Abstract
Th17 cells play a critical role in the pathogenesis of autoimmune diseases, including multiple sclerosis, rheumatoid arthritis, systemic lupus erythematosus, Sjogren’s syndrome, and inflammatory bowel disease. Despite the extensive investigation into this T cell lineage, little is understood regarding the role of Th17 lineage-specific lncRNAs (long non-coding RNAs) > 200 nt. lncRNAs may influence disease through a variety of mechanisms; their expression could be regulated by SNPs. lncRNAs can also affect the expression of neighboring genes or complementary miRNAs, and their expression may have lineage-specific patterns. In the system biology study presented here, the effective lncRNAs from different criteria were predicted for each autoimmune disease, and we then evaluated their expression levels in 50 MS patients compared to 25 controls using qRT-PCR. We identified changes in the expression levels of AL450992.2, AC009948.5, and RP11-98D18.3 as potential peripheral blood mononuclear cell (PBMC) biomarkers for MS among our studied lncRNAs in which co-expression analysis of AL450992.2 had the most AUCs, and the relationship to RORC was also assessed. We propose that the recurrently deregulated lncRNAs identified in this report could provide a valuable resource for studies aimed at delineating the relationship between functional lncRNAs and autoimmune disorders.
Keywords: autoimmune disease, biomarker, lncRNA
Introduction
Approximately 7% to 9% of people around the world suffer from autoimmune and immune-related diseases (AIDs) from a variety of heterogeneous disorders.1 Th17 cell subsets with specific developmental requirements and functions play critical roles in the pathogenesis of autoimmune diseases.2, 3 Although transcriptional regulation of Th17 cell differentiation has been extensively studied, there is little understanding of the post-transcriptional regulation of Th17 cells.
Recently, reports suggest that long non-coding RNAs (lncRNAs), i.e., non-coding RNAs > 200 nt, have more stability compared to protein-coding genes in body fluids and tissues, have tissue-specific patterns of expression, and are relatively easy to detect through various techniques in liquid biopsies. lncRNAs are attractive disease biomarkers and therapeutic targets, since they may modulate gene expression through several mechanisms.4, 5, 6, 7 In order to manifest the associated lncRNAs with the disease, several possibilities exist, including the possibility that they could be influenced by SNPs, affect the expression of neighboring genes or complementary microRNAs (miRNAs), or affect lineage-specific expression. In this paper, we address all of these influences.
The unique functions of each cell type are performed by its specialized gene expression. Therefore, regulating lineage-specific lncRNAs could affect cell fate and play a critical role in the pathology of disease.8 Roughly 50% of protein-coding genes within 50 kb were co-expressed with the respective lncRNA gene, and this correlation will decrease by each kilobase farther away from the genomic position of the protein-coding gene.9
Integration of AID-associated SNPs with genome functional region data demonstrate that ∼90% of SNPs are associated with regulatory DNA enhancer or promoter regions rather than with protein-coding regions.10, 11 Reported trait and/or disease-associated SNPs (TASs) were significantly overrepresented only in nonsynonymous sites and 5-kb promoter regions compared to SNPs randomly selected from genotyping arrays.12 Logistically, genomic regions containing SNPs associated with disease could include disease-associated genes like lncRNAs.
lncRNAs may have critical roles through targeting miRNA in a wide range of biological processes; accordingly, competing endogenous RNA theories could explain how lncRNAs regulate coding genes via competitively binding with their complementary miRNA.13, 14
Identifying influential biomarkers could potentially shed light on the underlying pathogenic mechanisms in AIDs. From a practical standpoint, the most useful autoimmune biomarkers will be those measurable in peripheral blood mononuclear cells (PBMCs) and serum. The utility of most of these markers is limited by their restriction to relatively inaccessible anatomic sites.
One emerging view is that lncRNAs could be novel molecules for disease diagnosis and therapy.15 In this system biology study, we aimed to deduce effective lncRNAs in several autoimmune diseases, including multiple sclerosis (MS), rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), Sjogren’s syndrome (SS), and inflammatory bowel disease (IBD) through several criteria, and we show that the expression levels of some are significantly elevated in MS.
Results
lncRNA Selection
Among the currently known lncRNAs, we selected 26 Th17 cell-lineage-specific lncRNAs that derived from previous RNA-sequencing (RNA-seq) data. These lncRNAs included 54 transcripts (Table S1).
lncRNA Selection from Adjustment to Th17 Cell Differentiation Genes
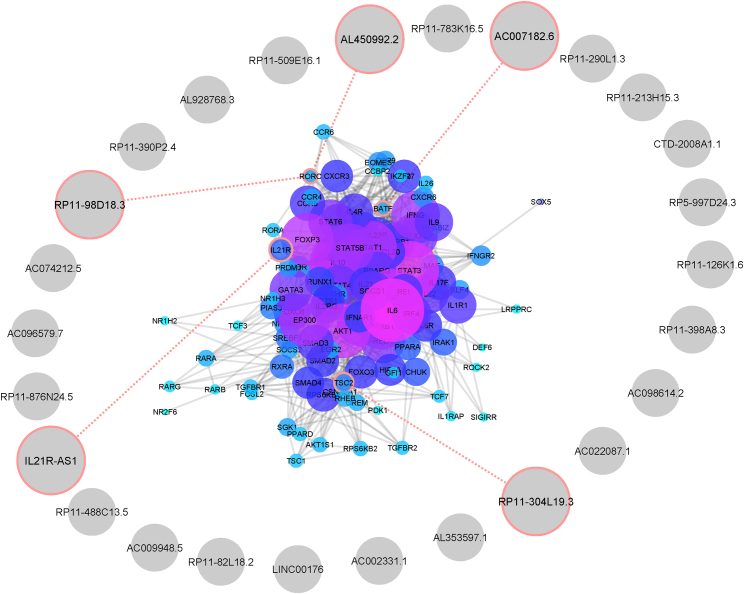
From mining the literature, we identified 116 genes encoding proteins involved in Th17 cell differentiation and categorized them according to their signaling pathway involvement (Table S2). From this list, we identified 5 lncRNAs that neighbor the genes involved in Th17 cell differentiation with a distance less than 50 kb and for which 4 of them were also co-expressed with IL21R, RORC, and BATF, which are also expressed in Th17 cell differentiation (Figure 1).
Figure 1.
