Summary
In amyotrophic lateral sclerosis (ALS) and frontotemporal degeneration (FTD), cytoplasmic aggregates of hyperphosphorylated TDP-43 accumulate and colocalize with some stressgranule components, but how pathological TDP-43 aggregation is nucleated remains unknown. In Drosophila, we establish that downregulation of Tankyrase, a poly (ADPribose) (PAR) polymerase, reduces TDP-43 accumulation in the cytoplasm and potently mitigates neurodegeneration. We establish that TDP-43 non-covalently binds to PAR via PAR-binding motifs embedded within its nuclear-localization sequence. PAR binding promotes liquid-liquid phase separation of TDP-43 in vitro and is required for TDP-43 accumulation in stress granules in mammalian cells and neurons. Stress-granule localization initially protects TDP-43 from disease-associated phosphorylation, but upon long-term stress, stress granules resolve leaving behind aggregates of phosphorylated TDP-43. Finally, small-molecule inhibition of Tankyrase in mammalian cells inhibits formation of cytoplasmic TDP-43 foci without affecting stress-granule assembly. Thus, Tankyrase inhibition antagonizes TDP-43-associated pathology and neurodegeneration and could have therapeutic utility for ALS/FTD.
Graphical Abstract
Introduction
In almost all cases of amyotrophic lateral sclerosis (ALS) and ~50% of frontotemporal degeneration (FTD-TDP), the normally nuclear RNA-binding protein TDP-43 forms phosphorylated aggregates in the cytoplasm of affected neurons and glia (Hasegawa et al., 2008; Mackenzie et al., 2007; Neumann et al., 2006). Increasing evidence suggests that stress pathways are central to these diseases. For example, TDP-43, along with other RNAbinding proteins associated with ALS/FTD, including FUS, Ataxin 2, TIA-1 and hnRNPA1, are components of stress granules (Li et al., 2013). Stress granules are cytoplasmic membraneless organelles that sequester RNA-protein complexes involved in the initiation of protein translation (Kedersha et al., 1999). In post-mortem tissue, phosphorylated TDP43 congregates with the stress-granule proteins eIF3, TIA-1 and PABPC-1 (Bentmann et al., 2012; Liu-Yesucevitz et al., 2010; McGurk et al., 2014). In cellular and animal models of ALS and FTD, stress-granule biogenesis may also contribute to the degenerative process, as downregulation of the stress-granule proteins Ataxin 2 and PolyA-binding protein (PABP) mitigates disease-associated toxicity (Becker et al., 2017; Elden et al., 2010; Kim et al., 2014; Zhang et al., 2018). Although mounting pathological and genetic evidence implicates stress pathways in ALS and FTD, how stress contributes to disease and to TDP-43associated pathology remains enigmatic.
Stress-granule assembly occurs via condensation of localized RNA-protein complexes into dynamic liquid droplets—a process called liquid-liquid phase separation (LLPS) (Hyman et al., 2014; Wippich et al., 2013). A remarkable feature of proteins that localize to stress granules is that they can undergo spontaneous LLPS in vitro (Lin et al., 2015; Mackenzie et al., 2017; Molliex et al., 2015; Murakami et al., 2015; Patel et al., 2015). Protein domains important for LLPS tend to be intrinsically-disordered regions, including the prion-like domains (PrLDs) of the ALS-associated proteins hnRNPA1, FUS, TIA1 and TDP-43, which drive LLPS as well as the formation of solid gel-like structures (Conicella et al., 2016; Han et al., 2012; Kato et al., 2012; Lin et al., 2015; Mackenzie et al., 2017; Molliex et al., 2015; Murakami et al., 2015; Patel et al., 2015; Ryan et al., 2018; Sun et al., 2011; Xiang et al., 2015). Although TDP-43 has been shown to phase separate it is unknown how LLPS of TDP-43 is regulated in health and disease. Thus, uncovering the regulatory mechanisms that link LLPS, stress-granule assembly and abnormal protein accumulation could illuminate important pathways relevant to disease.
A regulator of protein localization and liquid demixing in the cellular milieu is poly (ADPribose), or PAR (Krietsch et al., 2013; Leung, 2014). PAR is a negatively charged biopolymer that is covalently attached to target proteins by PAR polymerases (PARPs) (Gibson and Kraus, 2012). The PAR polymer is recognized by ‘reader’ proteins and in this way PAR ‘reading’ drives the assembly of protein complexes (Krietsch et al., 2013; Teloni and Altmeyer, 2016). PARP activity regulates a plethora of cellular processes (Caldecott, 2014; Fatokun et al., 2014; Hottiger, 2015; Hsiao and Smith, 2008), including stress-granule assembly (Catara et al., 2017; Isabelle et al., 2012; Leung et al., 2011). Here, we demonstrate that inhibition of the PARP, Tankyrase, reduces TDP-43-associated neurodegeneration in vivo. We show that Tankyrase regulates the formation of stressinduced cytoplasmic TDP-43 foci and that PAR binding to TDP-43 regulates stress-granule recruitment. These studies provide insight into the liquid demixing of TDP-43 and suggest that small-molecule inhibitors of Tankyrase could be developed as therapeutics for ALS/FTD.
Results
Tankyrase modulates TDP-43-associated toxicity in Drosophila
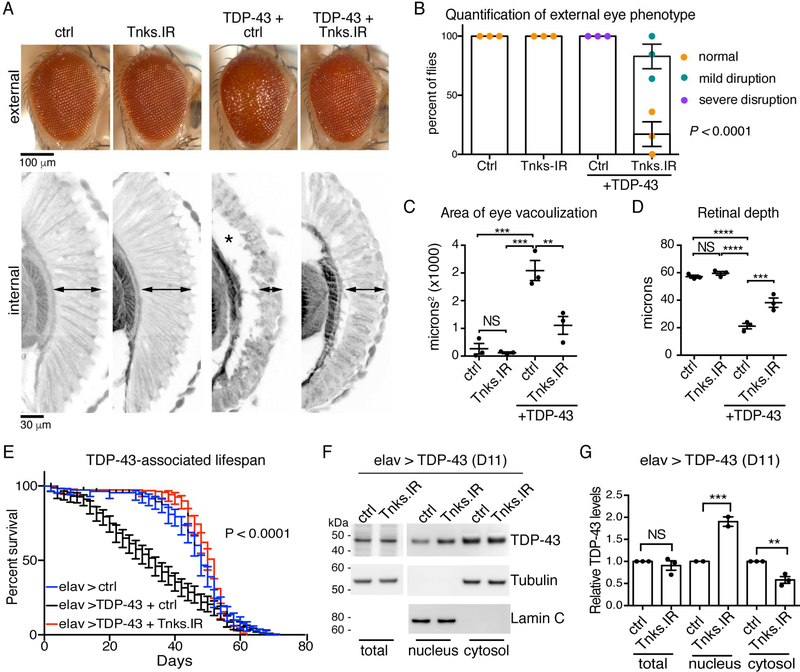
To identify novel pathways relevant to TDP-43-associated toxicity we initiated a geneticmodifier screen in the fly. Selective expression of the human TDP-43 protein in the fly eye using the glass multiple repeat-GAL4 (gmr-GAL4) driver leads to external and internal degeneration (Figure 1A). In the screen, we discovered that downregulation of Tankyrase (Tnks, CG4719), a PAR polymerase (PARP) (Smith et al., 1998), significantly reduced degeneration of the eye (Figure 1A-D). We confirmed that expression of the inverted repeat reduced Tnks mRNA levels (0.5±0.1 fold (±s.e.m.) of normal levels) (Figure S1A) and that reduced Tnks had no effect on total TDP-43 protein levels or the levels of a control protein (β-galactosidase) (Figure S1B and S1C), indicating that downregulation of Tnks did not affect the GAL4 expression system. Additionally, reduction of Tnks had no effect on the Alzheimer’s and FTD-associated protein Tau (Figure S1D), thus the interaction was selective for TDP-43-associated degeneration. Finally, upregulation of Tnks enhanced TDP43-associated toxicity in the eye without having an effect on the eye morphology on its own (Figure S1E) or on the GAL4 expression system (Figure S1F). Together these data implicate Tnks as a novel dose-sensitive regulator of TDP-43-associated degeneration.
Figure 1:
PARylation modulates TDP-43 toxicity in the fly.
(A) Compared to control (ctrl), human TDP-43 (TDP-43+ ctrl) disrupted the external eye (upper panel) and internal retina (lower panel, arrowheads:retinal width and asterisk: vacuolization). Reduction of Tnks (Tnks.IR) mitigates the degeneration of TDP-43 (TDP-43 + Tnks.IR) and has no effect on its own (Tnks.IR).
(B) Reduction of Tnks improved the external eye of TDP-43 flies. Mean (±s.e.m.), n=3, and a two-way ANOVA (P < 0.0001).
(C) Reduction of Tnks reduced vacuolization of the TDP-43 retina. Mean (±s.e.m.), n=3, a one-way ANOVA (P < 0.0001) and a Tukey’s test. Asterisk: significant, NS: not significant.
(D) Reduction of Tnks increased the retinal width of TDP-43 flies. Mean (±s.e.m.), n=3, a one-way ANOVA (P < 0.0001) and a Tukey’s test. Asterisk: significant, NS: not significant.
(E) Reduction of Tnks (Tnks.IR) mitigated the lifespan defect of flies expressing TDP-43 in the brain. More than 190 male flies were followed per genotype. Log-rank test for trend, P < 0.0001.
(F) Reduction of Tnks (Tnks.IR) had no effect on total TDP-43 levels, but increased nuclear and decreased cytoplasmic TDP-43.
(G) TDP-43 protein levels relative to control protein (F). Mean (±s.e.m), one-way ANOVA (P < 0.001) and a Tukey’s test. Asterisks: significant, NS: not significant.Genotypes:
(A-D) ctrl (control) is UAS-mCD8-GFP/+; gmr-GAL4 (III)/+: Tnks.IR is: UAS-Tnks.IR/+; gmr (III)-GAL4 (III) /+: TDP-43 + ctrl is UAS-TDP-43(M)/UAS-mCD8-GFP; gmr-GAL4 (III)/+ and TDP-43+Tnks-IR is UAS-TDP-43 (M)/UAS-Tnks-IR4179R−4; gmr-GAL4 (III)/+.
(E-G) elav > ctrl (control): UAS-mCD8-GFP/+; elav3A-GAL4/+: elav > TDP-43 + ctrl is UASmCD8-GFP/+; elav3A-GAL4, UAS-TDP-43 (S)/ + and elav > TDP-43+Tnks.IR is UAS-TnksIR4179R−4/+; elav3A-GAL4, UAS-TDP-43 (S)/+.
See Figure S1.
We extended these studies to the fly central nervous system. Neuronal expression of TDP-43 with elav-GAL4 driver resulted in early death (Figure 1E). Downregulation of Tnks significantly restored the lifespan defects of the TDP-43 flies, while having no effect on the lifespan of controls (Figure 1E and Figure S1G-H). Downregulation of Tnks in the nervous system had no effect on the total protein levels of TDP-43 (Figure 1F-G). However, lowered Tnks levels led to a significant increase in nuclear TDP-43 (1.9±0.1 (s.e.m.)) and a significant decrease in cytoplasmic TDP-43 (0.6±0.1 (s.e.m.)) compared to the control (Figure 1F-G and S1I-J). Taken together, these data indicate that the downregulation of Tankyrase in the nervous system correlates with increased nuclear TDP-43 and mitigates TDP-43-induced degeneration of the nervous system.
TDP-43 binds to PAR via the nuclear-localization sequence
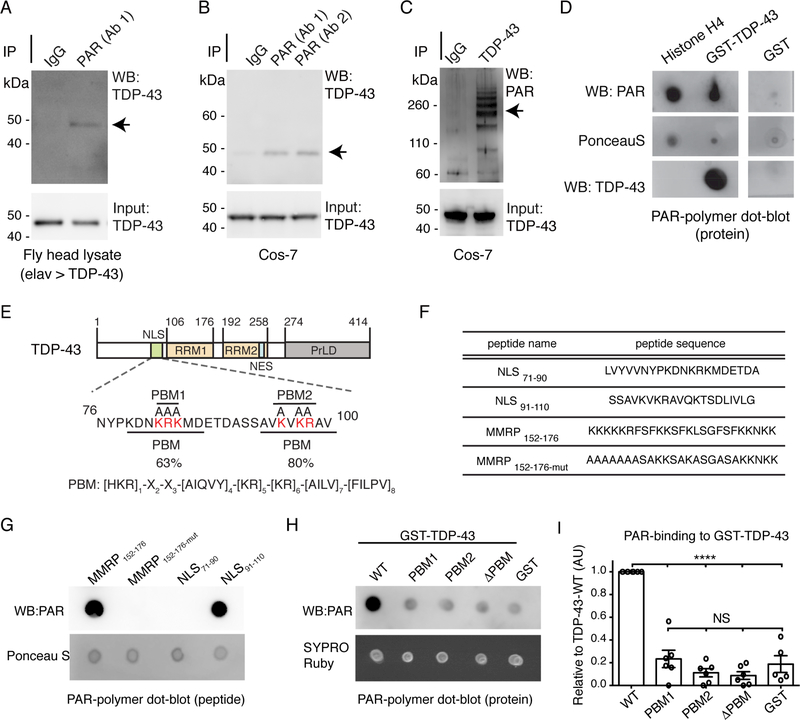
A previous proteomic screen identified TDP-43 as one of many proteins that coimmunoprecipitated with a PAR glycohydrolase (PARG) (Gagne et al., 2012), suggesting that TDP-43 could be a PAR-binding protein. Given that Tankyrase genetically interacted with TDP-43 in the fly, we determined whether TDP-43 and PAR may associate in the same complex. Human TDP-43 was expressed in the fly nervous system and, under nondenaturing conditions, we observed PAR to co-immunoprecipitate with TDP-43 from fly protein lysate (Figure 2A). Furthermore, PAR and endogenous TDP-43 co-immunoprecipitated from mammalian cells (Figure 2B-C). These data indicate that TDP-43 and PAR can co-exist in the same protein complex in fly and mammalian protein lysate, and raise the possibility that TDP-43 non-covalently interacts with PAR.
