Abstract
Background
Airway type 2 inflammation is usually corticosteroid sensitive, but the role of type 2 inflammation as a mechanism of asthma in patients receiving high-dose inhaled corticosteroids (ICSs) is uncertain.
Objective
We sought to determine whether airway type 2 inflammation persists in patients treated with ICSs and to evaluate the clinical features of patients with steroid-resistant airway type 2 inflammation.
Methods
We used quantitative PCR to generate a composite metric of type 2 cytokine gene expression (type 2 gene mean [T2GM]) in induced sputum cells from healthy control subjects, patients with severe asthma receiving ICSs (n = 174), and patients with nonsevere asthma receiving ICSs (n = 85). We explored relationships between asthma outcomes and T2GM values and the utility of noninvasive biomarkers of airway T2GM.
Results
Sputum cell T2GM values in asthmatic patients were significantly increased and remained high after treatment with intramuscular triamcinolone. We used the median T2GM value as a cutoff to classify steroid-treated type 2–low and steroid-resistant type 2–high (srT2-high) subgroups. Compared with patients with steroid-treated type 2–low asthma, those with srT2-high asthma were older and had more severe asthma. Blood eosinophil cell counts predicted srT2-high asthma when body mass index was less than 40 kg/m2 but not when it was 40 kg/m2 or greater, whereas blood IgE levels strongly predicted srT2-high asthma when age was less than 34 years but not when it was 34 years or greater.
Conclusion
Despite ICS therapy, many asthmatic patients have persistent airway type 2 inflammation (srT2-high asthma), and these patients are older and have more severe disease. Body weight and age modify the performance of blood-based biomarkers of airway type 2 inflammation.
Keywords: Severe asthma, type 2 inflammation, steroid resistance, biomarkers
Asthma affects 300 to 400 million persons worldwide,1 and the 5% to 10% of asthmatic patients with severe disease account for much of the public health burden of asthma.2 Airway type 2 inflammation mediated by type 2 cytokines (IL-4, IL-5, and IL-13) is a key pathologic mechanism of asthma,3 but the role of type 2 inflammation in patients with severe asthma is not fully understood. Inhaled corticosteroids (ICSs) are a mainstay treatment for asthma because they decrease airway type 2 inflammation,4 but many patients continue to experience asthma exacerbations, asthma symptoms, and airflow obstruction despite ICS treatment.5 One possibility is that some patients have persistent airway type 2 inflammation despite ICS therapy. Alternatively, some patients can have a type 2–low asthma endotype that is not effectively treated by ICSs. There are few studies to guide thinking about which of these 2 possibilities is operating in patients with severe asthma.
Recently, we have shown that it is feasible to quantify airway type 2 inflammation using PCR-based measures of expression of type 2 cytokines (IL4, IL5, and IL13) in induced sputum cells.6 We also proposed the type 2 gene mean (T2GM) as a composite metric of gene expression for IL4, IL5, and IL13 in sputum cells.6 Sputum cell T2GM provides a sensitive and specific measure of airway type 2 inflammation, and it allows quantitative analysis of type 2 inflammation in large patient cohorts.6 The Severe Asthma Research Program (SARP) cohort includes patients with nonsevere and severe forms of asthma who undergo detailed characterization studies and collection of biospecimens, including induced sputum.7,8 The majority of patients in the cohort are treated with ICSs, usually in high doses.8 To determine whether airway type 2 inflammation persists in patients with asthma treated with ICSs and the clinical features of the subgroup with steroid-resistant airway type 2 inflammation, we measured T2GM values in induced sputum in the SARP cohort in an analysis restricted to patients receiving ICSs.
METHODS
Study design
Subjects studied included a reference (healthy) cohort and a cohort of adult asthmatic patients recruited by 7 clinical centers in the National Health and Blood Institute’s SARP-3.7 The SARP protocol is an ongoing, 6-visit, 3-year, longitudinal cohort study in which 60% of participants have severe asthma, as defined by the European Respiratory Society/American Thoracic Society criteria.5 The visit structure of the SARP protocol is outlined in Fig E1 in this article’s Online Repository at www.jacionline.org. The first baseline visit included completion of medical history and asthma control questionnaires, spirometry, measurement of fraction of exhaled nitric oxide (FENO), immunologic tests (ImmunoCAP), and biospecimen collection (induced sputum and blood). In addition, maximum bronchodilator reversibility tests (spirometry before and after 4–8 puffs of albuterol) were performed on baseline visits 2 and 3, and participants also underwent a systemic corticosteroid response test. The systemic corticosteroid response test involved an intramuscular injection of triamcinolone acetonide (40 mg) on baseline visit 2 and repeat characterization (including maximum bronchodilator reversibility tests, sputum induction, and blood draw) on visit 3 (2–4 weeks later).
Healthy subjects
Healthy control subjects (n = 57) were recruited between November 1, 2012, and October 1, 2015. All healthy control subjects had no history of pulmonary disease, allergic rhinitis, or any other atopic disease. Each of the 7 centers was required to recruit a minimum of 7 healthy control subjects. The healthy control subjects recruited across all centers were required to be greater than 55% female, greater than 40% older than 45 years of age, and greater than 20% of African American race to ensure that the demographic characteristics of the healthy control group resembled those of the asthmatic patients. The recruitment strategy for the healthy control cohort was continuously monitored by using the SARP data coordinating center to ensure appropriate recruitment. In pulmonary function tests all healthy control subjects had normal airway responses to inhaled methacholine and normal FEV1 (FEV1 percent predicted >80%). Four healthy control subjects were excluded for having evidence of significant atopy (>5 positive test results for common aeroallergens on Immuno-CAP immunologic testing).
Asthmatic patients
Five hundred twenty-eight adult (age ≥18) asthmatic patients were recruited by SARP-3 centers between November 1, 2012, and October 1, 2015. The SARP protocol included 2 baseline visits, with a 40-mg triamcinolone steroid injection at the second visit and a follow-up visit 2 weeks later. At each visit, patients underwent detailed disease characterization and provided samples of blood and induced sputum.7 Of the 528 adult subjects enrolled in the SARP-3 protocol, 55 were not taking ICSs. We restricted our analysis to the subgroup of asthmatic patients receiving ICSs (n = 473). Additional details about recruitment methods, subject enrollment, study measurements, and study procedures are provided in the Methods section in this article’s Online Repository at www.jacionline.org.
Sputum induction and processing
Sputum induction and processing were performed by using a standard procedure in all subjects.9 Briefly, subjects inhaled nebulized 3% saline through a mouthpiece for 12 minutes, as previously described.9 Subjects interrupted inhalation at 2-minute intervals to spit saliva into a saliva cup and induced sputum into a sputum cup. Saliva was discarded, and induced sputum was processed. A 10% solution of Sputolysin (EMD Millipore, Burlington, Mass) was added at a 1:1 gram per milliliter (sputum weight/Sputolysin) ratio to the induced sputum, mixed with a serologic pipette, and placed in a 37°C shaking water bath for 15 minutes. Samples were removed at 5-, 10-, and 15-minute intervals for additional mixing with the pipette, and a portion of this sample was used to determine total and differential cell counts, as previously described.9 The sample was then centrifuged in the cold (4°C) at 2000 rpm for 10 minutes. The cell pellet was then resuspended in 1 mL of Qiagen RNAprotect Saliva Reagent (Qiagen Hilden, Germany). All pellets were stored at −80°C.
