Abstract
Purpose
A common variant of the melatonin receptor 1B (MTNR1B) gene has been related to increased signaling of melatonin, a hormone previously associated with body fatness mainly through effects on energy metabolism. We examined whether the MTNR1B variant affects changes of body fatness and composition in response to a dietary weight loss intervention.
Methods
The MTNR1B rs10830963 variant was genotyped for 722 overweight and obese individuals, who were randomly assigned to one of four diets varying in macronutrient composition. Anthropometric and body composition measurements (DXA scan) were collected at baseline and at 6 and 24 months of follow-up.
Results
Statistically significant interactions were observed between the MTNR1B genotype and low-/high-fat diet on changes in weight, body mass index (BMI), waist circumference (WC) and total body fat (p interaction=0.01, 0.02, 0.002 and 0.04, respectively), at 6 months of dietary intervention. In the low-fat diet group, increasing number of the sleep disruption related G allele was significantly associated with a decrease in weight (p=0.004), BMI (p=0.005) and WC (p=0.001). In the high-fat diet group, carrying the G allele was positively associated with changes in body fat (p=0.03). At 2 years, the associations remained statistically significant for changes in body weight (p=0.02), BMI (p=0.02) and WC (p=0.048) in the low-fat diet group, although the gene-diet interaction became less significant.
Conclusions
The results suggest that carriers of the G allele of the MTNR1B rs10830963 may have a greater improvement in body adiposity and fat distribution when eating a low-fat diet.
Keywords: Melatonin receptor 1B, gene-diet interaction, high-fat diet, weight-loss intervention, adiposity
INTRODUCTION
It has long been recognized that circadian system is implicated in the regulation of energy balance, and subsequently affecting body fatness [1]. In humans beings, disruption of circadian rhythms by means of shift work, social jet lag, sleep deprivation, timed feeding, and consumption of a high-fat diet, among others, has been related to obesity and metabolic disturbances such as type 2 diabetes and cardiovascular disease [2–4]. One of the most important chronobiotics is melatonin, a hormone secreted mainly by the pineal gland and a key mediator used by the central master clock to synchronize the circadian system [5].
Recent genome-wide association studies (GWAS) have identified common variants in the Melatonin receptor 1B (MTNR1B) gene, which encodes one of the two high-affinity receptors for melatonin [6, 7], associated with fasting plasma glucose and the risk of type 2 diabetes [8–10]. Among them, the MTNR1B rs10830963 risk allele has been related to melatonin signaling [11]. The sleep disruption G allele has also been associated with adiposity measures such as body mass index (BMI) and waist circumference (WC) as well as with body weight loss [12–15]. Notably, evidence has indicated that melatonin also plays a key role in the regulation of adipocyte biology (lipolysis, lipogenesis), the activation of brown adipose tissue, the participation in the browning process of white adipose tissue, and the maintenance of an adequate energy balance acting on the regulation of energy expenditure and energy intake [5, 16, 17]. Therefore, we hypothesized that the MTNR1B genotype might affect changes in body fatness and composition in response to dietary interventions.
In the current study, we investigated the effect of the MTNR1B rs10830963 genetic variant on changes in body fatness and body composition in response to weight-loss diets varying in macronutrient contents in the Preventing Overweight Using Novel Strategies (POUNDS Lost) trial.
METHODS
Study population
The POUNDS Lost trial is a 2-year randomized clinical trial (clinical trial reg. no. NCT00072995) conducted from October 2004 through December 2007 at two sites: Harvard School of Public Health and Brigham and Women’s Hospital in Boston, MA, and the Pennington Biomedical Research Center of Louisiana State University System, Baton Rouge, LA. Details of the study design and methods have been described elsewhere [18]. This 2-year study included 811 overweight and obese (BMI 25 – 40 kg/m2) individuals who were randomly assigned to one of four diets in which calories were restricted by 750 kcal/day. The target percentages of energy derived from fat, protein, and carbohydrate in the four diets were: 20, 15 and 60%; 20, 25 and 55%; 40, 15 and 45%; and 40, 25 and 35%, respectively. Thus, the 4 diets constituted a 2-by-2 factorial design: two diets were low fat (20%), two diets were high fat (40%), two diets were average protein (15%), and two diets were high protein (25%). At baseline, 6 months and 2 years of the intervention anthropometric and body composition measurements, as well as blood samples (serum lipids, glucose and insulin levels) were collected under fasting conditions. Moreover, 24-hour urine samples, blood pressure, and measurement of resting metabolic rate were obtained. Major exclusion criteria in this trial were the presence of diabetes or unstable cardiovascular disease, the use of medications that affect body weight, and insufficient motivation [18]. The study was approved by the institutional review board (Reference 15-852312, Tulane University) and by a data and safety monitoring board appointed by the National Heart, Lung and Blood Institute. All participants provided written informed consent.
