Abstract
The transient receptor potential (TRP) channel TRPV3 plays a crucial role in skin physiology and pathophysiology. Mutations in TRPV3 are associated with various skin diseases, including Olmsted syndrome, atopic dermatitis and rosacea. Here we present cryo-EM structures of full length mouse TRPV3 in the closed and agonist-bound open states. The agonist binds three allosteric sites distal to the pore. Channel opening is accompanied by conformational changes in both the outer pore and the intracellular gate. The gate is formed by the pore-lining S6 helices that undergo local α-to-π helical transitions, elongate, rotate, and splay apart in the open state. In the closed state, the shorter S6 segments are entirely α-helical, expose their non-polar surfaces to the pore, and hydrophobically seal the ion permeation pathway. These findings further illuminate TRP channel activation and can aid in design of drugs for the treatment of inflammatory skin conditions, itch and pain.
Introduction
Cation-selective transient receptor potential (TRP) channels play important roles in numerous physiological processes and are known as polymodal sensors that integrate a wide range of physical and chemical stimuli in cellular signaling1. TRPV3, a representative of the vanilloid subfamily of TRP channels2–4, is predominantly expressed in skin keratinocytes and is implicated in cutaneous sensation including thermo-sensation, nociception and itch, in addition to maintenance of the skin barrier, wound healing, and hair growth3,5–7.
The amino acid sequence of TRPV3 is ~30–40% similar to other temperature-sensitive vanilloid subtype TRP channels: TRPV1, 2 and 4. In contrast to TRPV1 and TRPV2, which are activated by noxious heat (>40°C), TRPV3 is activated at innocuous temperatures (~33°C)2,4,8,9, similar to TRPV4. TRPV3 is also activated by natural compounds such as camphor, carvacrol, eugenol and thymol, as well as by the small synthetic compound 2-aminoethoxydiphenyl borate (2-APB)10–16, which has recently been shown to suppress tumor growth and invasiveness17. Electrophysiological studies have demonstrated that 2-APB can regulate numerous TRP channels; 2-APB activates TRPV1, TRPV3, TRPA1, TRPM6, a TRPM2-like ion channel from Nematostella vectensis18–21, and inhibits TRPV6, TRPM7, TRPM2, TRPC3, TRPC6, and TRPC722–25. TRPV3 is inhibited by the lipid PIP2, an important regulatory lipid present in cell membrane, which also inhibits the closely related ion channels TRPV1 and TRPV2 26,27, activates TRPV5 and TRPV6, and facilitates the activation of TRPV428–30. Unlike TRPV1, 2 and 4, which all desensitize upon successive heat or agonist applications, TRPV3 undergoes sensitization14,15, which is a property that has been attributed to the reduced Ca2+-dependent inactivation of TRPV331.
The dysfunction of TRPV3 channels, often a result of genetic mutations, is associated with numerous human skin diseases including a genodermatosis known as Olmsted syndrome, atopic dermatitis, and rosacea32,33. Furthermore, overexpression of TRPV3 is implicated in the development and progression of colorectal and lung cancer34. Therefore, TRPV3 represents an important therapeutic target for the treatment of a variety of devastating diseases12,16. Indeed, GRC15300, a small molecule inhibitor of TRPV3, is in clinical trials as an analgesic drug35,36. Efforts to better understand the role of TRPV3 in physiology and disease, and to target this ion channel with informed drug design, however, are hampered by a lack of TRPV3 structural data.
Here, we determine cryo-electron microscopy (cryo-EM) structures of the full-length mouse TRPV3 in the closed (apo) state and in an open state bound to the agonist 2-APB. Together, these structures shed light on the functional architecture of TRPV3 and its gating mechanism. More specifically, the mechanism by which a ligand can regulate the gating state of TRPV3 is markedly different from similar processes in the closely related ion channels TRPV1 and TRPV6.
Results and Discussion
Closed apo-state structure
We used Fluorescence-detection Size-Exclusion Chromatography (FSEC)37 to screen different TRPV3 orthologues and found mouse TRPV3 to be a promising target for our cryo-EM structural studies. We expressed full-length TRPV3 channels in HEK-293 cells and confirmed their function by Fura2 AM-based measurements of changes in intracellular calcium in response to the application of an agonist, 2-APB (Supplementary Fig. 1a).
First, we determined the structure of TRPV3 without the addition of ligand. The resulting apo-state cryo-EM reconstruction, refined in C4 symmetry to an overall resolution of 4.3 Å (Fig. 1 and Table 1), exhibited a higher local resolution in the core of the channel (~2.7 Å) when compared to its periphery (Supplementary Fig. 2). Our cryo-EM maps (Supplementary Fig. 3) allowed for the building of an accurate model of the TRPV3 homotetramer (residues 115 to 756), guided by a high-resolution structure of the isolated TRPV3 ankyrin repeat domain (ARD)38 and by cryo-EM reconstructions of TRPV1, 2, 4 and 639–42. The overall TRPV3 architecture (Fig. 1a,b,d,e) is reminiscent of other TRPV channels39–44 and includes the ion channel-forming transmembrane domain (TMD), which is composed of the S1-S4 and pore domains in a domain-swapped arrangement, and a large intracellular skirt domain that is primarily assembled of ankyrin repeat domains (ARDs) and encloses a large cytoplasmic cavity. Each ARD is connected to the TMD by a linker domain. Amphipathic TRP helices run nearly parallel to the membrane and interact with both the skirt and the TMD. A comparison of TRPV3 with other members of the TRPV subfamily highlights similarities and differences in their respective TMDs and ARDs (Supplementary Fig. 4).
Figure 1. 3D cryo-EM reconstruction and structure of TRPV3 in the apo state.

a,b, Top (a) and side (b) views of 3D cryo-EM reconstruction of TRPV3 in the apo state with subunits coloured green, yellow, violet and cyan and lipid in purple. The densities are shown at a threshold of 0.057 (UCSF Chimera). c, Expanded view of the two putative lipid densities shown as purple mesh at 4σ. d,e, Top (d) and side (e) views of the TRPV3 structure with the same coloring as in panels a-c. f, Expanded view of the boxed region in (e) displaying the interaction of the TRPV3 C-terminal loop domain with the ARD from the neighbouring subunit.
Table 1.