Figure 2:
TDP-43 binds to PAR via a PAR-binding region in the N-terminus.
(A) Human TDP-43 expressed in the fly brain co-immunoprecipitated with PAR (arrow). PAR antibody: clone 10H, BSA-free, Tulip Biolabs. Head lysate purified from the adult fly head at day 4. elav > TDP-43 is +/+; elav3A-GAL4, UAS-TDP-43 (S)/ +.
(B) PAR from Cos-7 cells co-immunoprecipitated with endogenous TDP-43 (arrow). Antibodies: Ab1: anti-PAR, clone 10H, BSA-free, Tulip Biolabs, Ab2: anti-PAR, ENZO.
(C) Endogenous TDP-43 from Cos-7 cells co-immunoprecipitated with PAR (arrows). PAR antibody: LP96, BD Pharmingen.
(D) GST-TDP-43-WT binds to PAR. Replica membranes stained for PonceauS and immunoblotted for TDP-43.
(E)TDP-43 domains. NLS: nuclear localization sequence, RRM: RNA recognition motif, NES: nuclear export sequence, PrLD: prion-like domain. The PAR-binding motifs (PBMs) in the NLS: PBM1 and PBM2. The basic amino acids in PBM1 and PBM2 (red) were mutated to alanine for this study.
(F) Peptides spanned either PBM1 (NLS71–90) or PBM2 (NLS91–110). MARCKS/MARCKSRelated Protein (MMRP) amino acids 152–176 (MMRP172–156) was the positive control and MMRP152–176-mut (Pleschke et al., 2000).
(G) PAR-binding was detected on MMRP152–176 and on NLS91–110. Replica membranes stained with PonceauS.
(H) Mutation of PBM1, PBM2 or both (PBM) in GST-TDP-43 diminished PAR binding. Replica membranes stained with SYPRO Ruby.
(I) PAR dot blots quantified for PAR immunoreactivity, relative to TDP-43. Mean (±s.e.m), n=6, one-way ANOVA (P < 0.0001) and a Tukey’s test is presented. Asterisks: significant, NS: not significant.
See Figure S2.
To ascertain if TDP-43 could bind to PAR, we performed a PAR-binding dot-blot. TDP-43 (GST-TDP-43-WT) was spotted onto a membrane with a canonical PAR-binding protein Histone H4 (Panzeter et al., 1993) and a GST negative control. Incubation with PAR polymer followed by immunoblotting with an antibody directed to PAR revealed that both Histone H4 and TDP-43 bound to PAR (Figure 2D). The most common PAR-binding module is the PAR-binding motif (PBM) (Teloni and Altmeyer, 2016). Alignment of the PBM ([HKR]1-X2-X3-[AIQVY]4-[KR]5-[KR]6-[AILV]7-[FILPV]8 (Gagne et al., 2008)) to TDP-43 revealed the region with highest similarity (80%) was in the NLS (Figures 2E and S2A), a second site with lower similarity (63%) was also identified within the NLS (Figures 2E and S2A). TDP-43 has a bipartite NLS typified by two clusters of basic amino acids, mutation of which leads to cytoplasmic accumulation of TDP-43 (Winton et al., 2008). Curiously, it was these clusters of basic residues that aligned to the PBM. Here we refer to these two regions as PBM1 and PBM2 (Figure 2E). These data raised the intriguing possibility that the NLS of TDP-43 may be involved in binding to PAR. To verify this possibility, we performed PARbinding dot-blots on peptides that spanned PBM1 or PBM2 (Figure 2F and2G). This revealed that PAR bound to the peptide corresponding to PBM2 of TDP-43 (Figure 2G) and not to the peptide corresponding to PBM1 (Figure 2G). These data suggest that the NLS, and PBM2, of TDP-43 is a region capable of binding to PAR.
We next determined whether mutation of the PBMs in the full-length TDP-43 protein altered PAR-binding and substituted the key lysine and arginine residues with alanine in PBM1, PBM2, or both (ΔPBM). Mutation of PBM1, PBM2 or both (ΔPBM) led to a significant reduction in PAR binding to TDP-43 (0.23 ±0.08-fold, 0.11±0.04-fold and 0.09±0.03-fold (±s.e.m.), respectively, of wild-type levels) (Figure 2H-I). These data indicate that although the PBM2 peptide, and not the PBM1 peptide, can directly bind to PAR, mutation of either PBM is sufficient to diminish the capacity of the full-length protein to bind to PAR. Taken together, these data implicate TDP-43 as a PAR-binding protein and establish the NLS as a region that mediates this interaction.
PAR promotes LLPS of TDP-43
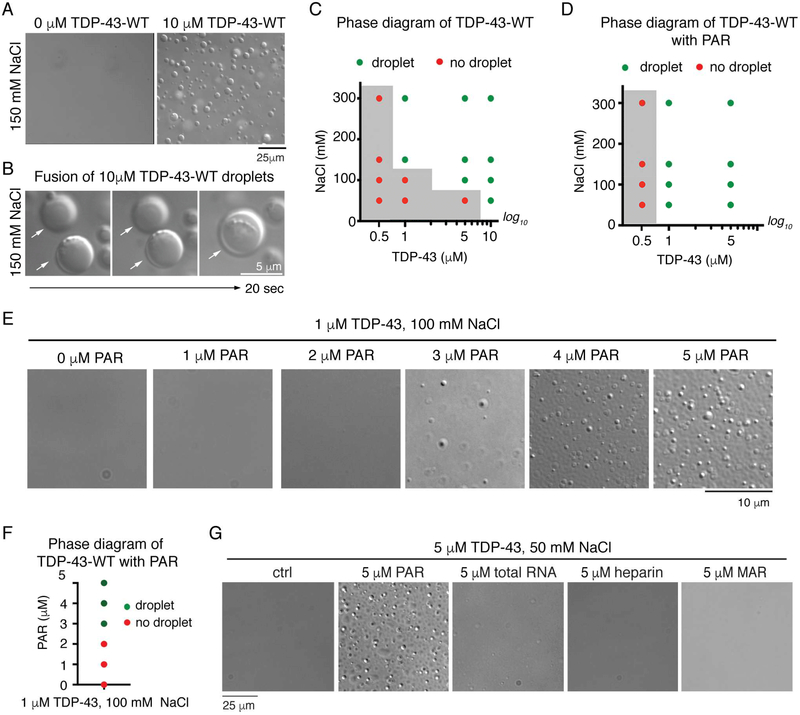
PAR seeds protein localization and liquid demixing in vivo (Leung, 2014; Teloni and Altmeyer, 2016). Liquid demixing is also an in vitro characteristic of proteins associated with ALS (Harrison and Shorter, 2017). Thus, we assessed whether TDP-43 could undergo liquid demixing (LLPS) in vitro and whether PAR could regulate this process. We used conditions that mimic the intracellular environment: we included dextran as a macromolecular crowding agent, buffer conditions of physiological salt (150mM NaCl) and pH (7.5), and we employed physiological (Ling et al., 2010) TDP-43 concentrations (110μM). Under these conditions, purified TDP-43 (His6-SUMO-TDP-43-WT) (Figure S2B) spontaneously formed dynamic spherical droplets that fused and increased in size indicating liquid-like properties (Figure 3A-B and Movie S1). LLPS of TDP-43 was induced at lower TDP-43 concentrations by increasing the salt (NaCl) concentration (Figure 3C). Thus, LLPS of full-length TDP-43 can be achieved under physiological conditions and is dependent upon TDP-43 and salt concentrations.
Figure 3:
PAR promotes LLPS of TDP-43 in vitro.
(A) Spontaneous LLPS of 10μM SUMO-TDP-43-WT in 150mM NaCl. Repeated more than five times on two preparations of protein.
(B) Fusion of two TDP-43-WT droplets. (white arrows).
(C) The presence (green) or absence (red) of TDP-43-WT droplets. The grey area indicates phase boundary. Confirmed in two independent protein preparations. Conditions either side of phase boundary were repeated three times.
(D) PAR reduced the phase boundary of TDP-43 LLPS. The presence (green) or absence (red) of protein droplets is plotted, grey area indicates phase boundary. Confirmed in two independent protein preparations. Conditions at phase boundary (grey side) were repeated three times. (E) Addition of PAR (1–5 μM mono ADP-ribose equivalents) promoted TDP-43-WT LLPS in a dose-dependent manner. This was performed on one protein preparation.
(F) The presence (green) or absence (red) of TDP-43 droplets.
(G) PAR promoted 5 μM TDP-43-WT LLPS in 50mM NaCL. Heparin, yeast total RNA, and mono (ADP-ribose) (MAR), at the same concentration had no effect. Repeated twice from 1 protein preparation. All at room temperature.
See Figure S2.
To evaluate if PAR could promote LLPS of TDP-43, we lowered the concentration of TDP-43 to 1μM and the salt concentration to 100mM; under these conditions TDP-43 did not undergo LLPS (Figure 3C). The addition of PAR (1–5μM ADP-ribose equivalents) at a chain length ranging from 2–300 ADP-ribose subunits, stimulated TDP-43 LLPS in a dose-dependent manner (Figure 3D-F). At lower protein and salt concentrations 5μM PAR promoted LLPS of TDP-43 (Figure 3D and S2E) and lowered the phase boundary. By contrast, the addition of heparin, total RNA, mono (ADP-ribose), or BSA did not stimulate TDP-43 LLPS, and PAR did not undergo LLPS in the absence of protein (Figure 3G and S2FJ). Finally,, we purified TDP-43 as a C-terminally tagged MBP-fusion protein (TDP-43-WTMBP-His6) (Figure S2C). Under physiological salt (150mM NaCl), and with dextran, TDP-43WT-MBP spontaneously formed liquid droplets (Figure S2K) and the addition of PAR to TDP-43-WT-MBP promoted LLPS (Figure S2L). Thus PAR-mediated LLPS of TDP-43 is a feature of TDP-43 and independent of the protein tag.
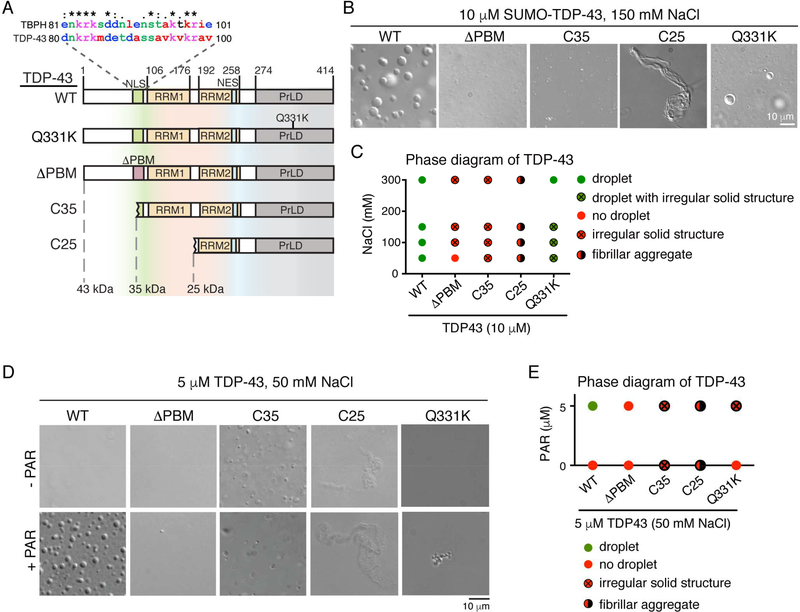
We next asked whether PAR-regulated LLPS of TDP-43 required the PAR-binding motifs in the NLS (see Figure 2). Under conditions in which TDP-43 phase separated, TDP-43 with mutation in both PAR-binding motifs (HIS6-SUMO-TDP-43-ΔPBM) (Figure S2B) formed irregular structures that were smaller in size and did not fuse, indicating an aberrant phase transition to solid-like states (Figure 4B-C). At a lower salt concentration (50mM NaCl), TDP-43-ΔPBM remained diffuse, indicating that transition into a solid state is salt dependent (Figure 4C). In support of our conclusion that the NLS harbors functional PARbinding motifs, the addition of PAR failed to promote LLPS of TDP-43-ΔPBM (Figure 4D-E and Figure S2M). Thus, the PAR-binding motifs are critical for normal LLPS of TDP-43.
Figure 4:
The PAR-binding region and the N-terminus are required for TDP-43 LLPS in vitro.
(A) The NLS of human TDP-43 and the Drosophila homologue TAR DNA-binding protein-43 homolog (TBPH), share 42.9% identity. The protein domains of TDP-43-WT, -Q331K, ΔPBM, -C35 (amino acids 85–414) and -C25 (amino acids 176–414) are shown. NLS: nuclear localization sequence, PBM: PAR-binding motif, RRM: RNA recognition motif, NES: nuclear export sequence and PrLD: prion-like domain.
(B) LLPS of 10 μM protein in 150mM NaCl. SUMO-TDP-43-WT underwent LLPS but SUMOTDP-43-ΔPBM and SUMO-TDP-43-C35 formed irregular-solid structures that did not fuse. SUMO-TDP-43-C25 formed fibrillar aggregates. TDP-43-Q331K formed liquid droplets and the occasional irregular-solid structure. Performed three independent times with two protein preparations of TDP-43-WT, C35 and C25 and twice from one protein preparation of TDP-43-Q331K.