In the present study, 722 subjects with baseline genotyped data of the MTNR1B rs10830963 variant, were included [19]. Among them, 79.3% were white, 15.7% were African American and 5.0% were Hispanic or other ethnic groups by self-report. For the analyses, body weight and WC data were available for 722 individuals at baseline, 648 and 645 individuals at 6 months, and 587 and 549 individuals at 2 years, respectively. A dual-energy X-ray absorptiometry (DXA) scan was carried out in a random sample of 50% of the total study population including 382 participants at baseline, 304 participants at 6 months and 222 participants at 2 years.
Measurements
Body weight and WC were measured in the morning before breakfast at baseline, 6 months and 2 years. Body weight was measured by calibrated hospital scales and WC was measured using a non-stretchable tape measure, 4 cm above the iliac crest. Height was measured at the baseline examination. BMI was calculated dividing weight (kg) by the square of height (m2). Body composition was analyzed by a DXA scan using a Hologic QDR 4500A (Hologic, Inc. Waltham, MA), after an overnight fast [20]. Total fat mass (kg), total lean mass (kg), the percentage of whole body fat mass, and percentage of trunk fat were obtained at baseline, 6 months, and 2 years of the intervention. To evaluate the adherence to the dietary intervention program, dietary intake was assessed in a random sample of 50% of the participants by a review of the 5 days diet record at baseline and by 24-h recall during a telephone interview on 3 nonconsecutive days at 6 months and 2 years of follow-up.
Genotyping
DNA was isolated from the buffy coat fraction of centrifuged blood using the QIAmp Blood Kit (Qiagen). The previously reported single nucleotide polymorphism (SNP) MTNR1B rs10830963 was successfully genotyped in 722 of 811 total participants using the OpenArray SNP Genotyping System (BioTrove) [19]. Replicated quality control samples (10%) were included and genotyped with >99% concordance. The genotype distribution was consistent with Hardy-Weinberg equilibrium (HWE) in all study participants or in the major ethnic group (whites) (p > 0.05).
Statistical analyses
The primary end points of this study were changes in body fatness (weight, BMI, WC) and composition measures (body fat, lean body mass, total fat mass percentage, and trunk fat percentage) over the course of the intervention. Chi-squared test for categorical variables, and general linear models for continuous variables, were performed for comparison of baseline characteristics across genotypes. To compare changes in body fatness and body composition measurements by genotypes according to low- or high-fat group at 6 months and at 2 years of the diet intervention, general linear models were used adjusted for covariates (Model 1 adjusted for age, gender, ethnicity, and the respective baseline variable; Model 2 adjusted for model 1 plus BMI at baseline). Gene-diet interactions were evaluated including the interaction term in the models (e.g., MTNR1B genotype x high-/low-fat diet group). Sensitivity analyses were performed among white individuals to evaluate the influence of potential population stratification. Additive genetic models were used in the analysis. Statistical analyses were performed using STATA/SE version 12.0 (StataCaorp, College Station, TX, USA). A p value < 0.05 was considered statistically significant.
RESULTS
Table 1 shows baseline characteristics of the participants according to the MTNR1B rs10830963 genotype. The minor allele frequency (G allele) was 0.27 in the study population. The genotype frequencies were similar between males and females and among diet groups. Nonetheless, the distribution of the SNP was different by ethnicity (p<0.001). Dietary intake and body fatness and compositional measurements were not related to the MTNR1B genetic variant at baseline examination. There were no associations of genotype with changes in body fatness and compositional measurements at 6 months and 2 years of diet intervention, after adjustment for age, sex ethnicity, BMI at baseline (if appropriate), high-/low-fat diet group, and baseline value for the according outcomes, were observed (data not shown) [19].
Table 1.
