesions. Cancer Discov. 2015;5:52–63. doi: 10.1158/2159-8290.CD-14-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pylayeva-Gupta Y, Lee KE, Hajdu CH, Miller G, Bar-Sagi D. Oncogenic Kras-induced GM-CSF production promotes the development of pancreatic neoplasia. Cancer Cell. 2012;21:836–47. doi: 10.1016/j.ccr.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Markowitz J, Brooks TR, Duggan MC, Paul BK, Pan X, Wei L, Abrams Z, Luedke E, Lesinski GB, Mundy-Bosse B, Bekaii-Saab T, Carson WE., 3rd Patients with pancreatic adenocarcinoma exhibit elevated levels of myeloid-derived suppressor cells upon progression of disease. Cancer Immunol Immunother. 2015;64:149–59. doi: 10.1007/s00262-014-1618-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vannini F, Kashfi K, Nath N. The dual role of iNOS in cancer. Redox biology. 2015;6:334–43. doi: 10.1016/j.redox.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burke AJ, Sullivan FJ, Giles FJ, Glynn SA. The yin and yang of nitric oxide in cancer progression. Carcinogenesis. 2013;34:503–12. doi: 10.1093/carcin/bgt034. [DOI] [PubMed] [Google Scholar]
- 41.Hong DS, Angelo LS, Kurzrock R. Interleukin-6 and its receptor in cancer: implications for translational therapeutics. Cancer. 2007;110:1911–28. doi: 10.1002/cncr.22999. [DOI] [PubMed] [Google Scholar]
- 42.Fisher DT, Appenheimer MM, Evans SS. The two faces of IL-6 in the tumor microenvironment. Seminars in immunology. 2014;26:38–47. doi: 10.1016/j.smim.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J, He P, Gaida M, Yang S, Schetter AJ, Gaedcke J, Ghadimi BM, Ried T, Yfantis H, Lee D, Weiss JM, Stauffer J, et al. Inducible nitric oxide synthase enhances disease aggressiveness in pancreatic cancer. Oncotarget. 2016;7:52993–3004. doi: 10.18632/oncotarget.10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li LM, Kilbourn RG, Adams J, Fidler IJ. Role of nitric oxide in lysis of tumor cells by cytokine-activated endothelial cells. Cancer research. 1991;51:2531–5. [PubMed] [Google Scholar]
- 45.Jiang H, Stewart CA, Fast DJ, Leu RW. Tumor target-derived soluble factor synergizes with IFN-gamma and IL-2 to activate macrophages for tumor necrosis factor and nitric oxide production to mediate cytotoxicity of the same target. Journal of immunology. 1992;149:2137–46. [PubMed] [Google Scholar]
- 46.Muerkoster S, Wegehenkel K, Arlt A, Witt M, Sipos B, Kruse ML, Sebens T, Kloppel G, Kalthoff H, Folsch UR, Schafer H. Tumor stroma interactions induce chemoresistance in pancreatic ductal carcinoma cells involving increased secretion and paracrine effects of nitric oxide and interleukin-1beta. Cancer research. 2004;64:1331–7. doi: 10.1158/0008-5472.can-03-1860. [DOI] [PubMed] [Google Scholar]
- 47.Liou GY, Bastea L, Fleming A, Doppler H, Edenfield BH, Dawson DW, Zhang L, Bardeesy N, Storz P. The Presence of Interleukin-13 at Pancreatic ADM/PanIN Lesions Alters Macrophage Populations and Mediates Pancreatic Tumorigenesis. Cell Rep. 2017;19:1322–33. doi: 10.1016/j.celrep.2017.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan X, Egami H, Ishikawa S, Sugita H, Kamohara H, Nakagawa M, Nozawa F, Abe M, Ogawa M. Involvement of matrix metalloproteinase-7 in invasion-metastasis through induction of cell dissociation in pancreatic cancer. International journal of oncology. 2005;26:1283–9. [PubMed] [Google Scholar]
- 49.Bera A, Zhao S, Cao L, Chiao PJ, Freeman JW. Oncogenic K-Ras and loss of Smad4 mediate invasion by activating an EGFR/NF-kappaB Axis that induces expression of MMP9 and uPA in human pancreas progenitor cells. PloS one. 2013;8:e82282. doi: 10.1371/journal.pone.0082282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knapinska AM, Estrada CA, Fields GB. The Roles of Matrix Metalloproteinases in Pancreatic Cancer. Progress in molecular biology and translational science. 2017;148:339–54. doi: 10.1016/bs.pmbts.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 51.Notari L, Miller A, Martinez A, Amaral J, Ju M, Robinson G, Smith LE, Becerra SP. Pigment epithelium-derived factor is a substrate for matrix metalloproteinase type 2 and type 9: implications for downregulation in hypoxia. Investigative ophthalmology & visual science. 2005;46:2736–47. doi: 10.1167/iovs.04-1489. [DOI] [PubMed] [Google Scholar]
