Abstract
Acute myeloid leukemia (AML) often harbors mutations in epigenetic regulators, and also has frequent DNA hypermethylation, including the presence of CpG island methylator phenotypes (CIMP). Although global hypomethylation is well-known in cancer, the question of whether distinct demethylator phenotypes (DMPs) exist remains unanswered. Using Illumina 450k arrays for 194 patients from The Cancer Genome Atlas we identified two distinct DMPs by hierarchical clustering: DMP.1 and DMP.2. DMP.1 cases harbored mutations in NPM1 (94%), FLT3 (71%) and DNMT3A (61%). Surprisingly, only 40% of patients with DNMT3A mutations were DMP.1, which has implications for mechanisms of transformation by this mutation. In contrast, DMP.2 AML was comprised of patients with t(8;21), inv(16), or t(15;17), suggesting common methylation defects connect these disparate rearrangements. RNA-seq revealed up-regulated genes functioning in immune response (DMP.1) and development (DMP.2). We confirmed these findings by integrating independent 450k datasets (236 additional cases), and found prognostic effects by DMP status, independent of age and cytogenetics. The existence of DMPs has implications for AML pathogenesis and may augment existing tools in risk stratification.
Keywords: AML, DNA methylation, CIMP, DMP, hypomethylation, demethylation, DNMT3A
Introduction
Epigenetic control of cell fate has long been studied in the context of organism development and cancer with non-random DNA methylation marks controlling various differentiation states 1, 2. Aberrant hypermethylation has been shown to affect tumor suppressor genes in cancer, and widespread hypermethylation defining a CpG island methylator phenotype (CIMP) can be observed in many tumor types 3–6. Many examples of specific DNA hypermethylation states have been shown to have clinical consequences in terms of therapy response and prognosis 6–8. Loss of DNA methylation in cancer has also been widely described for many years, however, the genomic targets, causes, and consequences of DNA demethylation remain unclear 9.
Acute myeloid leukemia is a heterogeneous and lethal disease in which studying the DNA methylome is a promising avenue for understanding cancer epigenetics and for clinically stratifying patients. While the overall somatic mutation burden in AML is low, some of the most frequently altered genes are epigenetic regulators. Approximately 25% of AML cases harbor mutations in the DNA methylation writer, DNMT3A 10. Mutations in the demethylase, TET2 have been reported in ~10% of cases, and IDH1/2 mutations occur in 10–15% of AML 10. Each of these genes has been reported to affect the leukemic methylome. DNMT3A has been shown to drive hypomethylation in the context of FLT3-ITD mutations 11, 12. TET2 has been reported to cause targeted DNA demethylation in some differentiation-related regions, and IDH1/2 mutations have been associated with oncometabolite formation causing hypermethylation via a TET-dependent mechanism 13. In addition, our group recently identified a TET2 associated (TET2-DMC-low) profile and a mutation-independent hypermethylation signature (A-CIMP) associated with favorable outcomes 6, 14.
In this study we sought to identify and characterize distinct DNA demethylator phenotypes (DMPs) in AML. To this end we studied TCGA AML samples profiled for DNA methylation, genetic mutations, gene expression, and clinical outcomes, and identified two distinct DMPs with important mechanistic and clinical implications in AML biology. We validated our findings in independent data and propose that DMPs may be used to augment existing clinical features for patient risk stratification and for understanding AML pathogenesis.
Methods
DNA methylation array data
DNA methylation data on the Illumina HumanMethylation 450k array platform were obtained for 194 AML patient samples from the TCGA data portal 10. Methylation data for 24 normal peripheral blood samples (GSE51388) were used to identify CpG sites which are normally methylated 15. Pre-processing of Level 1 data was done using functional normalization implemented by the minfi R package 16. We excluded CpG sites with NA values and were left with 375324 sites for analysis. In order to enrich for sites which lose methylation in cancer, we applied filtering criteria to obtain the subset of probes which show variable methylation in AML but high methylation in normal blood (average beta-value > 0.8 in normal blood and beta-value standard deviation > 0.2 in AML). To extend our analysis and increase statistical power for validation we merged multiple AML sample cohorts (TARGET, GSE62298, GSE58477, and GSE64934) interrogated with Illumina 450k arrays to compile a superset of 236 additional cases 10, 11, 17–19. We merged tables of beta values for these cases using R. To identify previously published epigenetic signatures we performed hierarchical clustering of the cases on methylation status of CpG sites for A-CIMP, I-CIMP, and TET2-DMC 6, 14. To test for the methylation status of differentiated leukocyte fractions we used data on the 450k array for both myeloid and lymphoid lineages (GSE35069) and selected for CpG sites of interest using R 20.
Characterizing hypomethylated CpG sites for protein binding and LINE-1 elements
Hypomethylated CpG sites were queried for protein binding using ChIP-seq peak data for CTCF in normal CD34+ hematopoietic progenitors, and SPI1 (PU.1) in HL-60 cells. Data were downloaded from the UCSC genome browser and peaks were overlapped with CpG sites interrogated by the Illumina 450k platform using hg19 probe coordinates in BED format with the intersection function in the Table Browser 21, 22. LINE-1 repetitive elements were mapped to CpG sites measured by the 450k array using RepeatMasker 23.
RNA-seq analysis
Level 3 RNA-seq data (per-gene read counts) were obtained for 176 available cases from the TCGA data portal 10. Count data were processed with the edgeR package to determine differential expression 24, 25. Genes identified as significantly up-regulated in demethylator AML clusters were queried for functional annotation enrichments using GeneCoDis 26–28. GeneCoDis annotations for Biological Process (BP), Molecular Function (MF), and Transcription Factors were analyzed with a false discovery rate (FDR)-corrected hypergeometric test. We filtered the annotations for those with at least 3 query genes present.