Baseline characteristics of study participants according to the MTNR1B rs10830963 genetic variant
| CC | CG | GG | p value | |
|---|---|---|---|---|
| n=393 | n=273 | n=56 | ||
| Age (years) | 50.8 (9.2) | 51.0 (9.1) | 53.0 (10.3) | 0.24 |
| Sex | 0.72 | |||
| Male | 149 (37.9) | 112 (41.0) | 22 (39.3) | |
| Female | 244 (62.1) | 161 (59.0) | 34 (60.7) | |
| Race or ethnic group | <0.001 | |||
| White | 279 (71.0) | 247 (90.5) | 49 (87.5) | |
| African American | 96 (24.4) | 12 (4.4) | 3 (5.4) | |
| Hispanic or other | 18 (4.6) | 14 (5.1) | 4 (7.1) | |
| Diet group | 0.69 | |||
| Low fat | 198 (50.4) | 134 (49.1) | 31 (55.4) | |
| High fat | 195 (49.6) | 139 (50.2) | 25 (44.6) | |
| Dietary intake per day a | ||||
| Energy (kcal) | 1945 (555) | 2016 (572) | 1884 (448) | 0.36 |
| Protein (%) | 18.0 (3.4) | 18.3 (3.0) | 18.2 (3.2) | 0.77 |
| Fat (%) | 36.9 (5.9) | 37.0 (6.1) | 37.6 (6.3) | 0.85 |
| Carbohydrate (%) | 45.1 (7.8) | 43.8 (7.6) | 44.9 (7.3) | 0.31 |
| Body weight (kg) | 93.7 (15.0) | 92.8 (16.0) | 92.0 (17.5) | 0.66 |
| BMI (kg/m2) | 32.9 (3.8) | 32.3 (3.9) | 32.5 (3.7) | 0.11 |
| WC (cm) | 103.9 (13.0) | 103.2 (13.2) | 103.9 (13.4) | 0.78 |
| Body composition b | ||||
| Total body fat (kg) | 35.3 (8.0) | 34.5 (8.1) | 34.6 (5.6) | 0.62 |
| Total body lean (kg) | 60.8 (12.8) | 60.4 (13.6) | 56.5 (13.1) | 0.26 |
| Total body fat mass (%) | 37.0 (7.1) | 36.6 (6.9) | 38.4 (6.5) | 0.44 |
| Trunk fat (%) | 38.2 (6.2) | 37.5 (6.1) | 38.6 (5.8) | 0.54 |
BMI, Body mass index; WC, Waist circumference Data are expressed as mean (SD) or n (%)
Data were available for 370 individuals (CC n=208, CG n=133, GG n=29)
Data were available for 382 individuals (CC n=204, CG n=149, GG n=29)
Although the targets of macronutrient intakes were not fully achieved, the reported dietary intake and changes in adherence biomarkers confirmed that participants modified their intakes of macronutrients in the direction of the intervention (Online Resource 1). There were no significant differences in mean values of macronutrient intakes and urinary protein at 6 months and 2 years across the MTNR1B rs10830963 genotype and group diet, except for fat intake at 24 months among participants in the high-fat group (p=0.04). Moreover, the G allele was significantly associated with a greater increase in respiratory quotient (RQ) in the low-fat diet group at 24 months, as has been previously reported [19].
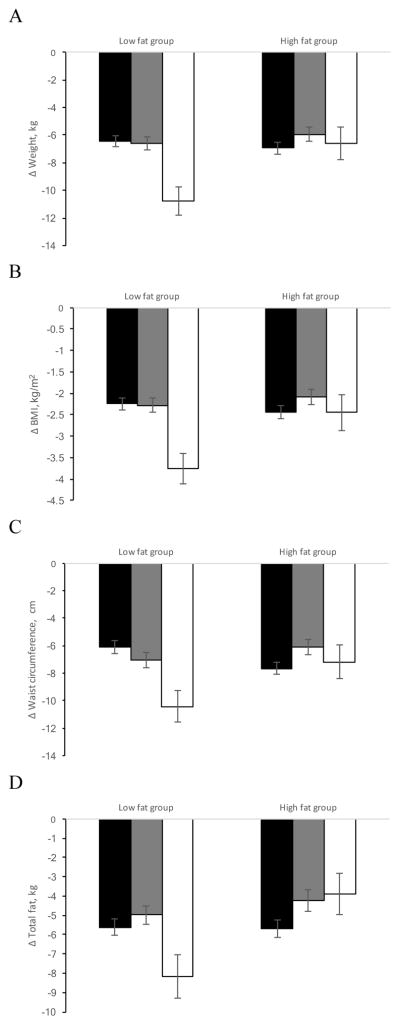
After adjustment for age, sex, ethnicity, BMI at baseline (if appropriate), and the respective baseline variable, significant interactions between the MTNR1B rs10830963 genotype and high-/low-fat diet on changes in weight, BMI, and WC were observed (Table 2 and Figure 1). An increasing number of the G allele of the MTNR1B genetic variant was significantly associated with a decrease in body weight, BMI, and WC at 6 months in response to the low-fat diet (p interaction=0.01, 0.02 and 0.002, respectively). When we applied the analyses among whites similar trends with significant interactions at 6 months on changes in body weight, BMI, and WC were found (all p interaction<0.05).
Table 2.