(C) The presence of liquid droplets, no liquid droplet, irregular solid structures, both liquid droplets and irregular solid structures and fibrillar aggregates is plotted.
(D) LLPS at 5 μM protein and 50mM NaCl. PAR at 5 μM equivalents to ADP-ribose promoted LLPS of TDP-43-WT and did not promote LLPS of TDP-43-ΔPBM, TDP-43-C35, and TDP-43-C25. PAR promoted the formation of irregular-solid structures of TDP-43Q331K. Performed three independent times with two independent protein preparations.
(E) The presence of liquid droplets, no liquid droplet, irregular-solid structures and fibrillar aggregates is plotted for each TDP-43 variant in the absence and presence of PAR.All at room temperature.
See Figure S2
Disease-associated TDP-43 has altered LLPS dynamics
In disease, TDP-43 can become N-terminally truncated (Igaz et al., 2009; Neumann et al., 2009; Neumann et al., 2006), raising the possibility that fragments of TDP-43 that accumulate in disease may have impaired LLPS properties. To assess this, we examined a 35 kDa isoform of TDP-43 identified in ALS spinal cord (Xiao et al., 2015) and a 25 kDa fragment of TDP-43 identified in ALS brain tissue (Kametani et al., 2016). The fragments were purified as His6-SUMO-tagged fusion proteins (His6-SUMO TDP-43-C35 and His6SUMO TDP-43-C25) (Figure 4A and S2B). Under conditions in which TDP-43-WT phase separated, TDP-43-C35, which retains PBM2, formed small, spherical structures that did not fuse, and the C25 fragment formed filamentous aggregates (Figure 4B-C), indicating they both lacked liquid properties. Moreover, PAR was ineffective at promoting LLPS of TDP-43-C35 or TDP-43-C25 at all salt and protein concentrations tested (Figure 4D-E and Figure S2M). Collectively these advances highlight that the N-terminal region and the PBMs are critical for TDP-43 liquid demixing.
To assess the impact of an ALS-linked mutation on PAR stimulated LLPS of TDP-43, we expressed and purified TDP-43 with an ALS-linked mutation Q331K (His6-SUMO-TDP-43-Q331K) (Figure S2D), previously shown to promote aggregation (Johnson et al., 2009). Surprisingly, the Q331K mutation did not completely prevent TDP-43 from forming protein droplets (Figure 4B-C), however the droplets were fewer in number and at lower NaCl concentrations the droplets coexisted with irregular solid structures (Figure 4B-C and S2M-N). At low NaCl concentrations, PAR promoted the aggregation of solid irregular structures of TDP-43-Q331K (Figure 4D-E) and was ineffective at higher salt concentrations (Figure S2M). Together these data suggest that the Q331K mutation is defective in LLPS and the propensity to aggregate can be stimulated by PAR.
The PBMs are required for TDP-43 stress-granule localization
We extended these findings to the intracellular milieu of mammalian cells. Under normal conditions PAR levels are low and TDP-43 is predominantly nuclear; however, upon stress, PAR and TDP-43 form foci in the cytoplasm (Figure S3A-C). PAR is a constituent of mammalian stress granules (Catara et al., 2017; Leung et al., 2011), hence, we co-labelled the cells with the stress-granules marker eIF3, TIAR, Ataxin-2 and PAR (Figure S3B-E) to reveal that TDP-43 and PAR co-exist in stress granules (see Figure 2A-C). To ascertain whether the PBMs in the NLS regulated TDP-43 localization to PAR-containing stress granules, we expressed TDP-43-YFP and immunostained for PAR. This revealed that mutation of the PBM reduced PAR-labeling of TDP-43-YFP from 93±2% to 17.6±4.9% (s.e.m.) (Figure S3F-H). These data indicate that mutation in the PBM reduces the ability of TDP-43 to interact with PAR in mammalian cells, in support of our data that the PBMs mediate binding to PAR (see Figure 2).
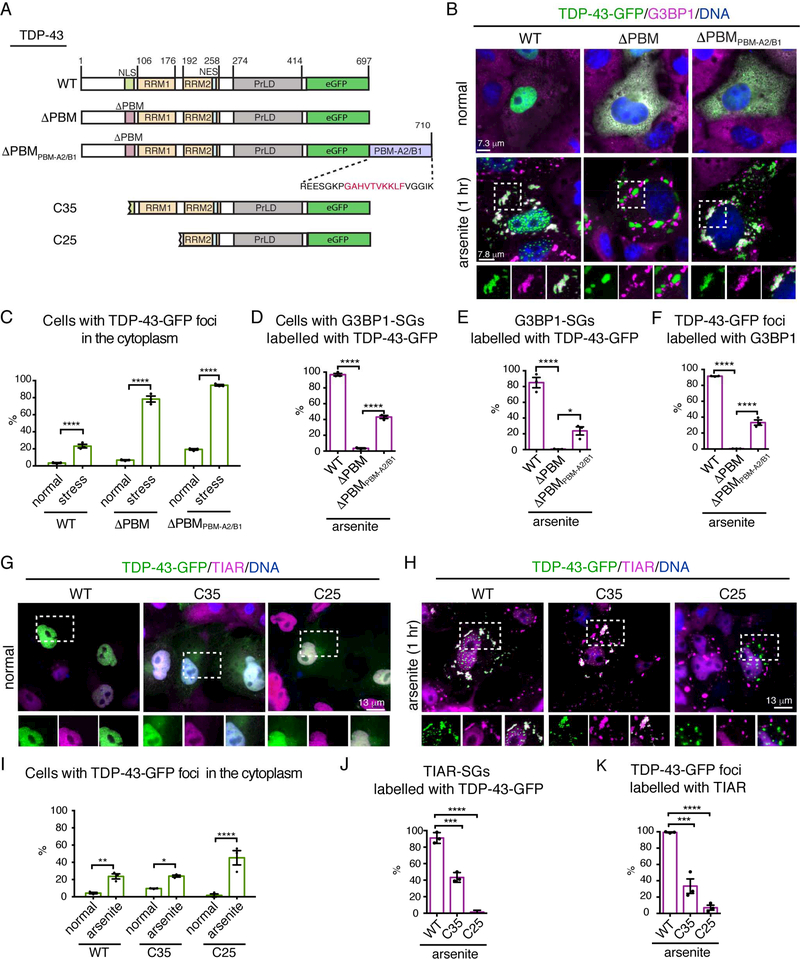
To determine if mutation in the PBM also reduced TDP-43 localization to stress granules we rigorously characterized TDP-43-GFP before and after stress. As anticipated, TDP-43-WT-GFP was predominantly nuclear (Figure 5B) while mutation in the PBMs (TDP-43ΔPBM-GFP), which is known to prevent nuclear localization (Winton et al., 2008), resulted diffuse cytoplasmic accumulation (Figure 5B). Upon stress, the percentage of cells with cytoplasmic foci of TDP-43-WT and TDP-43-ΔPBM increased to 23.7±3% and 89.7±4.9% (s.e.m.), respectively (Figure 5A-C and S3I-J); furthermore, the stress-induced foci of TDP43-WT, and not TDP-43-ΔPBM, co-labelled with the stress-granule markers G3BP1 and TIAR (Figure 5B, 5D-F and S3I-K). Staining with additional markers of stress granules (Ataxin-2 and eIF3) in mammalian cells, immunostaining with TIAR in rat primary neurons, and exposure to a different stress (heat shock) confirmed that TDP-43-WT was recruited to PAR-containing stress granules, whereas stress-induced foci of TDP-43-ΔPBM were excluded from these stress granules (Figure S4A-F). Together these data suggest that a functional PBM is required for TDP-43 to localize to stress granules.
Figure 5:
The PAR-binding region is required for the recruitment of TDP-43 to stress granules.
(A) Domain architecture of TDP-43-WT, -ΔPBM, -ΔPBMPBM-A2/B1, -C35 and -C25. The peptide sequence added to the C-terminus of TDP-43-ΔPBMPBM-A2/B1 is indicated. Red amino acids show the PBM of hnRNPA2/B1, with flanking amino acids of hnRNAPA2/B1 in black. NLS: nuclear localization sequence, PBM: PAR-binding motif, RRM: RNA recognition motif, NES: nuclear export sequence and PrLD: prion-like domain.
(B) Normally, TDP-43-WT was nuclear and TDP-43-ΔPBM and TDP-43-ΔPBMPBM-A2/B1 were cytoplasmic. Upon arsenite-treatment TDP-43-WT localized to stress granules, TDP-43ΔPBM was excluded from stress granules and TDP-43-ΔPBMPBM-A2/B1 localized to stress granules. Cells exposed to 0.5mM sodium arsenite 1 hr, labelled for G3BP1 and stained with Hoescht.
(C) The percentage of cells with cytoplasmic TDP-43-GFP foci. Mean (± s.e.m.), n=3, twoway ANOVA (P < 0.0001) and a Holm-Sidak’s test is presented Asterisks: significant.
(D) The percentage of cells with one or more G3BP1-labelled stress granule (SG) colabelled with TDP-43-GFP. Mean (±s.e.m.), n=3, one-way ANOVA (P < 0.0001) and a Tukey’s test is presented. Asterisks: significant.
(E) The percentage of G3BP1-labelled stress granules (SGs) co-labelled with TDP-43-GFP. Mean (±s.e.m.), n=3, one-way ANOVA (P < 0.0001) and a Tukey’s test is presented. Asterisks: significant.
(F) The percentage of TDP-43-GFP foci co-labelled with G3BP1. Mean (±s.e.m.), n=3, oneway ANOVA (P < 0.0001) and a Tukey’s test is presented. Asterisks: significant.
(G-H) Foci of TDP-43-C35 and TDP-43-C25-GFP are largely excluded from stress granules. Cells exposed to 0.5mM sodium arsenite 1 hr, labelled for TIAR and stained with Hoescht (DNA).
(I) The percentage of cells with cytoplasmic TFP-43-GFP foci. Mean (±s.e.m.), n=3. two-way ANOVA (P = 0.0042) and a Holm-Sidak’s test is presented. Asterisks: significant.
(J) The percentage of TIAR-labelled stress granules (SGs) co-labelled with TDP-43-GFP. Mean (±s.e.m.), n=3, two-way ANOVA (P < 0.0001) and a Tukey’s test is presented. Asterisks: significant, NS: not significant.
(K) The percentage of TDP-43-GFP foci co-labelled with TIAR. Mean (±s.e.m.), n=3, one-way ANOVA (P < 0.0001) and a Tukey’s test is presented. Asterisks: significant.
To determine if PAR binding was necessary for TDP-43 recruitment to stress granules, we added an exogenous PBM from hnRNPA2/B1 (Gagne et al., 2003) to TDP-43-ΔPBM (Figure 5A). The PBM of hnRNPA2/B1 is situated in the N-terminal domain between the two RRMs and is distinct form the NLS (Gagne et al., 2003). Under normal conditions TDP-43ΔPBMPBM-A2/B1 was diffusely enriched in the cytoplasm (Figure 5B), indicating that the PBM of hnRNPA2/B1 did not alter the cellular distribution of TDP-43-ΔPBM. The application of stress initiated the formation of cytoplasmic TDP-43-ΔPBM-PBMA2/B1 foci (Figure 5B-C). Strikingly, the exogenous PBM significantly increased the percentage of cells with TDP-43-ΔPBM localized to G3BP1-labeled stress granules, from 0.6±0.4% (s.e.m.) to 23.7±9% (Figure 5D-F). Thus, the addition of a functional PAR-binding motif is sufficient to restore stress-granule accumulation of TDP-43 defective in PAR binding (TDP-43-ΔPBM). We suggest that the localization of TDP-43 to stress granules is dependent upon PAR-binding.
We next addressed whether the C-terminal fragments of TDP-43, which lack the N-terminus and PBMs, could localize to stress granules. TDP-43-C35-GFP and TDP-43-C25GFP, which normally diffusely localized to the nucleus and cytoplasm formed cytoplasmic foci upon arsenite treatment (Figure 5FG-I). TDP-43-C35, retains PBM2, and localized to 43.5±3.4% (s.e.m.) of TIAR-labelled stress granules (Figure 5F-H). By contrast, TDP-43-C25 localized to 2.3±1.3% (s.e.m) of TIAR-labeled stress granules (Figure 5F-H). These data indicate that the N-terminal region of TDP-43 is important for the recruitment of the protein to stress granules, and suggest that, in disease, stress-granule recruitment of the C-terminal fragments of TDP-43 will be partially (C35) or almost fully (C25) impaired.