Statistics
Unsupervised hierarchical clustering was performed in R using Ward’s method implemented in the hclust function 29. Comparisons for binary variables (e.g. mutation status) across groups were tested using Fisher’s Exact Test. Continuous clinical variables (e.g. age, blast count) were compared using Student t-tests. Kaplan-Meier and Cox regression analyses were done using the survival package in R 30. Differences in Kaplan-Meier curves were compared using the log-rank test. Quantitative DNA methylation differences were defined as a difference in average beta-value across conditions greater than 0.1 and an FDR < 0.05. Differential expression analysis for RNA-seq data was done using the edgeR package and significance was defined as an FDR-corrected p-value < 0.05 with a fold-change of 2. Odds ratios for context-specific DNA hypomethylation (e.g. LINE-1 repeats, gene bodies, etc.) were calculated in R using the following formula:
Data availability
The TCGA and TARGET data used in this study – including Illumina 450k arrays, RNA-seq, and clinical annotations – are publicly available in the Genomic Data Commons (https://gdc.cancer.gov/). Other DNA methylation datasets used are available in the Gene Expression Omnibus (GEO): GSE51388, GSE62298, GSE58477, GSE35069, and GSE32251. ChIP-seq data for HL-60 and CD34+ cells are available on the UCSC Genome Browser (https://genome.ucsc.edu/).
Results
Distinct demethylator phenotypes in subsets of AML patients
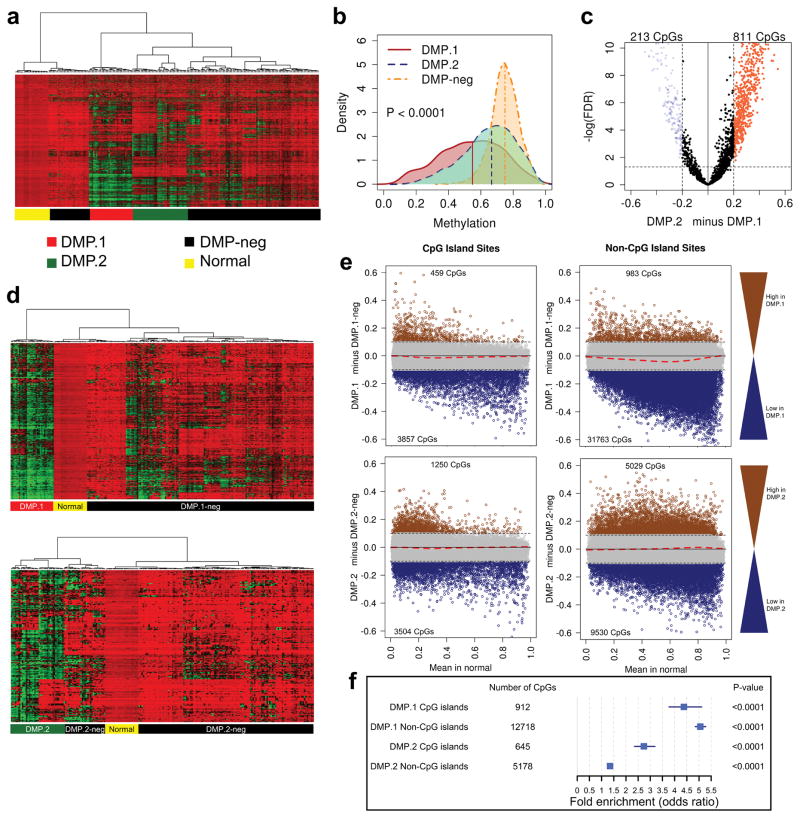
To study associations between DNA hypomethylation and AML biology we analyzed Illumina 450k arrays interrogating 194 TCGA leukemia samples. Patient characteristics are presented in Supplementary Table S1. To enrich for CpG sites that lose methylation (i.e. ‘demethylate’) in cancer, we selected for those with high methylation in normal peripheral blood, and variable methylation across the AML cohort (see methods). Hierarchical clustering of the samples based on these 2,606 sites revealed two distinct groups showing profound hypomethylation: DMP.1 and DMP.2 (Figure 1a). Analysis of the distributions of DNA methylation levels for these clusters showed a significant shift of density to lower values (Figure 1b). To rule-out potential cell of origin artifacts, we performed the same analysis using data obtained from bone marrow-derived hematopoietic stem cells as the normal comparator and obtained similar results (Supplementary Figure S1). For downstream exploration of DMP biology we refined a classifier of each DMP using differential methylation analysis (Figure 1c). From this, we identified signatures of 213 DMP.2 CpG target sites, and 811 DMP.1 target sites (FDR<0.05 and beta-value difference > 0.2; Supplementary Tables S2 and S3). Re-classifying the AML samples on the DMP.1 targets revealed a cluster of 31 DMP.1 patients, and the remaining 163 were DMP.1-negative (DMP.1-neg; Fig. 1d top). The same analysis done using the DMP.2 markers identified groups of 38 DMP.2 and 156 DMP.2-negative (DMP.2-neg) cases (Fig. 1d bottom). Importantly, DMP.2 and DMP.1 were mutually exclusive phenotypes (i.e. no cases were identified as both DMP.2 and DMP.1).
Figure 1. Identification of demethylator phenotypes in AML.
a) Hierarchical clustering of 194 AML cases and 24 normal peripheral blood controls based on 2,606 CpG sites with high methylation in normal blood (average beta-value > 0.8) and variable methylation in AML (beta-value standard deviation > 0.2). b) Density plot of distribution of DNA methylation levels in DMP.1, DMP.2, and DMP-neg leukemia. Vertical bars indicate median value. c) Volcano plot showing differentially methylated CpG sites between DMP.1 and DMP.2 cases from (a) which are used as a refined DMP classifier. d) Hierarchical clustering of the cases based on the CpG sites identified in (c) classify patients as DMP.1/DMP.1-neg (top), and DMP.2/DMP.2-neg (bottom). e) Differential methylation analysis comparing DMP.1 (top) and DMP.2 (bottom) to DMP.1-neg cases for CGI (left), and non-CGI (right) sites. The data show widespread demethylation (blue dots) and minimal hypermethylation (brown dots) in DMP.1, while there is less demethylation and more hypermethylation in DMP.2. Dashed red lines correspond to LOESS regression. f) Summary odds ratios of genomic region specificity of demethylation for DMP.1 and DMP.2 compared to DMP-neg. DMP.1 demethylation is more pronounced at both CGI and non-CGI sites compared to DMP.2, but neither compartment is more likely to hypomethylate. DMP.2 shows a relative preference for demethylation of CGI sites. Blue squares represent point estimates of odds ratios with blue lines representing 95% confidence intervals around the estimate.