Association of Th17 Cell Differentiation Genes and lncRNAs
Th17 cell differentiation genes were deduced through data mining, and their proximities to selected lncRNAs were evaluated through Python programming language. Desired lncRNAs with the appropriate distance are indicated by red circles.
lncRNA Selection from Adjustment to Differentially Expressed Genes in AIDs
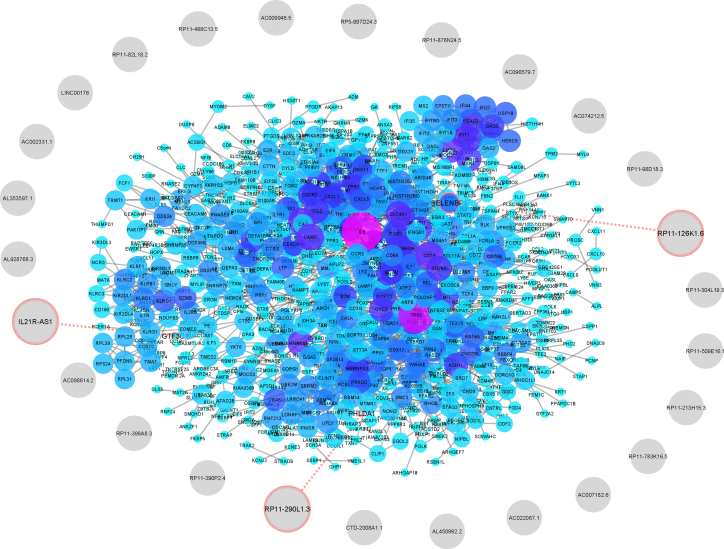
In the next phase, we analyzed array datasets of AID genes by comparing patient PBMC samples to controls and identified the most differentially expressed protein-coding genes (MS, 405; SLE, 102; SS, 55; RA, 23; and IBD, 238) (Table S3). We then examined the proximity of these genes to candidate lncRNA genes and recorded lncRNAs that were located less than 50 kb from protein-coding genes as associated or linked. We retrieved 1 lncRNA linked to MS and 3 linked to IBD genes with a potentially cis-regulatory role in neighboring genes, one of which was shared between these two groups (Figure 2).
Figure 2.
Association of lncRNAs and Array-Expressed Genes
Differentially expressed mRNAs in AID samples compared to controls were retrieved from the Gene Expression Omnibus (GEO) database, and their proximities to selected lncRNAs were analyzed by Python programming language, inputted to STRING-db, and visualized in Cytoscape. Appropriate lncRNAs in logical distance are indicated with red circles.
lncRNA Selection from Adjustment to the AID-Associated SNPs
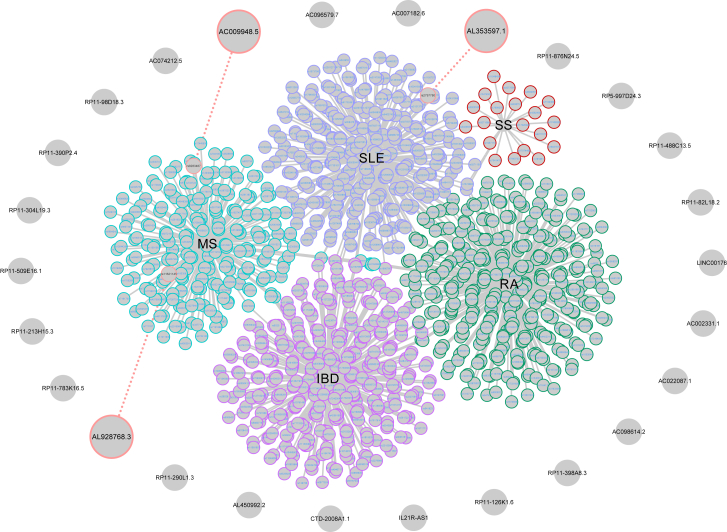
SNPs associated with each AID were retrieved from the Genome-Wide Association Studies (GWAS) catalog (MS, 210; SLE, 307; RA, 365; SS, 26; and IBD, 345) (Table S4). Accordingly, we predicted 3 lncRNAs related to AIDs via SNPs, including two associated with MS and one associated with SLE that was located less than 50 kb away from selected SNPs (Figure 3).
Figure 3.
SNP-lncRNA Association
SNPs associated with selected AIDs were determined, and their distance from each lncRNA locus was assessed by Python programming language and visualized via Cytoscape. Selected lncRNAs in this analysis were connected to the associated SNP, as indicated with red circles.
lncRNA-miRNA Interactions and Associated Diseases
Next, we inserted the sequence of every single exonic lncRNA transcript in FASTA format into the “LncRNAs Input” panel of LncDisease software, as shown in Table 1.
Table 1.
Deduced Potentially Associated lncRNAs according to miRNA Interaction Deduced from LncDisease Software
Putative lncRNAs Associated with MS
We selected 8 putative lncRNAs that, in addition to Th17 cell-lineage specificity, were also derived from at least one of the other criteria: 4 lncRNAs involved in Th17 cell-differentiation pathways, 1 lncRNA selected from an array expression criterion, 1 derived from LncDisease software, 1 lncRNA related to a SNP association criterion, and 1 derived from both SNP association and LncDisease software. AL450992.2 was associated with miRNA; therefore, we only evaluated this transcript for SNP association. This lncRNA was associated with MS (Table 2), so we explored its expression levels in a case-control study by real-time qPCR analysis.
Table 2.
Putative lncRNAs in MS Pathology Derived from System Biology Analyses
| Criterion | lncRNA |
|---|---|
| SNP association with AIDs | AC009948.5 |
| AL928768.3 | |
| miRNA interactions | AC009948.5-007 |
| RP11-290L1.3-001 | |
| Differentially expressed genes in AIDs | RP11-126K1.6 |
| Th17 cell differentiation genes | IL21R-AS1 |
| RP11-98D18.3 | |
| AL450992.2 | |
| AC007182.6 |
Patient Characteristics
The clinical and pathological data including sex, age, illness duration, and score on the Expanded Disability Status Scale (EDSS) have been recorded. Statistical analyses showed no significant differences between all samples in each group (Table 3).
Table 3.
Patient Clinical and Demographic Data
| Characteristics | RRMS Patients | Relapsing | Remitting | Control | p Value Comparison between Patients and Control/Relapsing, Remitting, and Control Samples |
|---|---|---|---|---|---|
| Sex: female/male | 38/12 | 19/6 | 19/6 | 17/8 | 0.46/0.76 |
| Age (and SD) | 34 (8.30) | 32.48 (7.07) | 35.52 (9.26) | 32.84 (6.06) | 0.49/0.23 |
| Duration of illness (and SD) | 3.08 (4.41) | 0 | 6.16 (4.47) | – | – |
| Disease severity (EDSS) | 1.47 | 1.56 | 1.38 | – | – |
EDSS, expanded disability status scale.