RNA extraction
RNAwas extracted from sputum cells with the RNeasy Qiagen kit (Qiagen) by using previously described methods.6 RNA quality was measured with the Agilent 2100 bioanalyzer (Biogen, Weston, Mass), which performs electrophoretic separations according to molecular weight. Each sample was assigned an RNA integrity number (RIN) based on the extent of RNA degradation. An RIN of 10 indicates intact RNA, whereas an RIN of 1 indicates totally degraded RNA.10 Samples with RIN values of less than 5 were excluded from analysis (see Fig E2 in this article’s Online Repository at www.jacionline.org).11 Purified RNA was placed in aliquots and stored at − 80°C.
Acceptable sputum sample
Samples were considered acceptable for PCR analysis if they had a squamous cell value of less than 80% and an RIN value of 5 or greater.
PCR-based measures of type 2 inflammation
By using previously described methods,6 real-time TaqMan-based quantitative PCR was performed on RNA extracted from induced sputum cell pellets. The genes for 3 type 2 cytokines (IL4, IL5, and IL13) were measured, and the genes for IFNG and IL17 were measured for comparison. Expression levels of 4 housekeeping genes (GAPDH, PPIA, YWHAZ, and PSMB2) were also measured.
Some reactions for the genes of interest did not yield a cycle threshold (CT) value after 40 cycles, and here we assigned a CT value equal to the highest CT value detected in other samples for that gene. All CT values were in the linear portion of the amplification curve. Gene expression data were normalized to the geometric mean of the 4 housekeeping genes. Gene expression data are displayed as log2 normalized. Details of the primers and probes are provided in Table E1 in this article’s Online Repository at www.jacionline.org. As a composite metric of airway type 2 gene expression, we averaged gene expression of IL4, IL5, and IL13 to generate the T2GM value, as previously described.6
Statistical methods
Statistical analyses were performed with the JMP 12 software package (SAS Institute, Cary, NC) and Stata 15.0 (StataCorp, College Station, Tex), and P values of less than .05 were taken as statistically significant. Two group comparisons for continuous variables were made by using the Student t test when normality assumptions were met or Wilcoxon rank sum tests when normality assumptions were not met. Two group comparisons for dichotomous variables were made by using a Pearson χ2 test. Tests for trends across ordered groups were performed by using a Wilcoxon-type test for trend.12
To assess the relationship between biomarkers of airway type 2 inflammation and the T2GM, we used ordinary least squares (OLS) linear regression models with T2GM as the dependent variable. In these models the predictor variables were log-transformed blood eosinophil cell counts, log-transformed sputum eosinophil percentages, log-transformed serum IgE levels, and FENO measures. Interaction terms were constructed between body mass index (BMI) and each of the biomarkers of airway type 2 inflammation on the dependent variable of the T2GM. This was repeated for the interaction of age and each of the biomarkers of airway type 2 inflammation. Age and BMI were divided into 5 categories for OLS regression models. Wald tests were performed by using the testparm command in STATA software to assess the statistical significance of interaction terms.13 Marginal effects for statistically significant interactions were assessed by using the margins command in STATA software.13 As a sensitivity analysis, we repeated all of the OLS linear regression models by using quantile regression models with robust SEs. Receiver operating characteristic (ROC) curves and tests of equality of the ROC area under the curve (AUC) were made by using the rocgold command in STATA software.13
RESULTS
Subjects
Acceptable sputum samples were available from 259 of the 473 adult asthmatic patients receiving ICSs at the baseline visit, 283 of the 473 adult asthmatic patients receiving ICSs at the poststeroid baseline visit, and 30 of the healthy control subjects (see Fig E2). Subjects’ demographics did not differ significantly between subjects with sputum samples compared with subjects without sputum samples (see Tables E2 and E3 in this article’s Online Repository at www.jacionline.org).
CT exclusion criteria for housekeeping genes
Gene expression for type 2 cytokines was 8 CT values greater on average than gene expression for the housekeeping genes. Based on this, we excluded data from sputum cell samples in which the CT for any 2 housekeeping genes was greater than 20 (see Fig E2). In our initial publication describing methods of quantitative PCR–based gene profiling in sputum cells from a smaller number of patients, we excluded data from sputum cell samples in which the CT for any 2 housekeeping genes was greater than 35.6 Based on data from the larger patient cohort studied here, we consider a CTof 20 to be better because it ensures that all measures of type 2 cytokine gene expression are in the linear portion of the amplification curve.
Refractory airway type 2 inflammation in a large subgroup of asthmatic patients treated with ICSs
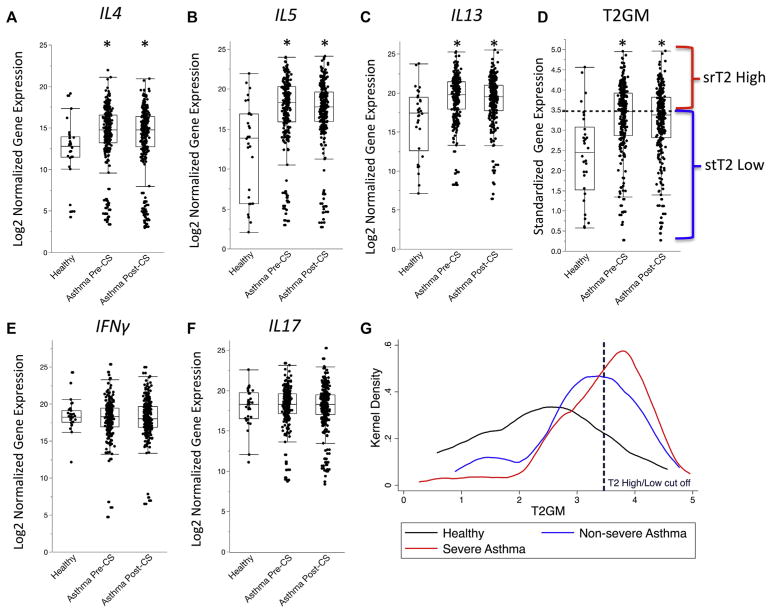
Mean gene expression levels in induced sputum cells for individual IL4, IL5, and IL13 cytokines was significantly greater in asthmatic patients than in healthy control subjects (Fig 1, A–C), and the T2GM value was also significantly increased (Fig 1, D). In contrast, mean gene expression levels in induced sputum cells for IFNG and IL17 were not significantly different in asthmatic patients than in healthy control subjects (Fig 1, E and F). Treatment with intramuscular triamcinolone did not significantly decrease gene expression of IL4, IL5, and IL13 cytokines (Fig 1, A–C). The triamcinolone injection had a minimal effect on the T2GM value, which decreased by 3% on average and did not reach the level of statistical significance (P =.19; Fig 1, D). Despite taking higher doses of ICSs, patients with severe asthma had significantly greater T2GM values than patients with nonsevere asthma (Fig 1, F). Mean gene expression levels in induced sputum cells for IFNG and IL17 were not significantly different in asthmatic patients than in healthy control subjects and did not change after the triamcinolone injection (Fig 1, F and G).
FIG. 1.
Sputum gene expression measures in 30 healthy control subjects, 259 asthmatic patients receiving ICSs before intramuscular injection of triamcinolone (Pre-CS), and 283 asthmatic patients receiving ICSs after a 40-mg intramuscular injection of triamcinolone (Post-CS). A–C, Measures of IL4, IL5, and IL13 are greater in asthmatic patients receiving ICSs compared with healthy control subjects and remain increased after triamcinolone injection. D, T2GM values (a summary metric of IL-4, IL-5, and IL-13 sputum gene expression) are significantly greater in asthmatic patients receiving ICSs than in healthy subjects at baseline and remains significantly greater after the triamcinolone injection. Asthmatic patients with values greater than the median T2GM value (Pre-CS) were classified as having srT2-high asthma, and patients with values of less than this median value were classified as having stT2-low asthma. E and F, Measures of IFNG and IL17 are no different in asthmatic patients compared with those in healthy control subjects. G, Kernel density plot demonstrating T2GM values are lower in healthy control subjects (black line) compared with patients with nonsevere asthma (blue line) or patients with severe asthma (red line; P < .01). T2GM measures are significantly greater in patients with severe asthma compared with those in patients with non-severe asthma (P < .01). *Significantly different from healthy control subjects: P < .05.
