Abstract
Introduction: Breast cancer, as one of the major causes of cancer death among women, is the central focus of this study. The recent advances in the development and application of computational tools and bioinformatics in the field of immunotherapy of malignancies such as breast cancer have emerged the new dominion of immunoinformatics, and therefore, next generation of immunomedicines .
Methods: Having reviewed the most recent works on the applications of computational tools, we provide comprehensive insights into the breast cancer incidence and its leading causes as well as immunotherapy approaches and the future trends. Furthermore, we discuss the impacts of bioinformatics on different stages of vaccine design for the breast cancer, which can be used to produce much more efficient vaccines through a rationalized time- and cost-effective in silico approaches prior to conducting costly experiments.
Results: The tools can be significantly used for designing the immune system-modulating drugs and vaccines based on in silico approaches prior to in vitro and in vivo experimental evaluations. Application of immunoinformatics in the cancer immunotherapy has shown its success in the pre-clinical models. This success returns back to the impacts of several powerful computational approaches developed during the last decade.
Conclusion: Despite the invention of a number of vaccines for the cancer immunotherapy, more computational and clinical trials are required to design much more efficient vaccines against various malignancies, including breast cancer.
Keywords: Bioinformatics, Cancer, Epitope-based vaccine design, Vaccine design
Introduction
Cancer is known as one of the leading causes of morbidity and mortality worldwide. The global occurrence of cancer is expected to increase up to 15 million with around 12 million deaths in the year 2020.1 Cancers, as large family of formidable diseases, are involved with the anomalous growth of the cells, which can potentially invade or spread to other tissues. The majority of cancer incidence (90%-95%) is due to the environmental factors while inherited genetics are considered as a minor factor (5%-10%).2 Risk of the cancer incidence is significantly increased with age and changes in lifestyle in the currently developing world.
Among various cancers, breast cancer is one of the most common malignancies and the leading cause of death among women worldwide. The incidence of this kind of cancer is increased over the last decade in countries like India and China around 30%, while it is doubled or even tripled in Korea, Japan and Singapore.3 According to the National Cancer Institute (NCI) of the United States, in 2013, there were 232 340 and 2240 new cases of breast cancer respectively in female and male, while the estimated death was about 39 620 in the female and 410 in the male.4 The number of breast cancer incidence is expected to grow up to 30%-40% by 2020.5 Such an increase requires more attention in terms of developing effective therapeutic and preventive methods for the breast cancer.
A vast number of efforts have been made to prevent, diagnose and treat cancer while several methods have been developed during the last decades regarding stage and type of cancer as well as the medical condition of patients. Chemotherapy, radiotherapy, surgery and biological therapies are the most common treatments that can be planned by physicians depending on different key factors such as patient's age, health, lifestyle, and the type and stage of cancer. The proposed methods have substantially improved cancer-associated morbidity and mortality in the western societies. However, there is a limited access to these advanced cancer therapy tools in the developing countries. Additionally, further improvements in the clinical outcome of the proposed treatment methods seem to be unlikely, in large part because of limitations such as the drug resistance. Altogether, despite substantial advancements in the early diagnosis and treatment of cancer, it remains as a major public health burden around the globe, and hence, necessitates the development of much more effective strategies for the prevention, diagnosis and therapy of cancer.
The use of vaccination for cancer treatment and prevention is highly attractive and innovative and has remained controversial in preclinical and clinical investigations. Despite experimental evidence showing a clinical benefit of the anticancer potentials of vaccines through induction of immune responses, their clinical applications are yet to be approved.6
Cancer vaccine can defiantly treat the malignancy through a dynamic activation of the individual's immune system, by which no/little side effects may occur, unlike the commonly used chemotherapies. This strategy can be developed for both cancer prevention and therapy in contrary to the classic concept of the vaccination against infectious diseases. However, designing a vaccine for cancer has confronted with various challenges, in part due to the existence of a large number of potential antigens (Ags) as the target for the immune system. Additionally, many of these Ags may arise during or after the tumorigenesis process. Despite encouraging successes in cancer immunotherapy, the field of vaccine design for the cancer therapy has been a challenging arena for many researchers in the recent decades.