Effect of the MTNR1B rs10830963 genetic variant on changes in body weight, BMI, WC and body composition in response to low-/high-fat diet at 6 months and 2 years of diet intervention
| 6 months | 2 years | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Low fat | High fat | p interaction | Low fat | High fat | p interaction | |||||
|
|
|
|||||||||
| β | p value | β | p value | β | p value | β | p value | |||
| Model1 | ||||||||||
| Δ Weigh t (kg) | −1.38 (0.48 ) | 0.004 | 0.55 (0.51 ) | 0.29 | 0.01 | −1.60 (0.66 ) | 0.02 | −0.05 (0.68 ) | 0.90 | 0.19 |
| Δ BMI (kg/m2 ) | −0.48 (0.17 ) −1.69 | 0.005 | 0.15 (0.18 ) 0.71 | 0.39 | 0.02 | −0.53 (0.23 ) −1.33 | 0.02 | −0.07 (0.23 ) 0.49 | 0.76 | 0.28 |
| Δ WC (cm) | (0.53) | 0.02 | (0.55) | 0.20 | 0.004 | (0.68) | 0.05 | (0.76) | 0.52 | 0.13 |
| Δ Total fat (g) | −430 (506) | 0.40 | 1063 (513) | 0.04 | 0.04 | −499 (791) | 0.53 | 100 (853) | 0.91 | 0.79 |
| Δ Total lean (g) | 62.5 (264) | 0.81 | 422 (284) | 0.14 | 0.41 | 201 (423) | 0.64 | 103 (413) | 0.81 | 0.70 |
| Δ Total fat mass (%) | −0.36 (0.36) | 0.33 | 0.63 (0.35) | 0.07 | 0.05 | −0.49 (0.51) | 0.34 | 0.12 (0.50) | 0.81 | 0.60 |
| Δ Trunk fat (%) | −0.37(0.48) | 0.44 | 0.81(0.47) | 0.09 | 0.08 | −0.51(0.65) | 0.43 | 0.24(0.65) | 0.71 | 0.65 |
|
| ||||||||||
| Model 2 | ||||||||||
| Δ WC (cm) | −1.67 (0.52 ) | 0.001 | 0.82 (0.54 ) | 0.13 | 0.002 | −1.31 (0.66 ) | 0.048 | 0.61 (0.75 ) | 0.42 | 0.09 |
| Δ Total fat (g) | −428 (508) | 0.40 | 1091 (513) | 0.03 | 0.04 | −453 (790) | 0.57 | 146 (858) | 0.87 | 0.77 |
| Δ Total lean (g) | 63.2 (262) | 0.81 | 416 (285) | 0.15 | 0.44 | 202 (425) | 0.63 | 99.5 (413) | 0.81 | 0.70 |
| Δ Total fat mass (%) | −0.30 (0.36 ) | 0.40 | 0.63 (0.34 ) | 0.07 | 0.07 | −0.47 (0.51 ) | 0.36 | 0.15 (0.49 ) | 0.77 | 0.61 |
| Δ Trunk fat (%) | −0.32 (0.48 ) | 0.51 | 0.76 (0.46 ) | 0.10 | 0.10 | −0.46 (0.65 ) | 0.48 | 0.21 (0.64 ) | 0.74 | 0.75 |
BMI, Body mass index; WC, Waist circumference
β represents changes in outcomes for the increasing number of G allele of the MTNR1B rs10830963 genetic variant
Model 1: Adjusted for age, gender, ethnicity and the respective baseline variable
Model 2: Model 1 plus BMI at baseline
Figure 1.
Effect of the MTNR1B rs10830963 genetic variant and fat diets on changes in body weight (A), BMI (B), WC (C) and body fat mass (D) at 6 months of diet intervention (black bars, CC genotype; gray bars, CG genotype, white bars, GG genotype). Data are means (SD) after adjusted for age, sex, ethnicity, BMI at the baseline (if appropriate) and value for the respective outcome trait at baseline
The interaction between the MTNR1B rs10830963 genotypes and dietary fat was also analyzed for the measurements of body composition including total fat, total lean body mass, percentage total fat mass, and percentage trunk fat at 6 months (Table 2 and Figure 1). Consistent with the observations of changes in body fatness measurements, a statistically significant interaction between the MTNR1B genetic variant and low-/high-fat diet was found on changes in total fat, adjusting for age, gender, ethnicity, BMI at baseline, and the respective baseline variable (p interaction=0.04). In response to the high-fat diet, an increasing number of the G allele was significantly associated with an increase of total fat (p=0.03). Similar trends were also found for percentage of total fat mass and percentage trunk fat although the gene-dietary fat interaction was no longer statistically significant (p interaction=0.07 and 0.10, respectively) after adjustment for covariates (model 2). Similar results were observed in the white population (data not were shown), although the interaction for body fat was not statistically significant (p interaction=0.07, model 2).