Genome-wide DNA methylation specificity in DMP+ AML
From a DNA methylation perspective, we next sought to explore the features of all interrogated genomic loci, not just those selected by the initial filtering criteria. To this end we performed differential methylation analysis for DMP.1 and DMP.2 for specific genomic compartments. Using an average beta-value difference threshold of 0.1 with an FDR < 0.05 we identified 3,857 sites in CGIs, and 31,763 sites in non-CGIs that lose methylation in DMP.1 AML compared to DMP.1-neg (Figure 1e top). There was also a relatively small number of CpG sites that gained methylation in DMP.1 (Figure 1e top). The same analysis for DMP.2 revealed 3,504, and 9,530 CGI and non-CGI sites, respectively, that lost methylation compared to DMP.2-neg (Figure 1e bottom). In contrast to DMP.1, however, DMP.2 leukemias demonstrated relatively more hypermethylation compared to DMP.2-neg (1,250 and 5,029 CpGs in CGI and non-CGI regions, respectively; Figure 1e bottom). We performed a genomic localization enrichment analysis using odds ratios to identify whether certain features (e.g. CGIs, non-CGIs) were more or less likely to demethylate in DMPs compared to DMP-neg leukemias (see methods). This analysis revealed that DMP.1 demethylation is more pronounced overall compared to DMP.2 by virtue of higher magnitude odds ratios, however, DMP.2 showed more specificity for CGI (versus non-CGI) demethylation (Figure 1f). An extended analysis of odds ratios for demethylation revealed no specificity for promoters, enhancers, gene bodies, LINE-1 repetitive elements, PU.1 binding sites, or CTCF binding sites for either DMP, but that the magnitude of enrichments at regulatory elements was generally higher for DMP.1 – i.e. the odds ratios were generally higher for DMP.1 sites compared to DMP.2 sites (Supplementary Figure S2).
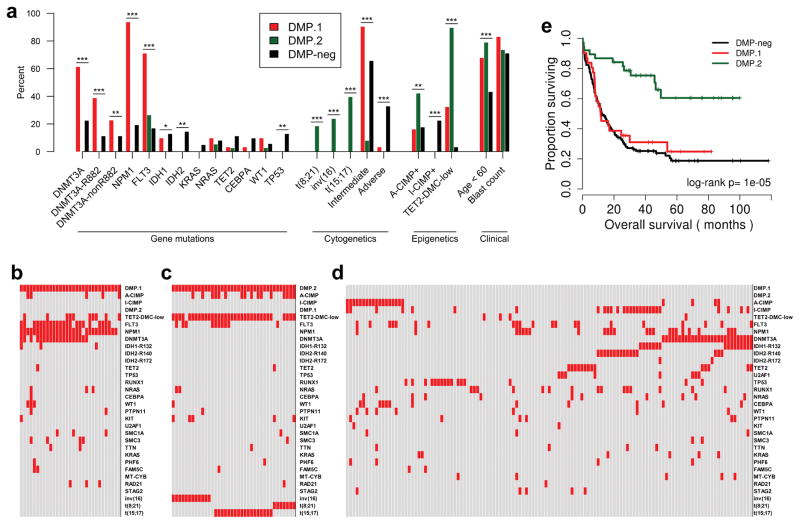
DMP status is associated with DNMT3A mutations and cytogenetic risk
We examined somatic alterations present in the DMPs and found that compared to DMP-neg, the DMP.1 cases were significantly enriched for mutations in DNMT3A, and FLT3 (61% vs. 22% and 71% vs. 17%, respectively, Fisher’s Exact P<0.001; Figure 2a). Strikingly, nearly all DMP.1 cases also harbored mutations in NPM1 (94% vs. 19%, Fisher’s Exact P<0.001; Figure 2a). In contrast, DMP.2 cases were characterized by a relative absence of somatic mutations, which was statistically significant for DNMT3A, NPM1, IDH1, IDH2, and TP53. (0% vs. 22%, 19%, 13%, 14%, and 13%; Fisher’s Exact P<0.001, <0.001, 0.04, <0.01, and <0.01, respectively; Figure 2a). Genomic rearrangements were also significantly different between DMPs, with DMP.1 cases showing significant enrichment for intermediate cytogenetic risk, and no cases harboring any favorable risk abnormality (Figure 2a). DMP.2, however, was characterized by all except one case harboring either t(8;21), inv(16), or t(15;17), suggesting a common epigenetic link between different good-risk cytogenetic aberrations (97% vs. 0%, Fisher’s Exact P<0.001; Figure 2a). Within DMP.2, the three cytogenetic abnormalities clustered separately by DNA methylation, with many CpG sites losing methylation in all cases, and a subset showing less demethylation in t(15;17) positive cases (Supplementary Figure S3a).
Figure 2. Genetic mutations, epigenetic signatures, and clinical outcomes in DMPs.
a) Enrichments for common gene mutations, cytogenetic abnormalities, previously published epigenetic profiles, and clinical features are plotted for DMP.1, DMP.2, and DMP-neg. Per-case (epi)genetic spectra are plotted for DMP.1 (b), DMP.2 (c), and DMP-neg (d); each column represents one patient with red boxes indicating positivity for the mutation/alteration. e) Kaplan-Meier analysis of overall survival stratified by DMP status. * P<0.05, ** P<0.01, *** P<0.001.
