Abstract
Eukaryotic nitrate reductase (NR) catalyzes the first step in nitrate assimilation and is regulated transcriptionally in response to external cues and intracellular metabolic status. NRs are also regulated post-translationally in plants by phosphorylation and binding of 14-3-3 proteins at conserved serine residues. 14-3-3 binding motifs have not previously been identified in algal NRs. A novel NR (NR2-2/2HbN) with a 2/2 hemoglobin domain was recently described in the alga Chattonella subsalsa. Here, a second NR (NR3) in C. subsalsa is described with a 14-3-3 binding motif but lacking the Heme-Fe domain found in other NRs. Transcriptional regulation of both NRs was examined in C. subsalsa, revealing differential gene expression over a diel light cycle, but not under constant light. NR2 transcripts increased with a decrease in temperature, while NR3 remained unchanged. NR2 and NR3 transcript levels were not inhibited by growth on ammonium, suggesting constitutive expression of these genes. Results indicate that Chattonella responds to environmental conditions and intracellular metabolic status by differentially regulating NR transcription, with potential for post-translational regulation of NR3. A survey of algal NRs also revealed the presence of 14-3-3 binding motifs in other algal species, indicating the need for future research on regulation of algal NRs.
Introduction
Nitrate and ammonium are the most abundant nitrogen sources that phytoplankton utilize for their growth and metabolism: approximately 40% of global primary production is the result of assimilating these two nitrogenous nutrients by phytoplankton1. Even so, nitrogen is a limiting nutrient for phytoplankton in many marine environments and its acquisition and assimilation is highly regulated. In order to utilize nitrate as a nitrogen source, phytoplankton reduce nitrate to ammonium by two sequential reactions that are catalyzed by nitrate reductase (NR), with electrons donated by NAD(P)H generated by the light reactions of photosynthesis, and nitrite reductase, with ferredoxin as an electron donor2,3. NR catalyzes the first enzymatic and also the rate-limiting step in nitrate assimilation, and plays a key role in coordinating nitrogen and carbon assimilation3,4.
NR gene expression and activity are regulated in response to environmental conditions and intracellular metabolic status5–9. The tight regulation of NR not only plays an important role for organisms to optimize nitrogen assimilation and adjust different metabolic pathways, but also prevents the accumulation of potentially toxic nitrite and formation of reactive oxygen and nitrogen species10,11. In general, both NR gene expression and activity respond to nitrogen source, temperature and light. Changes in NR transcript abundance typically reflect changes in NR protein abundance and activity12,13. In some species, NR gene expression is induced by the presence of nitrate, and repressed or inhibited completely by the presence of ammonium7. NR gene expression can also be constitutive, as in the raphidophyte, Heterosigma akashiwo14. Regulation of algal NR transcript abundance by temperature has been examined in diatoms, where increased transcription and NR activity in this group of algae may serve as a means to dissipate excess energy under high light and low temperature conditions4. NR gene expression also varies over a diel cycle15. These variations can be driven by an endogenous circadian rhythm, or in response to light- dark cycles, where NR gene expression is regulated by metabolic feedback controls15,16. For instance, Vergara et al. demonstrated that NR gene expression in the marine diatom Thalassiosira weissflogii was regulated by the photosynthetic organic carbon accumulated in the light period, indicating coordination with photosynthetic activity and light- dark cycle regulation instead of endogenous control by a circadian clock17.
NR enzyme activity is also regulated in higher plants at the post-translational level by reversible phosphorylation of a conserved serine (Ser) residue in the hinge 1 region, permitting binding of 14-3-3 proteins18,19. Binding of 14-3-3 proteins inhibits NR activity via obstruction of intra-molecular electron flow (i.e. the movement of electrons between domains)20,21. According to the published studies to date, the phospho-Ser residue required for the binding of 14-3-3 proteins in NRs of higher plants is absent from NRs of mosses and algae, suggesting alternative mechanisms for regulation of NRs in these species6,22. However, the lack of 14-3-3 binding motifs in NRs of algae were only examined in a few species from limited phylogenetic groups, including several green algal species (Chlamydomonas reinhardtii23, Volvox carteri23,24, Chlorella vulgaris23,25, and Dunaliella tertiolecta26), and diatoms27 (Phaeodactylum triconutum and Thalassiosira pseudonana).
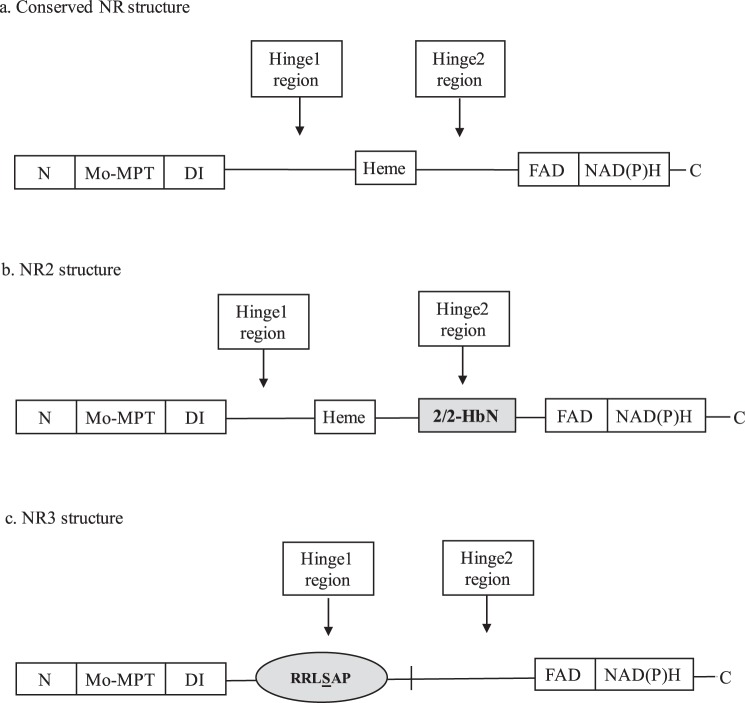
Despite differences in regulation of NRs among species, eukaryotic NRs share a conserved structure constituted by five domains3 (Fig. 1a): (a) molybdenum-molybdopterin cofactor domain (Mo-MPT); (b) dimer interface (DI); (c) cytochrome b5-binding domain containing Heme-Fe (Heme-Fe domain); (d) flavin adenine dinucleotide (FAD) domain; and (e) NAD(P)H domain. NR also includes three regions that function to regulate and stabilize the enzyme3: the N-terminal region, a hinge 1 region that is located between the DI and Heme-Fe domains, and a hinge 2 region between the Heme-Fe and FAD domains. Recently, a novel nitrate reductase sequence, NR2-2/2HbN (hereafter referred to as NR2), was identified in the harmful algal bloom species Heterosigma akashiwo and Chattonella subsalsa (Raphidophyceae)2 (Fig. 1b). NR2 has a 2/2 hemoglobin (2/2HbN) domain inserted in the hinge 2 region that was not present in the deduced amino acid sequence of H. akashiwo NR1 (HsNR1)14 or in any other eukaryotic NR to date12,26–28. Consistent with reports of NR sequences from other algae, a 14-3-3 binding site motif was not identified in the translated amino acid sequences of HsNR1 or the NR2 sequences for either H. akashiwo or C. subsalsa2.
Figure 1.
Alternative domain structures for nitrate reductase enzymes. (a) The conserved structure of nitrate reductase in higher plants, algae, and fungi, depicting five domains (MO-MPT, DI, Heme-Fe, FAD, and NADH domains) and 3 regions (N-terminal, hinge 1 and hinge 2 regions)3. (b) The structure of NR2 with a 2/2HbN domain (indicated by grey box) inserted in its Hinge 2 region of Heterosigma akashiwo and Chattonella subsalsa2. (c) The structure of NR3 lacking the Heme-Fe domain and with a 14-3-3 protein binding motif (indicated by grey circle) at a phospho-Ser residue (underlined in RRLSAP) in the Hinge 1 region of Chattonella subsalsa. N: N-terminal region; Mo-MPT: molybdenum-molybdopterin cofactor domain; DI: dimer interface; Heme: cytochrome b5-binding domain containing Heme-Fe; FAD: flavin adenine dinucleotide domain; NAD(P)H: NAD(P)H domain; C: C-terminal of NR enzyme.