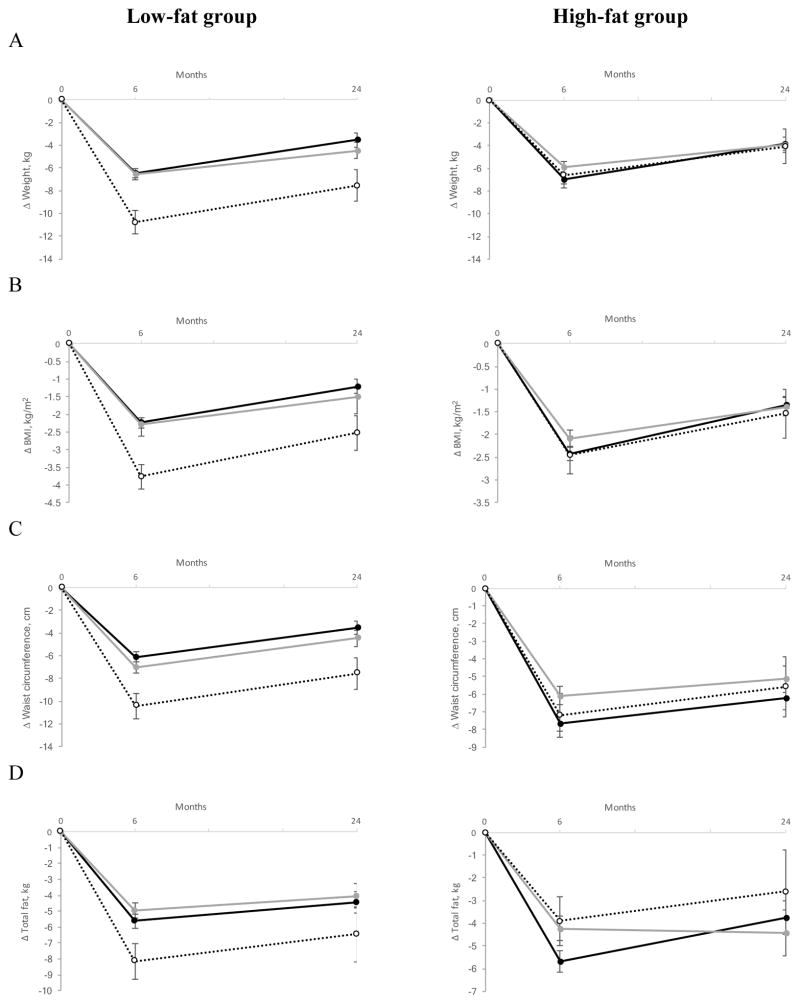
At 2 years participants, on average, regained body weight [18]. The associations with MTNRB rs10830963 genotypes remained statistically significant for weight (p=0.02), BMI (p=0.02), and WC (p=0.048) in the low-fat diet group, although the gene-diet interaction became non-significant (Table 2 and Figure 2).
Figure 2.
Effect of the MTNR1B rs10830963 genetic variant and fat diets on changes in body weight (A), BMI (B), WC (C) and body fat mass (D) at 6 months and 2 years of diet intervention (black circle and solid line, CC genotype; gray circle and gray solid line, CG genotype; white circle and dotted line, GG genotype). Data are means (SE) after adjusted for age, sex, ethnicity, BMI at baseline (if appropriate) and the value for the respective outcome trait at baseline
DISCUSSION
In this 2-year randomized dietary weight-loss intervention trial, we found significant interactions between the circadian rhythm genotype MTNR1B and dietary fat intake on changes in fatness, fat distribution and body composition measurements, especially at 6 months. In response to the low-fat diet, increasing number of the G allele was associated with a greater reduction in body weight, BMI, WC, and total body fat.
The circadian system has long been implicated in the regulation of body fatness due to its role in the control of energy balance [21]. Mice with mutations in the circadian rhythm gene Clock fed with a high-fat diet developed obesity at a young age as wells as a variety of metabolic and endocrine abnormalities consistent with the metabolic syndrome [22]. In addition, the circadian Clock mutant mice exhibited decreased expression of transcripts encoding selected hypothalamic peptides involved in energy balance. Melatonin is used by the central master clock as an internal synchronizer coordinating central and peripheral tissues [5]. Our findings are consistent with the biological role of melatonin in energy metabolism and energy balance [5]. On one hand, melatonin plays roles not only in the regulation of metabolic processes but also in the maintenance of their circadian organization [23]. On the other hand, the effect of melatonin on energy balance has been consistently observed [5]. Wolden-Hanson et al. demonstrated that melatonin supplementation therapy decreased body weight and intraabdominal fat, and increased the nocturnal locomotor activity and core body temperature [24]. While the precise mechanisms underlying our results remain largely unknown, several lines of evidence have implicated the MTNR1B rs10830963 genotype in regulating melatonin signaling [11]. Tuomi et al. ascertained that subjects carrying 1 and 2 MTNR1B rs10830963 G alleles showed a 2- and 4-fold increase in MTNR1B mRNA expression in human pancreatic islets, respectively, compared with the non-carriers [11]. Moreover, the authors demonstrated that the administration of melatonin to nondiabetic individuals inhibits insulin secretion in all subjects, and the effect was stronger among GG than those did not carry this allele. Another recent study reported that the common genetic variant was associated with the timing of the melatonin rhythm [25]. MTNR1B rs10830963 G allele carriers showed a later melatonin offset and a longer duration of elevated melatonin levels. The authors suggested that the disruption of melatonin rhythm among carriers of the risk allele may result in an increase of food intake to coincide with elevated melatonin levels in the morning leading to decreased glucose tolerance.