Though remarkable advances achieved in vaccine design, there is no generally accepted universal strategy/tool for rationally designing of vaccines. The procedure for vaccine design is still a time-consuming and costly empirical task. However, the computational methods can significantly be used to design vaccines with strongly reduced time and cost through in silico mapping of thousands of biological components. Recently, the impact of these tools on vaccine design has been highlighted from various aspects such as reverse vaccinology, structural vaccinology, system vaccinology and epitope prediction.7
Because of large incidence, the breast cancer is one of the important candidates for vaccine design.8 Furthermore, in the last 2 decades, there have been essential signs of progress in analyzing the molecular mechanism of breast tumorigenesis and developing immunologic therapies to fight the tumor-related Ags by means of antibodies (Abs). The growing attention to the immunotherapy of breast cancer has led researchers to work on vaccines to activate the immune system in order to prevent the host cells from the initiation of cancer and/or to destroy the developed cancer cells. The main objective of this review was to investigate the recent developments on the breast cancer vaccine and discuss the role of bioinformatics tools for the in-silico computational design of breast cancer vaccine.
Breast cancer
During initiation of the tumor, a normal cell is altered into a cancerous one through an evolutionary complex process, including several genetic and epigenetic variations. These variations are the basis of the initiation and progression for a tumor in the presence of enough growth signals with an insensitivity to the antigrowth signals, evasion from the programmed cell death, unlimited replicative potential, sustained angiogenesis, and finally the ability to invade and metastasize.3,9,10 Despite considerable attempts and impressive advances to understand the molecular mechanisms of cancerous cells, it remains a major challenge for biomedical scientists.
Breast cancer is generally acknowledged to be a molecularly heterogeneous disease. It consists of an extensive spectrum of molecular, pathogenic and clinical features with different prognostic and therapeutic consequences 11,12 Despite continuous study and analysis of these features and considerable achievements, physicians still trust on the traditional clinicopathologic features for tumor diagnosis, and hence, administration of treatment modalities. Recent researches show that a complete characterization of breast cancer can be accomplished by molecular classification as the gold standard.13,14 Within different schemes for molecular classification of the breast cancer, three special receptors on the outside of the cancer cells play a key role as tumor markers, including estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). Based on these special receptors, three important subtypes are considered for the breast cancer, including (i) hormone receptor-positive, (ii) HER2-positive, and (iii) triple-negative cancers. The breast cancer is called hormone receptor-positive if it expresses both the ER and PR receptors. The growth of tumor is depended on both receptors. About 60% to 75% of all breast cancer cases have ER and/or PR receptors. Furthermore, the breast cancer expressing HER2 is called HER2-positive and their growth is depended on this gene. About 20-25% of breast cancers encompass this gene. Finally, the tumors without ER, PR, and/or HER2 is called triple-negative and constitute about 15% of invasive breast cancers. These tumors may grow faster than the hormone receptor-positive tumors and more sensitive to the chemotherapy modalities. The above three receptors are routinely found in the breast cancers, which can serve as the reliable markers for making a decision on the treatment modalities.
A number of factors have been shown to increase the risk of breast cancer development, including genetic and non-genetic factors. Among them, around 20% of the breast cancer cases were caused by genetic risk factors.15 The inherited mutations in a set of 40 genes are significantly associated with the developing breast cancer. Among the genes involved with the breast cancer incidence, mutations in BRCA1, BRCA2, TP53, STK11, CDH1 and PTEN play critical roles in the development and progression of cancer.10 Mutations in BRIP1, CHEK2, PALB2 and ATM seem to have a moderate role, while mutations in the rest genes show a weak association. BRCA1, BRCA2 and TP53 are the most potent genes to predispose hereditary breast cancer development. Mutations in BRCA1 and BRCA2 appear to extremely affect the lifetime risk of mutation carriers by the age of 70 years with the incident rates of 46-87% and 26-84%, respectively. Mutations in TP53 also associate in the development of breast cancer approximately in 30% of the cases.16 Additionally, other susceptibility genes have been identified that associate with an increased risk of breast cancer.
An exciting opportunity has been provided through the widespread study of mutations in the breast cancer susceptibility genes to recognize people with a high risk of breast cancer development. However, investigating the impacts of mutations in these genes and translation of their clinical roles appears to be a striking issue for a large number of researchers. A number of online databases are available for breast cancer, including several scientific details related to breast cancer such as Breast cancer Information Core (BIC) database and Breast Cancer Linkage Consortium (BCLC).