Clinical features of steroid-treated type 2–low and steroid-treated type 2–high asthma
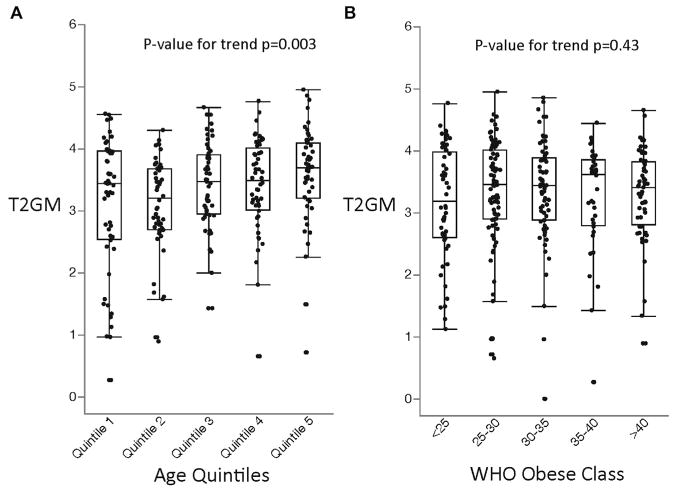
The median split value for T2GM in the asthma subgroup approximated the 90th percentile value for the T2GM value in the healthy subgroup (Fig 1, D), and we used the 50th percentile value in asthma as a cutoff to classify the steroid-treated type 2–low (stT2-low) and steroid-resistant type 2–high (srT2-high) subgroups. In effect, this means that 50% of the asthmatic patients had refractory type 2 inflammation in their airways despite ICS treatment. Subjects with srT2-high asthma were significantly older than those with stT2-low asthma (Table I). To further explore the effect of age on T2GM, we divided asthmatic patients into quintiles based on age (quintile 1, 18.3–33.6 years [n = 52]; quintile 2, 33.8–45.0 years [n = 52]; quintile 3, 45.3–52.9 years [n = 52]; quintile 4, 52.9–61.6 years [n = 52]; and quintile 5, 62.0–84.4 years [n = 51]). We found that T2GM values were significantly greater in the older age quintiles than in the younger age quintiles (Fig 2, A). In contrast to the positive relationships we found between age and T2GM values, we found that subgroups of patients with srT2-high and stT2-low asthma had similar BMIs (Table I) and that T2GM values did not increase significantly from low to high BMI classes, as defined by World Health Organization guidelines (Fig 2, B).14
TABLE I.
Clinical characteristics of stT2-low and srT2-high asthma
| Characteristic | Healthy subjects (n = 30) | Asthmatic patients | P value | |
|---|---|---|---|---|
|
|
|
|||
| stT2-low (n = 129) | srT2-high (n = 130) | srT2-high vs stT2-low | ||
| Age (y) | 38.0 (13.1) | 46.2 (13.7)* | 50.3 (14.0)*† | .02 |
|
| ||||
| Female sex, no. (%) | 18 (60) | 81 (63) | 96 (74) | .06 |
|
| ||||
| Race, no. (%)† | .01 | |||
|
| ||||
| American Indian and Alaska Native | 0 (0) | 1 (1) | 0 (0) | .01 |
|
| ||||
| Asian | 2 (7) | 1 (1) | 11 (9) | |
|
| ||||
| African American | 4 (13) | 25 (19) | 34 (26) | |
|
| ||||
| White | 20 (67) | 94 (73) | 76 (59) | |
|
| ||||
| Native Hawaiian or other Pacific Islander | 0 (0) | 0 (0) | 0 (0) | |
|
| ||||
| Mixed race | 4 (13) | 8 (6) | 9 (7) | |
|
| ||||
| BMI (kg/m2) | 26.7 (4.9) | 33.0 (8.8)* | 32.4 (8.3)* | .53 |
|
| ||||
| Percentage with BMI >30 kg/m2 | 6 (21) | 76 (59)* | 70 (54)* | .41 |
|
| ||||
| Spirometry | ||||
|
| ||||
| FEV1 (% predicted) | 97.5 (12.2) | 76.1 (19.2)* | 69.5 (17.9)*† | .005 |
|
| ||||
| FVC (% predicted) | 98.0 (14.6) | 85.9 (16.5)* | 84.1 (16.2)* | .36 |
|
| ||||
| FEV1/FVC ratio | 99.3 (4.6) | 87.5 (12.1)* | 81.7 (11.0)*† | <.001 |
|
| ||||
| FEV1 % <80, no. (%) | 0 (0) | 70 (54) | 93 (72) | .004 |
|
| ||||
| Blood cell counts (× 106/L) | ||||
|
| ||||
| Total white blood cells | 6.0 (1.5) | 7.5 (2.7) | 7.9 (2.6) | .21 |
|
| ||||
| Neutrophils | 3422 (1526) | 4585 (2269)* | 4632 (2050) | .86 |
|
| ||||
| Eosinophils | 126 (62) | 199 (157)* | 399 (319)*† | <.001 |
|
| ||||
| Serum IgE (IU/L) | 55 (59) | 265 (395)* | 468 (608)*† | <.001 |
|
| ||||
| FENO (ppm) | 19 (14) | 20 (16) | 35 (23)*† | <.001 |
|
| ||||
| No. of sIgEs (of 15)‡ | 0 (0–3) | 3 (1–6)* | 4 (1–8)* | .15 |
|
| ||||
| Sputum cell counts (%) | ||||
|
| ||||
| Eosinophils | 0.25 (0.1–0.7) | 0.4 (0.1–0.7) | 1.9 (0.6–7.2)† | <.001 |
|
| ||||
| Neutrophils | 64 (45–79) | 51 (34–73) | 53 (37–75) | .43 |
|
| ||||
| Sputum IL17 gene expression§ | 17.9 (2.4) | 17.9 (2.3) | 18.3 (2.4) | .14 |
|
| ||||
| Sputum IFNG gene expression§ | 18.4 (2.0) | 17.7 (2.5) | 18.5 (2.4) | .01 |
|
| ||||
| ACT score|| | 18 (14–21) | 17 (14–21) | .43 | |
|
| ||||
| Severity ATS/ERS criteria | .02 | |||
|
| ||||
| Mild | 26 (20) | 11 (9) | ||
|
| ||||
| Moderate | 25 (19) | 23 (18) | ||
|
| ||||
| Severe | 78 (61) | 96 (74) | ||
|
| ||||
| Receiving high-dose ICS, no. (%)¶ | 80 (62) | 98 (75) | <.001 | |
|
| ||||
| Receiving daily oral corticosteroids, no. (%) | 13 (10) | 19 (15) | .27 | |
|
| ||||
| Asthma exacerbations, no. (%) | ||||
|
| ||||
| No exacerbation in past year | 63 (49) | 41 (39) | .02 | |
|
| ||||
| One to 2 exacerbations in past year | 39 (31) | 51 (40) | ||
|
| ||||
| More than 2 exacerbations in the past year | 26 (20) | 36 (28) | ||
|
| ||||
| Necessitating hospitalization in past year, no. (%) | 16 (12) | 13 (10) | .54 | |
|
| ||||
| Necessitating ED visit in past year, no. (%) | 33 (26) | 30 (23) | .64 | |
|
| ||||
| Age of asthma onset (y) | 15.8 (13.7) | 18.1 (16.1) | .22 | |
|
| ||||
| Maximal BD response test (FEV1 %) | 9.7 (7.1) | 14.0 (8.6) | <.001 | |
|
| ||||
| SCRT (FEV1 %) | −0.06 (7.1) | 5.3 (8.9) | <.001 | |
Data are reported as means (SDs), unless otherwise indicated.