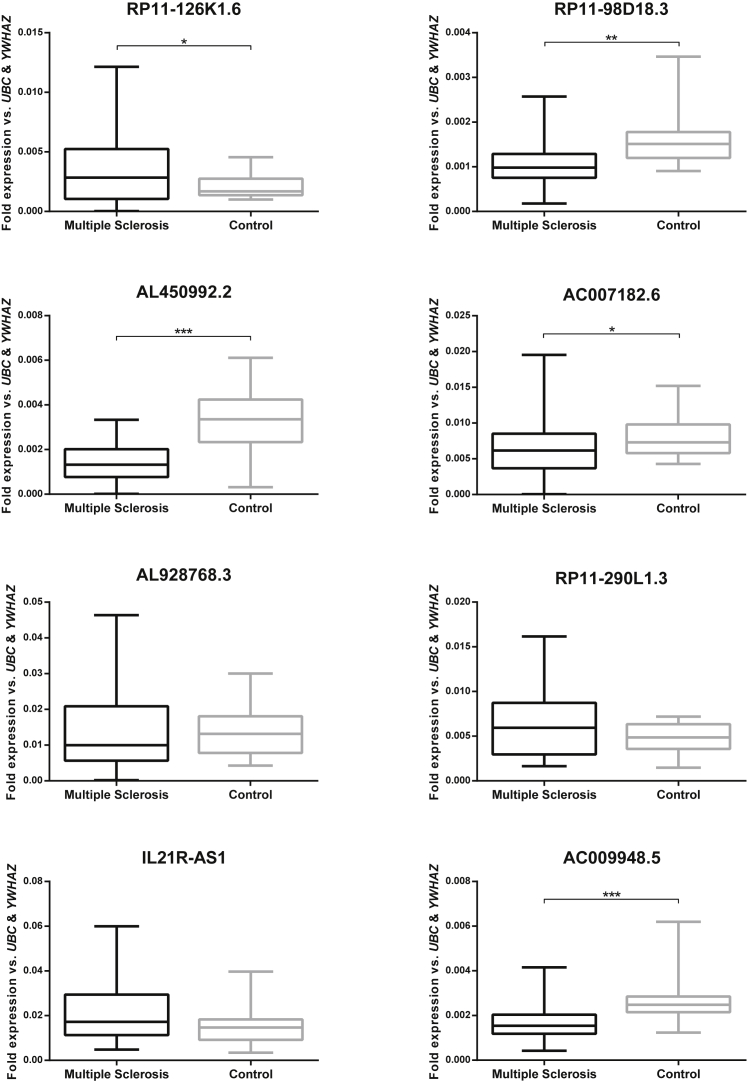
Significant Changes in Levels of Expression of Specific lncRNAs in MS Patients
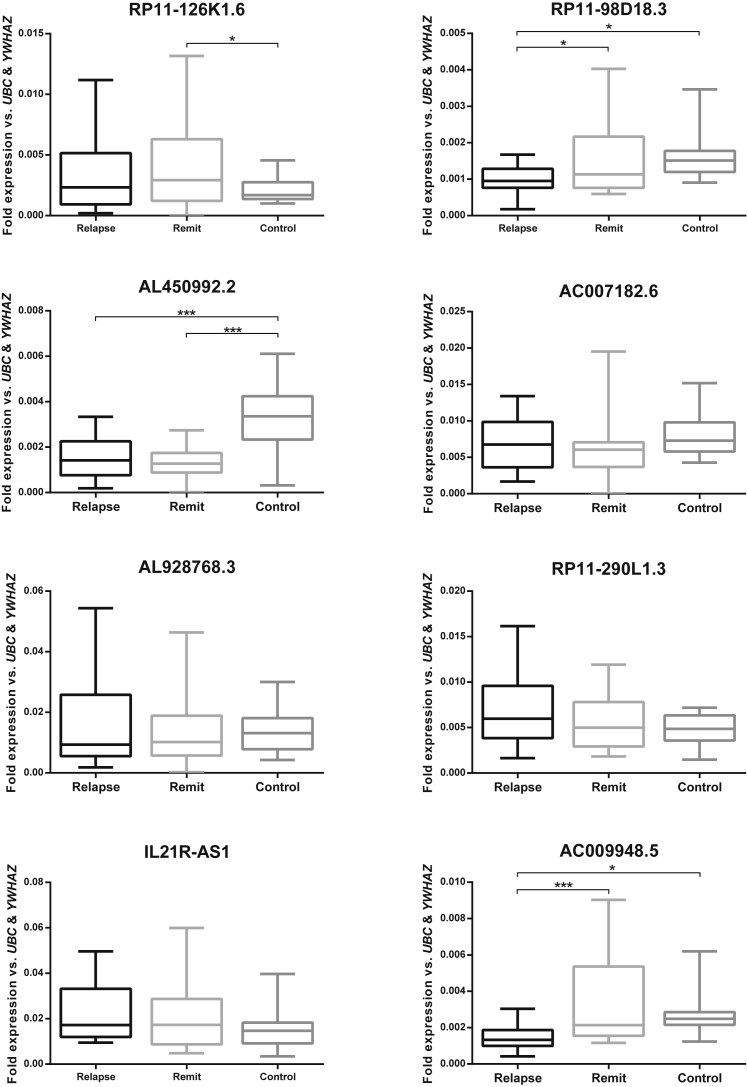
We evaluated the expression levels of selected lncRNAs in the PBMCs of MS patients compared to the control group. Real-time qPCR analysis showed a significant elevation of RP11-126K1.6 expression in the PBMCs of MS patients compared to those of the healthy controls (p = 0.023). Furthermore, RP1198D18.3, AL450992.2, and AC007182.6 lncRNAs associated with the Th17 cell differentiation genes, and AC009948.5—an lncRNA associated with miRNA and SNP—were downregulated in patients compared to the healthy volunteers (p = 0.001, p < 0.001, p = 0.041, and p < 0.001, respectively). In contrast, expression levels of these lncRNAs showed no statistically significant difference between the MS samples and control group: AL928768.3 as a SNP-related lncRNA, RP11-290L1.3 as a miRNA-associated lncRNA, and IL21R-AS1 as associated with a differentially expressed protein-coding gene (Figure 4).
Figure 4.
lncRNA Expression in MS compared to Controls
Boxplots of the expression levels of lncRNA genes in MS and control samples examined by real-time qPCR; values are given as mean normalized expression relative to UBC and WYHAZ; asterisks indicate significant differences to the controls (*p < 0.05; **p < 0.01; ***p < 0.001).
Significant Changes in Levels of Expression of Specific lncRNAs in Relapsing- and Remitting-Phase Patients
RP11-126K1.6, a lncRNA adjusted to array-expressed genes, had elevated expression levels in the remitting phase of disease compared to healthy volunteers (p = 0.039). RP1198D18.3 and AL450992.2, lncRNAs close to genes involved in Th17 cell differentiation, which were evaluated in relapsing-phase patients compared to controls using real-time qPCR, showed significantly decreased transcript levels (p = 0.030 and p < 0.001, respectively) and a decreased RP1198D18.3 expression level in relapsing-phase compared to remitting-phase patients (p = 0.016). Data indicated that AL450992.2 was downregulated in remitting-phase rather than healthy volunteers (p < 0.001). However, these values were not significant for IL21R-AS1 and AC007182.6 between the relapsing-phase, remitting-phase, and control groups.