A second novel NR gene, designated here as NR3, was revealed in the transcriptome of C. subsalsa as part of the Marine Microbial Eukaryote Transcriptome Sequencing Project29 (MMETSP) (Sample Names: MMETSP0947–0950; PI: Kathryn Coyne; https://www.imicrobe.us/#/combined_assemblies/21; accessed on 3/2/2018; sequence ID: CAMPEP_0187150166 in combined assembly peptides). A sequence orthologous to HsNR1 was not identified in the transcriptome of C. subsalsa. The deduced amino acid sequence of C. subsalsa NR3 (Fig. 1c) lacks the 2/2HbN domain found in CsNR2 as well as the conserved Heme-Fe domain present in all other eukaryotic NRs. In contrast to CsNR2 and other algal NRs described to date, the deduced amino acid sequence of NR3 also includes a proline- and serine-rich region in the hinge 1 region, similar to 14-3-3 protein binding domains in plant NRs. The regulation of NR2 and NR3 by environmental conditions and their relative impact on the competitive success of C. subsalsa in natural phytoplankton communities remain unknown.
The objectives of this investigation were to examine the hypothesis that NR2 and NR3 transcript abundances in C. subsalsa are differentially regulated in response to nitrogen source, temperature and diel changes in light. Mechanisms by which NR3 may retain function without the Heme-Fe domain are also discussed. Finally, a comprehensive survey was conducted to identify 14-3-3 binding motifs in other algal NRs.
Results
Sequence analysis of NR3 in C. subsalsa
The deduced amino acid sequences of NR3 cDNA clones in plasmids CsNR3-P2 and CsNR3-P10 were identical to the NR3 sequence (sequence ID: CAMPEP_0187150166 in combined assembly peptides) obtained from the Marine Microbial Eukaryote Transcriptome Sequencing Project29 (MMETSP), except that the sequence obtained from cloned cDNA included 7 sites that were undetermined in the original sequence, 1 additional amino acid at site 166, and 15 additional amino acids (HASSQEVFNNESTTA) within the hinge 1 region at site number 418 to 432 (Supplementary Data S1). A region of the sequence spanning from the DI domain to the hinge 2 region showed that the deduced amino acid sequence of NR3 lacked both the 2/2 HbN domain present in NR2 as well as the cytb-5 (Heme-Fe) binding domain found in other NRs (Fig. 2).
Figure 2.
Alignment of the hinge 1 region and adjacent regions of NR3 in C. subsalsa with NR2 in C. subsalsa, NR1 of Heterosigma akashiwo, NRs of four other representative algal species, Thalassiosira pseudonana, Phaeodactylum tricornutum, Dunaliella salina, and Chlamydomonas reinhardtii, as well as NRs in higher plants, Glycine max, Nicotina tabacum, and Zea mays. The single underlined sequences indicate the adjacent domains, showing the absence of the cytb-5 binding domain in NR3 (indicated by grey arrow). The double underlined sequences indicate the motif containing the serine phosphorylation site for 14-3-3 protein binding in NR3 and NRs in higher plants (indicated by black arrow). The identical residues in all sequences are marked by *; the highly and weakly conserved residues are marked by : and ., respectively.
Phylogenetic analysis
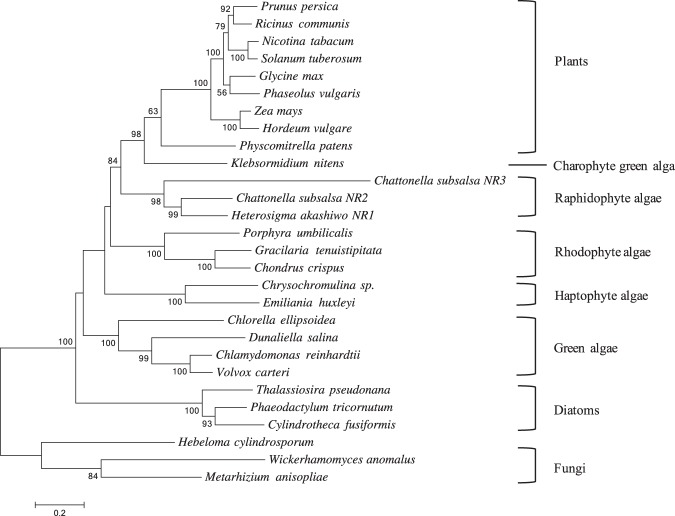
Phylogenetic analysis of NR3 in C. subsalsa revealed a close relationship to CsNR2 and HsNR1, with high bootstrap support (Fig. 3). Note that HsNR1 and HsNR2 are identical except for insertion of the 2/2 hemoglobin domain in HsNR2. Emiliania huxleyi NR was clustered with NR of its haptophyte relative Chrysochromulina, and Porphyra umbilicalis NR clustered with NR sequences of other rhodophyte species Gracilaria tenuistipitata, and Chondrus crispus, all with high bootstrap support. Interestingly, Klebsormidium nitens NR is more closely related to NR in terrestrial plants than with other algal species. Phylogenetic analysis suggests that Heterosigma and Chattonella NRs may be more similar to NRs in terrestrial plants, as noted for HsNR1 analysis by Coyne14, than to NRs found in other algal species, although the bootstrap support was low.
Figure 3.
Molecular phylogenetic analysis of NR amino acid sequences in plants and algae. The tree was generated using the Maximum Likelihood method based on the Le Gascuel 2008 model76. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites (+G) and allowed for some sites to be evolutionarily invariable (+I). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. All positions containing gaps and missing data were eliminated. There were a total of 568 positions in the final dataset. Evolutionary analyses were conducted in MEGA772. The tree was rooted by fungal NRs from Wickerhamomyces anomalus, Hebeloma cylindrosporum, and Metarhizium anisopliae. Bootstrap values less than 50 are not shown.
Evolutionary divergence between the deduced amino acid sequence of NR3 and NRs of other species indicates that NR3 is 49.82% and 49.65% identical to HsNR1 and CsNR2, respectively. Similar to results indicated by phylogenetic analysis, NR3 shares higher identity with Klebsormidium nitens NR (46.83%) and plant NRs (e.g. 44.19% identical to Glycine max NR) than to other algal NRs. When compared to algal NRs, for instance, NR3 is 37.68% and 40.32% identical to NR in Thalassiosira pseudonana (diatom) and Chlamydomonas reinhardtii (green alga), respectively.
Diel expression of NR genes under different light conditions
Treatments were sampled at five time points from C. subsalsa cultures growing in a 12:12 h light to dark cycle (L:D, Fig. 4) or constant light (L:L). There were no significant changes in gene expression for either NR2 or NR3 over the five sampling time points for cultures in L:L treatments (p > 0.05, data not shown), although the lowest point measured for NR2 gene expression coincided with that for the L:D treatment. NR2 gene expression was significantly (3–30 fold) higher than NR3 gene expression at all time points in each treatment (p < 0.05). Both NR2 and NR3 transcript levels exhibited diel expression patterns in the L:D treatment, but this pattern differed between NR2 and NR3. There was a significant difference in NR3 transcript abundance in samples collected in the light compared to the dark phase (p < 0.05), but no significant differences in NR2 gene expression between light and dark phases (p > 0.05). NR3 gene transcript abundance decreased significantly (p < 0.05) at one hour after lights on compared to the initial time point in the dark phase, and exhibited a trend (although not significant) toward increased expression of NR3 after entering the dark phase. In addition, there was a significant increase (p < 0.05) in NR3 transcript abundance at six hours after dark compared to six hours after lights on in the light phase. In contrast, there was no significant difference in NR2 transcript abundance between the first two time points, with the change from dark to light phase (p > 0.05). There was a significant decrease in NR2 transcript levels at six hours after lights on compared to 1 hour after lights on in the light phase (p < 0.05), and transcript abundance remained low at one hour after the start of the dark period before a trend to increasing expression at six hours after dark (not significant, p > 0.05).
Figure 4.
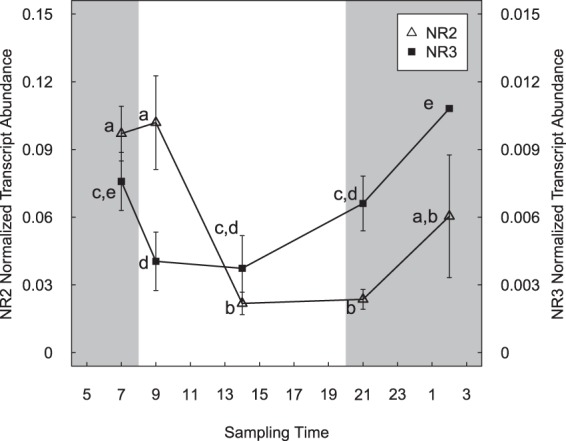
Transcript abundance of NR2 (y axis shown at left) and NR3 (y axis shown at right) normalized to actin in C. subsalsa cultured in a 12:12 h light: dark cycle. The shaded portion on the graph indicates the dark period. Different letters beside the symbols indicate significant differences in NR transcript abundance between each time point (p < 0.05). Error bars are standard deviation of three biological replicates.