Vaccine design for cancer prevention and treatment
The preventive and therapeutic roles of the immune system in the cancer development had remained controversial until the recent observation that confirmed the ability of the immune system to destroy many nascent cancers before they are clinically diagnosed.13 However, during the development of cancers, the cancerous cells can often evade the defensive mechanism of the immune system. Accordingly, much attention has been paid to the field of cancer immunotherapy by planning several research projects to design and develop cancer vaccines. For instance, Sipuleucel-T is the first cancer vaccine that has been approved in 2010 by the United States Food and Drug Administration (FDA) for the treatment of prostate cancer.14 However, the widespread acceptability and usage of vaccine for the prevention and treatment of cancer demand rationalized translational research approaches.
The human immune system defends the body against external invading agents through mounting a humoral or cellular response. During a humoral response, an interaction between B cells and an external antigen leads to additional separation of the B cells into plasma cells or memory cells in the presence of helper T cells. Particular Abs are secreted by the plasma cells against Ags. Furthermore, the memory cells are long-term cells with the ability to show a quick response to the same Ag. On the other hand, through the cellular response, T cells interact with non-native part of the external protein exposed to the surface of other cells by major histocompatibility (MHC) proteins.
A critical role of the immune system is the identification and elimination of tumors – a function so-called immunosurveillance. The tumor cells express Ags that are either under-expressed or are not expressed by the normal cells. The CD4+ and CD8+ T-cell responses are initiated by the activated professional Ag-presenting cells (APC) such as dendritic cells (DCs), to which tumor cells, in turn, respond by activating different signaling paths and expressing various aberrant Ags.17 These Ags are processed into the MHC class II-binding peptides by the endosome, or into the MHC class I-binding peptides by the proteasome.18 The MHC class I peptide epitopes are transferred to the endoplasmic reticulum by the transporter of antigen processing (TAP), and then, translocated to the surface of the cell through interaction with the MHC class I molecules. Concurrently, tumor antigens are presented to both CD4+ and CD8+ T cells by respectively the professional APC in the presence of MHC class II and MHC class I, with an efficient cross-priming the Ag-specific immune response.19 Fig. 1 shows the Ag-processing pathways within a cell. The CD8+ T-cell response is initiated and intensified by the activated CD4+ T cells explicitly by making co-stimulatory cytokines, and implicitly by regulating a set of co-stimulatory molecules on the APC providing accessory signals for the activation of T cells.17 In consequence, as shown in Fig. 2, the CD8+ T cells migrate towards the tumor sites and lyse the cancerous cells.20 The tumor cells may also accompany the tumor Ags towards CD8+ and/or CD4+ T cells specific for the tumor immunity. Accordingly, the primed and activated immune system can detect the tumor Ags, which are harmful to the nature of the ensuing response. Altogether, the activation of the immune system mechanism(s) against the cancer cells can be an effective strategy for the prevention and treatment of cancer.
Fig. 1.
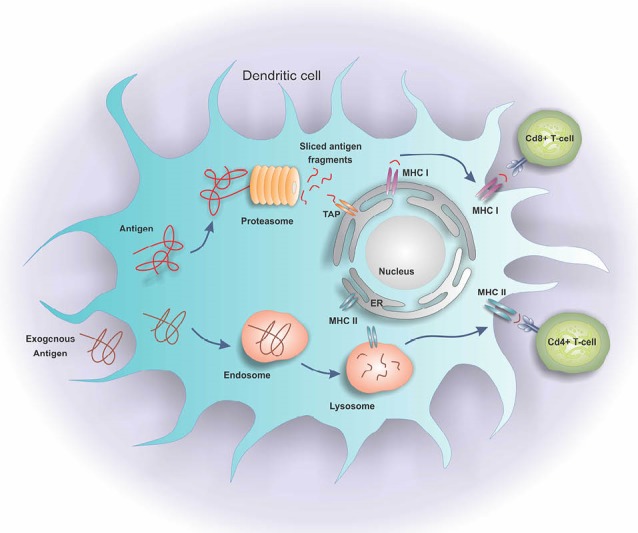
Antigen processing pathways in a dendritic cell. The dendritic cells process the Ags through the MHC class II-binding peptides by the endosomes, or the MHC class I-binding peptides by the proteasomes. The MHC class I peptide epitopes are transferred to the endoplasmic reticulum by transporter of antigen processing (TAP). Then, they are translocated to the surface of cell through interaction with the MHC class I molecules. Tumor Ags are presented to both CD4+ and CD8+ T cells in the presence of MHC II and I.