We next analyzed the data for the presence or absence of known epigenetic signatures; A-CIMP and TET2-DMC-low are favorable prognostic factors, while I-CIMP is a non-prognostic hypermethylation profile associated with IDH1/2 mutations (Supplementary Table S4–S6) 6, 14. In this analysis we found both DMPs to be I-CIMP-negative (0% in both vs. 22% in DMP-neg; Fisher’s Exact P<0.001), but DMP.2 cases were enriched for A-CIMP, and almost entirely overlapping with TET2-DMC-low (42% vs. 18%, and 89% vs. 3%; Fisher’s Exact P<0.01, <0.001, respectively; Figure 2a). The enrichment for A-CIMP+ leukemias in DMP.2 is consistent with the observed DNA methylation data (Figure 1e bottom); CpG sites which have low methylation in normal blood hypermethylate readily, while sites with high methylation in normal blood demethylate in this context, suggesting a widespread reprogramming phenomenon in CGI DNA methylation.
The observed co-occurrence and mutual exclusivity of different genetic mutations in DMP+ cases also revealed striking patterns. DMP.1 AML was enriched for co-occurrence of NPM1, FLT3, and DNMT3A mutations (Figure 2b). In contrast, nearly every DMP.2 case had one of three recurring favorable risk rearrangements. The sole outlier case was positive for A-CIMP, TET2-DMC-low, and a FLT3 mutation, but lacked other common alterations in AML (Figure 2c).
From a clinical perspective we also observed important differences between DMP.1, DMP.2, and DMP-neg. Bone marrow blast percentage did not differ by DMP status, however, both DMP.1 and DMP.2 patients were significantly younger compared to DMP-neg, with DMP.2 patients being the youngest (median age, years: DMP.1=55, DMP.2=49.5, DMP-neg=61; P<0.001; Figure 2a). In addition, Kaplan-Meier survival analysis demonstrated DMP.1 status was not associated with overall survival (OS) in this cohort (median OS, months: DMP.1=11.5, DMP.1-neg=19; log-rank P=0.51; Figure 2e), but DMP.2 leukemia patients had significantly improved OS (median OS, months: DMP.1=11.5, DMP.2=not reached, DMP-neg=12.4; log-rank P<0.001; Figure 2e). Because of the near-perfect overlap between methylation and cytogenetic risk, we could not assess whether their prognostic effects were independent in this dataset. When we examined whether there were differences between cytogenetic groups within DMP.2, we found that neither genetic mutations, nor overall survival varied by specific cytogenetic rearrangement (median OS, months: t(15;17)=not reached, t(8;21)=30.6, inv(16)=not reached, other=not reached; log-rank P=0.77; Supplementary Figure S3b). Furthermore, the subset of DMP.2 FLT3 mutation-positive cases did not have significantly different outcomes compared to FLT3 wild-type cases (median OS, months: FLT3-ITD=not reached, FLT3-TKD=not reached, Wild-type=not reached; log-rank P=0.75; Supplementary Figure S3c).
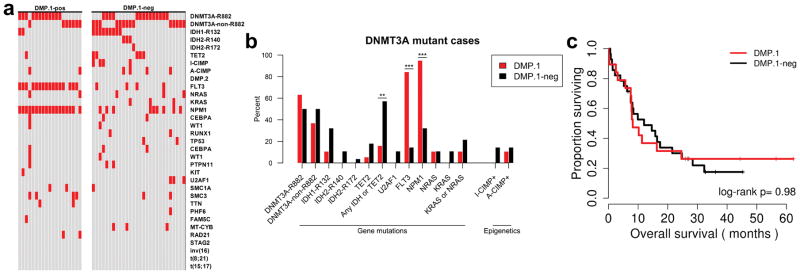
DNMT3A mutated AML is not always DMP.1
Given the observation that many DNMT3A mutant cases are DMP-neg, we sought to characterize this interesting subset (Figure 2d). We isolated the DNMT3A mutant leukemias and found that only 19/47 (40%) were DMP.1, with the rest being DMP.1-neg (Figure 3a). From a genetic perspective we found that the DMP.1-neg leukemias had a mix of the canonical DNMT3A R882 mutations, and non-R882 mutations (Figure 3a). Importantly, the DMP.1-neg cases also often had co-occurring mutations in IDH1, IDH2, or TET2 – three genes reported to cause aberrant hypermethylation – and relatively few FLT3 mutations (Figure 3a). We tested these observations statistically and found that the enrichments for IDH1, IDH2, and TET2 were not individually significant, but reached significance when aggregated (57% in DMP.1-neg vs. 16% in DMP.1; Fisher’s Exact P=0.006; Figure 3b). This analysis also revealed significantly fewer mutations in FLT3 and NPM1 in DMP.1-neg AML (14% in DMP.1-neg vs. 84% in DMP.1 and 32% in DMP.1-neg vs. 95% in DMP.1, respectively; Fisher’s Exact P<0.001 for each; Figure 3b). From a clinical perspective, the DNMT3A mutant cases did not differ in OS by DMP.1 status (median OS, months: DMP.1=8.2, DMP.1-neg=12.0; log-rank P=0.98; Figure 3c).
Figure 3. Genetic and epigenetic characteristics of DNMT3A-mutant AML.
a) Per-case (epi)genetic spectra are plotted for DNMT3A-mutant AML cases, and are divided by DMP.1 status. Red squares indicate presence of specified mutations/alterations. Each column represents data from a single patient. b) Barplot showing mutational enrichments for DNMT3A-mutant AML stratified by DMP.1 status. c) Kaplan-Meier analysis of overall survival for DNMT3A-mutant cases stratified by DMP.1 status. * P<0.05, ** P<0.01, *** P<0.001.