We assessed the expression levels of RP11-290L1.3 as a miRNA-associated lncRNA; however, the differences between the relapsing-phase, remitting-phase, and control groups were not as significant as for AL928768.5, a SNP-related lncRNA. However, there was a significant decrease in AC009948.5, a lncRNA associated with both miRNA and SNP, in the relapsing phase compared to the controls (p = 0.016) and in the relapsing phase compared to the remitting phase (p < 0.001) (Figure 5).
Figure 5.
lncRNA Expression in Relapsing and Remitting Phases compared to the Controls
Boxplots of differential expression levels of lncRNA genes in relapsing-phase, remitting-phase, and control samples (*p < 0.05; ***p < 0.001).
ROC Curve
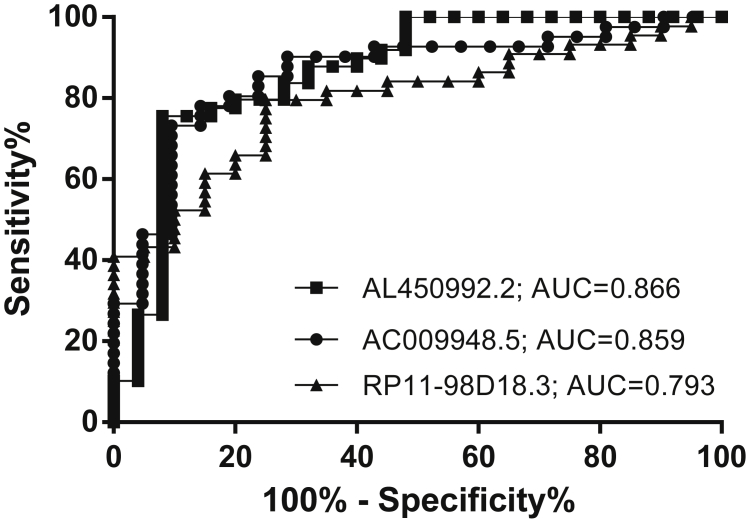
We performed a comparative analysis of the lncRNAs generated, and then we ranked them as biomarker candidates according to decreasing area under the receiver operating characteristic (ROC) curve (AUC) and graphed the results. The first three lncRNAs ranked by AUC are AL450992.2, with an AUC of 0.866 (95% confidence interval [CI] = 0.770–0.962, p < 0.001); AC009948.5, with an AUC of 0.859 (95% CI = 0.761–0.958, p < 0.001); and RP11-98D18.3, with an AUC of 0.793 (95% CI = 0.682–0.904, p < 0.001) (Figure 6).
Figure 6.
ROC Curve Analysis with Multiple Markers
Discriminatory power of the individual most differentially expressed lncRNAs, AL450992.2, AC009948.5, and RP11-98D18.3, for the diagnosis of MS patients and controls.
Correlation of lncRNAs Adjusted to RORC
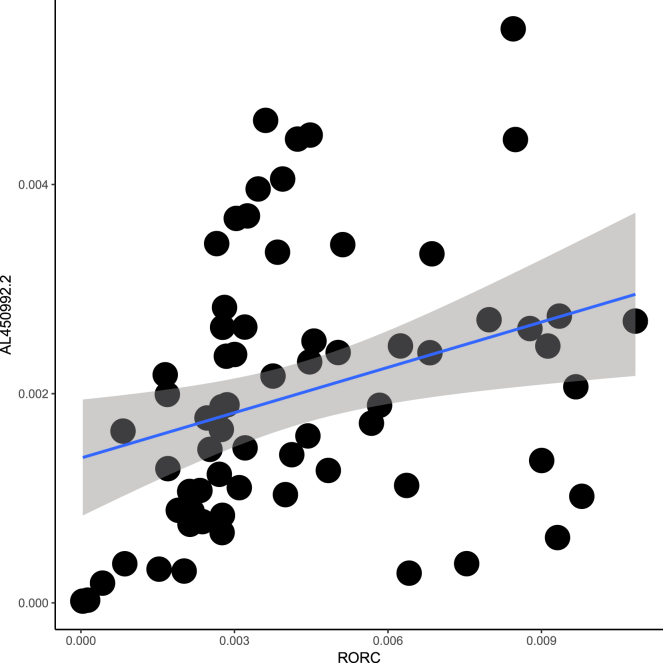
We assessed RORC expression levels in the PBMCs of patients compared to those of controls, which were adjusted to RP11-98D18.3 and AL450992.2. AL450992.2 relative expression fold change in PBMCs and RORC in MS patients showed a significant positive correlation (Pearson’s correlation = 0.306, p < 0.05) (Figure 7), while no correlation was observed between RP11-98D18.3 and RORC (Pearson’s correlation = 0.210, p = 0.11); moreover, there was no significant difference in RORC expression levels in MS patients compared to those of the controls.
Figure 7.
Correlation between AL450992.2 and RORC in MS Patients
Correlation between AL450992.2 relative expression fold change in PBMCs and RORC (Pearson’s correlation = 0.31, p < 0.05).
Discussion
To identify disease-associated lncRNAs, we applied an integrated method considering several criteria. Investigation of the chromosomal locations of obtained results of criteria, including Th17 cell differentiation genes, differentially expressed genes in AIDs; and AIDs-associated SNPs with lineage-specific lncRNAs indicated that ∼4.31% of Th17 cell differentiation genes were neighbors to the selected lncRNAs, while only ∼0.04% of genes retrieved from array expression were in association with lncRNA and only ∼0.02% of SNPs were in association with lncRNAs involved in disease suseptibility, which points to the fact that genes encoding Th17 cell differentiation are enriched in the genomic regions containing genes encoding lineage-specific lncRNAs. Hence, by applying these techniques to cell types involved in different diseases and protein-coding genes in specific cells, we could potentially identify more disease-associated lncRNAs.
Effector Th17 cell populations can cause CNS inflammation and demyelinating lesions.16 Accordingly, as demonstrated earlier, protein-coding genes associated with lncRNAs9 (RP11-98D18.3, AL450992.2, AC007182.6, and IL21R.AS1) were decreased in effector Th17 cells compared to primary cell cultures,9 and our investigations showed consistency with these results, as three of these lncRNAs (RP11-98D18.3, AL450992.2, and AC007182.6) were downregulated in MS patients compared to the controls.
Real-time qPCR of RP11-126K1.6 showed upregulation in MS patients compared to the controls, which was in accordance with its differentially expressed gene in MS samples, as this study evaluated expressed genes in MS patients compared to the controls with no discriminatory group in patients, consequently, in order to verify that lncRNAs associated with this criterion-specific sampling of patients on differentially expressed genes could influence the results.