ACT, Asthma Control Test; ATS, American Thoracic Society; BD, bronchodilator; ED, emergency department; ERS, European Respiratory Society; FVC, forced vital capacity; MBRT, maximal bronchodilator response test; SCRT, systemic corticosteroid response test.
Indicates statistically different from healthy subjects (P < .05).
Statistically different between srT2-high and stT2-low asthma (P < .05).
Total number of positive specific blood IgE test results (ImmunoCAP) of the 15 allergens tested.
Gene expression data are in log2 scale normalized to 4 housekeeping genes.
ACT scores range from 5 to 25, with lower scores indicating worse asthma control.
Definition of high-dose ICSs is greater than or equal to the equivalent of 1000 μg of fluticasone daily.
FIG. 2.
A, T2GM measures a gradual increase in older subjects. Age quintiles: 1, 18 to 33.7; 2, 33.8 to 45.1; 3, 45.3 to 52.9; 4, 52.9 to 61.6; and 5, 62.0 to 84.4. B, T2GM measures do not change in more obese subjects. WHO, World Health Organization.
Compared with patients with stT2-low asthma, patients with srT2-high asthma had more severe asthma, as evidenced by lower FEV1, higher frequency of asthma exacerbations in the prior year, and higher frequency of a severe asthma diagnosis (Table I). Subjects with srT2-high asthma also had higher blood and sputum eosinophil cell counts (Table I and see Fig E3 in this article’s Online Repository at www.jacionline.org), higher FENO measures, higher blood IgE measures (Table I), and slightly increased sputum cell gene expression for IFNG and a trend for higher IL17 levels (Table I).
Numbers of patients with srT2-high asthma differed among patients with nonsevere and severe asthma in the SARP cohort. Although 40% of the patients with nonsevere asthma had srT2-high asthma, 55% of the patients with severe asthma had srT2-high disease (P = .02). Although the majority of patients with severe asthma had srT2-high asthma, 45% of patients with severe asthma did not have an increase in their sputum cell T2GM values. Among this large stT2-low subgroup, we noted that 54% had an FEV1 of less than 80%, and 20% were exacerbation prone (>2 exacerbations in the past year, Table I).8 As a sensitivity analysis, we repeated these comparisons by using quantile regression models and found similar results (see Table E4 in this article’s Online Repository at www.jacionline.org).
Utility of noninvasive biomarkers as tests of refractory airway type 2 inflammation
Blood IgE levels, blood eosinophil counts, sputum eosinophil counts, and FENO values are noninvasive biomarkers of airway type 2 inflammation.15 To compare the performance of these tests as biomarkers of refractory airway type 2 inflammation, we used ROC analysis. Sputum and blood eosinophil counts outperformed blood IgE levels and FENO values for predicting srT2-high asthma (Fig 3). Specifically, AUC values for sputum eosinophil counts (0.79; 95% CI, 0.72–0.84) and blood eosinophil counts (0.76; 95% CI, 0.70–0.82) were statistically greater than the AUC values for IgE (0.62; 95% CI, 0.55–0.69; P <.002 for both comparisons) and trended toward being greater for FENO values (0.72; 95% CI, 0.66–0.78; P = .09 for comparison of FENO values with sputum eosinophil counts and P = .34 for comparison of FENO values with blood eosinophil counts; Fig 3).
FIG. 3.
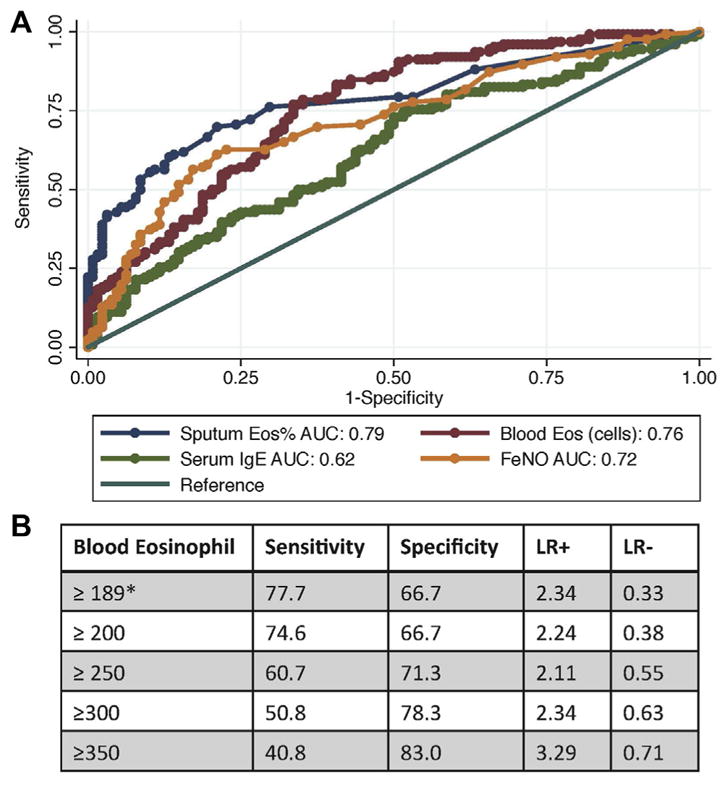
A, ROC curves for type 2 biomarkers for the outcome of srT2-high asthma. Eos, Eosinophils. B, Performance characteristics of blood eosinophil cell counts for predicting srT2-high asthma. *Blood eosinophil cell counts of 189 cells/μL or greater had the highest specificity and sensitivity for differentiating stT2-high asthma from stT2-low asthma. LR, Likelihood ratio.