Recruitment to stress-granules inhibits phosphorylation of TDP-43
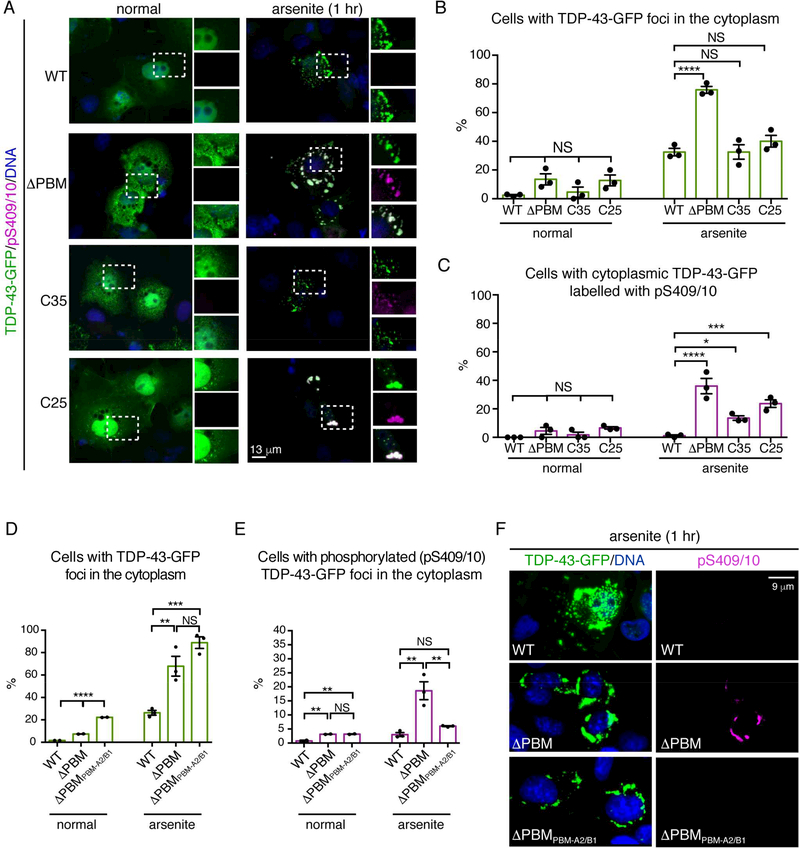
A longstanding notion is that TDP-43 recruited to stress granules becomes marked with the phosphorylation hallmark of disease (Li et al., 2013). To test this, we examined phosphorylation of TDP-43 foci that were localized to, or excluded from, stress granules. Surprisingly, staining for the disease-specific phosphorylation mark pS409/10 revealed that arsenite-induced and heat-induced foci that were excluded from stress granules (TDP-43-ΔPBM, C35 and C25) - and not TDP-43 localized to stress granules (TDP-43-WT) - were phosphorylated (Figure 6A-C and S5A-E). We then examined the phosphorylation status of TDP-43-ΔPBM with an exogenous PBM (TDP-43-ΔPBMPBM-A2/B1). Importantly, the percentage of cells with arsenite-induced cytoplasmic foci of TDP-43-ΔPBMPBM-A2/B1 that co-labelled with pS409/10 was indistinguishable from TDP-43-WT (Figure S6D-F), indicating that the exogenous PBM was sufficient to both localize TDP-43-ΔPBM to stress granules (see Figure 5) and to prevent stress-induced phosphorylation.
Figure 6:
Initial recruitment to stress granules protects TDP-43 from phosphorylation.
(A) Stress induced the formation of phosphorylated (pS409/10) foci of TDP-43-ΔPBM, TDP-43-C35 and TDP-43-C25 but not TDP-43-WT. Cells were stressed with 0.5 mM arsenite 1 hr, labelled for pS409/10 and Hoescht (DNA).
(B) The percentage of cells with cytoplasmic TDP-43-GFP foci. Mean (± s.e.m.), n=3, twoway ANOVA (P < 0.0001) and Tukey’s test is presented Asterisks: significant and NS: not significant.
(C) The percentage of cells with cytoplasmic TDP-43 foci co-labelled with pS409/10. Mean (±s.e.m.), n=3, one-way ANOVA (P <0.0001) and a Tukey’s test is presented. Asterisks: significant and NS: not significant.
(D) The percentage of cells with cytoplasmic TDP43-GFP (WT, ΔPBM or ΔPBMPBM-A2/B1) foci. Mean (± s.e.m.) n=3, one-way ANOVA and Dunnet’s test (P < 0.001) is presented. Asterisks: significant.
(E) The percentage of cells with cytoplasmic TDP-43-GFP (WT, ΔPBM or ΔPBMPBM-A2/B1) foci co-labelled with pS409/10. Mean (± s.e.m.), n=3, one-way ANOVA and Dunnet’s test is presented (normal P < 0.0001 and arsenite P = 0.0024).
(F) Stress-induced the phosphorylation of cytoplasmic foci of TDP-43-ΔPBM-GFP foci and not TDP-43-WT-GFP or TDP-43-ΔPBMPBM-A2/B1-GFP. Cells exposed to 0.5 mM arsenite (1 hr), were labelled for pS409/10 and Hoescht.
Next, we established a live-cell assay to examine the internal dynamics of stress-induced foci of TDP-43. Diffuse TDP-43-WT and TDP-43-ΔPBM in the cytoplasm recovered rapidly from photobleaching (Figure S5F-G), indicating that the two proteins were normally highly mobile. By contrast, stress-induced, cytoplasmic foci formed by both proteins were immobile (Figure S5F-G). However, the stress-induced foci of TDP-43-WT resolved after removal of stress, while the forms of the protein that aggregated outside of the stress granule persisted (Figure S5H). These data indicate that the localization of TDP-43 to the stress-granule environment is critical for preventing stress-induced phosphorylation and for the resolution of TDP-43 foci upon the recovery from stress.
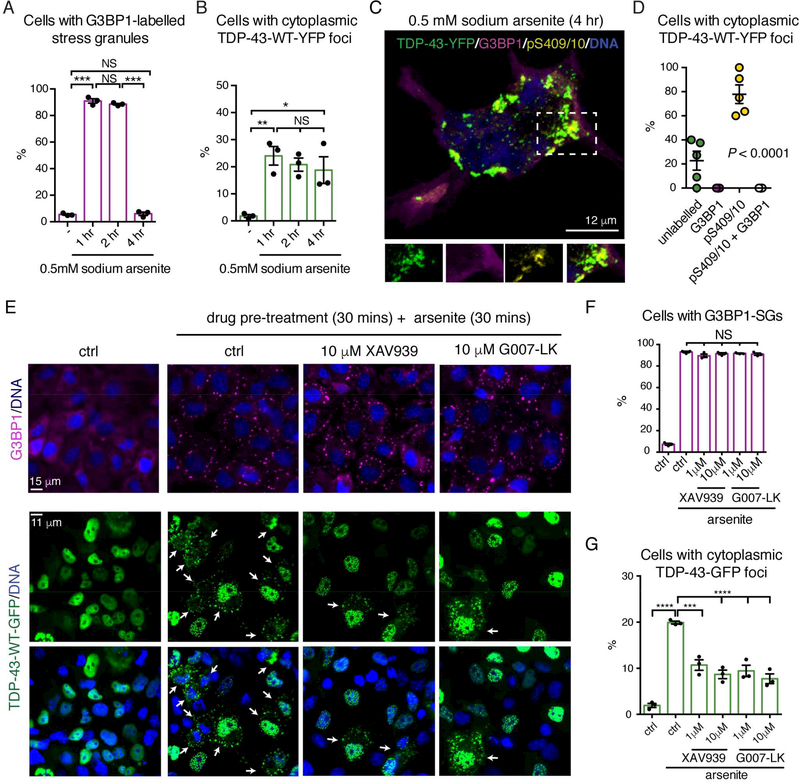
We next determined whether there were conditions of stress that could promote phosphorylation of TDP-43-WT. Intriguingly, low levels of prolonged stress (25 μM arsenite for 6 hr) led to an increase in the percentage of cells with cytoplasmic TDP-43-WT foci, the majority (81± 6% (s.e.m.)) of which were phosphorylated and devoid of the stressgranule protein eIF3 (Figure S6A-E). In addition, exposure to high levels of stress for a prolonged period (0.5 mM arsenite for 4 hr) led to the formation of, and the subsequent resolution of G3BP1-labeleld stress granules (Figure 7A). By contrast, stress-induced foci of TDP-43-WT were sustained over this time period (Figure 7B), and in 78±8% (s.e.m.) of cells, the TDP-43-WT foci were immunolabelled for pS409/10 and devoid of G3BP1 (Figure 7C-D). Thus, consistent with our finding that the stress-granule environment inhibits S409/410 phosphorylation, prolonged stress induces the formation of phosphorylated TDP-43-WT foci in the absence of stress granules.
Figure 7:
Prolonged stress leads to the phosphorylation of TDP-43 foci and TDP-43 foci formation can be mitigated with small molecule inhibitors of Tankyrase.
(A) G3BP1-labelled stress granules resolved after 4 hr exposure to 0.5 mM arsenite. Mean (± s.e.m), n=3, one-way ANOVA (P = 0.005) and a Tukey’s test is presented. Asterisks: significant, NS: not significant.
(B) Stress-induced foci of TDP-43-WT-YFP persisted after 4 hr exposure to 0.5 mM arsenite. Mean (± s.e.m.), n=3, one-way ANOVA (P < 0.0001) and a Tukey’s test is presented. Asterisks: significant and NS: not significant.
(C) ytoplasmic TDP-43-WT-YFP foci formed in 0.5 mM arsenite (4 hr) were co-labelled for pS409/10 and not G3BP1. Cells were labelled for G3BP1, pS409/10 and Hoescht, and imaged by confocal microscopy.
(D) Confocal images of cells with cytoplasmic TDP-43-WT-YFP foci were quantified. Mean (± s.e.m.), n=5 and one-way ANOVA (P < 0.0001) is presented.
(E) Cells untreated or treated with 0.5 mM arsenite (30 mins) in the absence or presence of XAV-939 or G007-LK. A 30 min pretreatment was required and lack of a dose-response is likely due to being above the IC50. (Figure S7H-I). Arrows: cells with cytoplasmic TDP-43WT-GFP foci. Cells were labelled for G3BP1 and Hoescht.
(F) Cells were quantified for the presence of G3BP1-labelled stress granules (SGs). Mean (± s.e.m.), n=3, one-way ANOVA (P < 0.0001) and a Tukey’s is presented. NS: not significant.
(H) Cells were quantified for the presence of cytoplasmic TDP-43-GFP foci. Mean (± s.e.m.), n=3, one-way ANOVA (P < 0.0001) and a Tukey’s test is presented. Asterisks: significant.
See Figure S7.
Small-molecule inhibition of Tankyrase inhibits stress-induced foci of TDP-43
Our studies were launched by discovery that lowered levels of Tankyrase mitigates TDP-43-elicited neurodegeneration in the fly. We thus examined the role of Tankyrase to modulate stress-induced foci of TDP-43 in mammalian cells. In mammals, there are two Tankyrase homologues, Tankyrase 1 and Tankyrase 2 (also known as PARP-5a and PARP5b), which share 82% sequence similarity, and are collectively detected as Tankyrase 1/2 (Citarelli et al., 2010). Under stress, Tankyrase 1/2 localized to 35±9% (s.e.m.) of Ataxin-2labelled stress granules and to 31±2% (s.e.m.) of TIAR-labelled stress granules (Figure S7A). Thus, Tankyrase 1/2 is not a constitutive stress-granule protein.
Tankyrase 1/2 inhibitors have been developed as anti-proliferative agents for cancer cells (Mariotti et al., 2017). We examined small-molecule inhibition of Tankyrase for an effect on stress-granule biogenesis and TDP-43-foci assembly. Treatment with the inhibitor XAV-939 (Huang et al., 2009), markedly reduced the formation of arsenite-induced cytoplasmic foci of TDP-43-WT-GFP without altering the percentage of cells with G3BP1-labelled stress granules (Figure 7E-G). Two additional inhibitors, G007-LK and IWR1-endo, with little to no effect on PARP1/2 activity (Huang et al., 2009; Voronkov et al., 2013) behaved similarly (Figure 7E, 7G and S7E-G). Thus, Tankyrase 1/2 activity regulates the formation of cytoplasmic TDP-43 foci, but not stress-granule formation. Together our data suggest that reduced Tankyrase activity mitigates ALS-associated degeneration by reducing the cytoplasmic localization of TDP43 and subsequent neurodegeneration.
DISCUSSION
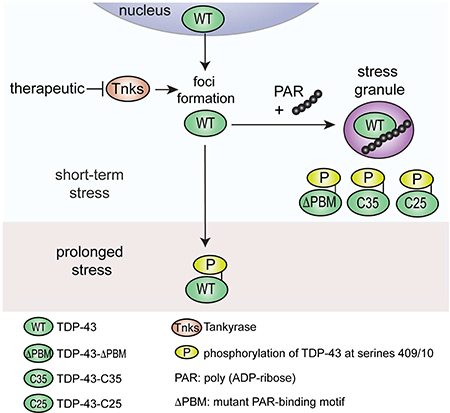
Our data lead to a two-step model for regulation of TDP-43 whereby Tankyrase regulates the formation of cytoplasmic foci of TDP-43 and PAR regulates the recruitment of TDP-43 to stress granules (Graphical Abstract). Under prolonged stress, stress-induced foci of TDP-43 become marked with the disease signature of phosphorylation. Importantly, small-molecule inhibitors of Tankyrase reduce the formation of TDP-43 foci in the cytoplasm. Thus, Tankyrase regulates the subcellular localization of TDP-43 and mitigates TDP-43-mediated neurodegeneration. Small-molecule inhibitors of Tankyrase may provide a therapeutic strategy for ALS/FTD.
The N-terminal domain and PBMs regulate TDP-43 LLPS and stress-granule recruitment
We have defined functional PBMs embedded in the TDP-43 NLS that are required for TDP-43 LLPS. Mounting evidence implicates the N-terminal domain of TDP-43 as critical for higher-order assemblies (Afroz et al., 2017; Chang et al., 2012; Jiang et al., 2017; Mompean et al., 2016; Romano et al., 2015; Zhang et al., 2013). Our data, and a report that that pinpoints serine 48 in the N-terminal domain (Wang et al., 2018), indicate that the Nterminal domain is also critical for TDP-43 phase separation in vitro. We show that negatively charged PAR can initiate LLPS of TDP-43 and suggest that this occurs via multivalent interactions with the PBM in the N-terminal domain.