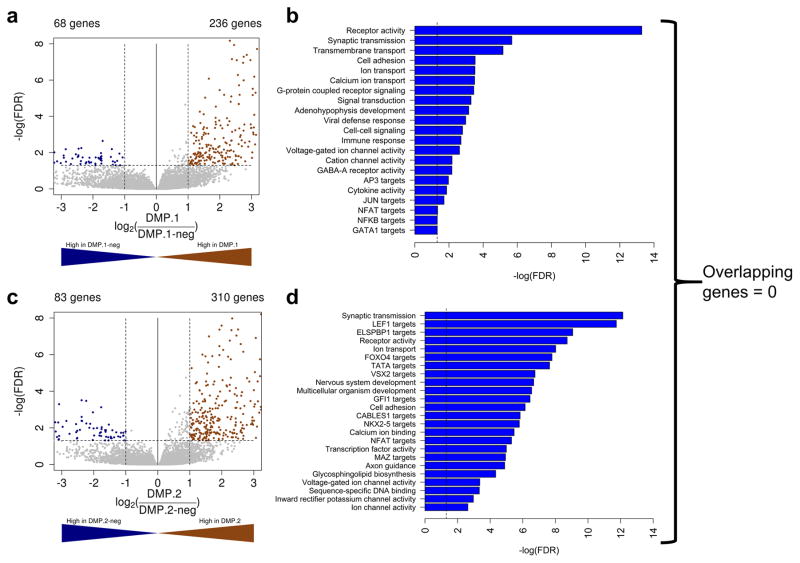
DMP.1 and DMP.2 AML have distinct gene expression signatures
To examine the transcriptomic differences between DMP.1 and DMP.2 we studied gene expression changes measured by RNA-seq. We performed differential expression analysis using edgeR (see methods) comparing DMP.1 to DMP.1-neg, and DMP.2 to DMP.2-neg and found there was generally more transcriptional up-regulation than down-regulation in both DMPs. In DMP.1 leukemias we identified 236 genes significantly up-regulated, and 68 down-regulated compared to DMP.1-neg (FDR<0.05; Fold-change>2; Figure 4a). For DMP.2 AML, 310 genes were up-regulated and 83, were down-regulated (FDR<0.05; Fold-change>2; Figure 4c). Importantly, the differentially expressed genes identified between the two DMPs were entirely mutually exclusive.
Figure 4. Distinct gene expression profiles in DMP+ AML.
Volcano plots showing differentially expressed genes for DMP.1 versus DMP.1-neg (a), and DMP.2 versus DMP.2-neg (c) leukemias. Gene set enrichment analysis was done using GeneCoDis for up-regulated genes in DMP.1 (b), and DMP.2 (d) AML.
Performing a gene set enrichment analysis for the up-regulated genes revealed that both DMPs had up-regulated genes functioning in ion transport, but that cell-cell signaling, immune response, JUN, and GATA1 target genes were specific to DMP.1, while gene ontology categories for development, FOXO4, LEF1, ELSPBP1, and other transcription factor targets were specific to DMP.2 AML (Figure 4b and d for DMP.1 and DMP.2, respectively).
We then tested whether there was a significant overlap between differentially methylated, and differentially expressed genes in this dataset. For DMP.1 AML 66 out of 236 up-regulated genes were also found to be hypomethylated compared to DMP.1-neg (Fisher’s Exact P=0.007). In contrast, only 16 of 310 up-regulated genes in DMP.2 leukemias were also significantly hypomethylated relative to DMP.2-neg (Fisher’s Exact P=0.48). This observation is consistent with the overall higher enrichments for DNA demethylation at regulatory elements for DMP.1 compared to DMP.2 (Supplementary Figure S2). Interestingly, gene set enrichment analysis of the subset of hypomethylated and up-regulated genes further confirmed the association between DMP.1 and immune response genes, and DMP.2 and developmental genes (Supplementary Tables S7 and S8 for DMP.1 and DMP.2, respectively). In addition, we queried DNA methylation status at DMP.1 and DMP.2 sites in multiple differentiated leukocyte fractions from healthy donors including NK cells, CD4+ T-cells, CD8+ T-cells, CD19+ B-cells, CD14+ monocytes, granulocytes, neutrophils, and eosinophils (Supplementary Figure S4). We found that the CpG sites demethylated in both DMPs are mostly methylated in normal leukocyte fractions, however, there are subsets of these sites that show low methylation levels, most markedly in the lymphoid fractions, further suggesting that DMP.2 demethylation defines a phenotype poised for differentiation (Supplementary Figure S4b). In contrast, the DMP.1 CpG sites with low methylation in healthy leukocyte fractions also tended to have low methylation in BM-HSCs (Supplementary Figure S4a).
Finally, we queried the RNA-seq data to explore gene expression levels of several known epigenetic regulators: DNMT1, DNMT3A, DNMT3B, TET1, TET2, and TET3 (Supplementary Figure S5). Although there were subtle differences in expression of DNMT1, and TET1 across DMPs (Supplementary Figure S5a and S5d), the most striking result was significant down-regulation of DNMT3A in DMP.1 leukemias with concurrent up-regulation of TET2 (Supplementary Figure S5b and S5e). The down-regulation of DNMT3A did not vary according to mutation status. We also observed a significant down-regulation of DNMT3B in DMP.2 AML (Supplementary Figure S5c). There was a significant up-regulation of MPO within this subset as well, consistent with data reported by Itonaga et al. suggesting that MPO is a key maturation marker of AML blasts (Supplementary Figure S5g) 31. TET3 expression was statistically identical across the DMPs.
Validation of DMPs in independent methylation data
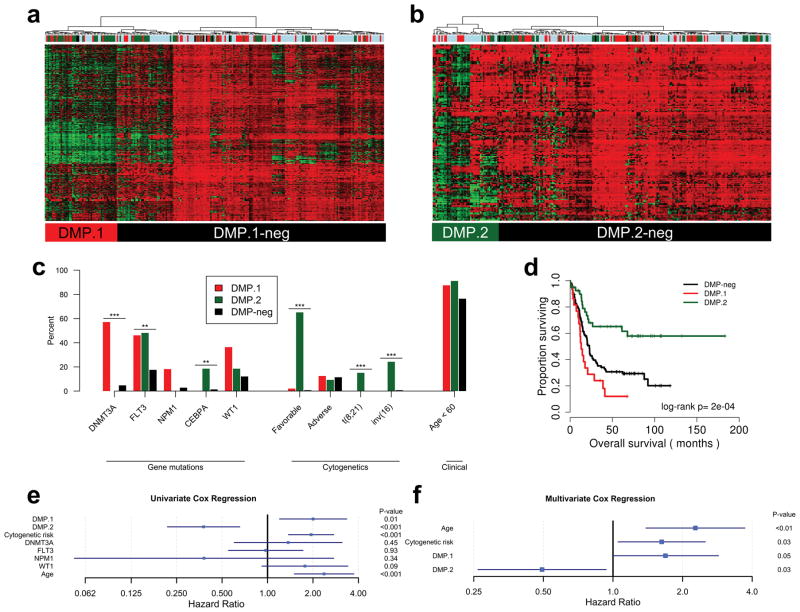
In order to validate our findings we queried additional AML datasets on the 450k platform, including TARGET, GSE62298, GSE58477, and GSE64934 (see methods). We integrated the validation datasets to form a superset consisting of 236 AML cases, and used our identified DMP signatures to classify the patient samples by hierarchical clustering. Based on DMP methylation status we found groups of 50, and 45 leukemias positive for DMP.1 and DMP.2, respectively (Figure 5a and b). Consistent with our previous analyses, these phenotypes were mutually exclusive.