Gene expression of NRs with nitrate or ammonium as a nitrogen source
Both NR2 and NR3 transcripts were detected in treatments with either NO3− or NH4+ as the sole nitrogen source (Fig. 5). NR2 had a significantly higher gene expression in treatments cultured with ammonium than with nitrate (p < 0.05), while gene expression of NR3 was not significantly affected by nitrogen source (p > 0.05).
Figure 5.
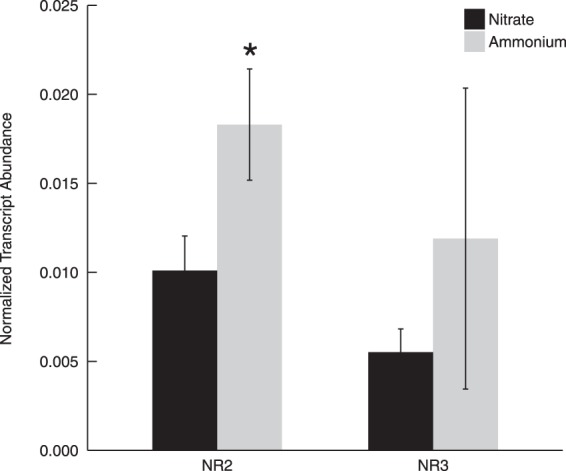
NR2 and NR3 transcript abundance normalized to actin in treatments cultured with nitrate or ammonium as the sole nitrogen source. Asterisk (*) reveals the significant difference between NR gene expression in nitrogen- treatments (p < 0.05). Error bars are standard deviation of three biological replicates.
Gene expression of NRs at different temperatures
NR2 gene expression was significantly (3.0- and 2.6- fold) higher in the cultures acclimated to 18 °C than those in 25° and 28 °C, respectively (p < 0.05; Fig. 6). There was no significant difference in NR2 gene expression for C. subsalsa cultured in 25° and 28 °C (p > 0.05). NR3 gene expression was not significantly affected by temperature (p > 0.05). Furthermore, NR2 gene expression values were significantly (4.5–10-fold) higher than NR3 gene expression under all temperature conditions (p < 0.05).
Figure 6.
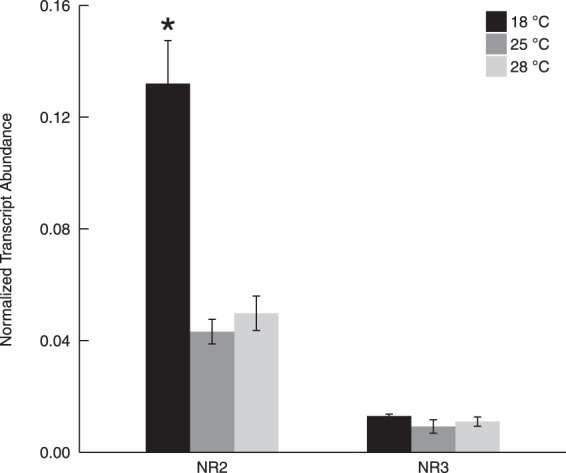
NR2 and NR3 transcript abundance normalized to actin at 18°, 25°, and 28 °C. Asterisk (*) indicates a significant difference in NR gene expression between treatments for each transcript (p < 0.05). Error bars represent standard deviation of three biological replicates.
Identification of 14-3-3 binding motifs
Alignment of the hinge 1 region of NR3 revealed the presence of a serine- and proline-rich region consistent with 14-3-3 binding motifs in plant NRs. The binding motif for 14-3-3 proteins in NR3 was predicted by Scansite 330 within the larger sequence SNRSRRLSAPALPLA, with the potential phosphorylation site at the Ser446 (underlined in the sequence) in the hinge 1 region of NR3 (Fig. 2). The search was rated in the top 0.147% of all sequence scores on Scansite 3. The predicted surface accessibility for this site within NR3 was 0.7828.
A total of 514 algal NR sequences in the NCBI database31 were downloaded and full-length algal NR sequences were selected for analysis (Supplementary Table S1). Among these 80 sequences, 12 were identified with at least one potential 14-3-3 binding motif as RXXSXP or KSXSXP. Full length algal NR sequences with putative 14-3-3 binding motifs were submitted for analysis using Scansite 330. Five algal NRs, in addition to C. subsalsa, were identified by Scansite 3 to include canonical 14-3-3 binding motifs (Supplementary Table S2). Three of these were identified with high stringency: the charophyte green algal species, Klebsormidium nitens (GAQ91960.1), the haptophyte coccolithophore, Emiliania huxleyi (DAA12507.1), and the rhodophyte macroalgal species, Porphyra umbilicalis (OSX81470.1).
Two potential 14-3-3 binding motifs were identified in Klebsormidium nitens NR, within the sequences PTMKRSVSQPRMAAV and RGGSRFLSAPTLDPL. These sequences were located in the hinge 1 and 2 regions with putative phosphorylation sites at S549 and S654 (underlined in the sequences above), respectively, and were rated in the top 0.094% and 0.068% of sequence scores, respectively. The putative 14-3-3 protein binding motif in the hinge 1 region had a surface accessibility of 1.60, while the motif in hinge 2 region had a low surface accessibility of 0.49 (Supplementary Table S2).
The 14-3-3 binding motif in NR of Emiliania huxleyi was identified within the sequence SAHLRAPSLPLGMRR with putative phosphorylation site at S639 (underlined) in the hinge 2 region. The search was rated in the top 0.068% on the site, and the surface accessibility was 0.50 (Supplementary Table S2).
The potential 14-3-3 binding motif identified in Porphyra umbilicalis included a phosphorylation site at S604 (underlined) in the hinge 1 region within the sequence ETLKRAKSAPQINKM. The putative 14-3-3 protein binding site was rated in the top 0.015% on the site, with a high surface accessibility of 1.68 (Supplementary Table S2).
In addition to the three sequences above identified by a high stringency search, putative 14-3-3 protein binding motifs were also found in the hinge 1 region of NR sequences in rhodophytes Gracilaria tenuistipitata (ACX31653.1) and Chondrus crispus (XP_005714488.1) using a medium stringency setting in Scansite 3. A 14-3-3 binding site was predicted within the large sequence of KSGSAPSLSTM at S523 in G. tenuistipitata, with a surface accessibility of 0.8526, and within the sequence of KSPSAPALSS in C. crispus at site S526, with a surface accessibility of 1.0043 (Supplementary Table S2).
Discussion
Blooms caused by the genus Chattonella have been responsible for massive fish kills in Japan, Korea, India, China, USA, and South Australia, with severe damage to fishery industries and economy32. Previous research identified temperature, salinity, irradiance, and nutrients as the most important factors to stimulate the growth and bloom formation of Chattonella spp.32. As both nitrate and ammonium contribute to growth of Chattonella33, the presence and relative concentrations of these nitrogen sources along with other environmental factors (such as temperature) that regulate NR likely play a key role in nitrogen assimilation and bloom formation by these species. In contrast to HsNR1 and HsNR2 in Heterosigma akashiwo, which are identical except for insertion of a 2/2HbN domain in HsNR22,14, analysis of NR amino acid sequences in C. subsalsa show that NR3 is distinct from CsNR2 (Fig. 2) and suggests that these enzymes may be differentially regulated. A primary goal of this study was to investigate the transcriptional regulation of these two novel nitrate reductase enzymes in C. subsalsa.
Regulation of NR gene expression by light and/ or the circadian rhythm has been well established for other plants and algae but differ in a species-specific manner. For instance, NR gene expression was regulated by diel changes in light but not the circadian rhythm in diatom Thalassiosira pseudonana34, while it was regulated by the endogenous rhythm or both in plants Arabidopsis thaliana35, Nicotiana tabacum L36. and Zea mays37, as well as the red alga Gracilaria tenuistipitata12. Studies show that regulation of NR gene expression by light was related to negative and positive feedback loops, where the pool of amino acids are thought to play an important role in this process16. In addition to metabolic feedback, NR may be under circadian control via the central clock, which allows organisms to predict daily (and seasonal) changes in their environment and metabolic needs in order to optimize their growth38.