Cryo-EM data collection, refinement and validation statistics
| TRPV3 (EMDB-8919) (PDB 6DVW) |
TRPV32-APB (EMDB-8920) (PDB 6DVY) |
TRPV3(Y564A)2-APB (EMDB-8921) (PDB 6DVZ) |
TRPV3(Y564A) (EMDB-8925) |
|
|---|---|---|---|---|
| Data collection and processing | ||||
| Magnification | 130,000x | 29,000x | 130,000x | 39,000 |
| Voltage (kV) | 300 | 200 | 300 | 300 |
| Electron exposure (e–/Å2) | 57 | 43 | 50 | 67 |
| Defocus range (μm) | −1.5 to −3.5 | −1.5 to −3.5 | 1.5 to −2.5 | −1.5 to −3.5 |
| Pixel size (Å) | 1.08 | 1.22 | 1.06/0.53 | 0.98 |
| Symmetry imposed | C4 | C4 | C4 | C4 |
| Initial particle images (no.) | 1,134,540 | 348,634 | 1,064,473 | 900,300 |
| Final particle images (no.) | 91,694 | 81,120 | 28,075 | 44,661 |
| Map resolution (Å) FSC threshold |
4.3 | 4.0 | 4.08 | 6.5 |
| Map resolution range (Å) | 2.6 to 7.0 | 2.6 to 7.0 | 2.6 to 7.0 | N/A |
| Refinement | ||||
| Initial model used (PDB code) | 3J5P, 6BO8 | This study | This study | |
| Model resolution (Å) FSC threshold |
4.3 | 4.0 | 4.08 | |
| Model resolution range (Å) | 2.6 to 7.0 | 2.6 to 7.0 | 2.6 to 7.0 | |
| Map sharpening B factor (Å2) | −298 | −211 | −204 | |
| Model composition | ||||
| Non-hydrogen atoms | 21,004 | 20,872 | 20,992 | |
| Protein residues | 2576 | 2564 | 2572 | |
| Ligands | N/A | 2 | 3 | |
| B factors (Å2) | ||||
| Protein | 76.87 | 75.9 | 80.5 | |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.01 | 0.01 | 0.01 | |
| Bond angles (°) | 1.16 | 1.15 | 1.13 | |
| Validation | ||||
| MolProbity score | 1.99 | 2.02 | 1.97 | |
| Clashscore | 5.69 | 7.02 | 7.23 | |
| Poor rotamers (%) | 0.87 | 0.88 | 0.97 | |
| Ramachandran plot | ||||
| Favored (%) | 83.72 | 86.23 | 82.63 | |
| Allowed (%) | 15.89 | 13.03 | 16.74 | |
| Disallowed (%) | 0.39 | 0.74 | 0.63 | |
Domain assembly and lipid-like densities
Following the C-terminal β3 strand of the 3-stranded β-sheet, the TRPV3 polypeptide forms a C-terminal loop domain, which has not been observed in other TRPV channels. It includes nineteen well-resolved residues (residues 737 to 756) that share low (<10%) sequence identity with other TRPV channels (Supplementary Fig. 5). The newly identified C-terminal loop domain extends along the corners of the TRPV3 intracellular skirt and forms an extensive network of interactions with ankyrin repeats 2–5 of the ARD (Fig. 1d-f). Two robust non-protein densities, presumably representing lipids, are observed in each subunit of the TRPV3 tetramer (Fig. 1a-e). The first density is sandwiched between the extracellular half of S4 of one subunit and the pore-domain of the adjacent subunit (Fig. 1c). Notably, this lipid binding site is analogous to the finger 1 binding site of the double-knot toxin (DkTx) in TRPV139 and has not been observed in other TRP channel structures. The second density is in a pocket formed by the intracellular half of the S1-S4 domain and the C-terminal portion of the TRP domain (Fig. 1c). An analogous pocket harbors putative lipids in other TRPV channels39–41,43, 2-APB in TRPV625 or menthol in TRPM845. Importantly, a lipid occupying site 1 would interact with the pore residues N647 and L657, which have been implicated in TRPV3 activation by temperature46. Thus, the site 1 lipid might be involved in TRPV3 gating and in its thermo-regulation in particular.
Closed pore of TRPV3
Similar to other tetrameric ion channels, the permeation pathway of TRPV3 is lined by the S6 helices and the extended portions of the pore loops (P-loops). A calculation of the radius of the TRPV3 pore in the apo-state (Fig. 2a-b) demonstrated that the narrowest pore constriction is formed by S6 methionines (M677), which project their side chains towards the center of the permeation pathway and form a hydrophobic seal. We conclude that M677 forms the intracellular gate and that the apo-state structure represents the closed non-conducting conformation of TRPV3. Indeed, the interatomic radius at the intracellular gate of TRPV3 (5.5 Å) is comparable to the corresponding radii in the closed state structures of other members of the TRPV subfamily. Similar to TRPV3, the intracellular gate is formed by methionine residues in TRPV240,44 (M643), TRPV442 (M714), TRPV5 (M578)43 and TRPV6 (M577)47. The corresponding methionine in TRPV1 is pointing away from the pore axis and the intracellular gate is formed by I67939,48.
Figure 2: Closed pore of TRPV3.
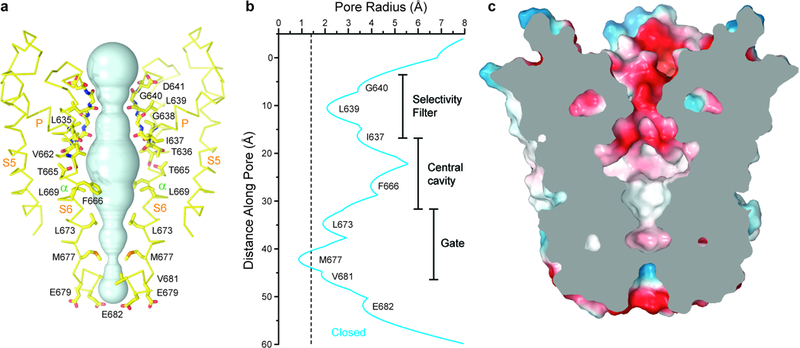
a, Pore-forming domain of TRPV3 in the apo state with residues lining the pore shown as sticks. Only two of four subunits are shown; the front and back subunits are omitted for clarity. The pore profile is shown as a space-filling model (cyan). b, Pore radius calculated using HOLE54. The vertical dashed line denotes the radius of a water molecule, 1.4 Å. c, Coronal section of the TRPV3 pore domain, with surface coloured by electrostatic potential.
The ion conduction pathway of TRPV3 in the closed state exhibits striking polarity; the outer pore module, including the selectivity filter, is lined exclusively by hydrophilic residues and backbone carbonyls, while the inner pore, including the central cavity and the intracellular gate region, is lined entirely by hydrophobic residues (Fig. 2a, c). The intracellular mouth of the TRPV3 pore is lined by a pair of negatively charged residues, E679 and E682, which were both shown to be important for intracellular magnesium block49. The selectivity filter of TRPV3 is defined by the residues 638GLGD641 in the P-loop extended region. The backbone carbonyl of L639 forms the selectivity filter’s narrowest constriction (7.5 Å) in the closed state (Fig. 2a-b). This constriction is comparable to that of TRPV1 in an open state48 (7.6 Å, formed by the G643 backbone carbonyls), but significantly larger than that of TRPV1 in the closed state (4.8 Å, formed by the G643 backbone carbonyls), TRPV240 (5.2 Å, formed by the G604 backbone carbonyls) and TRPV647 (4.2 Å, formed by the D541 side chains). The wide selectivity filter of TRPV3 in the closed state, which has a negatively charged surface, is large enough to permeate hydrated Na+, K+ or Ca2+ ions and suggests the absence of an upper gate. The TRPV3 gating mechanism, therefore, starkly contrasts that of TRPV1, which has been shown to have two, upper and lower, gates39,48.