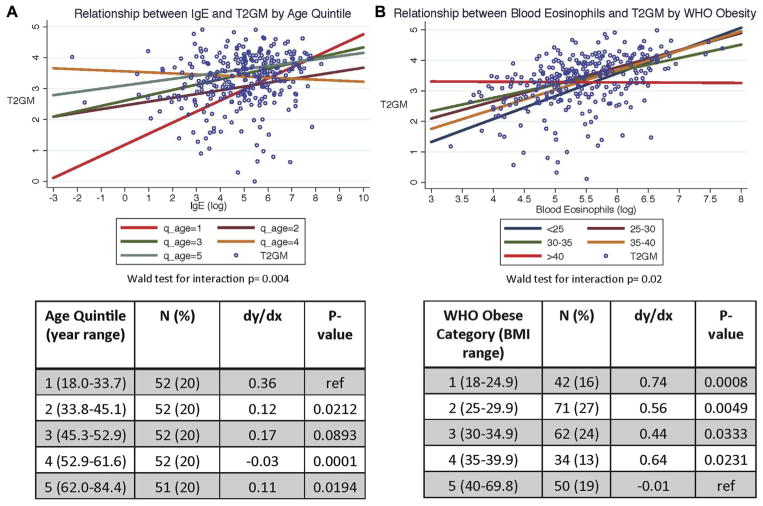
Because type 2 inflammation increased with age (above) and because prior reports show that obesity modifies the relationships between systemic and airway measures of type 2 inflammation in asthmatic patients,16 we explored whether age or BMI influenced the performance of noninvasive biomarkers as predictors of sputum cell T2GM values. For this analysis, we built linear regression models. In these models the outcome variable was the T2GM values, and the predictor variables were age (by age quintile) and each of 4 type 2 biomarkers (blood IgE levels, blood and sputum eosinophil counts, and FENO values). We included interaction terms for age and each of the 4 type 2 biomarkers, and we constructed linear regression models with predictor variables of BMI (by World Health Organization obesity categories) and each of the 4 type 2 biomarkers and interaction terms for BMI and each of the 4 type 2 biomarkers. We did not find any effect of age or BMI on the performance of sputum eosinophil cell percentages or FENO values as predictive biomarkers for the sputum cell T2GM (see Tables E5 and E6 in this article’s Online Repository at www.jacionline.org). However, we found a strong modifying effect of age on the relationship between blood IgE levels and sputum cell T2GM values (P value test for interaction = .004). Specifically, the correlation between blood IgE levels and sputum cell T2GM values was strong in patients in age quintile 1 (age <33.6 years) but decreased in the older patients in age quintiles 2 to 5 (Fig 4, A). In addition, we found a strong modifying effect of BMI on the relationship between blood eosinophil counts and sputum cell T2GM values (P value test for interaction = .02). Specifically, although blood eosinophil cell counts correlated well with sputum cell T2GM values in patients with a BMI of less than 40 kg/m2, the correlation decreased with increasing BMI and was very weak in morbidly obese patients (BMI >40 kg/m2; Fig 4, B).
FIG. 4.
A, Relationship between serum IgE levels and T2GM values is modified by age (Wald test for interaction, P = .004). Five linear regression lines demonstrate the relationship between log-transformed serum IgE levels and T2GM values subgrouped by age quintiles. Asthmatic patients in the youngest quintile have serum IgE levels that strongly correlate with airway T2GM measures (red line), whereas this relationship decreases in the older quintiles (green, blue, maroon, and orange lines). B, Relationship between blood eosinophil counts and T2GM values is modified by BMI (Wald test for interaction, P = .02). Five linear regression lines demonstrate the relationship between log-transformed blood eosinophil counts and T2GM values subgrouped by World Health Organization (WHO) BMI categories. Although asthmatic patients in BMI categories of less than 40 kg/m2 have blood eosinophil counts that correlate to airway T2GM measures (green, blue, maroon, and orange lines), this relationship decreases in patients with BMI of 40 kg/m2 or greater (red line). dy/dx, Marginal effect of serum IgE levels on T2GM values stratified by age quintiles or the marginal effect of blood eosinophil counts on T2GM values stratified by BMI WHO classification. P values are for (1) comparison of average marginal effects of the 4 oldest age quintiles compared with the youngest (reference) quintile and (2) comparison of average marginal effects of the 4 lowest BMI WHO categories compared with the highest (reference) BMI WHO category.
To illustrate this point more clearly, we used an ROC analysis. Although blood eosinophil counts are a strong predictor of stT2-high asthma in asthmatic patients with a BMI of less than 40 kg/m2 (AUC, 0.80), they are a poor predictor of patients with stT2-high asthma with a BMI of 40 kg/m2 or greater (AUC, 0.57; see Fig E4 in this article’s Online Repository at www.jacionline.org). As a sensitivity analysis, we repeated these OLS models using quantile regression models and found similar results (see Fig E5 in this article’s Online Repository at www.jacionline.org).
Treatment responses in patients with srT2-high and stT2-low asthma
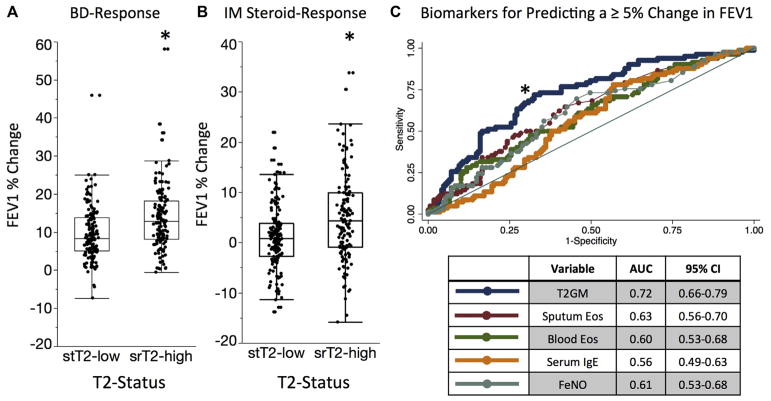
The treatment response to bronchodilators and systemic corticosteroids was better in patients with srT2-high asthma than in those with stT2-low asthma. Specifically, for the maximum bronchodilator reversibility test, which involved measurement of spirometry before and after 4 to 8 puffs of albuterol, the FEV1 response in patients with srT2-high asthma was significantly greater than in patients with stT2-low asthma (Fig 5, A). Similarly, for the systemic corticosteroid response test, which involved measurement of spirometry before and 2 to 4 weeks after an intramuscular injection of triamcinolone acetonide (40 mg), the FEV1 response in the patients with srT2-high asthma was significantly greater than in the patients with stT2-low asthma (Fig 5, B).
FIG. 5.
A, Maximal response to bronchodilators (BD-Response) is greater in patients with srT2-high asthma compared with those with stT2-low asthma. B, Absolute change in FEV1 percentage 2 weeks after a triamcinolone injection is greater in patients stT2-high asthma compared with those with stT2-low asthma. C, ROC curves demonstrating T2GM values as a superior predictor of a 5% absolute change in FEV1 percentage 2 weeks after a triamcinolone injection compared with other biomarkers of airway type 2 inflammation. *P < .05 for the T2GM AUC compared with other biomarkers. Eos, Eosinophils.
To compare the discriminating ability of the T2GM value to predict improvement in FEV1 with corticosteroid treatment, we again used an ROC analysis. We found that the sputum cell T2GM performed better than blood eosinophil cell counts for predicting a 5% absolute change in FEV1 percentage after triamcinolone (AUC, 0.71 vs 0.59; P =.001; Fig 5, C). The T2GM value also performed better than sputum eosinophil counts (AUC, 0.63; P =.01), FENO values (AUC, 0.61; P =.02), and blood IgE levels (AUC, 0.56; P =.003; Fig 5, C). As a sensitivity analysis, we also evaluated the discriminating ability of the T2GM value for predicting a 7.5% absolute change and a 10% absolute change in FEV1 percentage after the triamcinolone injection. In these analyses the T2GM value was also a superior predictor than the other type 2 biomarkers (see Fig E6 in this article’s Online Repository at www.jacionline.org).
Clinical features of persistently srT2-high asthma
One hundred ninety-six of the 259 asthmatic patients (76%) had paired T2GM measurements before and after the triamcinolone injection. We divided these subjects into 4 groups based on the change in T2GM value after triamcinolone injection. T2GM measurements were initially low and remained less than the T2-high threshold value after triamcinolone injection in 71 (36%) subjects (group 1); T2GM measurements were initially low and increased to greater than the T2-high threshold after triamcinolone in 25 (13%) subjects (group 2); T2GM measurements were initially high and decreased to less than the T2-high threshold after triamcinolone in 41 (21%) subjects (group 3), and T2GM measurements were initially high and remained at greater than the T2-high threshold after the triamcinolone injection in 59 (30%) of subjects (group 4, see Table E6). Compared with groups 1 to 3, subjects in group 4 (resistant T2-high subjects) were older and had more severe asthma (see Table E7 in this article’s Online Repository at www.jacionline.org).