In this study, both NR2 and NR3 gene expression in C. subsalsa exhibited significant changes over time when cultured in a 12 h:12 h L:D cycle (Fig. 4). This pattern was abolished when the cultures were exposed to continuous light in the L:L treatment, indicating that gene expression for both NRs in this species were not constrained by an endogenous rhythm. Gene expression patterns between NR2 and NR3 in the L:D treatments, however, show that NR3 gene expression was more responsive to changes in light, with a significant decrease in transcript abundance at one hour after lights on in the light phase compared to one hour before lights on in the dark phase. The slower response of NR2 to the change from dark to light in the L:D treatment suggests a lower level of sensitivity to negative feedback loops regulating the expression of this gene. At six hours after the start of the light phase, both NR2 and NR3 transcript levels were at their lowest level. This decrease in transcript abundance during the light phase is similar to gene expression patterns for NR observed in other algal species12,34, and is consistent with studies demonstrating direct or indirect feedback repression of NR transcription by products of nitrogen assimilation such as amino acids16. Strong repression of NR gene expression by glutamine, for example, has been reported in plant species Fagus sylvatica39 and Nicotiana plumbaginifolia40, as well as in the red alga Cyanidioschyzon merolae41. The lack of a circadian rhythm in C. subsalsa is in contrast to NR gene expression patterns observed in the raphidophyte, H. akashiwo, where preliminary experiments indicate that HsNR1 transcript abundance is under circadian control14. Overall, these results indicate multiple mechanisms regulating NR transcription between and among algal species.
NR gene expression in algal species typically increases with addition of nitrate, and can be strongly repressed by the presence of ammonium in some species6. NR gene expression was repressed or completely inhibited by ammonium in the green alga Dunaliella tertiolecta26, diatom Cylindrotheca fusiformis42, and red alga Cyanidioschyzon merolae43, for instance, although repression of NR gene expression by ammonium is not universal7. Zhang et al.33 found that C. subsalsa reached higher cell density in cultures supplied with ammonium as a nitrogen source compared to nitrate, but it was not clear if NR is repressed in this species by the presence of ammonium. In this study, NR2 and NR3 transcripts in C. subsalsa were detected in all treatments, even when ammonium was the sole nitrogen source (Fig. 5). The lack of repression by ammonium was also observed for NR1 transcription in H. akashiwo14. Interestingly, the transcript abundance of NR2 in C. subsalsa was significantly higher when growing with ammonium than with nitrate in this study, while NR3 gene expression was not significantly different with either nitrogen source. The increased transcript abundance for NR2 in ammonium cultures may reflect differences in cellular nutrient status associated with growth stage for these cultures compared to those provided with nitrate as a nitrogen source. Differential regulation of other genes involved in nitrogen metabolism by internal nitrogen status vs. external nitrogen source and concentrations has been observed in other algal species. The chloroplastic form of glutamine synthetase (GSII), for example, is stimulated by the presence of nitrate in the medium, while the mitochondrial form (GSIII) is expressed in response to ammonium derived from cellular processes7,44.
C. subsalsa is typically observed in Delaware’s inland bays at temperatures between 16 °C and 30 °C33. In this study, NR transcript abundance was measured after acclimation to temperatures that this species would experience in the field, and revealed a higher level of NR2 gene expression at 18 °C compared to 25 °C and 28 °C, while NR3 transcript abundance remained unchanged (Fig. 6). Parker and Armbrust4 suggested that photosynthesis and Calvin cycle efficiency in diatoms decrease at lower temperatures, and that increased rates of nitrate reduction under this condition may serve to dissipate excess energy in high light. Lomas and Glibert45 also suggested that diatoms took up nitrate in excess of nutritional requirements at lower temperatures, and used nitrate reduction as an electron sink when the harvesting and utilization of light energy were imbalanced. While we did not measure growth rates in this study, previous work by Zhang et al.33 found that C. subsalsa had a significantly reduced growth rate at 16 °C compared to 20 °C. The elevated NR2 transcript abundance at lower temperatures measured here, along with reduced growth rates reported by Zhang et al., suggests that nitrate reduction may act as an energy sink in C. subsalsa when activities of other enzymes are not sufficient to sustain growth at lower temperatures. This is in contrast to the gene expression of HsNR1 in H. akashiwo14, which was not significantly affected by the temperatures ranging from 18 ° to 28 °C, and maintained growth rates at 16 °C33, suggesting that nitrogen acquisition strategies may differ for these co-occurring species over this temperature range.
Proteins in the 14-3-3 family regulate widespread cellular processes in eukaryotes, such as intracellular signaling and control of cell cycles46–49. Under adverse conditions (e.g. low light), 14-3-3 proteins bind to the phosphorylated serine residue of plant NRs and block the electron flow from its NADH domain at the C-terminus to the Mo-MPT domain at the N-terminus, thus inhibiting NR activity and preventing the accumulation of toxic nitrite50. The 14-3-3 binding motif is highly conserved in plant NRs6,20. 14-3-3 proteins bind to a phosphorylated Ser residue within the motifs RXXpSXP as originally described by Muslin et al.51, or KSXpSXP in angiosperms52, where X in the sequence indicates any amino acid, and pS is the phospho-Ser residue53. Although the 14-3-3 binding motifs have not been reported in algal NR, evidence of indirect regulation of NR activity by 14-3-3 binding has been reported. In the green alga Chlamydomonas, for example, nitrogen assimilation is regulated by interaction of 14-3-3 proteins with glutamine synthetase I, indirectly affecting NR activity by feedback inhibition23,54.
Here, a putative 14-3-3 binding motif was found in the deduced amino acid sequence of NR3 in C. subsalsa. Regulation of NR3 by 14-3-3 binding proteins was also supported by the presence of a transcript encoding a 14-3-3 protein in the C. subsalsa transcriptome (ID: CAMPEP_0187155116). Putative 14-3-3 binding motifs were also identified in NRs of other algal species, Klebsormidium nitens, Emiliania huxleyi, and Porphyra umbilicalis, with a high stringency, and in NRs of Gracilaria tenuistipitata and Chondrus crispus with a medium stringency. Interestingly, 14-3-3 binding motifs identified in C. subsalsa and other algal species are all located in the hinge regions of these algae, consistent with 14-3-3 binding motifs in plant NRs. 14-3-3 binding motifs located within the hinge 1 region of C. subsalsa, K. nitens, P. umbilicalis, G. tenuistipitata, and C. crispus in particular, had surface accessibility scores indicating a higher probability of interactions with 14-3-3 binding proteins (Supplementary Table S1).
NRs with 14-3-3 binding motifs in the three rhodophyte species P. umbilicalis, G. tenuistipitata, and C. crispus are phylogenetically divergent from NRs in raphidophyte C. subsalsa, charophyte K. nitens, and haptophyte E. huxleyi (Fig. 3). If these NRs are regulated by 14-3-3 binding as in plants, this mode of regulation may have evolved independently among these algal species, or lost among other lineages over time. Interestingly, NR in K. nitens is closely related to NRs of terrestrial plants, consistent with a previous study showing that the K. nitens genome has a number of genes that were shared with land plants55. Although there have been no reported studies of NR regulatory mechanisms in P. umbilicalis and C. crispus, regulation of NR transcription in G. tenuistipitata12 and NR enzyme activity in both G. tenuistipitata56 and in Gracilaria chilensis57,58 have been investigated. NR activity in G. tenuistipitata was regulated by light and its circadian clock56. Intriguingly, NR in G. chilensis was regulated at the post-translational level by nitrogen source and light, involving phosphorylation and dephosphorylation57,58, providing for the possibility of NR regulation by 14-3-3 binding in this species. The amino acid sequence of NR in E. huxleyi and its gene expression responsive to nitrogen source were described by Bruhn et al.59. This enzyme was also characterized by Iwamoto and Shiraiwa60, showing that E. huxleyi NR is a hexamer and is NADH-specific. However, the post-translational regulation of E. huxleyi NR activity is not clear. Future investigations involving post-translational regulation of NRs in these species and the potential role of 14-3-3 binding motifs are required.