In the cellular milieu, TDP-43 is not a ubiquitous stress-granule protein and does not readily localize to the cytoplasm (Bentmann et al., 2012; Cohen et al., 2012; Dewey et al., 2011). Previous studies have focused on TDP-43 with mutation in the NLS without a comparison to the WT protein (Bentmann et al., 2012; Nonaka et al., 2009). Here, by successfully inducing the accumulation of TDP-43-WT in the cytoplasm we were able to uncover the essential role of the PAR binding and N-terminal regions for the recruitment of TDP-43 to stress granules. Importantly, the addition of the PAR-binding motif from hnRNPA2/B1 to TDP-43-ΔPBM was sufficient to re-localize TDP-43-ΔPBM back to stress granules. The PBM from hnRNPA1/B2 did not alter the cytoplasmic localization of TDP-43ΔPBM, suggesting that the lack of stress-granule recruitment was not a result of a loss of nuclear localization but was due to loss of PAR binding. Direct PARylation of specific stressgranule proteins assists in stress-granule formation (Leung et al., 2011) and we propose that non-covalent PAR-binding is an added means by which proteins enter stress granules. Interestingly, the C35 fragment retains PBM2 and is only partially localized to stress granules upon stress. The partial exclusion of C35 and the almost complete exclusion of C25 from stress granules could be due to a lack of binding to the PAR-scaffold or because these forms of TDP-43 undergo aberrant phase separation.
Recruitment to stress granules inhibits phosphorylation of TDP-43
Our studies refine the longstanding model that stress-granule localization of TDP-43 nucleates disease-associated pathology (Li et al., 2013). We present a model whereby TDP-43 foci excluded from stress granules readily transition into disease-like aggregates. Various kinases promote stress-granule biogenesis (Kedersha et al., 2013) and it is possible that TDP-43 aggregates excluded from stress granules are exposed to, and phosphorylated by these stress-responsive kinases. We suggest that an additional role of stress granules is to protect proteins from the activated kinases during cellular stress. In disease the phosphorylation and aggregation of C35 and C25 may arise from a failure to phase separate and localize to stress granules. TDP-43 that can localize to stress granules becomes phosphorylated upon long-term stress and in the absence of stress granules. This finding may explain only a subset of TDP-43 inclusions in the post-mortem spinal cord co-label with stress-granule components (Bentmann et al., 2012; Colombrita et al., 2009; McGurk et al., 2014). Stress-granule protein may become trapped in TDP-43 aggregates as the stress granules dissolves. Thus, in the spinal cord, TDP-43 may be recruited to stress granules and be protected, but may transition to a pathological aggregated state as a result of the stressgranule dissolution process.
Tankyrase as a potential therapeutic strategy in ALS/FTD
It has been reported that upregulation of Tankyrase 1 promotes stress-granule formation (Leung et al., 2011). Our studies uncover that Tankyrase is not a ubiquitous stress-granule protein and that Tankyrase inhibition does not alter the formation of G3BP1-labelled stress granules. It is possible that Tankyrase-mediated regulation of stress granules may be independent of enzymatic activity, or may only occur in select stress granules or on select proteins such as TDP-43. We hypothesize that Tankyrase acts upstream of TDP-43 recruitment to stress granules, and may regulate the transport of TDP-43 to stress granules. Although stress-granule localization is initially protective, prolonged stress leads to phosphorylated aggregates. A potential therapeutic strategy could be to target Tankyrase activity to inhibit the initial accumulation of TDP-43 in the cytoplasm. Our studies bring PARylation in ALS/FTD to the forefront, and specifically implicate Tankyrase inhibitors as potential therapeutics in ALS/FTD.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Requests for resources, reagents and further information should be directed to, and will be fulfilled by, Nancy M. Bonini (nbonini@sas.upenn.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila stocks and maintenance
All Drosophila stocks are described in the Resource Table. Briefly, transgenic lines for TDP-43 were described previously (Elden et al., 2010; Kim et al., 2014). The RNAi line to Tankyrase (4179R-4) was obtained from the Fly Stocks of National Institute of Genetics (NIG-Fly), Kyoto, Japan. UAS-mCD8-GFP (y1 w*; UAS-mCD8:GFP.L) obtained from the Bloomington stock center was backcrossed, six times, to w1118 line isogenic for chromosomes 1, 2 and 3 (5905). UAS-hTau R406W (Wittmann et al., 2001) was a kind gift from a gift of M. Feany (Harvard Medical School, Boston, MA, USA). All other stocks were obtained from the Bloomington Drosophila stock center, Indiana, USA. All fly experiments were carried out at 25oC in standard cornmeal molasses agar.
Mammalian cells and culture details
Cos-7 cells were originally purchased by ATCC and were kindly provided by Virginia M. Lee, University of Pennsylvania. Cells were authenticated by ATCC before purchase. The cell lines were not tested for mycoplasma contamination. No commonly misidentified cell lines were used. Cells were routinely cultured in Dulbecco’s modified Eagle’s medium (DMEM) with high glucose supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. Cells were grown at 37oC with 5% CO2, and a water bath was used for humidification. Cells were washed with dPBS without calcium and without magnesium and trypsinized in Trypsin with 0.25 % EDTA (ThermoFisher Scientific).
Rat cortical neuron cultures
Studies were approved by the Children’s Hospital of Pennsylvania Institutional Animal Care and Use Committee (CHOP IACUC), the assigned protocol number was 594. All studies adhered to all regulatory standards. Rat cortical neuron cultures were prepared from embryos isolated from female Sprague Dawley wild rats that were 16–18 days pregnant. Neurons were cultured as previously described (Mojsilovic-Petrovic et al., 2006). Briefly, tissue was dissociated with 5% trypsin and 1mg/ml DNase, cells were cultured in 12-well plates in conditioned neuralbasal medium with serum-free B-27 (ThermoFisher Scientific) and penicillin-streptomycin (ThermoFisher Scientific). After 3–4 days in vitro (DIV) cell division was stopped by the addition of 5microM cytosine arabinoside (AraC) (SigmaAldrich). Cells were maintained in preconditioned neuralbasal medium with serum-free B27 (ThermoFisher Scientific) and penicillin-streptomycin (ThermoFisher Scientific).
METHOD DETAILS
Drosophila external eye imaging and histology
External eyes and internal retinas were imaged in female flies aged 2–3d. For quantification, the integrity of the external eye was scored in ten female flies from three independent crosses for each genotype. Control and experimental crosses were propagated at the same time and incubated in the same incubator (25oC). The internal retina was quantified using imageJ (Rueden et al., 2017) from paraffin sections at the same anatomical position for all animals. Three sections per animal were imaged and quantified, this was repeated on 3–5 female heads from each genotype.
Drosophila lifespan
For lifespan analysis, more than 190 males (1d-old) for each genotype were separated into groups of 15–20 and aged in vials containing fresh standard cornmeal molasses agar. Every second day dead flies were scored and the survivors were tipped into a fresh food vial. Kaplan-Meir survival curves were plotted and a log-rank test for trend was performed using Graphpad prism 6. The lifespan analysis was repeated three independent times at different times throughout the year.
Nuclear Cytoplasmic fractionation
The NER-PER nuclear and cytoplasmic extraction reagents (Thermo Scientific) was used to fractionate the nuclear and cytoplasmic protein from fly heads. The instructions were followed with the following modifications: 10 male heads were homogenized in 110 microL of ice-cold CERI for 1 minute, the sample was vortexed on the highest setting for 30 seconds to fully suspend the cell pellet. The sample was incubated the on ice for 10 minutes and then 5.5 microL of ice-cold CER II was added. The sample was vortexed for 10 seconds on the highest setting, and then incubated on ice for 2 mins. The sample was vortexed on the highest setting for 15 seconds and then centrifuged at maximum speed for 10 mins at 4oC. The supernatant (cytoplasmic extract) was immediately transferred to a clean prechilled tube. The supernatant was spun again at maximum speed for 10 minutes at 4oC, the supernatant (cytoplasmic extract) was transferred to a pre-chilled tube. The nuclear pellet was washed in 50 microL of ice-cold CERI, and then centrifuged at maximum speed for 10 minutes at 4oC. The supernatant was discarded and the pellet was resuspended in 55 microL of ice-cold NER. The samples were vortexed on the highest setting for 10 secs every 10 minutes for a total of 40 mins. The samples were centrifuged at maximum speed for 10 mins at 4oC. The supernatant (nuclear fraction was transferred to pre-chilled tube. Samples for immunoblotting were made up in 4X LDS loading buffer, with β-mercaptoethanol, heat denatured at 95oC, chilled for 5 minutes and centrifuged at 5000 rpm for 5 mins at 4oC. For the total protein immunoblots that correspond to the nuclear and cytoplasmic fractionation, 10 male heads were homogenized in 100microL Laemelli buffer with βmercaptoethanol, heat denatured at 95oC, chilled for 5 minutes and centrifuged at 5000 rpm for 5 mins at 4oC. The equivalent of half of a fly head was loaded into each well for each sample type (nuclear, cytoplasmic and total). Protein was electrophoresed on 4–12% BisTrsi gel and transferred onto nitrocellulose by wet transfer (20volts for 75 mins). Blots were blocked in 5% milk in TBST. Primaries were rabbit anti-TDP-43 (1 in 3000, Proteintech), then mouse anti-Lamin A/C (1 in 1000, DSHB) and then anti-tubulin-HRP (1 in 1,000; Cell Signaling Technology). TDP-43 was always detected first, tubulin, migrates closely to TDP-43, and will interfere with TDP-43 signal detection. Secondary antibodies were anti-mouse HRP (1 in 5000, abcam) and anti-Rabbit-HRP (1 in 5000, EMD Millipore). TDP-43 was detected with ECL (GE healthcare Bio-Sciences) and tubulin and Lamin A/C were detected with ECL Prime (GE Healthcare Bio-Sciences). Quantification of protein blots was performed using Image J software (Rueden et al., 2017). All experiments were carried out on three or more biological replicates and statistical analysis was carried out using prism 6 software.
Immunoblotting of Drosophila total protein
For experiments where the transgene (TDP-43 or LacZ) was expressed in the eye with gmrGAL4, protein was extracted from 10 female heads in 100microL of 2X Laemelli buffer with 5% (v/v) β-mercaptoethanol, heat denatured at 95oC, chilled on ice for 5 mins and centrifuged at 5000 rpm for 5 mins at 4oC. The equivalent of half of a fly head was loaded into each well of a 4–12% Bis-Tris gel and transferred onto nitrocellulose by wet transfer (20 volts for 75 mins). Blots were blocked in 5% milk in TBST. Primary antibodies used: TDP-43 (1 in 3000; Proteintech), α-Tubulin-HRP (1 in 1,000; Cell Signaling Technology) and β−galactosidase (1 in 1,000; Promega). HRP-coupled secondary antibodies used: goat antibody to rabbit (1 in 5,000; EMD Millipore) and goat antibody to mouse (1 in 2,000; Jackson ImmunoResearch Laboratories, Inc.). All experiments were carried out on three or more biological replicates, blots were quantified with Image J (Rueden et al., 2017) and statistical analysis was carried out using prism 6 software.
Real-time PCR
20 third instar larvae from each genotype were homogenized in 250 microliters of Trizol. A further 250 microL of Trizol and 100 microliters of chloroform was added and the tube was vigorously shaken for 15 secs at room temperature. The samples were centrifuged at maximum speed for 10 minutes at 4oC and the upper-aqueous phase was transferred to a fresh and pre-chilled tube. The chloroform extraction was repeated on the aqueous phase. RNA was precipitated in 2 volumes of 100% ethanol and 1/10th the volume of 3M sodium acetate pH 5.2, and the RNA sample was left on ice for 25 mins. The samples were then centrifuged at maximum speed at 4oC for 10 mins. The RNA pellet was washed in 70% ethanol and centrifuged at maximum speed at 4oC for 10 mins. The pellet was air dried and the RNA was dissolved in 40 microL of DEPC-treated water. Genomic DNA in half of the RNA sample was digested with turbo DNase (Ambion) using the vigorous protocol. 500 ng of RNA was used for first-strand DNA synthesis by Superscript III (ThermoFisher Scientific) with random primers. Fast SYBR Green (ThermoFisher Scientific) was used for real-time analysis. Standard curves were used to test the efficiency of all the primers and to determine the amount of cDNA to add to the PCR reaction. Each experiment was carried out on larvae prepared from 4 independent fly crosses. Statistics were calculated using prism 6 software. Primers used were:
Tankyrase RT FP: 5’ ATCTCTGCGCATACCAGCTT 3’ Tankyrase RT RP: 5’ ATCTCTGCGCATACCAGCTT 3’ beta-tubulin FP: 5′CATCCAAGCTGGTCAGTG 3’ beta-tubulin RP: 5′ GCCATGCTCATCGGAGAT 3’
Co-immunoprecipitation
To co-immunoprecipitate PARylated protein and TDP-43 from male fly heads the Pierce coimmunoprecipitation kit (ThermoFisher Scientific) was used according to the manufacturer’s instructions. Briefly, 100microL (1mg/ml) of anti-PAR (clone 10H, BSA free, Tulip Biolabs) was diluted in 100microL of PBS and desalted using 2ml Zeba desalting columns (Thermo scientific). 33microgram of desalted antibody was immobilized to the agarose resin. Adult male flies, expressing TDP-43 with elav-GAL4, were aged to day 4, a total of no more than 22 heads were homogenized for 1 minute on ice in 200microL of IP Lysis/wash buffer (Thermo Scientific), that was supplemented with cOmplete Mini EDTAfree inhibitor (Roche) and ADP-HPD (Calbiochem) at a final concentration of 1 microMolar. A total of 20microL was removed for input. Samples were pre-cleared with agarose (1 hour with gentle shaking at 4oC). Precleared lysate was incubated with the antibody-agarose resin at 4oC for 14 hours, then washed 3 times in lysis buffer containing protease inhibitor and 1 microMolar ADP-HPD (10 minutes each with rotation at 4oC), and eluted in 50 microL of elution buffer. Samples for immunoblotting were prepared in 4X LDS sample buffer, heat denatured at 90oC for 6 minutes and electrophoresed on a NuPAGE 4–12% BisTris gel. TDP-43 was detected using anti-TDP-43 (1 in 250; Proteintech) and ECL Prime (GE Healthcare Bio-Sciences).