TRPV3 structure in complex with 2-APB
In order to determine the open state structure, we purified wild type TRPV3 in the presence of the agonist 2-APB. Surprisingly, the resulting cryo-EM reconstruction was identical to the apo state reconstruction (Supplementary Fig. 6, Table 1), suggesting that 2-APB did not open the ion channel. To shift the TRPV3 gating equilibrium towards the open state, we made a substitution of a conserved tyrosine (Y564A, Supplementary Fig. 5) in lipid binding site 2 that increases the apparent affinity of 2-APB ~20 fold25 (Supplementary Fig. 1b,d). The resulting cryo-EM reconstruction of TRPV3(Y564A)2-APB (Supplementary Fig. 7, Table 1) was significantly different from the closed state reconstructions (Supplementary Fig. 6) and contained three strong non-protein densities per subunit (Fig. 3a-b).
Figure 3: Structure of 2-APB-bound TRPV3(Y564A).
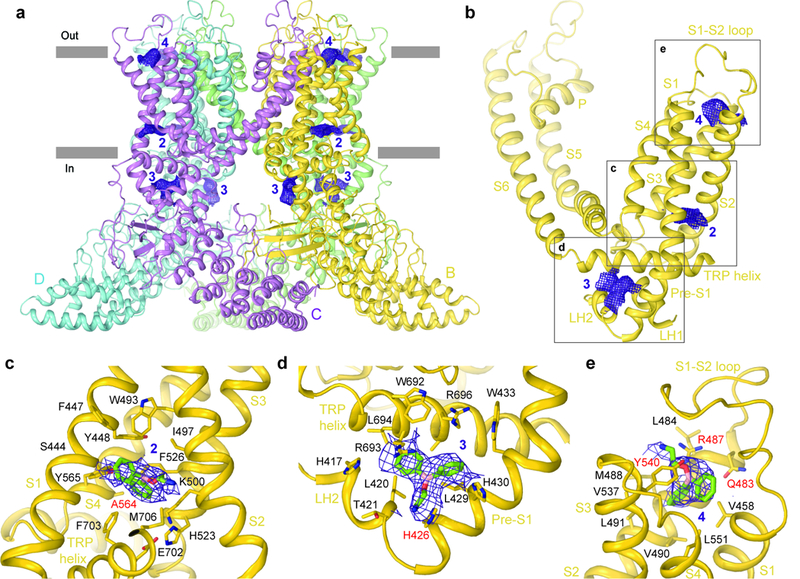
a, Side view of the TRPV3(Y564A)2-APB structure with each subunit coloured differently and non-protein densities shown as purple mesh at 4σ. b-e, Non-protein densities for one subunit (b) with expanded views of densities at site 2 (c), site 3 (d) and site 4 (e). 2-APB molecules in (c-e) and residues surrounding them are shown as sticks.
One density was observed in the same pocket as putative lipid density 2 in the closed state structure (cf. Figs. 1 and 3). We hypothesize that this site 2 density in TRPV3(Y564A)2-APB represents a molecule of 2-APB. Indeed, this site harbors the Y564A mutation, which increases the potency of 2-APB (Supplementary Fig. 1), it binds 2-APB in the analogous site in TRPV625, and the shape of TRPV3(Y564A)2-APB density 2 matches 2-APB. Correspondingly, the chemical environment of site 2 supports binding of 2-APB (Fig. 3c); S444 of S1 interacts with the boron atom of 2-APB, E501 of S2 and Y565 of S3 form polar interactions with the 2-APB central oxygen atom and the 2-APB tail amino group, respectively. Additionally, one of the two phenyl rings of 2-APB interacts with the imidazole group of H523 and with F526 of S3 while the other ring stacks hydrophobically with W493 of S2.
The density in site 3 is located at the nexus of the linker domain, pre-S1 helix and TRP domain, and also matches the shape of 2-APB (Fig. 3d). Binding of 2-APB to this site is supported by a previous mutagenesis-based study50. Indeed, the mutation H426A in site 3 completely eliminates TRPV3 activation by 2-APB but not by camphor (Supplementary Fig. 1c-d). Despite the drastic effect of the H426A mutation, binding of 2-APB to site 3 might not be the only requirement for TRPV3 activation by 2-APB. The Y564A mutation in site 2 restored activation of the H426A mutant and nearly completely recovered its apparent affinity for 2-APB (Supplementary Fig. 1d). At site 3, 2-APB binding is mediated primarily by polar residues, including H417 and T421 of the linker domain, H426 and H430 of the pre-S1 helix, and R693 and R696 of the TRP domain (Fig. 3d). The density at site 4 is nested in a cavity formed by the extracellular portions of helices S1-S4 (Fig. 3e), which in the closed state TRPV3 structure is occupied by the S1-S2 loop (Supplementary Fig. 8a-b). 2-APB binding at site 4 is mediated by both hydrophobic and hydrophilic residues including V458, Y540, R487, and Q483. Alanine substitutions of Y540, R487 or Q483 result in rightward shifted dose-response curves for TRPV3 activation by 2-APB (Supplementary Fig. 1d), supporting the role of these residues in 2-APB binding. Notably, all three putative 2-APB binding sites of TRPV3(Y564A)2-APB are distinct from the binding site at the S4-S5 interface that binds an activating lipid in TRPV641, agonists such as resiniferatoxin and capsaicin, and antagonists, such as capsazepine, in TRPV139, and the inhibitor econazole in TRPV551.
Ion conducting pore in the open state
A calculation of the pore radius confirmed that the TRPV3(Y564A)2-APB pore is wide open for conducting hydrated ions (Fig. 4). The entire ion conduction pathway is lined by polar or negatively-charged side chains, or backbone carbonyls (Fig. 4a), and it has an overall negative surface charge (Fig. 4c), which is likely critical for the cation selectivity of TRPV3. The pore’s most narrow constrictions at the gate region are defined by the side chains of E682 (interatomic distance of 8.2 Å) and I674 (interatomic distance of 8.7 Å). Despite their hydrophobic character, the I674 side chains do not introduce strong changes in the overall negatively-charged surface of the pore (Fig. 4c) and constrict the pore to a similar extent as the residues forming the narrowest pore constriction at the intracellular gate region of open TRPV148 (interatomic distance of 9.3 Å, measured between the side chains of I679) and TRPV641 (interatomic distance of 9.6 Å, measured between the side chains of I575). The diameter of the narrowest constriction in the selectivity filter (7.5 Å) is defined by the backbone carbonyl of G638. Although the narrowest constriction in the closed state is formed by the backbone carbonyl of a different residue (L639), its diameter is almost identical to the constriction in the open state (Fig. 4b), supporting the idea that this region of the TRPV3 pore does not form a gate and functions primarily to support ion selectivity and conductance.
Figure 4: Open pore of TRPV3.