In addition to the presence of a putative 14-3-3 binding motif in C. subsalsa NR3, the deduced amino acid sequence lacks the conserved Heme-Fe domain (cytochrome b5) found in all other NRs (Fig. 2). This domain is considered one of the three strictly conserved functional domains (Heme-Fe, FAD, and Molybdenum cofactor) in eukaryotic NRs6,61. These enzymes are members of sulfite oxidase (SO) family62,63, which differs in structure between plants and animals. Similar to eukaryotic NRs, SO in animals contains a Heme-Fe domain64. The deduced amino acid sequence of NR3 in C. subsalsa, however, is more similar to plant SO which lacks the Heme-Fe domain64, preventing intramolecular electron transfers between redox centers. Instead of utilizing Fe(III) and cytochrome c as electron acceptors as occurs in animal SOs, plant SO uses molecular oxygen as the terminal electron acceptor63,65. It has also been shown that eukaryotic NRs have partial activities with fragments of different domains, using alternative electron donors and acceptors15,66, lending support for the function of NR3 in C. subsalsa without requiring the Heme-Fe domain.
The structure of NR may also make it possible for NR3 to function without the Heme-Fe domain. NRs are homodimers where dimerization is necessary for function, and further formation of a tetramer is a tendency by NR homodimers67. NR3 may participate in tetramer formation with CsNR2, which includes both the Heme-Fe domain and a novel 2/2 hemoglobin domain. The additional heme-binding domain in CsNR2 may complement the loss of the Heme-Fe domain in NR3 after formation of the multimeric protein. In addition to nitrate reduction and energy dissipation, NRs have also been reported to have alternative functions5, including participating in nitric oxide metabolism2, signaling68, and ATP generation69. Fully elucidating the function of NR3 will require additional study.
In summary, results of this study demonstrate a range of transcriptional responses to diel light cycles, nitrogen source, and temperature that act to differentially regulate NR gene expression in C. subsalsa. Transcription of NR2 and NR3 were differentially regulated by diel changes in light intensity, while NR2 gene expression was also responsive to nitrogen source and temperature. Sequence analysis of the deduced amino acid sequence of CsNR3 revealed the existence of a putative 14-3-3 binding motif in the hinge 1 region, suggesting the potential for post-translational regulation of this enzyme, as well as the lack of the Heme-Fe (cytochrome b5) domain. Comparisons between NR3 and plant SO suggest that alternative electron acceptors, or heterotetramer formation with CsNR2, may permit this enzyme to function in the absence of Heme-Fe domain. To date, this is the first report of the presence of a putative 14-3-3 binding motif in algal NRs. A survey of NR sequences in other algal species, however, suggests that the presence of a putative 14-3-3 binding motif in CsNR3 is not unique and that post-translational mechanisms regulating NR activity in plants may be shared with some algal species. Future research efforts may be directed to identify these mechanisms for a better understanding of environmental factors affecting nitrogen assimilation in algae.
Materials and Methods
Stock culture conditions
Stock cultures of C. subsalsa (National Center for Marine Algae and Microbiota, CCMP2191) were maintained in f/2 medium70 -Si at a salinity of 20 with either f/2 levels of nitrate (882 µM) or 100 µM ammonium. Cultures were incubated at 25 °C and 100 µmol m−2s−1 irradiance with cool fluorescent lights, and on a 12:12 h light: dark cycle.
Identification and confirmation of NR3 sequence in C. subsalsa
The transcriptome of C. subsalsa (CCMP2191) was sequenced as part of the Marine Microbial Eukaryote Transcriptome Sequencing Project29 (MMETSP; Sample Name: MMETSP0947–0950; PI: Kathryn Coyne). A unique NR (designated NR3; ID: CAMPEP_0187150166; https://www.imicrobe.us/#/combined_assemblies/21) was identified and evaluated by BLASTX search71.
Plasmids designated CsNR3-P2 and CsNR3-P10 were prepared for sequencing to confirm NR3 sequence data obtained from MMETSP. Primers were designed to obtain overlapping segments of the entire NR3 cDNA sequence. RNA was extracted from stock cultures, and cDNA was prepared as described below. cDNA was diluted 1:20 with LoTE [3 mM Tris-HCl (pH 7.5), 0.2 mM EDTA] and used as template in PCR reactions. The PCR reactions included 1.0 µL diluted template, 0.2 mM dNTPs, 2.5 mM MgCl2, 1X Taq polymerase buffer (Sigma Chem. Co., St. Louis, MO, US), 0.25 units Jump-Start Taq Polymerase (Sigma Chem. Co.), and 0.25 µM of primers CsNR3 P2F and CsNR3 1228 R or CsNR3 1143 F and CsNR3 P10R. The PCR cycle consisted of 37 cycles of 30 s at 94 °C, 30 s at 58 °C, and 1.5 min at 72 °C. PCR products were cloned into pCR4 TOPO plasmid vector (Thermo Fisher Scientific Inc., Waltham, MA, US). Plasmids were prepared using QuickClean 5 M Miniprep Kit (GenScript Corporation, Piscataway, NJ, US) from liquid media following the manufacturer’s protocol.
Plasmids CsNR3-P2 and CsNR3-P10 were sequenced at Delaware Biotechnology Institute (University of Delaware, Newark, DE, US). The overlapping sequences were aligned with NR3 sequence from MMETSP using MEGA772 (Supplementary Data S1). A consensus sequence of NR3 was deposited in GenBank (accession number: MH708883).
Phylogenetic analysis
The deduced NR3 sequence was aligned with NR sequences of Klebsormidium nitens (GAQ91960.1), Emiliania huxleyi (DAA12507.1), Porphyra umbilicalis (OSX81470.1), Gracilaria tenuistipitata (ACX31653.1) and Chondrus crispus (XP_005714488.1), as well as other plant, fungal and algal NR sequences, including CsNR2 (AER70127.1), HsNR1 (ACS44801.1), NRs from plants [Prunus persica (BAB55002), Ricinus communis (AAG30576), Nicotina tabacum (CAA32216), Solanum tuberosum (BAB93534), Glycine max (AAA96813), Phaseolus vulgaris (P39866), Zea mays (AAD38068), and Hordeum vulgare (CAA40976)], moss [Physcomitrella patens (BAE19755)], haptophyte [Chrysochromulina sp. (KOO21257.1)], diatoms [Phaeodactylum tricornutum (AAV66996), Cylindrotheca fusiformis (AAY59538), and Thalassiosira pseudonana (EED88244.1)], green algae [Chlorella ellipsoidea (AAP32278), Dunaliella salina (AAP75705), Chlamydomonas reinhardtii (AAF17595), and Volvox carteri (AAA1114)], and fungi [Wickerhamomyces anomalus (AAF28059.1), Hebeloma cylindrosporum (CAB60010), and Metarhizium anisopliae (CAA04554)].
The sequences were aligned using Clustal W73 in Mega 772, and the evolutionary divergence between these sequences were computed using p- distance method with complete deletion of gaps. The sequences included Mo-MPT, DI, FAD, and NAD(P)H domains, as well as hinge 1 and N-terminal regions. To build a phylogenetic tree, the best model was computed and selected in Mega 772, then a phylogenetic tree was constructed using the LG + G + I model with a discrete Gamma distribution and 5 rate categories. The Maximum Likelihood method was used, and the tree was rooted by fungal NR sequences. Five hundred replicates were used to generate the bootstrap values.
Diel expression of NR genes under different light conditions
C. subsalsa was cultured as above and cells were counted under a light microscope with a Sedgewick-Rafter chamber and inoculated into six replicate cultures at 11,400 cells mL−1. Three replicate cultures were moved into constant light at 100 µmol photons m−2 s−1 for 24 hours before the initial sampling and remained under constant light for the entire sampling period of 19 hours. The other replicate cultures (N = 3) were maintained on a 12:12 h light:dark cycle. Samples were collected at five time points: 1 hour before lights on (7:00), 1 hour after lights on (9:00), 6 hours after lights on (14:00), 1 hour after dark (21:00), and 6 hours after dark (2:00). At each time point, cultures were gently filtered onto 3 µm pore size polycarbonate filters (EMD Millipore Corporation, Billerica, MA, USA) for RNA extraction, described below.
Gene expression of NRs with nitrate or ammonium as a nitrogen source
C. subsalsa was cultured as described under “Stock culture conditions” in f/2 medium with either 100 µM NO3− or 100 µM NH4+ (N = 3). Semi-continuous cultures were maintained in exponential phase for several weeks (diluted every 3 to four days), and harvested during exponential phase growth at 6 hours after the start of the light cycle. Samples were collected as described above for RNA extraction. RNA was extracted as described below.
Gene expression of NRs at different temperatures
C. subsalsa was cultured as described under “Stock culture conditions” at 18°, 25°, and 28 °C. Semi-continuous cultures (N = 3) were maintained in exponential phase for several weeks as described above before harvesting at 6 hours after the start of the light cycle. The samples were collected as above for RNA extraction. RNA was extracted as described below.