Cos-7 cells were lysed in IP Lysis buffer (Thermo Scientific) containing 1 microMolar ADPHPD (Calbiochem) and cOmplete Mini EDTA-free inhibitor (Roche). Lysate was removed for input analysis and the remaining cell lysate was pre-cleared in either Agarose-G or dynabeads™ protein G (both ThermoFisher Scientific) for 1 hr at 4oC. 5 micrograms of antiTDP-43 (Proteintech), 5 micrograms of anti-PAR (10H) BSA free (Tulip Biolabs) and 5 micrograms of anti-PAR (Enzo life sciences) were coupled to 50 microlitres of Agarose-G or dynabeads™ protein G and incubated with the precleared lysate. The beads were washed once in lysis buffer, once in lysis buffer supplemented with 300mM NaCl, and once more in lysis buffer (each 10 min with rotation at 4oC). Beads were denatured at 70oC for 10 minutes in 1X LDS buffer and electrophoresed on a NuPAGE 4–12% Bis-Tris gel. Antibodies used for detection were rabbit anti-TDP-43 (1 in 3000; Proteintech), rabbit anti-PAR LP-96 (1 in 500; BD Biosciences). HRP-coupled secondary antibodies used: goat antibody to rabbit (1 in 5,000; EMD Millipore) and goat antibody to mouse (1 in 2,000; Jackson ImmunoResearch Laboratories, Inc.).
Plasmids
Human TDP-43 in pDuet-GST was previously described (Johnson et al., 2009). Human TDP43-WT-YFP cloned in to pcDNA3.2 was described previously (Elden et al., 2010). Fulllength human TDP-43, TDP-43-C35 and TDP-43-C25 were subcloned into pE-SUMO (LifeSensors).
TDP-43-WT, TDP-43-ΔPBM, TDP-43-C35 (amino acids 85–414) and TDP-43-C25 (amino acids 176–414) were subcloned into pcDNA3-C-eGFP by GenScript. TDP-43-MBP-His6 in PJ4M/TDP-43, obtained from ADDGENE, was described previously (Wang et al., 2018). PBM1 and PBM2 mutations were generated in TDP-43-WT-YFP (pcDNA3.2), GST-TDP-43WT (pDuet-GST) and SUMO-TDP-43-WT (pE-SUMO) by performing site directed mutagenesis (QuikChange II XL) using the following primers:
PBM1: 5’GTTGTCAACTATCCAAAAGATAACGCAGCAGCAATGGATGAGACAGATGCT 3’
PBM2: 5’GATGCTTCATCAGCAGTGGCAGTGGCAGCAGCAGTCCAGAAAACATCGAT 3’
The Q331K mutation in TDP-43-WT (pE-SUMO) was generated by site directed mutagenesis (QuikChange II XL) using the following primer:
Q331K: 5” CGCCCAGGCAGCACTAATGAGCAGTTGGGG 3’
The DNA sequence corresponding to the hnRNPA1/B2 peptide, was cloned immediately upstream of the stop site in TDP-43-ΔPBM in pcDNA3-C-eGFP, by GenScript, to create TDP-
43ΔPBM-PBM-A2/B1. The DNA sequence subcloned was:
5’AGAGAGGAATCTGGAAAACCAGGGGCTCATGTAACTGTGAAGAAGCTGTTTGTTG
GCGGAATTAAA 3’
All plasmid inserts were sequenced and confirmed to be free of any errors.
Cell stress assays and immunofluorescence
Cos7 cells were seeded at an appropriate density onto glass coverslips coated with poly-Llysine (neuVitro) in 24 well plates. The following day each well was transfected with 500 ng of plasmid DNA, 1.75 microL lipofectamine LTX and 0.5 microL PLUS reagent in 100 microL of OPTIMEM I (all ThermoFisher Scientific). For phosphorylation of the TDP-43-WT, TDP-43-C35 and TDP-43-C25 cells were transfected with lipofectamine 2000 (ThermoFisher Scientific), all other transfections of Cos-7 cells used lipofectamine LTX. Again 500 ng of plasmid with 1.75 microL of lipofectamine 2000 were made up in 100 microL of OPTIMEM I and incubated at room temperature for 5 mins. All transfection mixtures were added to each well containing 400 microL of DMEM with high glucose supplemented with 10% FBS and no antibiotics. Transfection mixtures were left on overnight until the time of the experiment. All experiments begun 19–24 hrs posttransfection. Cells were exposed to oxidative stress by incubating cells at 37oC with 5% CO2 in either DMEM media supplemented with 10% FBS and antibiotics or DMEM media supplemented with 10% FBS, antibiotics and the indicated concentration of sodium arsenite. For stress-resolution experiments, cells were exposed to 0.25mM sodium arsenite for 1 hr and then washed with PBS twice and then incubated in pre-warmed DMEM with 10% FBS and antibiotics. See below for immunofluorescence details.
For drug inhibitor assays, Cos-7 cells were grown on glass coverslips in 24-well plates and transfected with lipofectamine LTX as described above. Transfection mixtures were not removed. Cells were incubated with 1 microM XAV-939 (SelleckChem), G007-LK (SelleckChem) in DMEM with high glucose and 10% FBS for 30 mins at 37oC with 5% CO2 before the application stress. For IWR1-endo (SelleckChem) experiments, cells were incubated in either 0.5 microM, 1 microM or 10 microM of drug for 30 mins at 37oC with 5% CO2 prior to stress. For all drugs, cells were exposed to 0.25mM sodium arsenite in DMEM with high glucose and 10% FBS and the indicated amount of drug for 30 mins at 37oC with 5% CO2. Experiments commenced 22–23 hrs after transfection. See below for immunofluorescence see details.
Primary cortical neurons, in 12-well plates, were cultured in neuralbasal medium with serum-free B-27 (ThermoFisher Scientific) and penicillin-streptomycin (ThermoFisher Scientific). Transfection was carried 6 DIV. Immediately before transfection the culture media was removed and cells were washed twice in pre-warmed PBS with calcium chloride and magnesium chloride (ThermoFisher). 1ml of pre-warmed neuralbasal medium supplemented with B27 was added to each well. Transfection reactions were made up in a total of 200 microL of OPTIMEM I (ThermoFisher Scientific) and contained 500ng or 1000ng (both work well) of plasmid DNA and 2 microL of lipofectamine 2000 (ThermoFisher Scientific). Transfection mixtures were left at room temperature for 5 minutes before being added to the well. The transfection media was removed after 6 hrs, each well was washed twice with pre-warmed PBS with calcium chloride and magnesium chloride, and then the cells were returned to pre-warmed and conditioned neuralbasal media supplemented with serum-free B-27 (ThermoFisher Scientific) and penicillin-streptomycin (ThermoFisher Scientific). The neurons were stressed 10 DIV (4 days post-transfection). Transected neurons were stressed in 0.5 mM sodium arsenite for 90 min. See below for immunofluorescence see details.
Neurons and Cos-7 cells were fixed for 15 minutes in 4% paraformaldehyde (made up in 50mM HEPES pH7.4, 150mM NaCl, 1mM MgCl and 1mM EGTA), permeablized by 3 sets of 3 mins in PEM-T buffer (100mM PIPES, 1mM MgCl2, 10mM EGTA pH 6.8, and 0.1% tritonX100), and blocked in 10% normal donkey serum (Sigma-Aldrich) made up in TBST. Primary antibodies were applied overnight at 4oC (see below for antibody details) in a humidified chamber, cells were washed 3 times in PEM-T buffer (3 mins each) and secondary antibody was applied for 45 mins (Cos-7 cells) or 1 hr (neurons) at room temperature and in the dark. Cells were washed 3 times in TBST (3 mins each), cells were stained with Hoescht for 5 mins and coverslips were washed in deionized H20 immediately before mounting in prolong diamond.
For detection of PAR, endogenous TDP-43 and Tankyrase in arsenite-induced stress granules, cells were washed twice in dPBS without magnesium and without calcium and extracted in 50 mM HEPES pH 6.8, 150 mM NaCl, 1 mM MgCl2, 1 mM EGTA and 0.1% Triton X-100 for 30 seconds before fixing. Cells were fixed for 15 minutes in 4% paraformaldehyde, permeabilized in PEM-T (3 incubations, 3 mins each), then in 100 mM sodium periodate (Sigma-Aldrich) made up in PEM-T for 5 mins, washed in PEM-T for 3 mins, 25 mM succinic dihydrazide (Sigma-Aldrich) made up in PEM-T (30 minutes), 3 washes in PEM-T (3 mins each) and then blocked in 10% normal donkey serum in TBST for 1 hr at room temperature.
Primary antibodies used for both Cos-7 and neurons: mouse anti-TIAR (1 in 500; BD Biosciences), mouse anti-Ataxin-2 (1 in 500; BD Biosciences), rabbit anti-G3BP1 (1 in 1000; Thermo Scientific), goat anti-eIF3 (1 in 50; Santa Cruz Biotechnology), mouse anti-eIF3 (1 in 500; Santa Cruz Biotechnology), rabbit anti-TDP-43 (1 in 300, Proteintech), mouse antiPAR (1 in 400: Tulip BioLabs), mouse anti PAR (1 in 400, ENZO), mouse pS409/10 (1 in 1000; Cosmo Bio USA) and rabbit pS409/10 (1 in 1000; Cosmo Bio USA). Alexa-Fluor −488, −568, −594 and −647 secondary antibodies were used for detection (all 1 in 500; Thermo Scientific).
All experiments were performed at least three independent times.
Image quantification and statistics
All imaging was performed on a Leica DMI6000 unless stated otherwise. To quantify the formation of stress granules in the cytoplasm we captured 4–8 independent images at 20X. The number of cells were quantified by ImageJ software (Rueden et al., 2017) and up to 900 cells per condition per experiment were quantified. The number of cells with 2 or more stress granules were counted. An overall percentage was obtained from three independent experiments and the mean (±s.e.m.) was calculated. To quantify TDP-43-foci formation 4 non-overlapping images at 20X were captured for the WT, C35 and C25 experiments and up to 66 transfected cells were quantified per condition per experiment. These experiments were repeated 3 independent times and a mean (±s.e.m.) was calculated. For the PBMhnRNPA2/B1 experiments 6 independent images at 20X were captured and up to 391 transfected cells were quantified. These experiments were repeated 3 independent times and a mean (±s.e.m.) was calculated.
To quantify the recruitment of TDP-43 to stress granules we captured 4–5 independent images at 40X of cells that had both TIAR-labelled stress granules (or one of the other stress granule markers) and cytosolic TDP-43 foci, up to 440 stress granules were quantified per condition per experiment. A mean (±s.e.m.) was calculated from three independent experiments. To quantify the percentage of TDP-43-GFP foci co-labelled with G3BP1 or TIAR, the same images were analyzed and up to 1419 TDP-43-GFP foci were quantified for the presence of TIAR or G3BP1. These experiments were repeated 3 independent times and a mean (±s.e.m.) was calculated.
To quantify the recruitment of TDP-43 to neuronal stress granules we imaged up to 6 transfected neurons and up to 40 stress granules per condition and quantified the number of stress granules localizing with TDP-43. A mean (±s.e.m.) was calculated from three independent experiments.
To quantify the phosphorylation of TDP-43-WT, C35 and C25, 4 non-overlapping images were captured at 20X magnification for each condition, up to 124 transfected cells were quantified per condition per experimental repeat. The percentage of cells with TDP-43 foci and phosphorylated foci was calculated for each condition. For the PBMhnRNPA2/B1 experiments 6 non-overlapping images were captured at 20X magnification for each condition, up to 700 transfected cells were quantified. These experiments were repeated 3 independent times and a mean (±s.e.m.) was calculated.
All triple labelled experiments were imaged using a Leica SP5 confocal. TFP-43-YFP and TDP-43-GFP alone were used as a control slide to optimize the settings for fluorescence bleed through. Cells with both G3BP1/TIAR/eIF3-labeled stress granules and cytoplasmic TDP-43-YFP or TDP-43-GFP foci were imaged at 40X magnification, 7–27 transfected cells were quantified per condition per experiment. Experiments were performed 3–5 independent times. Statistics for all experiments were performed using Graphpad prism 6 software.
Statistics (T tests, one-way ANOVA, two-way ANOVA and multiple comparison tests) for all experiments were performed using Graphpad prism 6 software, the details for each experiment are described in each legend.