Figure 5. Validation of DMPs in independent DNA methylation datasets.
Hierarchical clustering of AML cases from independent datasets based on markers for DMP.1 (a), and DMP.2 (b) confirmed the presence of both DMPs in a subset of patients. Color bars above each heatmap correspond to the different datasets-of-origin (see methods). c) Plots of enrichments for available genetic and demographic data in DMP.1, DMP.2, and DMP-neg. d) Kaplan-Meier analysis of the validation cohort revealed differences in overall survival based on DMP.1 and DMP.2 status. e) Univariate cox regression analysis for age, genetic features, and DMP status. f) Multivariate Cox regression for significant covariates from univariate analyses in (e). For Cox regression analyses, the point estimate for the hazard ratio is plotted along with the 95% confidence interval. * P<0.05, ** P<0.01, *** P<0.001.
We then sought to characterize these cases based on genetic mutations and clinical outcomes. We confirmed relative enrichments for DNMT3A and NPM1 mutations in DMP.1, however, unlike in the TCGA cohort, the majority of cases did not harbor NPM1 mutations (57% vs. 5% in DMP-neg and 18% vs. 3% in DMP-neg, respectively; Fisher’s Exact P<0.001, 0.06, for DNMT3A and NPM1, respectively; Figure 5c). We speculate this is due to the relative rarity of NPM1 mutations in the pediatric TARGET dataset, however, DMP.1 leukemias still carried the highest percentage of mutations compared to DMP.2 and DMP-neg. DMP.2 leukemias, in contrast, lacked DNMT3A mutations, and were significantly enriched for favorable cytogenetics, however, not all DMP.2 cases harbored these rearrangements (65% vs. 1% in DMP-neg; Fisher’s Exact P<0.001; Figure 5c). We also examined DNMT3A mutant cases in isolation and found that 80% were positive for DMP.1. We further considered a recent study published by Cauchy et al. which identified a FLT3-associated epigenetic signature 18. We sought to see whether the DMP.1 signature could recapitulate this FLT3 phenotype by hierarchical clustering of DNA methylation for the AML cases published in the Cauchy study, and found that demethylation of the DMP.1 sites was present in two patients, one of which harbored a FLT3 mutation (Supplementary Figure S6). Thus, it is likely that the FLT3 and DMP.1 DNA methylation signatures are independent. Importantly, within this external cohort we confirmed that DMP.1 demethylation does not occur in cases with IDH1/2 or TET2 mutations (Supplementary Figure S6).
In addition to validation of mutational signatures, we found that in this cohort DMP.1 patients had significantly inferior overall survival compared to DMP-neg, while patients with DMP.2 disease had significantly better outcomes (median OS, months: DMP.1=13.1, DMP.2=not reached, DMP-neg=22.8; log-rank P<0.001; Figure 5d). We also found that patients with intermediate or poor risk cytogenetic abnormalities who had DMP.2 disease had significantly better OS compared to DMP-neg (median OS, months: DMP.2=not reached, DMP-neg=20.3; log-rank P=0.03; Supplementary Figure S7).
Because both DMPs were associated with survival and neither was completely defined by gene mutations, we tested whether the DNA methylation effects were independent from cytogenetics and age in this cohort. First we performed univariate Cox regression analysis using available clinical covariates and gene mutations and found that only DMP status (DMP.1 HR=2.00, 95% CI: 1.20–3.35, P=0.008; DMP.2 HR=0.38, 95% CI: 0.22–0.65, P=0.001), age > 60 (HR=2.37, 95% CI: 1.50–3.73, P<0.001), and cytogenetic risk (HR=1.94, 95% CI: 1.38–2.74, P<0.001) were associated with outcome (Figure 5e). In multivariate Cox regression models both DMP.1 (HR=1.69, 95% CI: 0.99–2.86, P=0.05) and DMP.2 (HR=0.49, 95% CI: 0.26–0.93, P=0.03) were associated with survival independent of each other, age and cytogenetic risk (Figure 5f). These data suggest that DMP status may be useful to augment existing clinical covariates in stratifying patients with AML.
Discussion
Although numerous studies have investigated the complexities of aberrant hypermethylation in cancer, there is a relative gap in knowledge in the biology surrounding losses of DNA methylation. In this study we interrogated genome-wide DNA methylation data to identify demethylator phenotypes in AML. We found two distinct and non-overlapping DMPs: DMP.1 and DMP.2. Although both types of leukemia were characterized largely by demethylation of normally methylated non-CGI CpGs, the specific genomic targets of demethylation were non-random and were associated with gene expression, mutational, and clinical signatures. Interestingly, the identification of DMP-positive leukemia did not materially differ between using normal bone marrow-derived hematopoietic stem cells versus normal whole peripheral blood as controls. This suggests that DMP demethylation is cancer-specific as the same CpG sites that lose methylation in DMP-positive AML are methylated in both normal peripheral blood cells (differentiated) and hematopoietic stem cells (undifferentiated).