AC009948.5 expression results indicated that this putative lncRNA could, indeed, be associated with decreased level of MS pathogenesis, as it was identified from SNP (rs9283487) association and LncDisease criteria, while the reported risk SNP (rs11621145) from GWAS that is associated with the expression of AL928768.7 was not dysregulated, which suggests functional mechanisms underlying the findings from GWAS that are more complex than regulatory variants or expression levels of nearby lncRNA genes. Chromatin-looping models could explain how a contributing lncRNA is not necessarily the closest gene that is influenced by SNPs.17
The insignificant expression level of RP11-290L1.3 could be due to the limitations of LncDisease software for determining putative lncRNAs associated with miRNAs, as the exact mechanism by which miRNAs regulate lncRNAs is not clear, which results in the prediction that lncRNA targets of miRNAs will have high false positives and high false negatives.
The expression of lncRNAs is more disease and tissue specific than that of protein-coding genes and are more associated with their biological function.18 lncRNAs as regulators of diverse biological processes by immune cells and the molecular mechanism of autoimmunity might be associated with numerous autoimmune diseases. Even recently, microvesicles found to contain lncRNAs involved in AIDs have been reported.19, 20, 21 Previous studies demonstrated the therapeutic and biomarker roles of lncRNAs in various diseases such as RA, IBD, and SLE. These include the effect of large intergenic noncoding RNA (lincRNA)-p21 on nuclear factor κB (NF-κB) activity in RA, PlncRNA1 mediated the function of the intestinal epithelial barrier in IBD, and NEAT1 as a regulator of inflammatory pathway in SLE.22, 23, 24
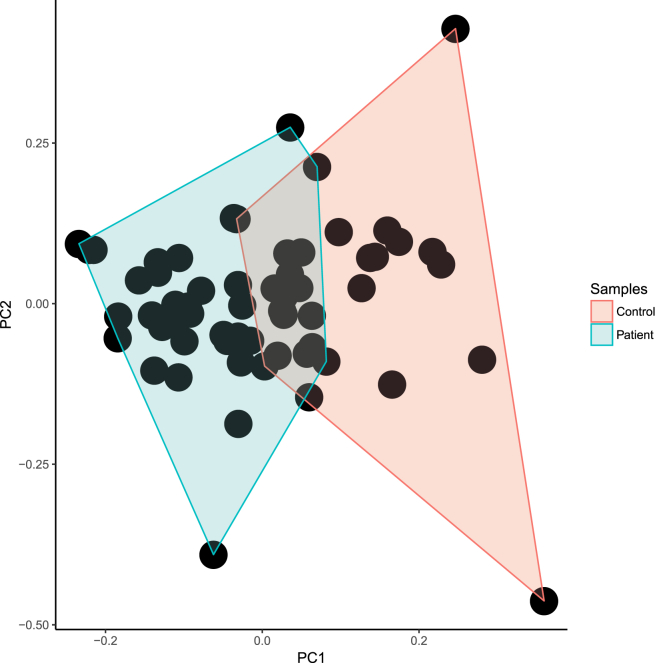
Arriving at more effective biomarker targets for AIDs seems a critical step, as Bernards declared that “a poor biomarker can be just as bad for the patient as a bad drug.”25 Improved clinical management of autoimmune diseases through biomarkers, which could be helpful for earlier and instant diagnosis, determining therapeutic strategies, and predicting outcome in this broad group of enervate disorders, is important.26 In the present study, we have identified that expression levels of AL450992.2, AC009948.5, and RP11-98D18.3 may potentially represent such PBMC biomarkers for MS among our studied lncRNAs. Moreover, the discriminatory power of these three lncRNAs was shown in the principal-component analysis (PCA) plot (Figure 8).
Figure 8.
PCA
The scatterplot of principal-component analysis using biomarker lncRNAs in order to indicate a distinct overlap of patients and controls.
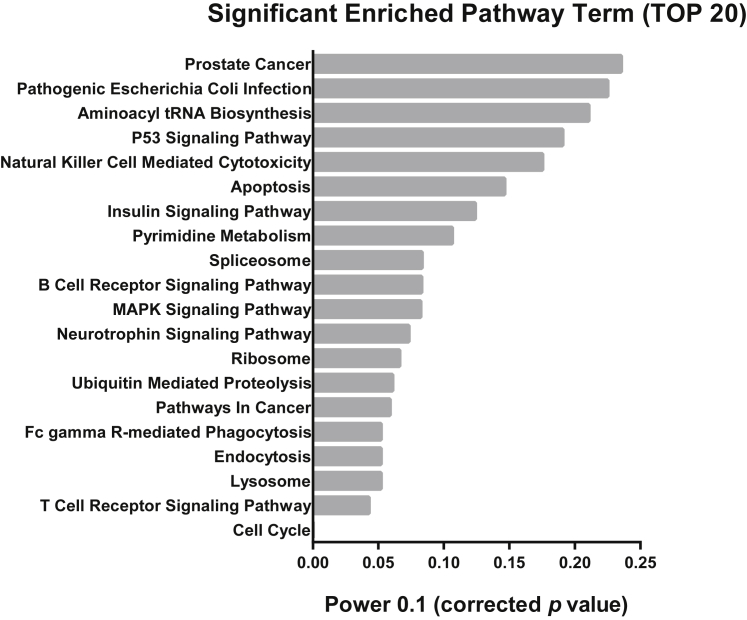
AL450992.2 and AC009948.5, as two potential MS biomarkers, are located adjacent to RORC, which is a key transcriptional regulator of Th17 lineage-specific lncRNAs and predominantly expressed in Th17 cells.9, 27 In accordance with the importance of RORC in the Th17 cell lineage, RORC correlation was assessed with the selected lncRNAs, revealing that AL450992.2 significantly correlated with RORC (Table S5). The possible role of AL450992.2 in different signaling pathways is determined through co-lncRNA and manifested through involvement with the cell cycle, T cell receptor signaling pathway, and many others (Figure 9).
Figure 9.
Enrichment Pathway Analysis
Top 20 pathways for the AL450992.2 lncRNA co-expressed genes based on Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways analysis of co-lncRNA. The most significant column is shown in the bottom of the graph, with a lower p value.
Materials and Methods
lncRNA Selection
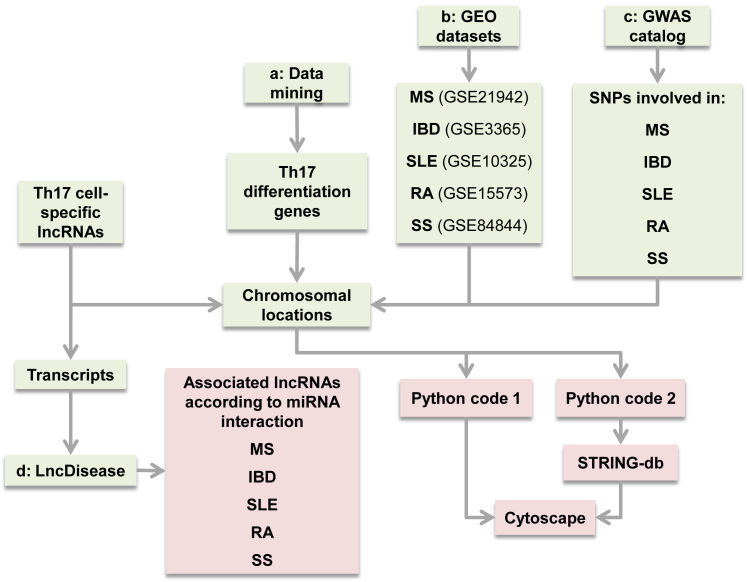
As the first step to the preliminary selection of Th17 lncRNAs, the study by Spurlock et al.9 on the whole-genome RNA-seq data of Th17 cell-lineage-specific lncRNAs was implemented. At the next step, the transcripts and chromosomal locations of each lncRNA were retrieved from Ensembl GRCh37 for further analyses to reach the defined list of potentially more effective lncRNAs based on the criteria and approach shown in the flowchart of the study in Figure 10, which is presented in detail.