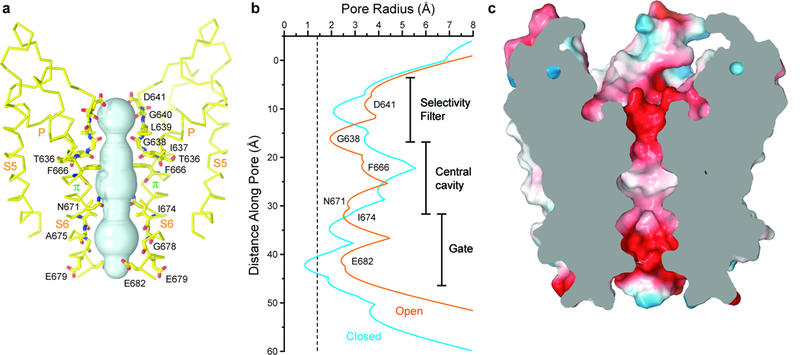
a, Pore-forming domain of TRPV3(Y564A)2-APB with residues lining the pore shown as sticks and the pore profile shown as a space-filling model (cyan). b, Pore radius calculated using HOLE54 for TRPV3 (blue) and TRPV3(Y564A)2-APB (orange). The vertical dashed line denotes the radius of a water molecule, 1.4 Å. c, Coronal section of the TRPV3(Y564A)2-APB pore-forming domain, with surface coloured by electrostatic potential.
Conformational changes in the pore during gating
Superposition of the closed and open TRPV3 structures reveals pronounced conformational changes in the pore-forming region (Fig. 5). For instance, upon channel opening, the pore helices tilt by 7° and their C-terminal ends, which point towards the center of the pore, move down by ~5 Å (Fig. 5a). The rearrangement in the upper pore results in different residues contributing their backbone carbonyls to the selectivity filter’s narrowest constriction in the closed (L639) and open (G638) states. Concurrently, each S6 helix undergoes a local α-to-π helical transition, similar to the one observed in TRPV641, which originates at T665 (Fig. 5a-c). T665 is highly conserved in TRPV1–4 and is analogous to the gating hinge alanine in TRPV641 and glycine in K+ channels52,53 (Supplementary Fig. 5). The formation of the short π-helix results in S6 splaying away from the pore by about ~11° and a ~100° rotation of its lower portion. The rotation exposes an entirely different set of S6 residues to the pore in the open and closed states. In the closed state, these are mostly hydrophobic residues that seal the pore and form the gate (Fig. 2). In the open state, however, these are primarily hydrophilic and negatively-charged residues that promote ion permeation (Fig. 4).
Figure 5: Structural changes associated with TRPV3 channel opening.
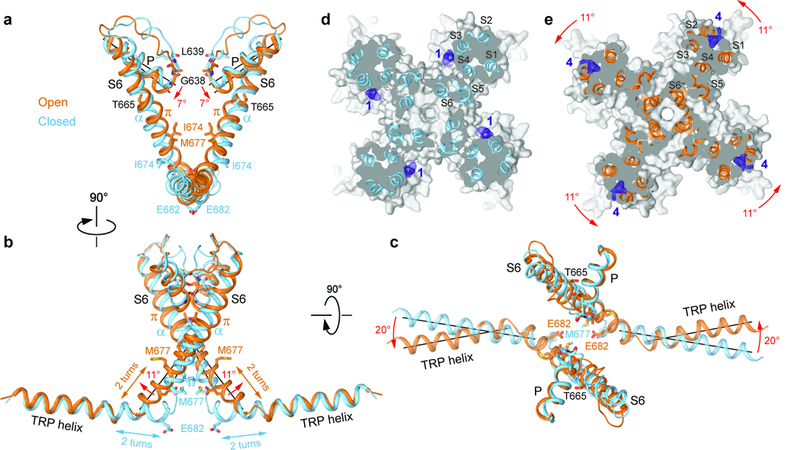
a-c, Superposition of the P-loop, S6 and TRP helices of TRPV3 (blue) and TRPV3(Y564A)2-APB (orange) viewed parallel to the membrane (a,b) or extracellulary (c). Only two of four subunits are shown for clarity. Residues lining the pore are shown as sticks. Rotations of the pore, S6 and TRP helices in TRPV3(Y564A)2-APB relative to TRPV3 are indicated by red arrows. d-e, Sections through the space filling models of TRPV3 (d, blue) and TRPV3(Y564A)2-APB (e, orange) made perpendicular to the pore axis and viewed extracellularly. Non-protein densities at site 1 (d) and site 4 (e) are shown as purple mesh at 4σ. The structures are aligned based on their pore domains. The 11° rotation of the S1-S4 domains in TRPV3(Y564A)2-APB relative to their position in TRPV3 is indicated by the red arrows.
In contrast to the gating rearrangements in TRPV139,48 and TRPV641, the conformational changes in S6 are propagated to the TRP helix and result in dramatic overall changes in TRPV3. In the open state, the S6 helices become longer by two helical turns (Fig. 5a-c) and the TRP helices become two helical turns shorter while rotating ~20° in the plane of membrane. As a result, the rest of the channel, including the S1-S4 domains and the intracellular skirt, rotate around the pore domain and the channel becomes ~5Å shorter upon opening (Fig. 6a-b). Such pronounced structural rearrangements have not been observed during activation of TRPV1 (Fig. 6c-d) or TRPV6 (Fig. 6e-f) and emphasize the substantially different gating mechanism of TRPV3. The rotation of the S1-S4 domain, with respect to the pore domain, reduces the distance between the extracellular halves of S4 and S6 of the neighboring subunit (Figs. 5d-e, Fig. 6a) and expels the lipid from site 1, which is only present in the closed state (Fig. 1). This lipid expulsion is consistent with previous observations that lipids can stabilize the resting state of TRPV3 and that transitioning from a closed to an open state can be accompanied by changes in lipid binding13.
Figure 6: Comparison of gating rearrangements in different TRPV channels.