The DMPs were genetically distinct with most DMP.2 patients harboring t(8;21), inv(16), or t(15;17). In our validation cohort, approximately one-third of DMP.2 patients did not have one of these rearrangements, and importantly, the DMP.2 methylation profile was associated with better survival independent of cytogenetic risk. This result suggests that a possible epigenetic mechanism may underlie the improved curability in patients harboring good-risk genomic rearrangements. This possibility is further alluded to by the observed up-regulation of genes enriched for organism development, and targets of specific hematopoietic transcription factors, including LEF-1 and GFI1 32, 33. The biological connection between the three cytogenetic abnormalities is likely a common differentiation “proneness”. This is further supported by the observation that some DMP.2 CpG sites are demethylated in certain differentiated leukocyte fractions derived from healthy individuals. We speculate that leukemias harboring demethylation at these sites may be epigenetically poised to differentiate regardless of the specific cytogenetic rearrangement that led to transformation.
In contrast to DMP.2, DMP.1 was associated with relatively poor clinical outcomes and was enriched for co-occurring DNMT3A, NPM1, and FLT3 mutations. Although the role of DNMT3A in writing DNA methylation marks is well-established, the strong association between NPM1 mutations and DNA demethylation may merit further investigation into a possible mechanistic relationship. In addition, we identified an intriguing subset of DNMT3A-mutation-positive leukemias which did not demethylate at DMP.1 CpG sites. These cases showed an enrichment for mutations in IDH1, IDH2, and TET2, all of which have been previously implicated in hypermethylation in cancer 6, 13, 34. Of equal interest, this same population of patients lacks hypermethylation at I-CIMP CpG sites, suggesting a possible co-dependency between DNMT3A and TET demethylases in regulating the methylome. RNA-seq data support this speculation by virtue of marked down-regulation of DNMT3A with up-regulation of TET2 in DMP.1 leukemias. The fact that DNMT3A mutant AML does not necessarily show global hypomethylation and that IDH1/2 mutant AML does not necessarily show hypermethylation raise questions as to methylation independent mechanisms of transformation by these mutations. Indeed, it was recently reported that a RAS signaling signature was upregulated in IDH and DNMT3A co-mutated leukemias, and that patient-derived primary cells were sensitive to MEK inhibition 35.
In addition to epigenetic regulators, the other key transcriptomic finding in DMP.1 AML was up-regulation of immune response genes. Based on analysis of transcriptomic data alone, immune activation and viral response were among the most significantly enriched functions, with specific genes including the chemokines CXCL9 and CXCL10, and interleukins IL-3 and IL-15. Analysis of genes which were both up-regulated and hypomethylated confirmed this functional enrichment. Thus the transcriptional programs of DMP.1 and DMP.2 are distinct, and may contribute to leukemia which is primed to proliferate via an immune response profile in the case of DMP.1, or primed to differentiate in the case of DMP.2, thereby contributing to their clinical differences 36, 37.
In summary we present evidence of two DNA demethylation signatures in AML which have mutually exclusive genetic backgrounds and clinical characteristics. Our data suggest an epigenetic link between different good-risk genomic rearrangements, and a methylation profile associated with DNMT3A mutations, and up-regulation of immune response genes. We found that both DMPs were prognostic - independent of age and cytogenetic risk - in an independent validation cohort. Future studies should validate the clinical relevance of these DMP signatures and attempt to integrate them with other established risk markers, and DNA methylation profiles in AML. In addition, the possible interactions between DNMT3A mutations and TET demethylases should be further explored from a molecular mechanistic perspective. More globally, the presence of demethylator phenotypes in cancer add to the complexity of epigenetic deregulation and should be examined across all cancer types in the same way CIMP was examined by the TCGA.
Supplementary Material
Acknowledgments
Funding: This work was supported by National Institutes of Health grants R01CA158112 and P50CA100632. J-PJI. is an American Cancer Society Clinical Research professor supported by a generous gift from the FM Kirby Foundation.
We would like to thank Drs. Humberto Ferreira and Manel Esteller for providing additional clinical data to supplement their 450k GEO submission.
Footnotes
Conflict of Interest
The authors declare that there are no competing financial interests with regard to the results presented in this study.
References
- 1.Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, et al. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982 Apr;10(8):2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Razin A, Riggs AD. DNA methylation and gene function. Science. 1980 Nov;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- 3.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999 Jul;96(15):8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toyota M, Ahuja N, Suzuki H, Itoh F, Ohe-Toyota M, Imai K, et al. Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype. Cancer Res. 1999 Nov;59(21):5438–5442. [PubMed] [Google Scholar]
- 5.Hill VK, Shinawi T, Ricketts CJ, Krex D, Schackert G, Bauer J, et al. Stability of the CpG island methylator phenotype during glioma progression and identification of methylated loci in secondary glioblastomas. BMC Cancer. 2014 Jul;14(1):506. doi: 10.1186/1471-2407-14-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelly AD, Kroeger H, Yamazaki J, Taby R, Neumann F, Yu S, et al. A CpG island methylator phenotype in acute myeloid leukemia independent of IDH mutations and associated with a favorable outcome. Leukemia. 2017 Jan; doi: 10.1038/leu.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van den Bent MJ, Gravendeel LA, Gorlia T, Kros JM, Lapre L, Wesseling P, et al. A hypermethylated phenotype is a better predictor of survival than MGMT methylation in anaplastic oligodendroglial brain tumors: a report from EORTC study 26951. Clin Cancer Res. 2011 Nov;17(22):7148–7155. doi: 10.1158/1078-0432.CCR-11-1274. [DOI] [PubMed] [Google Scholar]