DISCUSSION
In this study we report that airway type 2 inflammation is not suppressed in approximately half of asthmatic patients treated with ICSs, and we describe how patients with srT2–high asthma are older and have more severe asthma than patients with stT2–low disease. We also show that age and BMI influence the performance of blood- and breath-based biomarkers of refractory airway type 2 inflammation. Our data have implications for understanding the numbers of patients who are eligible for treatment with specific inhibitors of the type 2 inflammation pathway and for how biomarkers can be used to identify such patients.
The proportion of patients with srT2-high asthma was greater in those with severe asthma than in those with nonsevere asthma. This is consistent with the idea that type 2 airway inflammation is refractory to treatment with corticosteroids in a subgroup of asthmatic patients and that incompletely suppressed type 2 inflammation is a mechanism of severe asthma. An argument against this interpretation would be that patients with severe asthma might not be compliant with corticosteroids and that their disease could be brought under control with proper compliance or higher doses of corticosteroids. This argument is not supported by the fact that systemic corticosteroid treatment did not significantly decrease the sputum cell T2GM values in these patients. Thus it is more likely that corticosteroids do not fully suppress type 2 inflammation in many patients and that these patients stand to benefit from adjunctive treatment with specific inhibitors of type 2 inflammation, including inhibitors of type 2 cytokine or inhibitors of the prostaglandin D2 receptor 2.17–21
We found that treatment with intramuscular triamcinolone did not significantly decrease airway gene expression for type 2 cytokines in induced sputum cells. Although unexpected, our data are not the first to suggest that corticosteroids have limited effects on airway inflammation in patients with severe forms of asthma. For example, it has been reported that dexamethasone only weakly decreases IL-13 production in lung lymphocytes from patients with moderate-to-severe asthma, whereas it strongly inhibits IL-13 production in lung lymphocytes in patients with mild asthma.22 In addition, we have shown previously that systemic corticosteroids have limited effects on airway inflammation and lung function in the same SARP cohort studied here.23 For example, we showed previously that the effect of intramuscular triamcinolone on sputum eosinophil counts and FENO values in the SARP cohort (most of whom were taking inhaled or oral corticosteroids) was relatively modest. Specifically, the absolute triamcinolone-associated reduction in sputum eosinophil percentages was 0.2 and in FENO values was 2 ppm in adults with severe asthma. Furthermore, the effect of triamcinolone on lung function was also modest (only 20% of patients with severe asthma had an improvement in FEV1 of 10% or greater with parenteral corticosteroids, and the overall steroid-associated improvement in FEV1 was only 3%).23 Taken together, these data indicate that the effect of additional systemic corticosteroids is blunted in patients with severe asthma already receiving high-dose inhaled steroids and that airway type 2 inflammation can remain increased in asthmatic patients in spite of high doses of corticosteroids.
We found that steroid-resistant airway type 2 inflammation was significantly greater in older asthmatic patients compared with younger asthmatic patients, a finding that provides a clue for how corticosteroid resistance develops. One possibility is that immunosenescence results in decreased regulatory T-cell activity that renders older patients who are more susceptible to type 2 inflammation.24 Alternatively, prior work has suggested that lung lymphocyte production of type 2 cytokines is not suppressed by corticosteroid treatment in patients with severe asthma.22 Thus prolonged exposure to high doses of ICSs over time can render lymphocytes resistant to the suppressive effects of corticosteroid treatment.
In examining the utility of noninvasive biomarkers as tests of refractory airway type 2 inflammation, we found that sputum and blood eosinophil counts outperformed blood IgE measures and FENO values as predictors of refractory type 2–high asthma. However, we also noted that age and BMI were significant modifiers of the performance of noninvasive biomarkers of airway type 2 inflammation. Most importantly, blood eosinophil counts performed poorly as a biomarker in morbidly obese patients, and blood IgE levels performed surprisingly well in patients younger than 35 years. These data are highly relevant to decisions for when to treat patients with severe asthma with protein therapeutics. In particular, care is needed in ruling out adjunctive treatment with inhibitors of type 2 inflammation in morbidly obese patients with low eosinophil counts. According to our data, the blood eosinophil number might be a poor indicator of airway type 2 inflammation in this asthma subgroup.
Our data provide new insights about bronchodilator responsiveness and airway type 2 inflammation. Specifically, we show that the treatment response to albuterol is better in the srT2-high asthma subgroup than in the stT2-low asthma subgroup. We do not believe that this effect of type 2 inflammation has been shown before, although eosinophilic inflammation has been more strongly associated with reversibility than other measures of lung function.25 The mechanism of this effect is not clear but could be related to the role of IL-13 in inducing proliferation and hypercontractility of airway smooth muscle cells.26
Based on sputum cell T2GM values, 45% of patients with severe asthma did not have increased type 2 inflammation in their airways. However, many of them had abnormal FEV1 or were exacerbation prone. We propose that these patients are less likely to benefit from treatment with inhibitors of type 2 inflammation. In exploring other mechanisms of disease in this subgroup, we found no increases in gene expression for IL-17 or IFN-γ in their sputum cells, and we are unable to point to the mechanism of persistent airway disease in these patients.
Our study adds to the growing number of studies that have used gene expression data from sputum cells to explore patterns of airway inflammation in asthmatic patients.27–29 In general, these studies find a subgroup of asthmatic patients with high type 2 gene expression measures who are phenotypically distinct from patients with low type 2 gene expression,27–29 but not all studies report this.30
In summary, we conclude that many asthmatic patients treated with ICSs have increased type 2 inflammation in their airways and that these steroid-treated patients with type 2–high asthma are older and have more severe asthma. This asthma subgroup is eligible for adjunctive treatment with specific inhibitors of type 2 inflammation, but care needs to be exercised in identifying eligible patients by using blood- or breath-based biomarkers because age and BMI modify the performance of these biomarkers as indicators of refractory airway type 2 inflammation.
METHODS
SARP patients
Adult asthmatic patients were recruited to SARP from November 1, 2012, to October 1, 2015, by 7 clinical research centers (including UCSF) across the United States. Recruitment techniques varied by study site and included use of physician-to-physician letters, physician-to-patient letters, newspaper advertisements, direct mail, television advertising, poster flyers in the local community and clinics, mass e-mail to students/faculty, and use of recruitment databases.
The SARP protocol included 2 to 3 baseline characterization visits in which patients underwent detailed characterization and provided a sample of venous blood and induced sputum. Inclusion criteria mandated that at least 60% of patients met the American Thoracic Society/European Respiratory Society definition for severe asthma. The medication requirements for this definition included treatment with either continuous or near-continuous systemic corticosteroids or high-dose ICSs (>880 μg of fluticasone daily or equivalent) for at least half of the past year and continuously for the prior 3 months, plus a second controller medication (ie, a long-acting β-agonist or leukotriene modifier). In addition to the medication requirement, patients with severe asthma had to meet at least 1 minor criterion that indicated persistence of significant asthma impairment or report that their symptoms deteriorated immediately after a medication taper. The remaining 40% of patients with asthma did not meet the criteria for severe asthma and were classified as having nonsevere asthma. All patients were nonsmokers (<10 pack-years of tobacco use if >30 years of age; <5 pack-years if <30 years of age) and required to have evidence of bronchial hyperresponsiveness (defined as a PC20 methacholine value <16 mg/mL) or reversible airflow obstruction, as evidenced by an increase in FEV1 of 12% or greater after albuterol inhalation (≤720 μg), ipratropium bromide inhalation (136 μg), or both.