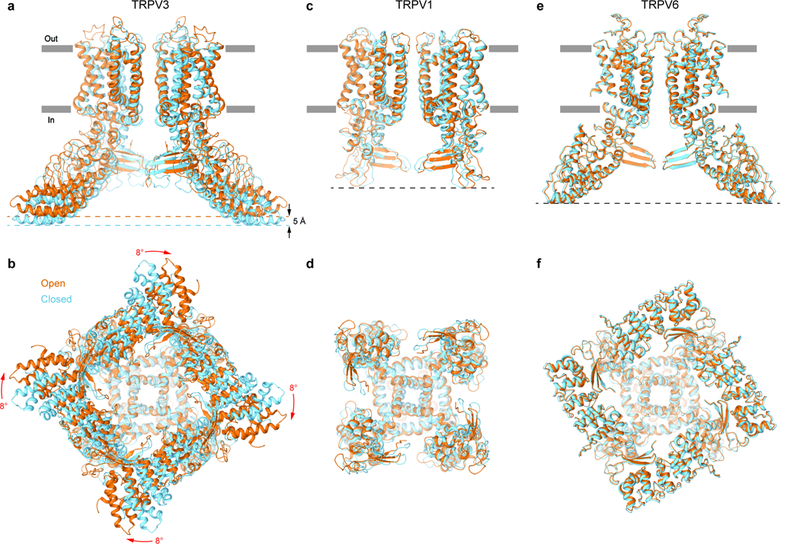
a-f, Superposition of structures in the open (orange) and closed (blue) states for TRPV3 (a-b, TRPV3(Y564A)2-APB and TRPV3, respectively), TRPV1 (c-d, PDB: 5IRX and PDB: 5IRZ, respectively) and TRPV6 (e-f, PDB: 6BO8 and PDB: 6BOA, respectively) viewed parallel to the membrane (a,c,e) or intracellularly (b,d,f). Only two of four subunits are shown in (a,c,e), with the front and back subunits omitted for clarity. The structures are aligned based on their pore domains. Note, the TRPV3 structure becomes shorter (black arrows) and its intracellular skirt undergoes substantial rotation (red arrows) during channel opening, while the overall architecture of TRPV1 and TRPV6 remains the same.
Mechanism of TRPV3 activation by 2-APB
By comparing the putative 2-APB binding sites 2 and 3 in the open and closed state structures of TRPV3, we discovered that they are almost identical and do not exhibit strong gating-associated conformational changes (Supplementary Fig. 8c). In contrast, dramatic structural rearrangements are observed at site 4 (Fig. 7a). We hypothesize that 2-APB binds at site 4, where it outcompetes the S1-S2 loop (Supplementary Fig. 8a-b). This binding seems to be important for channel opening because the closed state structure of TRPV32-APB has 2-APB molecules at sites 2 and 3 but not at site 4. As a result of 2-APB binding to site 4, the top of the S1-S4 bundle expands, the cleft between S4 and the neighboring S6 narrows, the lipid is squeezed out of site 1, and the S1-S4 and pore domain interface rearranges in a manner that supports channel opening (Fig. 7b). Binding of 2-APB to sites 2 and 3 is likely necessary for the relative stability of the pore, S1-S4 and skirt domains during channel opening and therefore is prerequisite for gating (Supplementary Fig. 1). Similar to TRPV6, two highly conserved residues in S5 (D586) and S6 (T680) form a hydrogen bond that stabilizes the open state (Fig. 7a). Further studies are necessary to decipher the energetics and domain dynamics during TRPV3 gating. The open and closed structures of TRPV3 presented here can therefore serve as a spring board for such studies as well as templates for the design of drugs targeting inflammatory skin conditions, itch, pain and cancer.
Figure 7: TRPV3 Gating Mechanism.
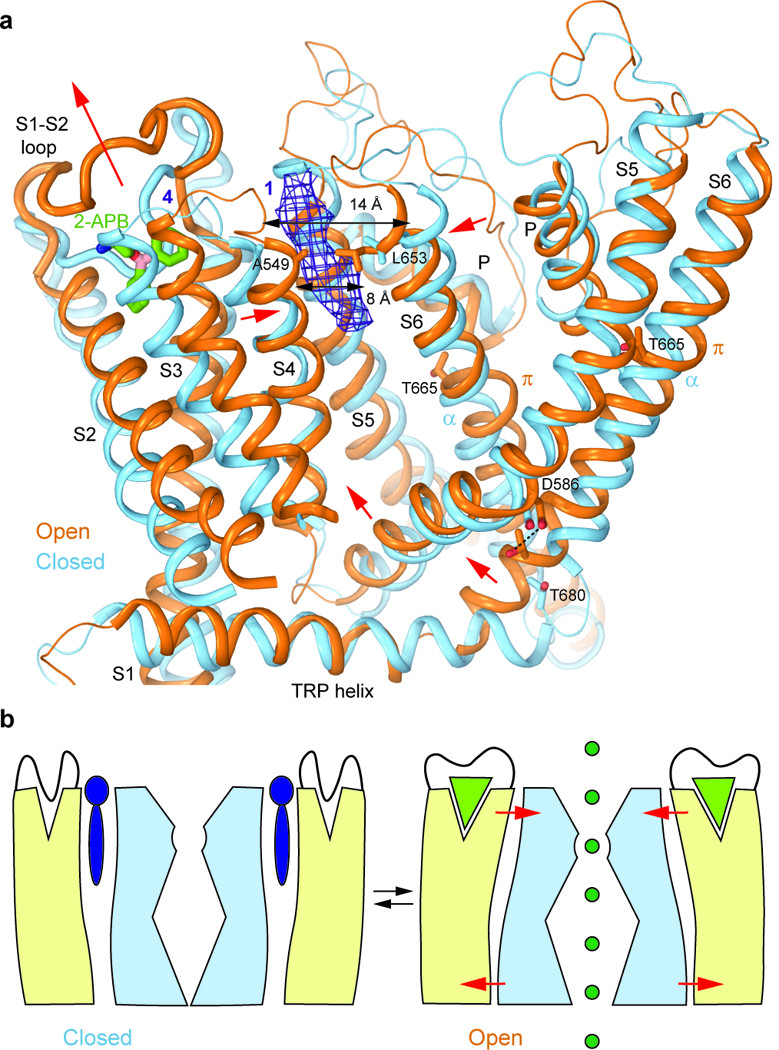
a, Superposition of the transmembrane domains and TRP helices of TRPV3 (blue) and TRPV3(Y564A)2-APB (orange) viewed parallel to the membrane. Only a single subunit and a fragment of a neighbouring subunit (S5 through S6) are shown for clarity. The putative molecule of 2-APB bound at site 4 in TRPV3(Y564A)2-APB is shown as sticks and the non-protein density at site 1 of TRPV3 is shown as purple mesh at 4σ. Red arrows indicate movements of domains in TRPV3(Y564A)2-APB relative to their positions in TRPV3. b, Cartoon illustrating gating in TRPV3. In the closed state (left), a lipid (purple) is bound between the S1-S4 (beige) and pore (light blue) domains and the pocket at the top of the S1-S4 domain is occupied by the S1-S2 loop. In the open state (right), 2-APB (light green triangle) binds to and expands the top of the S1-S4 domain, squeezes out the lipid and brings the S1-S4 and pore domains closer together. As a result, the S1-S4-pore domain interface rearranges, and S5 and S6 move away from the pore, opening it for permeation of ions (dark green).
Online Methods
Construct
The full-length mouse TRPV3 (residues 1–825) was introduced into a pEG BacMam vector37, with the C-terminal thrombin cleavage site (LVPRG) followed by the streptavidin affinity tag (WSHPQFEK). The H426A, Q483A, R487A, Y540A, and Y564A, mutations were introduced by conventional mutagenesis.