- 8.Hegi ME, Diserens AC, Godard S, Dietrich PY, Regli L, Ostermann S, et al. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res. 2004 Mar;10(6):1871–1874. doi: 10.1158/1078-0432.ccr-03-0384. [DOI] [PubMed] [Google Scholar]
- 9.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983 Jan;301(5895):89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013 May;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferreira HJ, Heyn H, Vizoso M, Moutinho C, Vidal E, Gomez A, et al. DNMT3A mutations mediate the epigenetic reactivation of the leukemogenic factor MEIS1 in acute myeloid leukemia. Oncogene. 2015 Oct [Google Scholar]
- 12.Yang L, Rodriguez B, Mayle A, Park HJ, Lin X, Luo M, et al. DNMT3A Loss Drives Enhancer Hypomethylation in FLT3-ITD-Associated Leukemias. Cancer Cell. 2016;29(6):922–934. doi: 10.1016/j.ccell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010 Dec;18(6):553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamazaki J, Taby R, Jelinek J, Raynal NJ, Cesaroni M, Pierce SA, et al. Hypomethylation of TET2 Target Genes Identifies a Curable Subset of Acute Myeloid Leukemia. J Natl Cancer Inst. 2016 Feb;108(2) doi: 10.1093/jnci/djv323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013 Jan;41(Database issue):D991–995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fortin JP, Labbe A, Lemire M, Zanke BW, Hudson TJ, Fertig EJ, et al. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 2014;15(12):503. doi: 10.1186/s13059-014-0503-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qu Y, Lennartsson A, Gaidzik VI, Deneberg S, Karimi M, Bengtzén S, et al. Differential methylation in CN-AML preferentially targets non-CGI regions and is dictated by DNMT3A mutational status and associated with predominant hypomethylation of HOX genes. Epigenetics. 2014 Aug;9(8):1108–1119. doi: 10.4161/epi.29315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cauchy P, James SR, Zacarias-Cabeza J, Ptasinska A, Imperato MR, Assi SA, et al. Chronic FLT3-ITD Signaling in Acute Myeloid Leukemia Is Connected to a Specific Chromatin Signature. Cell Rep. 2015 Aug;12(5):821–836. doi: 10.1016/j.celrep.2015.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farrar JE, Schuback HL, Ries RE, Wai D, Hampton OA, Trevino LR, et al. Genomic Profiling of Pediatric Acute Myeloid Leukemia Reveals a Changing Mutational Landscape from Disease Diagnosis to Relapse. Cancer Res. 2016;76(8):2197–2205. doi: 10.1158/0008-5472.CAN-15-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reinius LE, Acevedo N, Joerink M, Pershagen G, Dahlén SE, Greco D, et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One. 2012;7(7):e41361. doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004 Jan;32(Database issue):D493–496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013 Jan;41(Database issue):D56–63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smit A, Hubley RPG. RepeatMasker. at http://repeatmasker.org. [cited 2017; Available from.
- 24.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010 Jan;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012 May;40(10):4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nogales-Cadenas R, Carmona-Saez P, Vazquez M, Vicente C, Yang X, Tirado F, et al. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009 Jul;37(Web Server issue):W317–322. doi: 10.1093/nar/gkp416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 2007;8(1):R3. doi: 10.1186/gb-2007-8-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A. GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012 Jul;40(Web Server issue):W478–483. doi: 10.1093/nar/gks402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.R Core Development Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2014. [Google Scholar]
- 30.Therneau T. R package version 2.37–7. 2014. A Package for Survival Analysis in S. [Google Scholar]
- 31.Itonaga H, Imanishi D, Wong YF, Sato S, Ando K, Sawayama Y, et al. Expression of myeloperoxidase in acute myeloid leukemia blasts mirrors the distinct DNA methylation pattern involving the downregulation of DNA methyltransferase DNMT3B. Leukemia. 2014 Jul;28(7):1459–1466. doi: 10.1038/leu.2014.15. [DOI] [PubMed] [Google Scholar]
- 32.Hock H, Hamblen MJ, Rooke HM, Traver D, Bronson RT, Cameron S, et al. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity. 2003 Jan;18(1):109–120. doi: 10.1016/s1074-7613(02)00501-0. [DOI] [PubMed] [Google Scholar]
- 33.Skokowa J, Cario G, Uenalan M, Schambach A, Germeshausen M, Battmer K, et al. LEF-1 is crucial for neutrophil granulocytopoiesis and its expression is severely reduced in congenital neutropenia. Nat Med. 2006 Oct;12(10):1191–1197. doi: 10.1038/nm1474. [DOI] [PubMed] [Google Scholar]
- 34.Yamazaki J, Jelinek J, Lu Y, Cesaroni M, Madzo J, Neumann F, et al. TET2 Mutations Affect Non-CpG Island DNA Methylation at Enhancers and Transcription Factor-Binding Sites in Chronic Myelomonocytic Leukemia. Cancer Res. 2015 Jul;75(14):2833–2843. doi: 10.1158/0008-5472.CAN-14-0739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glass JL, Hassane D, Wouters BJ, Kunimoto H, Avellino R, Garrett-Bakelman FE, et al. Epigenetic Identity in AML Depends on Disruption of Nonpromoter Regulatory Elements and Is Affected by Antagonistic Effects of Mutations in Epigenetic Modifiers. Cancer Discov. 2017 Aug;7(8):868–883. doi: 10.1158/2159-8290.CD-16-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meazza R, Basso S, Gaggero A, Detotero D, Trentin L, Pereno R, et al. Interleukin (IL)-15 induces survival and proliferation of the growth factor-dependent acute myeloid leukemia M-07e through the IL-2 receptor beta/gamma. Int J Cancer. 1998 Oct;78(2):189–195. doi: 10.1002/(sici)1097-0215(19981005)78:2<189::aid-ijc12>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 37.Kittang AO, Hatfield K, Sand K, Reikvam H, Bruserud Ø. The chemokine network in acute myelogenous leukemia: molecular mechanisms involved in leukemogenesis and therapeutic implications. Curr Top Microbiol Immunol. 2010;341:149–172. doi: 10.1007/82_2010_25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The TCGA and TARGET data used in this study – including Illumina 450k arrays, RNA-seq, and clinical annotations – are publicly available in the Genomic Data Commons (https://gdc.cancer.gov/). Other DNA methylation datasets used are available in the Gene Expression Omnibus (GEO): GSE51388, GSE62298, GSE58477, GSE35069, and GSE32251. ChIP-seq data for HL-60 and CD34+ cells are available on the UCSC Genome Browser (https://genome.ucsc.edu/).