Fluorescence recovery after photobleaching
Cos-7 cells were cultured as described above. For live cell-culture experiments Cos-7 cells were grown in a poly-d-lysine coated glass bottom 35 mm culture dish (MatTek). Cells were transfected with using 7 microL lipofectamine LTX, 2 microL of plus reagent (both ThermoFisher Scientific) and 2000 ng of plasmid DNA in a total of 400 microL of OPTIMEM I (all ThermoFisher Scientific) in 1.6 ml DMEM with high glucose and 10% FBS. The transfection reagent was not removed and experiments were performed 21 hr after transfection. FRAP experiments were performed on a laser scanning Leica SP8 confocal with the FRAP module located at the CDB microscopy core, University of Pennsylvania. A cell-culture climate chamber was attached to the microscope stage. The temperature control of the chamber was previously calibrated with thermocouple measurements such that heating the outside of the chamber to 39oC gave an internal temperature of 37oC. An objective heating collar was attached to a 40X water immersion objective and the temperature was set to 33oC. The chamber was attached to a mixed gas chamber. The cellculture dish was placed in the chamber, the media was removed and rinsed in PBS and then incubated in DMEM with high glucose, 10% FBS and 1x penicillin-streptomycin in the presence or absence of 0.25 mM sodium arsenite
For FRAP experiments, images collected were 12 bit and at a resolution of 512 by 64. Scanning was unidirectional and at frequency of 1800 Hz with a zoom of 7.5 and a line average of 1. Care was taken to assure that the temperature of the microcopy room did not increase above 23oC during data acquisition. Conditions for diffuse TDP-43 were; prebleaching, 0.2% of the 488 nm laser, for 90 iterations at 76 ms; for bleaching, 100% of the 488 nm laser was used for 360 iterations at 76 ms; and for post-bleach, 0.2% of the 488 nm laser for 2500 iterations at 76 ms was used. Conditions for the TDP-43 foci for prebleaching, 0.2% of the 488 nm laser, for 50 iterations at 76ms; for bleaching, 100% of the 488 nm laser for 150 iterations at 76 ms to bleach and 0.2% of the 488 nm laser for 1750 iterations at 76 ms for the post-bleach. One GFP-positive granule was bleached per cell. Bleaching was focused on a circular region of interest (ROI) set to 1 micron in diameter. For early stress, cells were bleached from 30 mins to 1 hr and for late stress cells were bleached after 2 hr of treatment. 7 to 14 cells were bleached for each condition. Each experiment was more than performed 3 independent times on different days. After image capture, an ROI was applied to a non-bleached GFP-positive granule (to account for photobleaching). The intensity data of the ROIs were normalized and exported as a csv file.
To determine the rate of fluorescence recovery (FR) the fluorescence of the bleached ROI was normalized to the unbleached ROI at every time point (t) to take into account changes in fluorescence during image acquisition:
(Ft(bleached)/ Ft(unbleached)).
These data were then used to calculate the rate of fluorescence recovery (FR) for each time point (t) with the following formula:
FRt=(Ft(bleached) - Ft(initial bleach)) (Ft(prebleach)-Ft(initial bleach))
Each condition was repeated three times each on different days. The data from each experiment was plotted in Graphpad 6 and a one-phase decay curve was extrapolated from the data with 1000 iterations per time point. From the curve the half time (t1/2) and the plateau (mobile/immobile fraction) was calculated. A Fisher’s test was performed to identify significant differences.
GST-TDP-43 protein purification
N-terminally tagged GST-TDP-43-WT was overexpressed in BL21(DE3)RIL E. coli. The E.coli cells were then lysed by sonication on ice in 40 mM HEPES (pH 7.4), 5 mM EDTA, 300 mM KCl, 1 mM DTT, 5% glycerol, 5 microM pepstatin, 10mg/l RNase A and protease inhibitors (cOmplete, EDTA-free, Roche Applied Science). The protein was purified over Glutathione Sepharose 4 Fast Flow beads (GE Healthcare) and eluted from the beads using 40 mM HEPES (pH 7.4), 150 mM NaCl, 5% glycerol, and 20 mM reduced glutathione.
PAR polymer
Free PAR (commercially obtained from TREVIGEN) polymer was synthesized from PARP-1 and ranged in size from 2–300 ADP-ribose subunits. Molar equivalencies were calculated by TREVIGEN, briefly, the Absorbance of the PAR at 258 nm was divided by the extinction coefficient of ADP-ribose (13, 500 cm−1 M−1) (Schultheisz et al., 2009; Shah et al., 1995).
PAR-binding dot blot assay
PAR-binding motifs were identified by aligning the consensus to TDP-43 using the PATTINPROT search engine (Combet et al., 2000). For dot-blot analysis of recombinant protein, 0.25–0.5 microgram of protein, diluted in TBS with 0.1% tween (TBST), was blotted onto 0.45 micron nitrocellulose membrane. Peptides, synthesized by GenScript (details are in the Resource Table and Figure 2), were spotted onto 0.1 micron nitrocellulose at 0.25–0.5 microgram. Membranes were left to dry for 60–90 mins and then incubated in TBST for 30 mins without agitation. The membrane was sealed in a hybridization bag (KPL) containing 0.4 ml of TBST containing 50nM PAR polymer (Trevigen) and incubated for 1 hr with rocking and rotation at room temperature. The membrane was washed 5 times in TBST (5 mins each) and blocked in TBSMT (5% milk in TBST) for 1 hr then incubated in primary antibody in TBST for 1 hour. After 5 washes in TBSMT (5 minutes each), the membrane was incubated with secondary antibody in TBSMT for 30 minutes. The membrane was washed 3 times in TBSMT, 2 times in TBST and 2 times in TBS (all 5 minutes each). ECL prime was used to detect secondary antibody. Antibodies used were anti-PAR (10H) (1 in 250; Tulip Biolabs and 1 in 500; ENZO) anti-TDP-43 (1 in 3000; Proteintech). HRP-coupled secondary antibodies used: goat antibody to rabbit (1 in 5,000; EMD Millipore) and goat antibody to mouse (1 in 2,000; Jackson ImmunoResearch Laboratories, Inc. and 1 in 5000; abcam). For SYPRO Ruby staining, blotted and dried membrane were submerged in TBS and then incubated in 7% acetic acid and 10% methanol for 15 minutes, the membrane was washed 4 times (5 mins each) in deionized H2O with gentle rotation, the membrane was incubated in SYPRO Ruby blot stain (ThermoFisher Scientific for 30 minutes in the dark, washed 4 times in deionized H2O (5 mins each) and imaged on a Safe Imager (ThermoFisher Scientific). The dot blot was carried out 6 independent times and in addition was repeated on independent preparation of proteins. Spot intensities were measured using ImageJ software (Rueden et al., 2017) and statistics were performed using Graphpad prism 6 software.
Purification of Recombinant His6-SUMO-TDP-43
His6-SUMO N-terminally tagged TDP-43-WT was overexpressed in BL21(DE3)RIL E. coli. The E.coli cells were then lysed by sonication on ice in 50 mM HEPES (pH 7.5), 2% TritonX100, 500 mM NaCl, 30 mM imidazole, 5% glycerol, 2 mM β-mercaptoethanol, and protease inhibitors (cOmplete, EDTA-free, Roche). The protein was purified over Ni-NTA agarose beads (Qiagen) and eluted from the beads using 50 mM HEPES (pH 7.5), 500 mM NaCl, 300 mM imidazole, 5% glycerol, and 5 mM DTT. The protein was subsequently buffer exchanged into 50 mM HEPES (pH 7.5), 500 mM NaCl, 5% glycerol, and 5 mM DTT, flash frozen in liquid N2, and stored as aliquots in −80°C until use. His6-SUMO N-terminally tagged TDP-43-ΔPBM and TDP-43-Q331K were purified using the protocol described above. His6-SUMO N-terminally tagged. Note: for C25, the expression was induced with 1 mM IPTG for 2 hours at 37oC whereas for all the other proteins it was with 1 mM IPTG for 16 hours at 15oC.
Purified His6SUMO-TDP-43 was thawed and centrifuged at 16,100 X g for 10 minutes at 4oC to remove preformed aggregates. Protein concentration was determined by Bradford assay (Bio-Rad). The purity of the expressed proteins was confirmed on a 4–20% polyacrylamide gel.
Purification of Recombinant TDP-43-MBP-His6
The plasmid encoding TDP-43-MBP-His6 was purchased from addgene (plasmid #104480) and expressed in BL21 (DE3) RIL E. coli. following the conditions reported previously (Wang et al., 2018). Cells were harvested by centrifugation, resuspended in TDP‐43 binding buffer (20 mM Tris–Cl pH 8.0, 1 M NaCl, 10 mM imidazole, 10% (v/v) glycerol, 1 mM DTT) supplemented with complete EDTA‐free protease inhibitor cocktail, and lysed using an Emulciflex C-3. The protein was purified over Ni-NTA agarose beads (Qiagen) and eluted from the beads using 20 mM Tris–Cl pH 8.0, 1 M NaCl, 300 mM imidazole, 10% (v/v) glycerol, 1 mM DTT. The eluate was further purified over amylose resin (NEB) and eluted from the amylose resin using 20 mM Tris–Cl pH 8.0, 1 M NaCl, 10mM maltose, 10% (v/v) glycerol, 1 mM DTT. The protein was concentrated, flash frozen in liquid N2, and stored as aliquots in 80°C until use.
Liquid-droplet formation
His6-SUMO-TDP-43 was prepared in 50 mM HEPES-NaOH (pH 7.5), 5% (v/v) glycerol, 5 mM DTT, and 100 mg/mL dextran from Leuconostoc spp. (Sigma, St. Louis, MO) with the indicated concentrations of NaCl. For TDP-43-MBP-His6, the protein was first buffer exchanged into 20 mM HEPES, pH7.4, 150 mM NaCl, 1 mM DTT and reactions were prepared in 20 mM HEPES, pH7.4, 1mM DTT, 100 mg/mL dextran from Leuconostoc spp. (Sigma, St. Louis, MO) with the appropriate concentration of NaCl. Protein was always added last to each LLPS reaction. Protein samples were spotted onto a coverslip and imaged by differential interference contrast (DIC) microscopy. All movies and images were captured within ten minutes of reaction set-up. Details of experimental repeats are in the associated legends.
To assess the effect of PAR on TDP-43 phase separation, SUMO-TDP-43 was prepared as before but with the addition of 5 microM mono-ADP-ribose equivalents of PAR (Trevigen) or equivalent volume of PAR buffer (10 mM Tris, pH8 and 1mM EDTA). For SUMO-TDP-43 and PAR experiments, all movies and images were captured within ten minutes of reaction set up. For TDP-43-MBP-His6 with PAR or PAR buffer control, reactions were incubated at room temperature (~19 −22oC) for 1 hr prior to imaging.
The PAR nucleation studies of TDP-43 LLPS were repeated twice at the phase boundary. The comparison between full-length TDP-43 and TDP-43 with mutation in the PAR-binding motifs has been tested three times on two or more different protein preps. The comparison between full-length TDP-43 and the C-terminal fragments has been repeated on two independent protein preps. Testing the effect of PAR on the TDP-43-WT, TDP-43-ΔPBM and the two C-terminal fragments has been repeated on two independent protein preps. Experiments examining the phase boundary of TDP-43-MBP were performed 3–5 times and experiments analyzing PAR nucleation three times using one protein prep. Further details are in each associated legend.
QUANTIFICATION AND STATISTICAL ANALYSIS
All data points in each graph are mean (±s.e.m.) and the n is a biological repeat. T Tests, one-way ANOVA, two-way ANOVA and multiple comparison tests were performed, and are described in each figure legend. Significance was set at P < 0.05. All statistical analyses were carried out using Graphpad prism6 software.
Highlights
The NLS contains PAR-binding motifs that localize TDP-43 to stress granules
PAR promotes TDP-43 liquid-liquid phase separation
Stress granules protect TDP-43 from disease-associated phosphorylation
Tankyrase regulates cytoplasmic accumulation of TDP-43
Supplementary Material
Acknowledgements
We thank Yanlan Huang, Jeffrey Nirschl and Yongqing Zhu for technical assistance. We thank Lindsey D. Goodman for reading the manuscript and for insightful comments, the CDB Microscopy core at the University of Pennsylvania for the use of their Leica TCS SP8 confocal, and Nicolas L Fawzi (Brown University, USA) for the TDP-43-MBP-His6 plasmid DNA. This work was funded by grants from the Ellison Medical Foundation (LG), American Federation for Aging Research (LG), Alzheimer’s Association (LG), Life Extension Foundation (JS), ALS Association (JS), Department of Biochemistry and Biophysics Pilot Grant (JS), Target ALS (JS and NMB), the Glenn Foundation (NMB), the Robert Packard Center for ALS Research at Johns Hopkins (JS), and the NIH R01 NS095746–01 (RGK), R21NS093439 (RGK), 5R21NS087077–02 (RGK), R01GM099836 (JS), R21NS090205 (JS), 5R01NS073660 (NMB), R35NS097275 (NMB).
Footnotes
Declaration of Interests
The authors declare no competing interests.