Expression and purification
All TRPV3 constructs were expressed and purified as was done previously for TRPV6cryst47,55 with minor differences. Bacmids and baculoviruses were produced as described in the literature37. Briefly, baculovirus was produced in Sf9 cells (Thermo Fisher Scientific, mycoplasma test negative, GIBCO #12659017) and was added to the suspension of HEK 293S cells lacking N-acetyl-glucosaminyltransferase I (GnTI-, mycoplasma test negative, ATCC #CRL-3022) that were grown in Freestyle 293 media (Gibco-Life Technologies #12338–018) supplemented with 2% FBS. The cells were cultured at 37°C and 5% CO2 until ~10 h after transduction, at which point 10 mM sodium butyrate was added and the temperature was reduced to 30°C. 48–72 hours post-transduction, the cells were pelleted by centrifugation at 5471 g for 15 min using a Sorvall Evolution RC Centrifuge (Thermo Scientific), washed in phosphate buffer saline (PBS) pH 8.0, and pelleted again by centrifugation at 3202 g for 10 min using an Eppendorf Centrifuge 5810. After resuspension in buffer (50 ml per 800 ml of culture) containing 150 mM NaCl, 20 mM Tris-HCl (pH 8.0), 1 mM βME (beta-mercaptoethanol) and protease inhibitors (0.8 μM aprotinin, 2 μg/ml leupeptin, 2 μM pepstatin A and 1 mM phenylmethysulfonyl fluoride) the cells were subjected to sonication with a Misonix sonicator (12 × 15 s, power level 8). After sonication, the cell lysate was cleared by slow speed centrifugation and the resulting supernatant was subjected to ultracentrifugation in a Beckman Coulter ultracentrifuge using Beckman Coulter Type Ti-45 rotor at 186,000 g for 1 h to pellet the membranes. The membranes were mechanically homogenized and solubilized for ~2 h in buffer containing 150 mM NaCl, 20 mM Tris-HCl pH 8.0, 1% digitonin and 1 mM BME. The mixture was subjected to ultracentrifugation for 1 h at 186,000 g to remove any remaining insoluble material. The supernatant was added to streptavidin-linked resin and rotated for 10–14 h at 4°C. The resin was washed with 10 column volumes of wash buffer containing 150 mM NaCl, 20 mM Tris-HCl pH 8.0, 1 mM BME and 0.05% digitonin and the protein was eluted with the same buffer to which 2.5 mM D-desthiobiotin was added prior. All constructs were purified by size-exclusion chromatography using a Superose 6 column equilibrated in 150 mM NaCl, 20 mM Tris-HCl pH 8.0, 1mM BME and 0.05% digitonin for TRPV3 and 0.01% GDN, rather than digitonin, for TRPV3(Y564A)2-APB. Tris(2-carboxyethyl)phosphine (TCEP, 10 mM) was added to the tetrameric peak fractions which were pooled together and concentrated to ~3.6 mg/ml.
Cryo-EM sample preparation and data collection
Au/Au grids were prepared as described in the literature56. Briefly, grids were prepared by first coating C-flat (Protochips, Inc., Morrisville, NC) CF-1.2/1.3–2Au mesh holey carbon grids with ~50 nm of gold using an Edwards Auto 306 evaporator. Subsequently, an Ar/O2 plasma treatment (4 min, 50 watts, 35.0 sccm Ar, 11.5 sccm O2) was used to remove the carbon with a Gatan Solarus (model 950) Advanced Plasma Cleaning System. The grids were again plasma treated (H2/O2, 20 s, 10 watts, 6.4 sccm H2, 27.5 sccm O2) prior to sample application in order to make their surfaces hydrophilic. A Vitrobot Mark V1 was used to plunge-freeze the grids after application of 3-µl protein solution with 100% humidity at 5˚C, a blot time of 2 s, blot force set to 3, and a wait time of 20 s.
The TRPV3 data was collected on a Titan Krios TEM (FEI) operating at 300 kV equipped with a Gatan K2 Summit direct electron detection (DED) camera (Gatan, Pleasanton, CA, USA) and using a post-column GIF Quantum energy filter. 4715 micrographs were collected in counting mode with a pixel size of 1.08 Å across a defocus range of −1.0 µm to −3.5 µm. The total dose, ~57 e-Å−2, was attained by using a dose rate of ~8.0 e-pixel−1s−1 across 40 frames for 8 s total exposure time.
The TRPV3(Y564A)2-APB data were collected on a Titan Krios TEM operating at 300 kV equipped with a Gatan K2 Summit DED camera using Leginon57. 1447 micrographs were collected in super resolution mode with a pixel size of 0.53 Å and 2404 micrographs were collected in counting mode with a pixel size of 1.06 Å. A defocus range of −1.0 µm to −2.5 µm was used and the total dose, ~50 e-Å−2, was attained by using a dose rate of 4.7 by e-pixel−1s−1 across 40 frames for 12 s total exposure time.
The TRPV32-APB data was collected on a Tecnai F20 TEM operating at 200 kV equipped with a Gatan K2 Summit DED camera using Leginon57. 1032 micrographs were collected in counting mode with an image pixel size of 1.22 Å. A defocus of range of −1.5 µm to −3.5 µm was used and the total dose, ~47 e-Å−2, was attained by using a dose rate of ~8.0 e-pixel−1s−1 across 40 frames for 8 s total exposure time.
The TRPV3(Y564A) data was collected on a Tecnai F30 Polara TEM operating at 300 kV equipped with a Gatan K2 Summit DED camera using Leginon57. 2734 micrographs were collected in counting mode with an image pixel size of 0.98 Å. A defocus of range of −1.5 µm to −3.5 µm was used and the total dose, ~67 e-Å−2, was attained by using a dose rate of ~8.0 e-pixel−1s−1 across 40 frames for 8 s total exposure time.
Image processing
Frame alignment for all data was done using MotionCor258 and micrographs that were collected using the Gatan K2 in super resolution mode were binned 2 × 2. CTF correction, using Gctf59, was performed on non-dose-weighted micrographs while subsequent data processing was done on dose-weighted micrographs. All other data processing, unless stated otherwise, was performed using Relion 2.060.
As a representative of our image processing work flow, the TRPV3 dataset was processed as follows. Initially, ~1000 particles were manually picked to generate 2D classes that were subsequently used to automatically pick 1,134,540 particles. The particle images were subjected to 3D classification using the open state structure of TRPV6 as a reference (PDB: 6BO8). One class, composed of ~200,000 particles, was refined with C4 symmetry and post-processed. The resulting ~4.7 Å structure was used a reference for the next round of 3D classification (with C4 symmetry) in which the ~200,000 particles were separated into 10 classes. 3 of the resulting classes, composed of 91,694 particles were refined with C4 symmetry and post-processed to generate the final 4.3 Å map.
Each data set was processed in a similar workflow to that described above and the reported resolutions were estimated using the Fourier shell correlation (FSC)=0.143 criterion61 on masking-effect-corrected FSC curves calculated between two independent half-maps62. The local resolutions were estimated with unfiltered half maps using ResMap63 and EM density visualization was done in UCSF Chimera64.