Fig. 2.
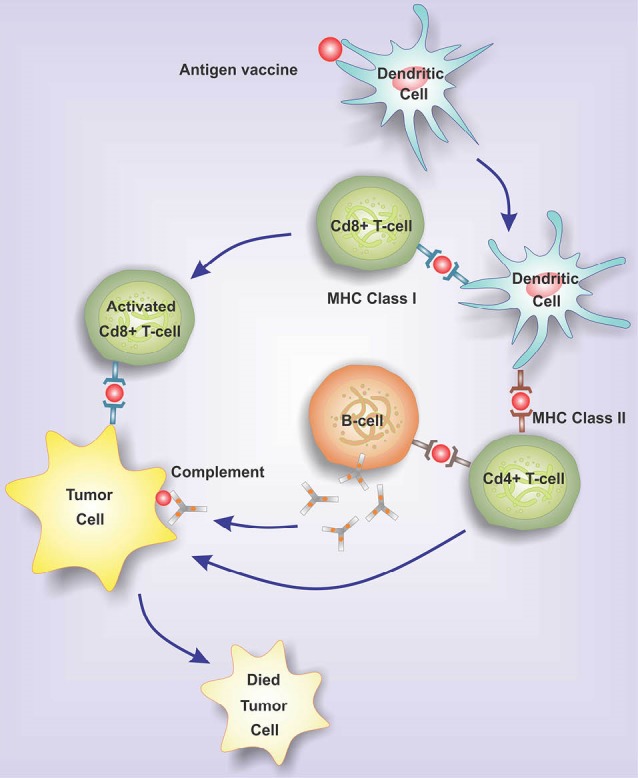
The migration of activated CD8+ T cells to tumor sites and lyse tumor cells. Once dendritic cells processed the tumor antigens and presented them to the lymphocytes, the CD8+ T cells migrate towards the tumor sites. Accordingly, the primed and activated immune system cells, CD8+ and/or CD4+ T cells, can detect the tumor Ags.
Tumor immunity can be activated through the interaction of vaccines with the tumor Ags in various platforms. The vaccine platform is generally designed based on the functions of B cells, T cells, or professional APC.21 These platforms may target Ags through peptide, protein or engineered plasmid DNA, or target cells such as DCs, autologous tumor cells or even tumor cell lysates derived from a patient. The active platforms under clinical development are peptide plus adjuvant,21 plasmid DNA,22 recombinant virus and bacteria,17 dendritic cell vaccines,23 tumor cell vaccines,24 heat-shock protein,25 and exosome-based vaccines.26 A detailed review of these platforms and their advantages and disadvantages have previously been described.17 In general, a vaccine can directly induce the T-cell immunity in 2 different ways, including genetical modification of tumor cells to express co-stimulatory molecules for the direct presentation of Ags, or modification of professional APC to express tumor Ags by gene transfer or direct loading of Ags.27
Vaccine design for breast cancer
Although the design and development of a vaccine for particular tumor cell Ags appear to be a straightforward approach, there are several potential challenges that make some limitations.28 For example, as a major issue, only low levels of an Ag may be expressed by the tumor cells even if the Ag associates with a specific kind of tumor cell. The Ags may be localized in a cryptic position or secured form. The antigenic profile of tumors may also be modified by growing the tumor or expression of the Ags by a fraction of the tumor cells. Furthermore, the MHC proteins may be expressed in a very low level by the tumor cells, and therefore, a more robust immune response is required to be generated.29 Another issue seems to be associated with the inaccessibility of cells deep within the solid tumors. Despite the existence of the above potential challenges in the immunotherapy of solid tumors, many attempts have been done to design and construct efficient vaccines for the treatment of breast cancer. Table 1 shows a summary of the constructed vaccines for the breast cancer therapy using different Ags expressed in the normal tissues and overexpressed or mutated in tumor cells. Table 2 shows the list of breast cancer vaccines that have been patented.