- 52.Hase R, Miyamoto M, Uehara H, Kadoya M, Ebihara Y, Murakami Y, Takahashi R, Mega S, Li L, Shichinohe T, Kawarada Y, Kondo S. Pigment epithelium-derived factor gene therapy inhibits human pancreatic cancer in mice. Clin Cancer Res. 2005;11:8737–44. doi: 10.1158/1078-0432.CCR-05-1323. [DOI] [PubMed] [Google Scholar]
- 53.Becerra SP, Notario V. The effects of PEDF on cancer biology: mechanisms of action and therapeutic potential. Nat Rev Cancer. 2013;13:258–71. doi: 10.1038/nrc3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cui R, Yue W, Lattime EC, Stein MN, Xu Q, Tan XL. Targeting tumor-associated macrophages to combat pancreatic cancer. Oncotarget. 2016;7:50735–54. doi: 10.18632/oncotarget.9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure: 5: Association of ductal invasion with higher proportion of pro-tumorigenic (M2 subtype) macrophage infiltration. (a) Subsequent sections of the 29 human PDA specimens were co-stained with CK 19 and CD 68 (pan-macrophage marker); the image quantification revealed a significant positive correlation between the two markers (r=0.8, p<0.01). (b) Representative 20× magnification images of slides co-stained with CK 19/CD 68 (top panel) with corresponding areas on progressive sections from the same sample co-stained with CK19/CD 163 (bottom panel) showing that as CK 19 expression increases (left to right) so does macrophage staining for CD68 (pan macrophage, top panel), most of which are CD 163 (M2 subtype macrophage, bottom panel). (c) This was quantified by calculating the CD 68 index and CD 163 index in each imaged field (see the results text) followed by calculating the correlation between the two indices; A significant positive correlation between the CD 68 index and CD 163 index (r=0.71) was observed, suggesting that ductal invasion is associated with a higher proportion of M2 macrophage infiltration.
Supplementary Figure 6: Correlation between gene expressions of CK19 (KRT19) with PEDF (SERPINF1) in 186 human pancreas adenocarcinomas. Data and graph were queried from The Cancer Genome Atlas (TCGA).
Supplementary Table 1. Reagents used for organoid culture in this study.
Supplementary Table 2. Primers used for gene expression analysis in this study.
Supplementary Figure 1: Comparison of CK19 and CFTR expressions in WT and mutant KRAS carrying organoids, bar graphs show means ± SE of n=3 experiments, asterisk indicates p<0.05.
Supplementary Figure 2: The fold change in gene expression of Amylase (acinar marker) and CFTR (Ductal marker) in WT and mutant KRAS carrying organoids after 48 h incubation with RAW 264.7 macrophages, compared to control (no macrophage incubation). D-A ratios increased in both pancreatic organoids after macrophage incubation, bar graphs show means ± SE of n=3 experiments, asterisk indicates p<0.05.
Supplementary Figure 3: Relative gene expression of Amylase and CFTR and Ductal-Acinar (D-A) ratios in WT organoids after 48 hours incubation with RAW 264.7 which was pre-treated with KRAS carrying organoids relative to RAW 264.7 pre-treated with WT organoids, bar graphs show means ± SE of n=3 experiments.
Supplementary Figure 4: Correlation between gene expression of CK19 (KRT19) with (a) iNOS (NOS1) and (b) IL6R as part of IL6 signaling pathways, in 186 human pancreas adenocarcinomas. Data and graph was queried from The Cancer Genome Atlas (TCGA).