Model building
To build the closed state and open state models of TRPV3 in COOT65, we used the TRPV1/2/6 cryo-EM structures39–41 as guides. The resulting models were refined against unfiltered half maps (work) in real space with constraints using PHENIX66. The refined models were tested for overfitting (Supplementary Fig. 2, 7) by shifting their coordinates by 0.5 Å (using shake) in PHENIX and generating densities in Chimera64 from the shaken models. FSC was calculated between the densities from the shaken models, the half maps used in PHENIX refinement (work), the second half maps (free), and the unfiltered sum maps, using EMAN267. The local resolutions in the transmembrane regions of our TRPV3, TRPV32-APB and TRPV3(Y564A) 2-APB maps all exhibited higher resolution details in their TM domains than in their soluble domains; this observation was supported by local resolution predictions made using ResMap63. These resolutions allowed us to unambiguously define the conformation of S6 in the open and closed state structures as well as the existence of the π-helix in the middle of S6 in the open state. Structures were visualized and figures were prepared in Pymol66.
Fura 2-AM measurements
Wild type TRPV3, TRPV3(H426A), TRPV3(Q483A), TRPV3(R487A), TRPV3(Y540A), TRPV3(Y564A) and TRPV3(H426A-Y564A) fused to C-terminal strep tags were expressed in HEK 293S cells. 50–60 h after viral transduction, cells were harvested by centrifugation at 600 g for 5 min. The cells were resuspended in pre-warmed modified HEPES buffered saline (HBS) (118 mM NaCl, 4.8 mM KCl, 1 mM MgCl2, 5 mM D-glucose, 2.5 mM CaCl2 and 10 mM HEPES pH 7.4) containing 5 μg/ml of Fura2-AM (Life Technologies) and incubated at 37°C for 45 min25,47. The loaded cells were then centrifuged for 5 min at 600 g, resuspended in prewarmed, modified HBS and incubated again at 37°C for 25–35 min in the dark. The cells were subsequently pelleted and washed twice, then resuspended in modified HBS for experiments. After preparing the cells, they were kept on ice and in the dark prior to taking an aliquot for a fluorescence recording. If the experiment lasted for greater than two hours, the cells were re-washed before taking further recordings to ensure that measurements were only taken from healthy cells. Fluorescence measurements were conducted using spectrofluorometer QuantaMaster™ 40 (Photon Technology International) at room temperature in a quartz cuvette under constant stirring. Intracellular Ca2+ was measured in response to different concentration of 2-APB or camphor, by taking the ratio at two excitation wavelengths (340 and 380 nm) and at one emission wavelength (510 nm). The excitation wavelength was switched with 1-s intervals.
Supplementary Material
Acknowledgements:
We thank H. Kao for computational support, R. Grassucci and F. Acosta-Reyes for assistance with microscope operation and U. Baxa and T. Edwards for help with data collection. L.L.M. is supported by the NIH (T32 GM008224). A.I.S. is supported by the NIH (R01 CA206573, R01 NS083660), the Amgen Young Investigator and Irma T. Hirschl Career Scientist Awards. Data were collected at the Frederick National Laboratory for Cancer Research National Cryo-EM Facility (NIH) and at the Simons Electron Microscopy Center and National Resource for Automated Molecular Microscopy (New York Structural Biology Center) supported by grants from the Simons Foundation (349247), NYSTAR and the NIH (GM103310).
Accession Codes: Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers EMD-8919 for TRPV3, EMD-8920 for TRPV32-APB, EMD-8921 for TRPV3(Y564A)2-APB and EMD-8925 for TRPV3(Y564A). Model coordinates have been deposited in the Protein Data Bank (PDB) under accession numbers 6DVW for TRPV3, 6DVY for TRPV32-APB and 6DVZ for TRPV3(Y564A)2-APB.
Footnotes
Data Availability
Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers EMD-8919 for TRPV3, EMD-8920 for TRPV32-APB, EMD-8921 for TRPV3(Y564A)2-APB and EMD-8925 for TRPV3(Y564A). Model coordinates have been deposited in the Protein Data Bank (PDB) under accession numbers 6DVW for TRPV3, 6DVY for TRPV32-APB and 6DVZ for TRPV3(Y564A)2-APB. All other data are available from the corresponding author upon request.
References
- 1.Clapham DE TRP channels as cellular sensors. Nature 426, 517–524 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Xu HX et al. TRPV3 is a calcium-permeable temperature-sensitive cation channel. Nature 418, 181–186 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Peier AM et al. A heat-sensitive TRP channel expressed in keratinocytes. Science 296, 2046–2049 (2002). [DOI] [PubMed] [Google Scholar]
- 4.Smith GD et al. TRPV3 is a temperature-sensitive vanilloid receptor-like protein. Nature 418, 186–190 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Cheng X. et al. TRP channel regulates EGFR signaling in hair morphogenesis and skin barrier formation. Cell 141, 331–343 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aijima R. et al. The thermosensitive TRPV3 channel contributes to rapid wound healing in oral epithelia. FASEB J 29, 182–192 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Imura K. et al. Influence of TRPV3 mutation on hair growth cycle in mice. Biochem Biophys Res Commun 363, 479–483 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Liu B, Yao J, Zhu MX & Qin F. Hysteresis of gating underlines sensitization of TRPV3 channels. J Gen Physiol 138, 509–520 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrer-Montiel A. et al. Advances in modulating thermosensory TRP channels. Expert Opinion on Therapeutic Patents 22, 999–1017 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Sherkheli MA, Vogt-Eisele AK, Weber K. & Hatt H. Camphor modulates TRPV3 cation channels activity by interacting with critical pore-region cysteine residues. Pak J Pharm Sci 26, 431–438 (2013). [PubMed] [Google Scholar]
- 11.Vogt-Eisele AK et al. Monoterpenoid agonists of TRPV3. British Journal of Pharmacology 151, 530–540 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Broad LM et al. TRPV3 in Drug Development . Pharmaceuticals 9, 55 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doerner JF, Hatt H & Ramsey IS Voltage- and temperature-dependent activation of TRPV3 channels is potentiated by receptor-mediated PI(4,5)P-2 hydrolysis. Journal of General Physiology 137, 271–288 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung MK, Lee H, Mizuno A, Suzuki M & Caterina MJ 2-aminoethoxydiphenyl borate activates and sensitizes the heat-gated ion channel TRPV3. Journal of Neuroscience 24, 5177–5182 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moqrich A. et al. Impaired thermosensation in mice lacking TRPV3, a heat and camphor sensor in the skin. Science 307, 1468–1472 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Wang G & Wang K. The Ca(2+)-Permeable Cation Transient Receptor Potential TRPV3 Channel: An Emerging Pivotal Target for Itch and Skin Diseases. Mol Pharmacol 92, 193–200 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Nelson AM et al. 2-APB arrests human keratinocyte proliferation and inhibits cutaneous squamous cell carcinoma in vitro. bioRxiv (2018). doi: 10.1101/249821 [DOI] [Google Scholar]