Figure 10.
In Silico Workflow of the Study
Lineage-specific lncRNAs were obtained from RNA-seq data. In step a, SNPs associated with each AID were retrieved from the GWAS catalog for analysis of their proximities to lncRNAs through Python programming language and were then visualized in Cytoscape. In step b, differentially expressed mRNAs in AID patients were obtained from corresponding GEO datasets. After chromosomal localization, their distances from selected lncRNAs were checked via Python programming language and, after inputting to STRING-db, visualized in Cytoscape. In step c, Th17 cell differentiation genes were selected through data mining. Their neighboring to the selected lncRNAs was determined and, after inputting to STRING-db, visualized in Cytoscape. In step d, lncRNAs’ transcripts were deduced and inputted to the LncDisease software, which checked lncRNA-miRNA interaction and outputted the diseases related to the predicted miRNAs.
lncRNAs Adjusted to Th17 Cell Differentiation Genes
Proteins and respective genes involved in Th17 cell differentiation were deduced through manual data mining,28 in which keywords such as Th17 cell, differentiation, and signaling pathway were used to filter the outcomes. Systematic literature mining was performed on papers that were published until April 2017 from several databases, including PubMed, Science Direct, and Web of Science. Next, chromosomal locations of selected genes were determined in Ensembl GRCh37. Proximities of previously selected lncRNAs to these genes were analyzed using Python programming language (v3.6.0) (Python Script S1). In this step, Python programming language code categorizes the co-localized protein-coding genes and individual lncRNAs located in the same chromosome; then their distances were checked on each side of each lncRNA up to a distance of 50 kb away. Meanwhile, in order to consider more effective lncRNAs, we only selected lncRNAs that were co-expressed with protein-coding genes in Th17 cell differentiation.9
lncRNAs Adjusted to Differentially Expressed Genes in AIDs
To identify differentially expressed mRNAs in AID PBMCs, we used investigative analysis of array datasets that were retrieved from the Gene Expression Omnibus (GEO) database: for IBD, GEO: GSE3365 (Platform: GPL96; Affymetrix Human Genome U133A Array; 127 samples); for MS, GEO: GSE21942 (Platform: GPL570; Affymetrix Human Genome U133 Plus 2.0 Array; 29 samples); for RA, GEO: GSE15573 (Platform: GPL6102; Illumina Human-6 v2.0 expression beadchip; 33 samples), for SS, GEO: GSE84844 (Platform: GPL570; Affymetrix Human Genome U133 Plus 2.0 Array; 60 samples), for SLE, GEO: GSE10325 (Platform: GPL96; Affymetrix Human Genome U133A Array; 67 samples). These were utilized for identification of differentially expressed mRNAs with the GEO2R analyzer between patient samples and controls. We considered differentially expressed genes with adjusted p values < 0.05 and absolute log fold changes > 1. Next, each protein-coding gene’s proximity to lncRNAs was checked via Python programming language in order to select adjusted lncRNAs (a similar code as that described in the immediately preceding section).
lncRNAs Adjusted to Autoimmune Diseases Associated with SNPs
SNPs (p ≤ 5 × 10−8) associated with selected AIDs were extracted from the GWAS catalog29 and were chromosomally localized in Ensembl GRCh37. The distance of each lncRNA locus from selected SNPs was assessed with Python programming language (v3.6.0) (Python Script S2). To do such analysis, SNPs relevant to each lncRNA categorized on the same chromosome and their positions were evaluated to reach the SNPs with less than 5-kb nt from selected lncRNAs.
lncRNA-Associated Diseases through miRNA Interactions
In order to predict lncRNA-associated diseases, a sequence-based bioinformatics tool, LncDisease, was used to identify potentially associated lncRNAs with autoimmune diseases.14 TargetScan and miRanda criteria were used to perform such analysis in this software. Each individual sequence of the lncRNA transcripts was input to LncDisease to predict the interacting miRNAs by using TargetScan and miRanda criteria. LncDisease then used the Human microRNA Disease Database (HMDD) for further analysis on the predicted miRNAs according to the TAM method. Finally, potential disease-associated lncRNAs through miRNAs interactions were listed.
PBMC Isolation of Patients and Control
In this step, we performed a case-control association study including 75 adults, divided into three groups. 10 mL peripheral blood was obtained from 25 patients recently diagnosed as having MS, according to the revised McDonald’s criteria,30 and 25 in the remitting phase, and both groups had no history of other diseases of the CNS, tumor(s) and systemic hematologic diseases, recent infection, and concomitant use of antineoplastic or immunomodulating therapies prior to blood sampling. Blood samples were gathered in EDTA-containing tubes from the Alzahra Hospital, Isfahan, Iran. Both groups were between 18 and 60 years old. Patients were also characterized according to the EDSS method of calculating and understanding disability in MS patients.31 Moreover, 25 age- and gender-matched blood samples were collected from healthy volunteers, and written informed consent was obtained. The human subject protocol was approved by an institutional review board of Royan Institute (Project ID no. 91000573). All study protocols were carried out in accordance with the approved guidelines. Human PBMCs were isolated with the Lymphoprep density gradient medium (STEMCELL Technologies, Cambridge, MA, USA).
RNA Extraction and cDNA Synthesis
To isolate total RNA, TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was used according to the manufacturer’s supplied instructions. The RNA was quantified, and the quality was evaluated according to a ratio of absorbance of 260/280 nm using a Nanodrop spectrophotometer (Nanodrop 1000, Thermo Scientific, Waltham, MA, USA). To eliminate any potential contaminating DNA, total RNA samples were treated with RNase-free DNase (Thermo Scientific, Waltham, MA, USA) prior to real-time qPCR. cDNA was synthesized from the total RNA using a RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA). Afterward, cDNA samples were stored at −80°C until further use.
Real-Time qPCR
Transcription levels were measured in triplicate by real-time qPCR using SYBR Green Master Mix: SYBR Premix Ex TaqII (TaKaRa, Tokyo, Japan) and were carried out using specific primer pairs in the Step One Plus Real-Time PCR thermal cycler (Applied Biosystems, Foster City, CA, USA). UBC and YWHAZ were used as the most stably expressed reference genes, which were in accordance with gene expression analysis of PBMCs between healthy volunteers, relapsing-remitting MS (RRMS) and RRMS-interferon (IFN)-β patients.32 Primer pairs used in these reactions are listed in Table 4.