Patients were excluded if they were pregnant or breast-feeding during the initial characterization period, had a history of premature birth (<35 weeks gestation), or had a diagnosis of any other chronic pulmonary disorder. SARP patients younger than 18 years of age were excluded from analysis.
Patients completed comprehensive phenotypic characterization similar to patients recruited at UCSF, including a physician-directed history, Asthma Control Test, spirometry, maximum bronchodilator reversibility (see below), complete blood count with cell differential, induced sputum cell counts, serum IgE measurements, and FENO measurements. In addition, patients completed extensive questionnaires that characterized asthma symptoms, quality of life, medication use, and health care use. The SARP protocol included 2 baseline visits in which patients underwent detailed characterization studies and provided samples of venous blood and induced sputum. Data reported here are from these 2 baseline visits. All patients signed an informed consent form approved by their local institutional review board.
Procedures for withholding asthma and allergy medications
SARP asthmatic patients were also asked to hold their bronchodilator medications before spirometric testing. The medication holds for SARP were as follows: short-acting β-agonists, 4 hours; short-acting anticholinergics, 6 hours; long-acting β-agonists, 12 hours; long-acting muscarinic antagonists, 24 hours; and leukotriene modifiers, 24 hours.
Maximum bronchodilator reversibility test (SARP)
Patients were evaluated for baseline spirometry, after which 4 puffs of albuterol (360 μg) were administered. Spirometry was then repeated after 15 minutes. An additional 2 puffs of albuterol (180 μg) were then administered, and spirometry was repeated again after 15 minutes. If the change in FEV1 from the spirometric maneuver performed after 4 puffs and after 6 puffs was greater than 5%, an additional 2 puffs of albuterol were administered with repeat spirometry after an additional 15 minutes. If the change was less than 5%, the procedure was stopped, and the last maneuver was taken to be the highest achievable measure. No more than 8 puffs of albuterol were administered as part of the maximum bronchodilator reversibility procedure.
Supplementary Material
Key messages.
ICSs or systemic corticosteroids do not fully suppress airway type 2 inflammation in a large subgroup of patients with severe asthma.
Patients with steroid-resistant T2-high asthma are characterized by older age and more severe disease.
Body weight and age modify the performance of blood-based biomarkers of airway type 2 inflammation.
Acknowledgments
Supported by the National Institutes of Health (PO1 HL107201, R01 HL080414, U19, AI077439, U10 HL109146, U10 HL109164, U10 HL109172, U10 HL109086, U10 HL109250, U10 HL109168, U10HL109257, U10 HL109152, K23 HL138303, and K12 HL119997-04) and by grants from the Parker B. Francis Foundation.
We thank Patricia Noel and Robert Smith (Division of Lung Diseases, National Heart, Lung, and Blood Institute, Bethesda, Md) for their support and leadership of SARP.
Abbreviations used
- AUC
Area under the curve
- BMI
Body mass index
- CT
Cycle threshold
- FENO
Fraction of exhaled nitric oxide
- ICS
Inhaled corticosteroid
- OLS
Ordinary least squares
- RIN
RNA integrity number
- ROC
Receiver operating characteristic
- SARP
Severe Asthma Research Program
- srT2-high
Steroid-resistant type 2–high
- stT2-low
Steroid-treated type 2–low
- T2GM
Type 2 gene mean
Footnotes
Disclosure of potential conflict of interest: M. C. Peters reports consultancy fees from Merck and Genentech. P. G. Woodruff reports consultancy fees from AstraZeneca, Theravance, Regeneron, Sanofi, Genentech, Novartis, and Janssen. B. D. Levy reports institutional National Institutes of Health (NIH) funding and consultancy fees from AstraZeneca, Merck, Pieris Pharmaceuticals, and Sanofi. E. Israel reports personal fees from AstraZeneca, Novartis, Philips Respironics, Regeneron Pharmaceuticals; fees from Research in Real Life (RiRL); personal fees and other from TEVA Specialty Pharmaceuticals; grants from Genentech; nonfinancial support from Boehringer Ingelheim, GlaxoSmithKline, Merck, Sunovion, and TEVA; grants from Sanofi; personal fees from Bird Rock Bio, Nuvelution Pharmaceuticals, and Vitaeris; grants from Boehringer Ingelheim; nonfinancial support from TEVA Specialty Pharmaceuticals; personal fees from Sanofi, Merck, Entrinsic Health Solutions, and Glaxo-SmithKline; other funds from Vorso; and personal fees from Pneuma Respiratory outside the submitted work. D. T. Mauger reports institutional grant funding from the NIH. S. C. Erzurum reports institutional NIH funding and has pending NIH grants and is Chair of the ABIM Pulmonary Disease Board and reports travel support from them. M. W. Johansson’s institution reports grant funding from the NIH, and he has grants pending from Hoffman-LaRoche, is a member of the Genentech Advisory Board, and received speaking fees from them. N. N. Jarjour reports consulting fees from Teva, Daiichi Sankyo, and AstraZeneca, and his institution reports NIH grant funding. A. M. Coverstone reports institutional NIH grants (U10 HL109257 and UL1 TR00448), and she has grant funding from Orbex (5U01HL3004502) and the Inner City Asthma Consortium (5UM1Al11427104 and 5UM1Al11427103). M. Castro reports grants from the NIH and ALA during the conduct of the study; personal fees from Aviragen, Boehringer-Ingelheim, Boston Scientific, Elsevier, Genentech, GlaxoSmithKline, Holaira, and Teva; and grants from Amgen, Boehringer-Ingelheim, Genentech, Gilead, GlaxoSmithKline, Invion, Medimmune, Sanofi-Aventis, and Vectura, all outside the submitted work. A. T. Hastie reports grants from the National Heart, Lung and Blood Institute (NHLBI) during the conduct of the study. E. R. Bleecker reports undertaking clinical trials through his employer, Wake Forest School of Medicine, and the University of Arizona and has also served as a paid consultant outside the submitted work. S. E. Wenzel has received consultancy fees from AstraZeneca (Anti-IL-5R/TSLP), Sanofi (anti–IL-4R), Genentech, and Teva (anti–IL-5) and her institution has grant funding pending from GlaxoSmithKline (anti–IL-33), Boehringer Ingelheim (anti–IL-23), and received funds from AstraZeneca (anti–IL-5R trials), GlaxoSmithK-line (anti–IL-5 trials), Sanofi (anti–IL-4R trials), and Novartis (CRTH2 antagonist trial). J. V. Fahy reports consultant fees from Boehringer Ingelheim, Dynavax, Medimmune, Theravance, Pieris, and Entrinsic Health Solutions; his institution has received NIH grant funding and has grants from NHLBI, Pfizer, Genentech, and Vitaeris, and the institution also received biomedical patents in which he was the named inventor (for the National Heart, Lung and Blood Institute Severe Asthma Research Program 3). The rest of the authors declare that they have no relevant conflicts of interest.