RNA extraction and cDNA synthesis
RNA was extracted from filtered cells using the RNeasy Mini Kit (Qiagen, Chatswort, CA, US). Contaminating DNA was digested in 10 µl reaction volumes containing 300 to 1000 ng RNA and 1 µl DNase (100 units, Amplification Grade, Thermo Fisher Scientific Inc.) in 1X DNase I Reaction Buffer (Thermo Fisher Scientific Inc.) for 15 min at room temperature. The reaction was stopped by addition of EDTA (pH 8.0, Thermo Fisher Scientific Inc.), and the mixture was incubated at 65 °C for 15 min to degrade the DNase. The purified RNA was stored at −80 °C until use.
First strand cDNA was synthesized using the Superscript III First Strand Supermix Kit (Thermo Fisher Scientific Inc.) according to manufacturer’s directions using random hexamers to prime the reaction. The reaction was stopped by incubation at 85 °C for 5 min. The cDNA was stored at −80 °C until PCR analysis.
Plasmid preparation
Plasmids were prepared from amplified cDNA to use as standards in qPCR analysis of NR2 and NR3. Diluted cDNA was used as template in PCR reactions with primers targeting C. subsalsa NR2 and NR3 (Table 1). PCR reactions were prepared as above with 0.075 µM CsNR3 1143 F and CsNR3 1228 R primers for CsNR3 amplification, or CsNR Glob 1005 F and CsNR Glob 1240 R for CsNR2 amplification. The PCR cycle consisted of 37 cycles of 30 s at 94 °C, 30 s at 53 °C, and 1 min at 72 °C.
Table 1.
Sequence of C. subsalsa- specific primers for nitrate reductase and actin. F, forward primer; R, reverse primer.
| Primers | Sequence (5′-3′) | Gene Targeted |
|---|---|---|
| CsNR Glob 1005 F | GAAGCTGCAATCCAAGCTGTAGT | NR2 |
| CsNR Glob 1240 R | CCAACTCAGTCAACGTGTTCA | NR2 |
| CsNR3 1143 F | CTGGACACAGCTGAGTTACA | NR3 |
| CsNR3 1228 R | GCAATGCGATGAGTTGGAG | NR3 |
| CsNR3 P2F | ATGGACGCATTCACATCCAGG | NR3 |
| CsNR3 P10R | GAAAGTGAAGCACATTTCACGC | NR3 |
| Cs Actin163F | GTGGGAGATGAGGCACAGTC | Actin |
| Cs Actin 294 R | TGCCACTCGAAGCTCATTGT | Actin |
qPCR analysis of NR gene expression
NR2 and NR3 gene expression was measured relative to actin transcript levels using quantitative real-time PCR (qPCR) on an ABI Prism 7500 Sequence Detection System (Thermo Fisher Scientific Inc.). Triplicate reactions included 5 µl Power SYBR® Green PCR Master Mix (Thermo Fisher Scientific Inc.), 0.3 µM forward primer (CsNR Glob 1005 F for NR2, and CsNR3 1143 F for NR3), 0.3 µM reverse primer (CsNR Glob 1240 R for NR2, and CsNR3 1228 R for NR3), and 1 µl diluted (1:20) template cDNA. Primer sequences are in Table 1. Ten-fold dilutions of plasmids for CsNR2 and CsNR3 were used to make a standard curve of known copy numbers. Negative controls (without template) were run simultaneously. The cycling parameters were 50 °C for 2 min, and 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, 56 °C for 30 s, and 60 °C for 1 min. The specificity of each reaction was evaluated by measuring the change in fluorescence with dissociation of duplex PCR products during a stepwise increase in temperature from 60 °C to 95 °C. The transcript copy numbers of NR2 and NR3 for each sample were determined by linear regression analysis, and then normalized to gene expression of actin (CAMPEP_ 0187146040) as an endogenous reference to produce a relative expression value for each NR in that sample.
Statistical analysis
Repeated measures ANOVA was used to test if there were significant differences in gene expression over the entire sampling period. If the effect was significant (p < 0.05), then another repeated measures ANOVA was used to test the significant difference in transcript abundance in light vs. in dark (in L:D treatments), and paired t-test was conducted to analyze the significance of the differences between the values at each time point. Additionally, paired t-test was used to analyze the difference between NR2 and NR3 gene expression at each time point.
One-way ANOVA was used to test if nitrogen source and temperature had a significant effect on NR gene expression. If the effect was significant, Tukey’s HSD test was used to analyze the difference in all possible pairs. All statistical analyses were performed in R74 (V.3.2.4 Revised).
Identification of 14-3-3 binding motifs
The translated amino acid sequence of NR3 was submitted to Scansite 330 (http://scansite3.mit.edu; accessed on 2/28/2018) to identify potential 14-3-3 binding motifs in this sequence. Scansite is a computational tool to predict interactions between proteins and sites of phosphorylation. The parameters used in this search are listed here: group: phosphoserine/threonine binding; motif: 14-3-3 mode 1; and stringency: high.
NR sequences from other algal species were identified in the NCBI31 database (accessed on 11 Sept, 2017) and downloaded for analysis. Potential 14-3-3 binding sites including the motifs RXXSXP and KSXSXP were searched in 80 full-length algal NR sequences using MATLAB75 (V. R2016a). Sequences with putative 14-3-3 binding motifs were submitted to Scansite 330 (http://scansite3.mit.edu) for further analysis, using parameters described above for CsNR3 with high and medium stringency.
Electronic supplementary material
Acknowledgements
We thank Douglas Miller (University of Delaware (UD)) and Julie Rose (US National Oceanic and Atmospheric Administration (NOAA)) for assistance with statistical analysis, and Anna Schutschkow (UD) for assistance with sample processing. This research was funded in part by the Center for Sponsored Coastal Ocean Research, NOAA (Grant number NA15NOS4780182 to Mark Warner, Jon Cohen and Kathryn Coyne; ECOHAB publication number 920) and the Gordon and Betty Moore Foundation through Grant #2637 to the National Center for Genome Resources. Samples MMETSP0947, MMETSP0948, MMETSP0949 and MMETSP0950 were sequenced at the National Center for Genome Resources.
Author Contributions
Y.W. conducted the research and wrote the manuscript, J.N.B. contributed to analysis of results, K.J.C. conceived of the project and contributed to manuscript preparation.
Data Availability Statement
All data generated during this study are available upon request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-31735-5.
References
- 1.Malerba ME, Connolly SR, Heimann K. An experimentally validated nitrate-ammonium-phytoplankton model including effects of starvation length and ammonium inhibition on nitrate uptake. Ecol. Modell. 2015;317:30–40. doi: 10.1016/j.ecolmodel.2015.08.024. [DOI] [Google Scholar]
- 2.Stewart JJ, Coyne KJ. Analysis of raphidophyte assimilatory nitrate reductase reveals unique domain architecture incorporating a 2/2 hemoglobin. Plant Mol. Biol. 2011;77:565–575. doi: 10.1007/s11103-011-9831-8. [DOI] [PubMed] [Google Scholar]
- 3.Campbell WH. Nitrate reductase structure, function and regulation: bridging the gap between biochemistry and physiology. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;50:277–303. doi: 10.1146/annurev.arplant.50.1.277. [DOI] [PubMed] [Google Scholar]
- 4.Parker MS, Armbrust EV. Synergistic effects of light, temperature, and nitrogen source on transcription of genes for carbon and nitrogen metabolism in the centric diatom Thalassiosira pseudonana (Bacillariophyceae) J. Phycol. 2005;41:1142–1153. doi: 10.1111/j.1529-8817.2005.00139.x. [DOI] [Google Scholar]
- 5.Young, E. B. & Berges, J. A. Nitrate reductase: a nexus of disciplines, organisms, and metabolism. Aquat. Microb. Ecol. Biogeochem. A Dual Perspect. 105–116 (2016).