Table 4.
Primer Pairs Used in This Study
| Primer | Sequence (5′–3′) | GC% | 3′ΔG (kcal/mol) | Amplicon Size (bp) | Ta (°C) |
|---|---|---|---|---|---|
| AC007182.6 F | GTAAGTCAGCATGTAGCAC | 47.7 | −7.5 | 86 | 50 |
| AC007182.6 R | GCTCCATCAATAACATCTTCAT | 36.4 | −5.7 | ||
| AC009948.5.007 F | ATCAAGTTACAGAGCAGAG | 4.1 | −64 | 83 | 50 |
| AC009948.5.007 R | AACATTACCGAGGACAAC | 44.4 | −6.5 | ||
| AL450992.2.001 F | TTAGACTCTCCTTGACCAT | 42.1 | −6.6 | 113 | 52 |
| AL450992.2.001 R | TTCTCCTTCTGTGCTTTC | 44.4 | −5.7 | ||
| AL928768.3 F | ACAGGGAGGAAGTGTGGAG | 57.9 | −6.9 | 93 | 56 |
| AL928768.3 R | GTGAGTAAGGGCGGGTC | 64.7 | −7.4 | ||
| IL21R-AS1 F | CTCCGACCACTCATTCAG | 55.6 | −6.1 | 119 | 56 |
| IL21R-AS1 R | CTTATCACCTTGCCGTCTG | 52.6 | −6.6 | ||
| RP11.126K1.6 F | AACAGTCGCCTTCCAACAC | 52.6 | −6.5 | 118 | 62 |
| RP11.126K1.6 R | TCCATCACTCCTACCCATCATT | 45.5 | −5.7 | ||
| RP11-98D18.3 F | AGGCTCAGTCACCTTTCC | 55.6 | −6.2 | 81 | 54 |
| RP11-98D18.3 R | CCTTCTCTGTGACCGTCGA | 57.9 | −7.4 | ||
| RP11-290L1.3 F | GCGAGTGCGGCTCGTGATCTC | 66.7 | −5.8 | 168 | 64 |
| RP11-290L1.3 R | CCGGTCAAGCTCAAGGAACTGC | 59.1 | −7.5 | ||
| RORC F | TGCCAGAATGACCAGATTGTGCTT | 45.8 | −7.1 | 132 | 60 |
| RORC R | GAACAGCTCCATGCCACCGTA | 57.1 | −7.1 | ||
| UBC F | GGATTTGGGTCGCAGTTCTTG | 52.4 | −6.1 | 135 | 58 |
| UBC R | TGCCTTGACATTCTCGATGG | 50 | −6.5 | ||
| YWHAZ F | ACTTTTGGTACATTGTGGCTTC | 40.9 | −6.9 | 94 | 62 |
| YWHAZ R | CCGCCAGGACAAACCAGTA | 57.9 | −5.8 |
F and R, forward and reverse primers, respectively; 3′ΔG stands for Gibbs energy for binding of the 3′ part of the primer with the template. Ta, annealing temperature of PCR reactions.
Statistical Analysis
We performed analyses using a standard two-tailed Student’s t test and a one-way ANOVA to assess differences, followed by pairwise comparisons using Tukey’s correction. Pearson correlation was also performed for lncRNA-mRNA co-expression in this study. The discriminatory power of biomarker panels was assessed by ROC analysis between the controls and the MS patients. The aforementioned statistical analyses were performed using SPSS 20 software (SPSS, Chicago, IL, USA) and GraphPad Prism (v6; GraphPad software). For all analyses, p < 0.05 was considered statistically significant. Clustering of samples was performed using PCA with the package ggfortify (v0.4.1) in R software (v3.1.1).33
Network Construction
The protein-protein interactions of significant genes were assessed by STRING-db34 and visualized by Cytoscape 3.6.0 software. Additionally, Cytoscape supplies a basic set of features for data integration, analysis, and visualization for complicated networks. Briefly, we transformed mined data into STRING-db to assess interactions and STRING-db results in Cytoscape. Next, to construct the network graph, we used methods available in Cytoscape to visualize and analyze the network by the number of direct edges. Co-LncRNA, a web-based computational tool that provides an overview of the relevant pathways of expressed protein-coding genes with inputted lncRNAs was also used in this study.35
Conclusions
In this study, most of the predicted lncRNAs of MS were derived from association with two criteria: Th17 cell differentiation genes and differentially expressed genes in AIDs. Most of the dysregulated lncRNAs were derived from these criteria. We propose that recurrently deregulated lncRNAs identified in this report could provide a valuable resource for studies aimed at delineating the relationship between functional lncRNAs and AIDs.
Author Contributions
S.T. and A.H. both participated in designing research studies, conducting experiments, acquiring data, analyzing data, providing reagents, and writing the manuscript; A.R. participated in designing research studies, conducting experiments, and acquiring data; K.G. participated in acquiring and analyzing data and writing the manuscript; E.G. carried out designing research studies, data interpretation, and manuscript writing; M.E. participated in acquiring data, data interpretation, manuscript writing, and giving final approval of the manuscript; M.H.N.-E. participated in designing research studies, data interpretation, manuscript writing, and giving final approval of the manuscript; and T.L.M. performed data interpretation, manuscript writing, and giving final approval of the manuscript.
Conflict of Interests
We declare no competing financial interests.
Acknowledgments
A part of this project was supported by the National Institute for Medical Research Development (NIMAD’s project no. 942792) and also partly by the Royan Institute for Biotechnology. The authors are grateful to Dr. Jamshid Hosseini (Marquette University, Milwaukee, WI, USA) for revising the text of the manuscript.
Footnotes
Supplemental Information includes five tables and two Python scripts and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.05.022.
Contributor Information
Kamran Ghaedi, Email: kamranghaedi@royaninstitute.org.
Mohammad Hossein Nasr-Esfahani, Email: mh.nasr-esfahani@royaninstitute.org.
Timothy L. Megraw, Email: timothy.megraw@med.fsu.edu.