References
- 1.To T, Stanojevic S, Moores G, Gershon AS, Bateman ED, Cruz AA, et al. Global asthma prevalence in adults: findings from the cross-sectional world health survey. BMC Public Health. 2012;12:204. doi: 10.1186/1471-2458-12-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaga M, Zervas E, Chanez P. Update on severe asthma: what we know and what we need. Eur Respir Rev. 2009;18:58–65. doi: 10.1183/09059180.00001009. [DOI] [PubMed] [Google Scholar]
- 3.Fahy JV. Type 2 inflammation in asthma—present in most, absent in many. Nat Rev. 2015;15:57–65. doi: 10.1038/nri3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woodruff PG, Modrek B, Choy DF, Jia G, Abbas AR, Ellwanger A, et al. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med. 2009;180:388–95. doi: 10.1164/rccm.200903-0392OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung KF, Wenzel SE, Brozek JL, Bush A, Castro M, Sterk PJ, et al. International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. Eur Respir J. 2014;43:343–73. doi: 10.1183/09031936.00202013. [DOI] [PubMed] [Google Scholar]
- 6.Peters MC, Mekonnen ZK, Yuan S, Bhakta NR, Woodruff PG, Fahy JV. Measures of gene expression in sputum cells can identify TH2-high and TH2-low subtypes of asthma. J Allergy Clin Immunol. 2014;133:388–94. doi: 10.1016/j.jaci.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peters MC, McGrath KW, Hawkins GA, Hastie AT, Levy BD, Israel E, et al. Plasma interleukin-6 concentrations, metabolic dysfunction, and asthma severity: a cross-sectional analysis of two cohorts. Lancet Respir Med. 2016;4:574–84. doi: 10.1016/S2213-2600(16)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denlinger LC, Phillips BR, Ramratnam S, Ross K, Bhakta NR, Cardet JC, et al. Inflammatory and comorbid features of patients with severe asthma and frequent exacerbations. Am J Respir Crit Care Med. 2017;195:302–13. doi: 10.1164/rccm.201602-0419OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gershman NH, Wong HH, Liu JT, Mahlmeister MJ, Fahy JV. Comparison of two methods of collecting induced sputum in asthmatic subjects. Eur Respir J. 1996;9:2448–53. doi: 10.1183/09031936.96.09122448. [DOI] [PubMed] [Google Scholar]
- 10.Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med. 2006;27:126–39. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Cuzick J. A Wilcoxon-type test for trend. Stat Med. 1985;4:87–90. doi: 10.1002/sim.4780040112. [DOI] [PubMed] [Google Scholar]
- 13.StataCorp. Stata Statistical Software: Release 12.1. College Station (TX): Stata-Corp LP; 2014. [Google Scholar]
- 14.Obesity: preventing and managing the global epidemic. Report of a WHO consultation. World Health Organ Tech Rep Ser. 2000;894:i–xii. 1–253. [PubMed] [Google Scholar]
- 15.Peters MC, Nguyen M-LT, Dunican EM. Biomarkers of airway type-2 inflammation and integrating complex phenotypes to endotypes in asthma. Curr Allergy Asthma Rep. 2016;16:71. doi: 10.1007/s11882-016-0651-4. [DOI] [PubMed] [Google Scholar]
- 16.Desai D, Newby C, Symon FA, Haldar P, Shah S, Gupta S, et al. Elevated sputum interleukin-5 and submucosal eosinophilia in obese individuals with severe asthma. Am J Respir Crit Care Med. 2013;188:657–63. doi: 10.1164/rccm.201208-1470OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wenzel S, Ford L, Pearlman D, Spector S, Sher L, Skobieranda F, et al. Dupilumab in persistent asthma with elevated eosinophil levels. N Engl J Med. 2013;368:2455–66. doi: 10.1056/NEJMoa1304048. [DOI] [PubMed] [Google Scholar]
- 18.Ortega HG, Liu MC, Pavord ID, Brusselle GG, FitzGerald JM, Chetta A, et al. Mepolizumab treatment in patients with severe eosinophilic asthma. N Engl J Med. 2014;371:1198–207. doi: 10.1056/NEJMoa1403290. [DOI] [PubMed] [Google Scholar]
- 19.Bel EH, Wenzel SE, Thompson PJ, Prazma CM, Keene ON, Yancey SW, et al. Oral glucocorticoid-sparing effect of mepolizumab in eosinophilic asthma. N Engl J Med. 2014;371:1189–97. doi: 10.1056/NEJMoa1403291. [DOI] [PubMed] [Google Scholar]
- 20.Corren J, Lemanske RF, Hanania NA, Korenblat PE, Parsey MV, Arron JR, et al. Lebrikizumab treatment in adults with asthma. N Engl J Med. 2011;365:1088–98. doi: 10.1056/NEJMoa1106469. [DOI] [PubMed] [Google Scholar]
- 21.Gonem S, Berair R, Singapuri A, Hartley R, Laurencin MFM, Bacher G, et al. Fevipiprant, a prostaglandin D2 receptor 2 antagonist, in patients with persistent eosinophilic asthma: a single-centre, randomised, double-blind, parallel-group, placebo-controlled trial. Lancet Respir Med. 2016;4:699–707. doi: 10.1016/S2213-2600(16)30179-5. [DOI] [PubMed] [Google Scholar]
- 22.Kaur M, Reynolds S, Smyth LJ, Simpson K, Hall S, Singh D. The effects of corticosteroids on cytokine production from asthma lung lymphocytes. Int Immunopharmacol. 2014;23:581–4. doi: 10.1016/j.intimp.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 23.Phipatanakul W, Mauger DT, Sorkness RL, Gaffin JM, Holguin F, Woodruff PG, et al. Effects of age and disease severity on systemic corticosteroid responses in asthma. Am J Respir Crit Care Med. 2017;195:1439–48. doi: 10.1164/rccm.201607-1453OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jagger AT, Shimojima Y, Goronzy JJ, Weyand CM. T regulatory cells and the immune aging process. Gerontology. 2014;60:130–7. doi: 10.1159/000355303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Modena BD, Tedrow JR, Milosevic J, Bleecker ER, Meyers DA, Wu W, et al. Gene expression in relation to exhaled nitric oxide identifies novel asthma phenotypes with unique biomolecular pathways. Am J Respir Crit Care Med. 2014;190:1363–72. doi: 10.1164/rccm.201406-1099OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Risse P-A, Jo T, Suarez F, Hirota N, Tolloczko B, Ferraro P, et al. Interleukin-13 inhibits proliferation and enhances contractility of human airway smooth muscle cells without change in contractile phenotype. Am J Physiol Lung Cell Mol Physiol. 2011;300:L958–66. doi: 10.1152/ajplung.00247.2010. [DOI] [PubMed] [Google Scholar]
- 27.Baines KJ, Simpson JL, Wood LG, Scott RJ, Gibson PG. Transcriptional phenotypes of asthma defined by gene expression profiling of induced sputum samples. J Allergy Clin Immunol. 2011;127:153–69. doi: 10.1016/j.jaci.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 28.Baines KJ, Simpson JL, Wood LG, Scott RJ, Fibbens NL, Powell H, et al. Sputum gene expression signature of 6 biomarkers discriminates asthma inflammatory phenotypes. J Allergy Clin Immunol. 2014;133:997–1007. doi: 10.1016/j.jaci.2013.12.1091. [DOI] [PubMed] [Google Scholar]
- 29.Rossios C, Pavlidis S, Hoda U, Kuo C-H, Wiegman C, Russell K, et al. Sputum transcriptomics reveal upregulation of IL-1 receptor family members in patients with severe asthma. J Allergy Clin Immunol. 2018;141:560–70. doi: 10.1016/j.jaci.2017.02.045. [DOI] [PubMed] [Google Scholar]
- 30.Seys SF, Scheers H, Van den Brande P, Marijsse G, Dilissen E, Van Den Bergh A, et al. Cluster analysis of sputum cytokine-high profiles reveals diversity in T(h)2-high asthma patients. Respir Res. 2017;18:39. doi: 10.1186/s12931-017-0524-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.