- 6.Sanz-Luque, E., Chamizo-Ampudia, A., Llamas, A., Galvan, A. & Fernandez, E. Understanding nitrate assimilation and its regulation in microalgae. Front. Plant Sci. 6, (2015). [DOI] [PMC free article] [PubMed]
- 7.Glibert PM, et al. Pluses and minuses of ammonium and nitrate uptake and assimilation by phytoplankton and implications for productivity and community composition, with emphasis on nitrogen-enriched conditions. Limnol. Oceanogr. 2016;61:165–197. doi: 10.1002/lno.10203. [DOI] [Google Scholar]
- 8.Young EB, Dring MJ, Berges JA. Distinct patterns of nitrate reductase activity in brown algae: light and ammonium sensitivity in Laminaria digitata is absent in Fucus species. J. Phycol. 2007;43:1200–1208. doi: 10.1111/j.1529-8817.2007.00403.x. [DOI] [Google Scholar]
- 9.Davison IR, Davison JO. The effect of growth temperature on enzyme activities in the brown alga Laminaria saccharina. Br. Phycol. J. 1987;22:77–87. doi: 10.1080/00071618700650101. [DOI] [Google Scholar]
- 10.Meyer C, Lea US, Provan F, Kaiser WM, Lillo C. Is nitrate reductase a major player in the plant NO (nitric oxide) game? Photosynth. Res. 2005;83:181–189. doi: 10.1007/s11120-004-3548-3. [DOI] [PubMed] [Google Scholar]
- 11.Lillo C, et al. Signalling cascades integrating light-enhanced nitrate metabolism. Biochem. J. 2008;415:11–9. doi: 10.1042/BJ20081115. [DOI] [PubMed] [Google Scholar]
- 12.Falcão VR, Oliveira MC, Colepicolo P. Molecular characterization of nitrate reductase gene and its expression in the marine red alga Gracilaria tenuistipitata (Rhodophyta) J. Appl. Phycol. 2010;22:613–622. doi: 10.1007/s10811-010-9501-2. [DOI] [Google Scholar]
- 13.Lea US, Leydecker M-T, Quilleré I, Meyer C, Lillo C. Posttranslational regulation of nitrate reductase strongly affects the levels of free amino acids and nitrate, whereas transcriptional regulation has only minor influence. Plant Physiol. 2006;140:1085–94. doi: 10.1104/pp.105.074633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coyne KJ. Nitrate reductase (NR1) sequence and expression in the harmful alga Heterosigma akashiwo (raphidophyceae) J. Phycol. 2010;46:135–142. doi: 10.1111/j.1529-8817.2009.00781.x. [DOI] [Google Scholar]
- 15.Solomonson LP, Barber MJ. Assimilatory nitrate reductase: functional properties and regulation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1990;41:225–253. doi: 10.1146/annurev.pp.41.060190.001301. [DOI] [Google Scholar]
- 16.Lillo C, Meyer C, Ruoff P. The nitrate reductase circadian system. The central clock dogma contra multiple oscillatory feedback loops. Plant Physiol. 2001;125:1554–7. doi: 10.1104/pp.125.4.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vergara JJ, Berges JA, Falkowski PG. Diel periodicity of nitrate reductase activity and protein levels in the marine diatom Thalassiosira weissflogii (Bacillariophyceae) Jounal Phycol. 1998;34:952–961. doi: 10.1046/j.1529-8817.1998.340952.x. [DOI] [Google Scholar]
- 18.Tischner R. Nitrate uptake and reduction in higher and lower plants. Plant, Cell Environ. 2000;23:1005–1024. doi: 10.1046/j.1365-3040.2000.00595.x. [DOI] [Google Scholar]
- 19.Huber JL, Huber SC, Campbell WH, Redinbaugh MG. Reversible light/dark modulation of spinach leaf nitrate reductase activity involves protein phosphorylation. Arch. Biochem. Biophys. 1992;296:58–65. doi: 10.1016/0003-9861(92)90544-7. [DOI] [PubMed] [Google Scholar]
- 20.Lambeck IC, et al. Molecular mechanism of 14-3-3 protein-mediated inhibition of plant nitrate reductase. J. Biol. Chem. 2012;287:4562–4571. doi: 10.1074/jbc.M111.323113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bachmann M, et al. Identification of Ser-543 as the major regulatory phosphorylation site in spinach leaf nitrate reductase. Plant Cell. 1996;8:505–517. doi: 10.1105/tpc.8.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medina-Andrés R, Lira-Ruan V. In silico characterization of a nitrate reductase gene family and analysis of the predicted proteins from the moss Physcomitrella patens. Commun. Integr. Biol. 2012;5:19–25. doi: 10.4161/cib.18534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pozuelo M, MacKintosh C, Galván A, Fernández E. Cytosolic glutamine synthetase and not nitrate reductase from the green alga Chlamydomonas reinhardtii is phosphorylated and binds 14-3-3 proteins. Planta. 2001;212:264–269. doi: 10.1007/s004250000388. [DOI] [PubMed] [Google Scholar]
- 24.Gruber H, Goetinck SD, Kirk DL, Schmitt R. The nitrate reductase-encoding gene of Volvox carteri: map location, sequence and induction kinetics. Gene. 1992;120:75–83. doi: 10.1016/0378-1119(92)90011-D. [DOI] [PubMed] [Google Scholar]
- 25.Dawson HN, Pendleton LC, Solomonson LP, Cannons AC. Cloning and characterization of the nitrate reductase-encoding gene from Chlorella vulgaris: structure and identification of transcription start points and initiator sequences. Gene. 1996;171:139–145. doi: 10.1016/0378-1119(96)00063-7. [DOI] [PubMed] [Google Scholar]
- 26.Song B, Ward BB. Molecular characterization of the assimilatory nitrate reductase gene and its expression in the marine green alga Dunaliella tertiolecta (Chlorophyceae) J. Phycol. 2004;40:721–731. doi: 10.1111/j.1529-8817.2004.03078.x. [DOI] [Google Scholar]
- 27.Allen AE, Ward BB, Song B. Characterization of diatom (Bacillariophyceae) nitrate reductase genes and their detection in marine phytoplankton communities. J. Phycol. 2005;41:95–104. doi: 10.1111/j.1529-8817.2005.04090.x. [DOI] [Google Scholar]
- 28.Fernandez E, et al. Isolation and characterization of the nitrate reductase structural gene of Chlamydomonas reinhardtii (restriction fragment length polymorphisms/regulatory mutant/transformation) Biochemistry. 1989;86:6449–6453. doi: 10.1073/pnas.86.17.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keeling PJ, et al. The marine microbial eukaryote transcriptome sequencing project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 2014;12:e1001889. doi: 10.1371/journal.pbio.1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–41. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 44, D7–19 (2016). [DOI] [PMC free article] [PubMed]
- 32.Imai I, Yamaguchi M. Life cycle, physiology, ecology and red tide occurrences of the fish-killing raphidophyte Chattonella. Harmful Algae. 2012;14:46–70. doi: 10.1016/j.hal.2011.10.014. [DOI] [Google Scholar]
- 33.Zhang Y, Fu FX, Whereat E, Coyne KJ, Hutchins DA. Bottom-up controls on a mixed-species HAB assemblage: a comparison of sympatric Chattonella subsalsa and Heterosigma akashiwo (Raphidophyceae) isolates from the Delaware Inland Bays, USA. Harmful Algae. 2006;5:310–320. doi: 10.1016/j.hal.2005.09.001. [DOI] [Google Scholar]
- 34.Brown KL, Twing KI, Robertson DL. Unraveling the regulation of nitrogen assimilation in the marine diatom Thalassiosira pseudonana (bacillariophyceae): diurnal variations in transcript levels for five genes involved in nitrogen assimilation. J. Phycol. 2009;45:413–426. doi: 10.1111/j.1529-8817.2009.00648.x. [DOI] [PubMed] [Google Scholar]
- 35.Pilgrim ML, Caspar T, Quail PH, Robertson Mcclung C, McClung CR. Circadian and light-regulated expression of nitrate reductase in Arabidopsis. Plant Mol. Biol. 1993;23:349–364. doi: 10.1007/BF00029010. [DOI] [PubMed] [Google Scholar]
- 36.Deng MD, Moureaux T, Leydecker MT, Caboche M. Nitrate-reductase expression is under the control of a circadian rhythm and is light inducible in Nicotiana tabacum leaves. Planta. 1990;180:257–261. doi: 10.1007/BF00194005. [DOI] [PubMed] [Google Scholar]
- 37.Lillo C, Ruoff P. An unusually rapid light-induced nitrate reductase mRNA pulse and circadian oscillations. Sci. Nat. 1989;76:526–528. doi: 10.1007/BF00374129. [DOI] [PubMed] [Google Scholar]