Supplemental Information
References
- 1.Cooper G.S., Bynum M.L., Somers E.C. Recent insights in the epidemiology of autoimmune diseases: improved prevalence estimates and understanding of clustering of diseases. J. Autoimmun. 2009;33:197–207. doi: 10.1016/j.jaut.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peters A., Lee Y., Kuchroo V.K. The many faces of Th17 cells. Curr. Opin. Immunol. 2011;23:702–706. doi: 10.1016/j.coi.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong C. TH17 cells in development: an updated view of their molecular identity and genetic programming. Nat. Rev. Immunol. 2008;8:337–348. doi: 10.1038/nri2295. [DOI] [PubMed] [Google Scholar]
- 4.Geisler S., Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qi P., Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod. Pathol. 2013;26:155–165. doi: 10.1038/modpathol.2012.160. [DOI] [PubMed] [Google Scholar]
- 6.Tong Y.-K., Lo Y.M. Diagnostic developments involving cell-free (circulating) nucleic acids. Clin. Chim. Acta. 2006;363:187–196. doi: 10.1016/j.cccn.2005.05.048. [DOI] [PubMed] [Google Scholar]
- 7.Ayers D. Long non-coding RNAs: novel emergent biomarkers for cancer diagnostics. J. Cancer Res. Treatment. 2013;1:31–35. [Google Scholar]
- 8.Seim I., Ma S., Gladyshev V.N. Gene expression signatures of human cell and tissue longevity. NPJ Aging Mech. Dis. 2016;2:16014. doi: 10.1038/npjamd.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spurlock C.F., 3rd, Tossberg J.T., Guo Y., Collier S.P., Crooke P.S., 3rd, Aune T.M. Expression and functions of long noncoding RNAs during human T helper cell differentiation. Nat. Commun. 2015;6:6932. doi: 10.1038/ncomms7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ricaño-Ponce I., Wijmenga C. Mapping of immune-mediated disease genes. Annu. Rev. Genomics Hum. Genet. 2013;14:325–353. doi: 10.1146/annurev-genom-091212-153450. [DOI] [PubMed] [Google Scholar]
- 12.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen G., Wang Z., Wang D., Qiu C., Liu M., Chen X., Zhang Q., Yan G., Cui Q. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41:D983–D986. doi: 10.1093/nar/gks1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J., Ma R., Ma W., Chen J., Yang J., Xi Y., Cui Q. LncDisease: a sequence based bioinformatics tool for predicting lncRNA-disease associations. Nucleic Acids Res. 2016;44:e90. doi: 10.1093/nar/gkw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wapinski O., Chang H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Baron J.L., Madri J.A., Ruddle N.H., Hashim G., Janeway C.A., Jr. Surface expression of alpha 4 integrin by CD4 T cells is required for their entry into brain parenchyma. J. Exp. Med. 1993;177:57–68. doi: 10.1084/jem.177.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ricaño-Ponce I., Zhernakova D.V., Deelen P., Luo O., Li X., Isaacs A., Karjalainen J., Di Tommaso J., Borek Z.A., Zorro M.M., BIOS Consortium. Lifelines Cohort Study Refined mapping of autoimmune disease associated genetic variants with gene expression suggests an important role for non-coding RNAs. J. Autoimmun. 2016;68:62–74. doi: 10.1016/j.jaut.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brunner A.L., Beck A.H., Edris B., Sweeney R.T., Zhu S.X., Li R., Montgomery K., Varma S., Gilks T., Guo X. Transcriptional profiling of long non-coding RNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol. 2012;13:R75. doi: 10.1186/gb-2012-13-8-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buzas E.I., György B., Nagy G., Falus A., Gay S. Emerging role of extracellular vesicles in inflammatory diseases. Nat. Rev. Rheumatol. 2014;10:356–364. doi: 10.1038/nrrheum.2014.19. [DOI] [PubMed] [Google Scholar]
- 20.Sigdel K.R., Cheng A., Wang Y., Duan L., Zhang Y. The emerging functions of long noncoding RNA in immune cells: autoimmune diseases. J. Immunol. Res. 2015;2015:848790. doi: 10.1155/2015/848790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu G.-C., Pan H.-F., Leng R.-X., Wang D.-G., Li X.-P., Li X.-M., Ye D.Q. Emerging role of long noncoding RNAs in autoimmune diseases. Autoimmun. Rev. 2015;14:798–805. doi: 10.1016/j.autrev.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Wang J., Peng H., Tian J., Ma J., Tang X., Rui K., Tian X., Wang Y., Chen J., Lu L. Upregulation of long noncoding RNA TMEVPG1 enhances T helper type 1 cell response in patients with Sjögren syndrome. Immunol. Res. 2016;64:489–496. doi: 10.1007/s12026-015-8715-4. [DOI] [PubMed] [Google Scholar]
- 23.Spurlock C.F., 3rd, Tossberg J.T., Matlock B.K., Olsen N.J., Aune T.M. Methotrexate inhibits NF-κB activity via long intergenic (noncoding) RNA-p21 induction. Arthritis Rheumatol. 2014;66:2947–2957. doi: 10.1002/art.38805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang F., Wu L., Qian J., Qu B., Xia S., La T., Wu Y., Ma J., Zeng J., Guo Q. Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus. J. Autoimmun. 2016;75:96–104. doi: 10.1016/j.jaut.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 25.Bernards R. It’s diagnostics, stupid. Cell. 2010;141:13–17. doi: 10.1016/j.cell.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 26.Gibson D.S., Banha J., Penque D., Costa L., Conrads T.P., Cahill D.J., O’Brien J.K., Rooney M.E. Diagnostic and prognostic biomarker discovery strategies for autoimmune disorders. J. Proteomics. 2010;73:1045–1060. doi: 10.1016/j.jprot.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 27.Hamzaoui K., Borhani Haghighi A., Ghorbel I.B., Houman H. RORC and Foxp3 axis in cerebrospinal fluid of patients with neuro-Behçet’s disease. J. Neuroimmunol. 2011;233:249–253. doi: 10.1016/j.jneuroim.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 28.Võsa U., Vooder T., Kolde R., Vilo J., Metspalu A., Annilo T. Meta-analysis of microRNA expression in lung cancer. Int. J. Cancer. 2013;132:2884–2893. doi: 10.1002/ijc.27981. [DOI] [PubMed] [Google Scholar]
- 29.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polman C.H., Reingold S.C., Banwell B., Clanet M., Cohen J.A., Filippi M., Fujihara K., Havrdova E., Hutchinson M., Kappos L. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann. Neurol. 2011;69:292–302. doi: 10.1002/ana.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurtzke J.F. Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS) Neurology. 1983;33:1444–1452. doi: 10.1212/wnl.33.11.1444. [DOI] [PubMed] [Google Scholar]
- 32.Oturai D.B., Søndergaard H.B., Börnsen L., Sellebjerg F., Christensen J.R. Identification of suitable reference genes for peripheral blood mononuclear cell subset studies in multiple sclerosis. Scand. J. Immunol. 2016;83:72–80. doi: 10.1111/sji.12391. [DOI] [PubMed] [Google Scholar]
- 33.Tang Y., Horikoshi M., Li W. ggfortify: unified interface to visualize statistical results of popular R packages. R J. 2016;8:478–489. [Google Scholar]
- 34.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Z., Bai J., Wu A., Wang Y., Zhang J., Wang Z., Li Y., Xu J., Li X. Co-lncRNA: investigating the lncRNA combinatorial effects in GO annotations and KEGG pathways based on human RNA-seq data. Database (Oxford) 2015;2015:bav082. doi: 10.1093/database/bav082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.