In line with our previous results, we found that the relations of the MTNR1B rs10830963 genetic variant with changes in fatness and body composition measurements were significantly modified by dietary fat intake [19]. Notably, a high-fat diet has been found to alter the circadian molecular clock, both centrally and peripherally, leading to a state of chronodisruption [1, 26]. Interestingly, several previous studies reported that the expression levels of clock genes and also the rhythmic mRNA expression were influenced by a high-fat diet consumption, in different animal tissues [27, 28]. Moreover, it has been demonstrated that a significant alteration of circadian rhythmicity of different hormones is related to obesity such as pineal melatonin, leptin, ghrelin, and adiponectin among others, in rats fed a high-fat diet [29–31]. Taken together, these data lent support to potential interplays between the MTNR1B genotype and dietary fat.
Our findings of body composition analysis suggested that the MTNR1B genotype might affect total body fat composition, instead of specific fat compositions (trunk fat). According to our results, a previous study showed that MTNR1B was expressed in rodent inguinal and epididymal adipocytes [32]. In this sense, it should be highlighted, that the effect of melatonin on adipocyte biology is not only mediated by means of the receptors in adipocytes but also through the action of the sympathetic nervous system [16]. In humans, Staiger et al. found an association between the MTNR1B rs4753426 genetic variant and total body fat [33]. More studies are needed to further verify our findings.
In the current study, the gene-diet interactions on changes in body fatness and composition measurements were attenuated at 2 years of follow-up. On the one hand, this might be due in part to decreasing adherence to the diet that occurred between 6 months and 2 years in the POUNDS Lost trial, similar to other weight-loss interventions [18, 34–36]. On the other hand, the statistical power might also be diminished because of more dropouts at 2 years of diet intervention than at 6 months. Even though, similar relations of the MTNR1B rs10830963 genotype with a decrease in body weight, BMI, and WC in the low-fat diet group were observed at 2 years, indicating the genetic effects were stable and for up to 2 years.
As far as we know, this is the first study to date to report significant interactions between the MTNR1B rs10830963 genetic variant and dietary fat intake on changes in fatness and body compositional measurements, in a large and long-term randomized weight-loss dietary intervention trial. Nonetheless, we acknowledge several limitations. First, we did not measure clock parameters such as melatonin and MTNR1B expression which limited our ability to explore plausible underlying mechanisms. Second, the power to detect long-term genotypic effect in response to a high-/low-fat diet was reduced due to the decline of adherence after 6 months of diet intervention, as has been reported by other authors [18, 34–36]. Third, because low-fat intake is usually characterized by high-carbohydrate intake, it is difficult to determine which macronutrient would best explain our results. Finally, around 80% of the study participants were white, and the genotype distribution differed across ethnicity, so further studies are required to generalize our findings to other ethnic groups.
In conclusion, the results of the present study showed that the level of fat intake might modify the effect of the circadian rhythms related MTNR1B rs10830963 genetic variant on changes in body fatness and composition. Subjects with the GG genotype might have a better response to a weight-loss dietary intervention by choosing a low-fat diet.
Supplementary Material
Dietary intake and biomarkers of adherence according to the MTNR1B rs10830963 genetic variant and diet group at 6 months and 2 years
Acknowledgments
The authors thank all participants of the study for their dedication and contribution to the research. The study is supported by grants from the National Heart, Lung, and Blood Institute (HL-071981, HL-034594, HL-126024), the National Institute of Diabetes and Digestive and Kidney Diseases (DK-091718, DK-100383, DK-078616), the Boston Obesity Nutrition Research Center (DK-46200), and United States-Israel Binational Science Foundation Grant 2011036. L.G. is a recipient of a pre-doctoral and a mobility grant from the Spanish Ministry of Education, Culture and Sport. Y.H. is a recipient of a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science. L.Q. was a recipient of the American Heart Association Scientist Development Award (0730094N).
Footnotes
Ethical standards
All procedures of the present study were in accordance with the ethical standards laid down the 1964 Helsinki declaration and its later amendments.
The study was approved by the human subjects committee at each institution and by a data and safety monitoring board appointed by the National Heart, Lung and Blood Institute.