- 38.Farré EM, Weise SE. The interactions between the circadian clock and primary metabolism. Current Opinion in Plant Biology. 2012;15:293–300. doi: 10.1016/j.pbi.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 39.Stoelken G, Simon J, Ehlting B, Rennenberg H. The presence of amino acids affects inorganic N uptake in non-mycorrhizal seedlings of european beech (Fagus sylvatica) Tree Physiol. 2010;30:1118–1128. doi: 10.1093/treephys/tpq050. [DOI] [PubMed] [Google Scholar]
- 40.Vincentz M, Moureaux T, Leydecker MT, Vaucheret H, Caboche M. Regulation of nitrate and nitrite reductase expression in Nicotiana plumbaginifolia leaves by nitrogen and carbon metabolites. Plant J. 1993;3:315–324. doi: 10.1111/j.1365-313X.1993.tb00183.x. [DOI] [PubMed] [Google Scholar]
- 41.Imamura S, et al. R2R3-type MYB transcription factor, CmMYB1, is a central nitrogen assimilation regulator in Cyanidioschyzon merolae. Proc. Natl. Acad. Sci. USA. 2009;106:12548–12553. doi: 10.1073/pnas.0902790106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poulsen N, Kröger N. A new molecular tool for transgenic diatoms. FEBS J. 2005;272:3413–3423. doi: 10.1111/j.1742-4658.2005.04760.x. [DOI] [PubMed] [Google Scholar]
- 43.Imamura S, et al. Nitrate assimilatory genes and their transcriptional regulation in a unicellular red alga Cyanidioschyzon merolae: genetic evidence for nitrite reduction by a sulfite reductase-like enzyme. Plant Cell Physiol. 2010;51:707–717. doi: 10.1093/pcp/pcq043. [DOI] [PubMed] [Google Scholar]
- 44.Hockin NL, Mock T, Mulholland F, Kopriva S, Malin G. The response of diatom central carbon metabolism to nitrogen starvation is different from that of green algae and higher plants. Plant Physiol. 2012;158:299–312. doi: 10.1104/pp.111.184333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lomas MW, Glibert PM. Temperature regulation of nitrate uptake: a novel hypothesis about nitrate uptake and reduction in cool-water diatoms. Limnol. Oceanogr. 1999;44:556–572. doi: 10.4319/lo.1999.44.3.0556. [DOI] [Google Scholar]
- 46.Hutchins JR, Dikovskaya D, Clarke PR. Dephosphorylation of the inhibitory phosphorylation site S287 in Xenopus Cdc25C by protein phosphatase-2A is inhibited by 14-3-3 binding. FEBS Lett. 2002;528:267–271. doi: 10.1016/S0014-5793(02)03327-6. [DOI] [PubMed] [Google Scholar]
- 47.Cotelle V, et al. 14-3-3s regulate global cleavage of their diverse binding partners in sugar-starved Arabidopsis cells. EMBO J. 2000;19:2869–2876. doi: 10.1093/emboj/19.12.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mackintosh C. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem. J. 2004;342:329–342. doi: 10.1042/BJ20031332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Denison FC, Paul AL, Zupanska AK, Ferl RJ. 14-3-3 Proteins in Plant Physiology. Semin. Cell Dev. Biol. 2011;22:720–7. doi: 10.1016/j.semcdb.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 50.Kaiser WM, Huber SC. Post‐translational regulation of nitrate reductase: mechanism, physiological relevance and environmental triggers. J. Exp. Bot. 2001;52:1981–1989. doi: 10.1093/jexbot/52.363.1981. [DOI] [PubMed] [Google Scholar]
- 51.Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/S0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 52.Nemie-Feyissa D, et al. Post-translational control of nitrate reductase activity responding to light and photosynthesis evolved already in the early vascular plants. J. Plant Physiol. 2013;170:662–667. doi: 10.1016/j.jplph.2012.12.010. [DOI] [PubMed] [Google Scholar]
- 53.Li Z, et al. Identification of 14-3-3 proteins phosphopeptide-binding specificity using an affinity-based computational approach. PLoS One. 2016;11:e0147467. doi: 10.1371/journal.pone.0147467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Montaigu A, Sanz-Luque E, Galván A, Fernández E. A soluble guanylate cyclase mediates negative signaling by ammonium on expression of nitrate reductase in Chlamydomonas. Plant Cell. 2010;22:1532–1548. doi: 10.1105/tpc.108.062380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hori K, et al. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 2014;5:3978. doi: 10.1038/ncomms4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lopes PF, et al. Diurnal fluctuation of nitrate reductase activity in the marine red algae Gracilaria tenuistipitata (Rhodophyta) J. Phycol. 1997;231:225–231. doi: 10.1111/j.0022-3646.1997.00225.x. [DOI] [Google Scholar]
- 57.Chow F, De Oliveira MC, Pedersén M. In vitro assay and light regulation of nitrate reductase in red alga Gracilaria chilensis. J. Plant Physiol. 2004;161:769–776. doi: 10.1016/j.jplph.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 58.Chow F, De Oliveira MC. Rapid and slow modulation of nitrate reductase activity in the red macroalga Gracilaria chilensis (Gracilariales, Rhodophyta): influence of different nitrogen sources. J. Appl. Phycol. 2008;20:775–782. doi: 10.1007/s10811-008-9310-z. [DOI] [Google Scholar]
- 59.Bruhn A, LaRoche J, Richardson K. Emiliania Huxleyi (Prymnesiophyceae): nitrogen-metabolism genes and their expression in response to external nitrogen sources. J. Phycol. 2010;46:266–277. doi: 10.1111/j.1529-8817.2010.00809.x. [DOI] [Google Scholar]
- 60.Iwamoto K, Shiraiwa Y. Characterization of NADH: nitrate reductase from the coccolithophorid Emiliania huxleyi (Lohman) Hay & Mohler (Haptophyceae) Mar. Biotechnol. (NY). 2003;5:20–26. doi: 10.1007/s10126-002-0051-8. [DOI] [PubMed] [Google Scholar]
- 61.Zhou J, Kleinhofs A. Molecular evolution of nitrate reductase genes. J. Mol. Evol. 1996;42:432–442. doi: 10.1007/BF02498637. [DOI] [PubMed] [Google Scholar]
- 62.Stolz JF, Basu P. Evolution of nitrate reductase: molecular and structural variations on a common function. ChemBioChem. 2002;3:198–206. doi: 10.1002/1439-7633(20020301)3:2/3<198::AID-CBIC198>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 63.Kappler U, Enemark JH. Sulfite-oxidizing enzymes. J. Biol. Inorg. Chem. 2015;20:253–264. doi: 10.1007/s00775-014-1197-3. [DOI] [PubMed] [Google Scholar]
- 64.Schrader N, et al. The crystal structure of plant sulfite oxidase provides insights into sulfite oxidation in plants and animals while animal SOs contain a Moco and a heme domain. Structure. 2003;11:1251–1263. doi: 10.1016/j.str.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 65.Hänsch R, et al. Plant sulfite oxidase as novel producer of H2O2: combination of enzyme catalysis with a subsequent non-enzymatic reaction step. J. Biol. Chem. 2006;281:6884–6888. doi: 10.1074/jbc.M513054200. [DOI] [PubMed] [Google Scholar]
- 66.Hyde GE, Campbell WH. High-level expression in Escherichia coli of the catalytically active flavin domain of corn leaf NADH:nitrate reductase and its comparison to human NADH:cytochrome B5 reductase. Biochem. Biophys. Res. Commun. 1990;168:1285–1291. doi: 10.1016/0006-291X(90)91168-R. [DOI] [PubMed] [Google Scholar]
- 67.Campbell WH. Structure and function of eukaryotic NAD(P)H:nitrate reductase. Cell. Mol. Life Sci. 2001;58:194–204. doi: 10.1007/PL00000847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Desikan R, Griffiths R, Hancock J, Neill S. A new role for an old enzyme: nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 2002;99:16314–16318. doi: 10.1073/pnas.252461999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kamp A, Stief P, Knappe J, De Beer D. Response of the ubiquitous pelagic diatom Thalassiosira weissflogii to darkness and anoxia. PLoS One. 2013;8:e82605. doi: 10.1371/journal.pone.0082605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guillard RRL, Ryther JH. Studies of marine planktonic diatoms I. Cyclotella nana Hustedt, and Detonula confervasea (Cleve) Gran. Can. J. Microbiol. 1962;8:229–239. doi: 10.1139/m62-029. [DOI] [PubMed] [Google Scholar]
- 71.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Team, R. C. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. (2015).
- 75.MathWorks, Inc. MATLAB: the language of technical computing. Desktop tools and development environment, version 7. MathWorks; (2005).
- 76.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated during this study are available upon request.