DATA AVAILIBILIY
Raw data sets are available in Mendeley Data and are available at https://data.mendeley.com/datasets/rh7zv7w8c8/draft?a=ad24c605–62f4–46fe-888717e9035a78cc.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afroz T, Hock EM, Ernst P, Foglieni C, Jambeau M, Gilhespy LAB, Laferriere F, Maniecka Z, Pluckthun A, Mittl P, et al. (2017). Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation. Nat Commun 8, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker LA, Huang B, Bieri G, Ma R, Knowles DA, Jafar-Nejad P, Messing J, Kim HJ, Soriano A, Auburger G, et al. (2017). Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature 544, 367–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentmann E, Neumann M, Tahirovic S, Rodde R, Dormann D, and Haass C (2012). Requirements for stress granule recruitment of fused in sarcoma (FUS) and TAR DNAbinding protein of 43 kDa (TDP-43). The Journal of biological chemistry 287, 23079–23094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott KW (2014). Protein ADP-ribosylation and the cellular response to DNA strand breaks. DNA Repair (Amst) 19, 108–113. [DOI] [PubMed] [Google Scholar]
- Catara G, Grimaldi G, Schembri L, Spano D, Turacchio G, Lo Monte M, Beccari AR, Valente C, and Corda D (2017). PARP1-produced poly-ADP-ribose causes the PARP12 translocation to stress granules and impairment of Golgi complex functions. Scientific reports 7, 14035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CK, Wu TH, Wu CY, Chiang MH, Toh EK, Hsu YC, Lin KF, Liao YH, Huang TH, and Huang JJ (2012). The N-terminus of TDP-43 promotes its oligomerization and enhances DNA binding affinity. Biochemical and biophysical research communications 425, 219–224. [DOI] [PubMed] [Google Scholar]
- Citarelli M, Teotia S, and Lamb RS (2010). Evolutionary history of the poly(ADP-ribose) polymerase gene family in eukaryotes. BMC Evol Biol 10, 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen TJ, Hwang AW, Unger T, Trojanowski JQ, and Lee VM (2012). Redox signalling directly regulates TDP-43 via cysteine oxidation and disulphide cross-linking. The EMBO journal 31, 1241–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombrita C, Zennaro E, Fallini C, Weber M, Sommacal A, Buratti E, Silani V, and Ratti A (2009). TDP-43 is recruited to stress granules in conditions of oxidative insult. Journal of neurochemistry 111, 1051–1061. [DOI] [PubMed] [Google Scholar]
- Combet C, Blanchet C, Geourjon C, and Deleage G (2000). NPS@: network protein sequence analysis. Trends Biochem Sci 25, 147–150. [DOI] [PubMed] [Google Scholar]
- Conicella AE, Zerze GH, Mittal J, and Fawzi NL (2016). ALS Mutations Disrupt Phase Separation Mediated by alpha-Helical Structure in the TDP-43 Low-Complexity C-Terminal Domain. Structure 24, 1537–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey CM, Cenik B, Sephton CF, Dries DR, Mayer P 3rd, Good SK, Johnson BA, Herz J, and Yu G (2011). TDP-43 is directed to stress granules by sorbitol, a novel physiological osmotic and oxidative stressor. Molecular and cellular biology 31, 1098–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, et al. (2010). Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466, 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatokun AA, Dawson VL, and Dawson TM (2014). Parthanatos: mitochondrial-linked mechanisms and therapeutic opportunities. Br J Pharmacol 171, 2000–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JP, Hunter JM, Labrecque B, Chabot B, and Poirier GG (2003). A proteomic approach to the identification of heterogeneous nuclear ribonucleoproteins as a new family of poly(ADP-ribose)-binding proteins. The Biochemical journal 371, 331–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, Dawson TM, and Poirier GG (2008). Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res 36, 6959–6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JP, Pic E, Isabelle M, Krietsch J, Ethier C, Paquet E, Kelly I, Boutin M, Moon KM, Foster LJ, et al. (2012). Quantitative proteomics profiling of the poly(ADP-ribose)related response to genotoxic stress. Nucleic Acids Res 40, 7788–7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson BA, and Kraus WL (2012). New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nature reviews. Molecular cell biology 13, 411–424. [DOI] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, et al. (2012). Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149, 768–779. [DOI] [PubMed] [Google Scholar]
- Harrison AF, and Shorter J (2017). RNA-binding proteins with prion-like domains in health and disease. The Biochemical journal 474, 1417–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Arai T, Nonaka T, Kametani F, Yoshida M, Hashizume Y, Beach TG, Buratti E, Baralle F, Morita M, et al. (2008). Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Annals of neurology 64, 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottiger MO (2015). Nuclear ADP-Ribosylation and Its Role in Chromatin Plasticity, Cell Differentiation, and Epigenetics. Annu Rev Biochem 84, 227–263. [DOI] [PubMed] [Google Scholar]
- Hsiao SJ, and Smith S (2008). Tankyrase function at telomeres, spindle poles, and beyond. Biochimie 90, 83–92. [DOI] [PubMed] [Google Scholar]
- Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, et al. (2009). Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461, 614–620. [DOI] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, and Julicher F (2014). Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol 30, 39–58. [DOI] [PubMed] [Google Scholar]
- Igaz LM, Kwong LK, Chen-Plotkin A, Winton MJ, Unger TL, Xu Y, Neumann M, Trojanowski JQ, and Lee VM (2009). Expression of TDP-43 C-terminal Fragments in Vitro Recapitulates Pathological Features of TDP-43 Proteinopathies. The Journal of biological chemistry 284, 8516–8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabelle M, Gagne JP, Gallouzi IE, and Poirier GG (2012). Quantitative proteomics and dynamic imaging reveal that G3BP-mediated stress granule assembly is poly(ADP-ribose)dependent following exposure to MNNG-induced DNA alkylation. J Cell Sci 125, 4555–4566. [DOI] [PubMed] [Google Scholar]
- Jiang LL, Xue W, Hong JY, Zhang JT, Li MJ, Yu SN, He JH, and Hu HY (2017). The N-terminal dimerization is required for TDP-43 splicing activity. Scientific reports 7, 6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BS, Snead D, Lee JJ, McCaffery JM, Shorter J, and Gitler AD (2009). TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. The Journal of biological chemistry 284, 20329–20339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kametani F, Obi T, Shishido T, Akatsu H, Murayama S, Saito Y, Yoshida M, and Hasegawa M (2016). Mass spectrometric analysis of accumulated TDP-43 in amyotrophic lateral sclerosis brains. Scientific reports 6, 23281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. (2012). Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Ivanov P, and Anderson P (2013). Stress granules and cell signaling: more than just a passing phase? Trends Biochem Sci 38, 494–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, and Anderson P (1999). RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. The Journal of cell biology 147, 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Raphael AR, LaDow ES, McGurk L, Weber RA, Trojanowski JQ, Lee VM, Finkbeiner S, Gitler AD, and Bonini NM (2014). Therapeutic modulation of eIF2alpha phosphorylation rescues TDP-43 toxicity in amyotrophic lateral sclerosis disease models. Nature genetics 46, 152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krietsch J, Rouleau M, Pic E, Ethier C, Dawson TM, Dawson VL, Masson JY, Poirier GG, and Gagne JP (2013). Reprogramming cellular events by poly(ADP-ribose)-binding proteins. Mol Aspects Med 34, 1066–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK (2014). Poly(ADP-ribose): an organizer of cellular architecture. The Journal of cell biology 205, 613–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Vyas S, Rood JE, Bhutkar A, Sharp PA, and Chang P (2011). Poly(ADPribose) regulates stress responses and microRNA activity in the cytoplasm. Molecular cell 42, 489–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YR, King OD, Shorter J, and Gitler AD (2013). Stress granules as crucibles of ALS pathogenesis. The Journal of cell biology 201, 361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DS, Rosen MK, and Parker R (2015). Formation and Maturation of PhaseSeparated Liquid Droplets by RNA-Binding Proteins. Molecular cell 60, 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling SC, Albuquerque CP, Han JS, Lagier-Tourenne C, Tokunaga S, Zhou H, and Cleveland DW (2010). ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc Natl Acad Sci U S A 107, 13318–13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bilgutay A, Zhang YJ, Vanderweyde T, Citro A, Mehta T, Zaarur N, McKee A, Bowser R, Sherman M, et al. (2010). Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PloS one 5, e13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Bigio EH, Ince PG, Geser F, Neumann M, Cairns NJ, Kwong LK, Forman MS, Ravits J, Stewart H, et al. (2007). Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Annals of neurology 61, 427–434. [DOI] [PubMed] [Google Scholar]
- Mackenzie IR, Nicholson AM, Sarkar M, Messing J, Purice MD, Pottier C, Annu K, Baker M, Perkerson RB, Kurti A, et al. (2017). TIA1 Mutations in Amyotrophic Lateral Sclerosis and Frontotemporal Dementia Promote Phase Separation and Alter Stress Granule Dynamics. Neuron 95, 808–816 e809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariotti L, Pollock K, and Guettler S (2017). Regulation of Wnt/beta-catenin signalling by tankyrase-dependent poly(ADP-ribosyl)ation and scaffolding. Br J Pharmacol 174, 4611–4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGurk L, Lee VM, Trojanowksi JQ, Van Deerlin VM, Lee EB, and Bonini NM (2014). Poly-A binding protein-1 localization to a subset of TDP-43 inclusions in amyotrophic lateral sclerosis occurs more frequently in patients harboring an expansion in C9orf72. Journal of neuropathology and experimental neurology 73, 837–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojsilovic-Petrovic J, Jeong GB, Crocker A, Arneja A, David S, Russell DS, and Kalb RG (2006). Protecting motor neurons from toxic insult by antagonism of adenosine A2a and Trk receptors. The Journal of neuroscience : the official journal of the Society for Neuroscience 26, 9250–9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, and Taylor JP (2015). Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mompean M, Romano V, Pantoja-Uceda D, Stuani C, Baralle FE, Buratti E, and Laurents DV (2016). The TDP-43 N-terminal domain structure at high resolution. FEBS J 283, 1242–1260. [DOI] [PubMed] [Google Scholar]
- Murakami T, Qamar S, Lin JQ, Schierle GS, Rees E, Miyashita A, Costa AR, Dodd RB, Chan FT, Michel CH, et al. (2015). ALS/FTD Mutation-Induced Phase Transition of FUS Liquid Droplets and Reversible Hydrogels into Irreversible Hydrogels Impairs RNP Granule Function. Neuron 88, 678–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Kwong LK, Lee EB, Kremmer E, Flatley A, Xu Y, Forman MS, Troost D, Kretzschmar HA, Trojanowski JQ, et al. (2009). Phosphorylation of S409/410 of TDP-43 is a consistent feature in all sporadic and familial forms of TDP-43 proteinopathies. Acta neuropathologica 117, 137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. (2006). Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133. [DOI] [PubMed] [Google Scholar]
- Nonaka T, Arai T, Buratti E, Baralle FE, Akiyama H, and Hasegawa M (2009). Phosphorylated and ubiquitinated TDP-43 pathological inclusions in ALS and FTLD-U are recapitulated in SH-SY5Y cells. FEBS letters 583, 394–400. [DOI] [PubMed] [Google Scholar]
- Panzeter PL, Zweifel B, Malanga M, Waser SH, Richard M, and Althaus FR (1993). Targeting of histone tails by poly(ADP-ribose). The Journal of biological chemistry 268, 17662–17664. [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. (2015). A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 162, 1066–1077. [DOI] [PubMed] [Google Scholar]
- Pleschke JM, Kleczkowska HE, Strohm M, and Althaus FR (2000). Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. The Journal of biological chemistry 275, 40974–40980. [DOI] [PubMed] [Google Scholar]
- Romano V, Quadri Z, Baralle FE, and Buratti E (2015). The structural integrity of TDP43 N-terminus is required for efficient aggregate entrapment and consequent loss of protein function. Prion 9, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, and Eliceiri KW (2017). ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan VH, Dignon GL, Zerze GH, Chabata CV, Silva R, Conicella AE, Amaya J, Burke KA, Mittal J, and Fawzi NL (2018). Mechanistic View of hnRNPA2 Low-Complexity Domain Structure, Interactions, and Phase Separation Altered by Mutation and Arginine Methylation. Molecular cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheisz HL, Szymczyna BR, and Williamson JR (2009). Enzymatic synthesis and structural characterization of 13C, 15N-poly(ADP-ribose). J Am Chem Soc 131, 14571–14578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah GM, Poirier D, Duchaine C, Brochu G, Desnoyers S, Lagueux J, Verreault A, Hoflack JC, Kirkland JB, and Poirier GG (1995). Methods for biochemical study of poly(ADP-ribose) metabolism in vitro and in vivo. Anal Biochem 227, 1–13. [DOI] [PubMed] [Google Scholar]
- Smith S, Giriat I, Schmitt A, and de Lange T (1998). Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282, 1484–1487. [DOI] [PubMed] [Google Scholar]
- Sun Z, Diaz Z, Fang X, Hart MP, Chesi A, Shorter J, and Gitler AD (2011). Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS biology 9, e1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teloni F, and Altmeyer M (2016). Readers of poly(ADP-ribose): designed to be fit for purpose. Nucleic Acids Res 44, 993–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronkov A, Holsworth DD, Waaler J, Wilson SR, Ekblad B, Perdreau-Dahl H, Dinh H, Drewes G, Hopf C, Morth JP, et al. (2013). Structural basis and SAR for G007-LK, a lead stage 1,2,4-triazole based specific tankyrase 1/2 inhibitor. Journal of medicinal chemistry 56, 3012–3023. [DOI] [PubMed] [Google Scholar]
- Wang A, Conicella AE, Schmidt HB, Martin EW, Rhoads SN, Reeb AN, Nourse A, Ramirez Montero D, Ryan VH, Rohatgi R, et al. (2018). A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. The EMBO journal 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winton MJ, Igaz LM, Wong MM, Kwong LK, Trojanowski JQ, and Lee VM (2008). Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. The Journal of biological chemistry 283, 13302–13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wippich F, Bodenmiller B, Trajkovska MG, Wanka S, Aebersold R, and Pelkmans L (2013). Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 152, 791–805. [DOI] [PubMed] [Google Scholar]
- Wittmann CW, Wszolek MF, Shulman JM, Salvaterra PM, Lewis J, Hutton M, and Feany MB (2001). Tauopathy in Drosophila: neurodegeneration without neurofibrillary tangles. Science 293, 711–714. [DOI] [PubMed] [Google Scholar]
- Xiang S, Kato M, Wu LC, Lin Y, Ding M, Zhang Y, Yu Y, and McKnight SL (2015). The LC Domain of hnRNPA2 Adopts Similar Conformations in Hydrogel Polymers, Liquid-like Droplets, and Nuclei. Cell 163, 829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Sanelli T, Chiang H, Sun Y, Chakrabartty A, Keith J, Rogaeva E, Zinman L, and Robertson J (2015). Low molecular weight species of TDP-43 generated by abnormal splicing form inclusions in amyotrophic lateral sclerosis and result in motor neuron death. Acta neuropathologica 130, 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Daigle JG, Cunningham KM, Coyne AN, Ruan K, Grima JC, Bowen KE, Wadhwa H, Yang P, Rigo F, et al. (2018). Stress Granule Assembly Disrupts Nucleocytoplasmic Transport. Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Caulfield T, Xu YF, Gendron TF, Hubbard J, Stetler C, Sasaguri H, Whitelaw EC, Cai S, Lee WC, et al. (2013). The dual functions of the extreme N-terminus of TDP-43 in regulating its biological activity and inclusion formation. Human molecular genetics 22, 3112–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.