- 18.Hu HZ et al. 2-aminoethoxydiphenyl borate is a common activator of TRPV1, TRPV2, and TRPV3. Journal of Biological Chemistry 279, 35741–35748 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Hinman A, Chuang HH, Bautista DM & Julius D. TRP channel activation by reversible covalent modification. Proc Natl Acad Sci U S A 103, 19564–19568 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li M, Jiang J. & Yue L. Functional characterization of homo- and heteromeric channel kinases TRPM6 and TRPM7. J Gen Physiol 127, 525–537 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn FJP, Mathis W, Cornelia K, Hoffmann DC & Luckhoff A. Modulation of activation and inactivation by Ca(2+) and 2-APB in the pore of an archetypal TRPM channel from Nematostella vectensis. Sci Rep 7, 7245 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Togashi K, Inada H & Tominaga M. Inhibition of the transient receptor potential cation channel TRPM2 by 2-aminoethoxydiphenyl borate (2-APB). Br J Pharmacol 153, 1324–1330 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lievremont JP, Bird GS & Putney JW Jr. Mechanism of inhibition of TRPC cation channels by 2-aminoethoxydiphenylborane. Mol Pharmacol 68, 758–762 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Chokshi R, Fruasaha P. & Kozak JA 2-Aminoethyl diphenyl borinate (2-APB) inhibits TRPM7 channels through an intracellular acidification mechanism. Channels 6, 362–369 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh AK, Saotome K, McGoldrick LL & Sobolevsky AI Structural bases of TRP channel TRPV6 allosteric modulation by 2-APB. Nature Communications 9, 2465 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chuang HH et al. Bradykinin and nerve growth factor release the capsaicin receptor from PtdIns(4,5)P2-mediated inhibition. Nature 411, 957–62 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Mercado J, Gordon-Shaag A, Zagotta WN & Gordon SE Ca2+-Dependent Desensitization of TRPV2 Channels Is Mediated by Hydrolysis of Phosphatidylinositol 4,5-Bisphosphate. Journal of Neuroscience 30, 13338–13347 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Elias A. et al. Phosphatidylinositol-4,5-biphosphate-dependent rearrangement of TRPV4 cytosolic tails enables channel activation by physiological stimuli. Proc Natl Acad Sci U S A 110, 9553–9558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee J, Cha SK, Sun TJ & Huang CL PIP2 activates TRPV5 and releases its inhibition by intracellular Mg2+. J Gen Physiol 126, 439–451 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thyagarajan B, Lukacs V. & Rohacs T. Hydrolysis of phosphatidylinositol 4,5-bisphosphate mediates calcium-induced inactivation of TRPV6 channels. J Biol Chem 283, 14980–14987 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao R. et al. Calcium plays a central role in the sensitization of TRPV3 channel to repetitive stimulations. Journal of Biological Chemistry 283, 6162–6174 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin ZM et al. Exome Sequencing Reveals Mutations in TRPV3 as a Cause of Olmsted Syndrome. American Journal of Human Genetics 90, 558–564 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ni C. et al. A novel mutation in TRPV3 gene causes atypical familial Olmsted syndrome. Scientific Reports 6, 21815 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li XL et al. Overexpression of TRPV3 Correlates with Tumor Progression in Non-Small Cell Lung Cancer. International Journal of Molecular Sciences 17, 437 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang SM & Chung MK Targeting TRPV3 for the Development of Novel Analgesics. Open Pain J 6, 119–126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reilly RM & Kym PR Analgesic Potential of TRPV3 Antagonists. Current Topics in Medicinal Chemistry 11, 2210–2215 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Goehring A. et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat Protoc 9, 2574–2585 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi DJ, Ye S, Cao X, Zhang R. & Wang K. Crystal structure of the N-terminal ankyrin repeat domain of TRPV3 reveals unique conformation of finger 3 loop critical for channel function. Protein Cell 4, 942–950 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao Y, Cao EH, Julius D. & Cheng YF TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 534, 347–351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zubcevic L. et al. Cryo-electron microscopy structure of the TRPV2 ion channel. Nature Structural & Molecular Biology 23, 180–186 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGoldrick LL et al. Opening of the human epithelial calcium channel TRPV6. Nature 553, 233–237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng ZQ et al. Cryo-EM and X-ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms. Nature Structural & Molecular Biology 25, 252–260 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hughes TET et al. Structural basis of TRPV5 channel inhibition by econazole revealed by cryo-EM. Nature Structural & Molecular Biology 25, 53–60 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huynh KW et al. Structure of the full-length TRPV2 channel by cryo-EM. Nature Communications 7, 11130 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin Y. et al. Structure of the cold- and menthol-sensing ion channel TRPM8. Science 359, 237–241 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grandl J. et al. Pore region of TRPV3 ion channel is specifically required for heat activation. Nature Neuroscience 11, 1007–1013 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saotome K, Singh AK, Yelshanskaya MV & Sobolevsky AI Crystal structure of the epithelial calcium channel TRPV6. Nature 534, 506–511 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao E, Liao M, Cheng Y. & Julius D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature 504, 113–118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo J, Stewart R, Berdeaux R & Hu H. Tonic inhibition of TRPV3 by Mg2+ in mouse epidermal keratinocytes. J Invest Dermatol 132, 2158–2165 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu HZ, Grandl J, Bandell M, Petrus M & Patapoutian A. Two amino acid residues determine 2-APB sensitivity of the ion channels TRPV3 and TRPV4. Proceedings of the National Academy of Sciences of the United States of America 106, 1626–1631 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hughes TET et al. Structural basis of TRPV5 channel inhibition by econazole revealed by cryo-EM. Nat Struct Mol Biol 25, 53–60 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang Y. et al. X-ray structure of a voltage-dependent K+ channel. Nature 423, 33–41 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Jiang Y. et al. Crystal structure and mechanism of a calcium-gated potassium channel. Nature 417, 515–522 (2002). [DOI] [PubMed] [Google Scholar]
- 54.Smart OS, Neduvelil JG, Wang X, Wallace BA & Sansom MS HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph 14, 354–360, 376 (1996). [DOI] [PubMed] [Google Scholar]
- 55.Singh AK, Saotome K. & Sobolevsky AI Swapping of transmembrane domains in the epithelial calcium channel TRPV6. Sci Rep 7, 10669 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Russo CJ & Passmore LA Electron microscopy: Ultrastable gold substrates for electron cryomicroscopy. Science 346, 1377–1380 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suloway C. et al. Automated molecular microscopy: the new Leginon system. J Struct Biol 151, 41–60 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kimanius D, Forsberg BO, Scheres SHW & Lindahl E Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife 5, e18722 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scheres SH & Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods 9, 853–854 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen S. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kucukelbir A, Sigworth FJ & Tagare HD Quantifying the local resolution of cryo-EM density maps. Nat Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Emsley P, Lohkamp B, Scott WG & Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Afonine PV et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang G. et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 68.Swint-Kruse L. & Brown CS Resmap: automated representation of macromolecular interfaces as two-dimensional networks. Bioinformatics 21, 3327–3328 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