All participants provided written informed consent.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Laermans J, Depoortere I. Chronobesity: Role of the circadian system in the obesity epidemic. Obes Rev. 2016;17:108–125. doi: 10.1111/obr.12351. [DOI] [PubMed] [Google Scholar]
- 2.Garaulet M, Ordovás JM, Madrid JA. The chronobiology, etiology and pathophysiology of obesity. Int J Obes (Lond) 2010;34:1667–1683. doi: 10.1038/ijo.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qian J, Scheer FAJL. Circadian system and glucose metabolism: Implications for physiology and disease. Trends Endocrinol Metab. 2016;27:282–293. doi: 10.1016/j.tem.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott EM. Circadian clocks, obesity and cardiometabolic function. Diabetes, Obes Metab. 2015;17:84–89. doi: 10.1111/dom.12518. [DOI] [PubMed] [Google Scholar]
- 5.Cipolla-Neto J, Amaral FG, Afeche SC, et al. Melatonin, energy metabolism, and obesity: A review. J Pineal Res. 2014;56:371–381. doi: 10.1111/jpi.12137. [DOI] [PubMed] [Google Scholar]
- 6.Dubocovich ML, Delagrange P, Krause DN, et al. Nomenclature, classification, and pharmacology of G Protein-Coupled Melatonin Receptors. Pharmocological Rev. 2010;62:343–380. doi: 10.1124/pr.110.002832.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gamble KL, Berry R, Frank SJ, Young ME. Circadian clock control of endocrine factors. Nat Rev Endocrinol. 2014;10:466–475. doi: 10.1038/nrendo.2014.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prokopenko I, Langenberg C, Florez JC, et al. Variants in MTNR1B influence fasting glucose levels. Nat Genet. 2009;41:77–81. doi: 10.1038/ng.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manning AK, Hivert M-F, Scott RA, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet. 2012;44:659–69. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaulton KJ, Ferreira T, Lee Y, et al. Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat Genet. 2015;47:1415–25. doi: 10.1038/ng.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tuomi T, Nagorny CLF, Singh P, et al. Increased melatonin signaling is a risk factor for type 2 diabetes. Cell Metab. 2016;23:1067–1077. doi: 10.1016/j.cmet.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Stancáková A, Kuulasmaa T, Paananen J, et al. Association of 18 confirmed susceptibility loci for type 2 diabetes with indices of insulin release, proinsulin conversion, and insulin sensitivity in 5,327 nondiabetic Finnish men. Diabetes. 2009;58:2129–2136. doi: 10.2337/db09-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong X, Zhang X, Xing X, et al. The association of type 2 diabetes loci identified in genome-wide association studies with metabolic syndrome and its components in a Chinese population with type 2 diabetes. PLoS One. 2015;10:1–21. doi: 10.1371/journal.pone.0143607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grotenfelt NE, Wasenius NS, Rönö K, et al. Interaction between rs10830963 polymorphism in MTNR1B and lifestyle intervention on occurrence of gestational diabetes. Diabetologia. 2016;59:1655–1658. doi: 10.1007/s00125-016-3989-1. [DOI] [PubMed] [Google Scholar]
- 15.Goni L, Cuervo M, Milagro FI, Martínez JA. Gene-gene interplay and gene-diet interactions involving the MTNR1B rs10830963 variant with body weight loss. J Nutrigenet Nutrigenomics. 2014;7:232–242. doi: 10.1159/000380951. [DOI] [PubMed] [Google Scholar]
- 16.Szewczyk-Golec K, Wo niak A, Reiter RJ. Inter-relationships of the chronobiotic, melatonin, with leptin and adiponectin: Implications for obesity. J Pineal Res. 2015;59:277–291. doi: 10.1111/jpi.12257. [DOI] [PubMed] [Google Scholar]
- 17.Barrenetxe J, Delagrange P, Martínez JA. Physiological and metabolic functions of melatonin. J Physiol Biochem. 2004;60:61–72. doi: 10.1007/BF03168221. [DOI] [PubMed] [Google Scholar]
- 18.Sacks FM, Bray GA, Carey VJ, et al. Comparison of weight-loss diets with different compositions of fat, protein, and carbohydrates. N Engl J Med. 2009;360:859–873. doi: 10.1056/NEJMoa1208410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mirzaei K, Xu M, Qi Q, et al. Variants in glucose- and circadian rhythm-related genes affect the response of energy expenditure to weight-loss diets: the POUNDS LOST Trial. Am J Clin Nutr. 2014;99:392–399. doi: 10.3945/ajcn.113.072066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Souza RJ, Bray GA, Carey VJ, et al. Effects of 4 weight-loss diets differing in fat, protein, and carbohydrate on fat mass, lean mass, visceral adipose tissue, and hepatic fat: Results from the POUNDS LOST trial. Am J Clin Nutr. 2012;95:614–625. doi: 10.3945/ajcn.111.026328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Summa KC, Turek FW. Chronobiology and obesity: Interactions between circadian rhythms and energy regulation. Adv Nutr. 2014;5:312S9S. doi: 10.3945/an.113.005132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turek FW, Joshu C, Kohsaka A, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science (80- ) 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar Jha P, Challet E, Kalsbeek A. Circadian rhythms in glucose and lipid metabolism in nocturnal and diurnal mammals. Mol Cell Endocrinol. 2015;418:74–88. doi: 10.1016/j.mce.2015.01.024. [DOI] [PubMed] [Google Scholar]
- 24.Wolden-Hanson T, Mitton DR, McCants RL, et al. Daily melatonin administration to middle-aged male rats suppresses body weight, intraabdominal adiposity, and plasma leptin and insulin independent of food intake and total body fat. Endocrinology. 2000;141:487–497. doi: 10.1210/en.141.2.487. [DOI] [PubMed] [Google Scholar]
- 25.Lane JM, Chang AM, Bjonnes AC, et al. Impact of common diabetes risk variant in MTNR1B on sleep, circadian, and melatonin physiology. Diabetes. 2016;65:1741–1751. doi: 10.2337/db15-0999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oosterman JE, Kalsbeek A, la Fleur SE, Belsham DD. Impact of nutrients on circadian rhythmicity. Am J Physiol - Regul Integr Comp Physiol. 2015;308:R337–R350. doi: 10.1152/ajpregu.00322.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yanagihara H, Ando H, Hayashi Y, et al. High-fat feeding exerts minimal effects on rhythmic mRNA expression of clock genes in mouse peripheral tissues. Chronobiol Int. 2006;23:905–914. doi: 10.1080/07420520600827103. [DOI] [PubMed] [Google Scholar]
- 28.Hsieh M-C, Yang S-C, Tseng H-L, et al. Abnormal expressions of circadian-clock and circadian clock-controlled genes in the livers and kidneys of long-term, high-fat-diet-treated mice. Int J Obes (Lond) 2010;34:227–239. doi: 10.1038/ijo.2009.228. [DOI] [PubMed] [Google Scholar]
- 29.Cano P, Jiménez-Ortega V, Larrad Á, et al. Effect of a high-fat diet on 24-h pattern of circulating levels of prolactin, luteinizing hormone, testosterone, corticosterone, thyroid-stimulating hormone and glucose, and pineal melatonin content, in rats. Endocrine. 2008;33:118–125. doi: 10.1007/s12020-008-9066-x. [DOI] [PubMed] [Google Scholar]
- 30.Cano P, Cardinali DP, Rios-Lugo MJ, et al. Effect of a high-fat diet on 24-hour pattern of circulating adipocytokines in rats. Obes (Silver Spring) 2009;17:1866–1871. doi: 10.1038/oby.2009.200. oby2009200 [pii]\r. [DOI] [PubMed] [Google Scholar]
- 31.Cardinali DP, Cano P, Jiménez-Ortega V, Esquifino AI. Melatonin and the metabolic syndrome: Physiopathologic and therapeutical implications. Neuroendocrinology. 2011;93:133–142. doi: 10.1159/000324699. [DOI] [PubMed] [Google Scholar]
- 32.Zalatan F, Krause JA, Blask DE. Inhibition of isoproterenol-induced lipolysis in rat inguinal adipocytes in vitro by physiological melatonin via a receptor-mediated mechanism. Endocrinology. 2001;142:3783–3790. doi: 10.1210/en.142.9.3783. [DOI] [PubMed] [Google Scholar]
- 33.Staiger H, Machicao F, Schäfer SA, et al. Polymorphisms within the novel type 2 diabetes risk locus MTNR1B determine β-cell function. PLoS One. 2008;3:e3962. doi: 10.1371/journal.pone.0003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foster GD, Wyatt HR, Hill JO, et al. A randomized trial of a low-carbohydrate diet for obesity. N Engl J Med. 2003;348:2082–2090. doi: 10.1056/NEJMoa022207. [DOI] [PubMed] [Google Scholar]
- 35.Dansinger ML, Gleason JA, Griffith JL, et al. Comparison of the Atkins, Ornish, Weight Watchers, and Zone Diets for weight loss and heart disease risk reduction: A randomized trial. J Am Med Assoc. 2005;293:43–53. doi: 10.1097/00008483-200505000-00012. [DOI] [PubMed] [Google Scholar]
- 36.Shai I, Schwarzfuchs D, Henkin Y, et al. Weight loss with a low-carbohydrate, Mediterranean, or low-fat diet. N Engl J Med. 2008;359:229–241. doi: 10.1056/NEJMoa1208410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dietary intake and biomarkers of adherence according to the MTNR1B rs10830963 genetic variant and diet group at 6 months and 2 years