Table 1. List of antigens used in vaccine construction for breast cancer immunotherapy .
| Application | Tool | Method | Type | Description |
| Molecular docking | Autodock | Protein–ligand docking | Software | Predicts bound conformations and binding energies of ligands with macromolecular targets using a grid-based technique |
| Gold | Flexible ligand. Partial flexibility for protein | Software | Calculates the docking modes of small molecules in protein binding sites and docking of protein-ligand using an evolutionary-based algorithm for optimization of the result | |
| ZDOCK | Rigid-body protein-protein docking | Server, Software | Predicts structures of protein-protein complexes and symmetric multimers-based on the rigid-body docking | |
| RosettaDock | Local protein-protein docking | Server | Analyses protein–protein interaction using a multi-scale and multi-start technique based on Monte Carlo algorithm | |
| EPIDOCK | A molecular docking-based tool | Server | Predicts MHC binding peptides | |
| Structure prediction | MODELLER | Comparative Protein Structure Modeling | Software | Models protein 3D structure through homology or comparative modeling |
| Phyre2 | Protein homology/analogy recognition engine | Server | Predicts and analyzes protein structure, function and mutations | |
| I-TASSER | Iterative Threading ASSEmbly Refinement | Server | Predicts protein structure and function based on a hierarchical approach from sequence to structure to function paradigm | |
| SWISS-MODEL | Protein structure homology modelling | Server | Builds a protein model based on homology models at different levels of complexity | |
| Robetta | Full-chain protein structure prediction server | Server | Generates structural models through either comparative modeling or de novo structure prediction techniques | |
| Structure Evaluation | ERRAT | Verifies protein structure | Server | Assesses quality of a structure for nonbonded atomic interactions |
| Verify 3D | A protein model assessment based on its 3D profile | Server | Assesses the compatibility of a 3D model with its primary sequence | |
| ProCheck | Protein structure quality assessment based on stereochemical characteristics | Server | Investigates the backbone conformation using a Psi/Phi Ramachandran plot | |
| Molecular Modeling | CHARMM | Chemistry at Harvard Macromolecular Mechanics | Software | Simulates and analysis the molecular dynamics associated with widely used set of force fields |
| GROMACS | GROningen MAchine for Chemical Simulations | Software | Simulates the molecular dynamics of proteins, lipids and nucleic acids | |
| Amber | Assisted Model Building with Energy Refinement | Software | Simulates the molecular dynamics of biomolecules via a family of force fields |
Table 2. List of breast cancer vaccines registered as patents .
| Patent number | Owner | Antigen | Description | Date of Patent |
| US9370560 B2 | University of the Health Science | HER2/neu | This invention proposes a novel technique to induce and preserve an immune response to HER2/neu expressing tumor cells. E75 peptide induces defensive immunity in patients with the HLA-A2 and -A3 haplotype through associating with MHC HLA-A2 and A3. | Jun. 21, 2016 45 |
| US20100210714A1 | Pangenomics Co., LTD | HER2/neu | This invention presents the DNA vaccine with the capability of usage as a therapeutic vaccine to reduce metastasis after tumor surgery. It can be also used for persons with genetic high risk as a prophylactic vaccine. | Aug. 19, 2010 46 |
| US9114099B2 | University of the Health Science | HER2/neu | Breast cancer recurrence can be prevented by this proposed method which induces and maintains a protective cytotoxic T-lymphocyte response to a peptide of the HER/neu oncogene, GP2. In this method, an amount of vaccine containing a pharmaceutically acceptable carrier an adjuvant such as GM-CSF, and the GP2 peptide are prescribed to the patient. | Aug. 25, 201547 |
| US 7674456 B2 | St. Vincent Medical Center | HER2/neu | A new SV-BR cancer cell lines is proposed by this invention that further makes relation to therapeutic usage of the novel cell lines as cancer vaccines. | Mar. 9, 201048 |
| US 9125848 B2 | The Cleveland Clinic Foundation | α-Lactalbumin (LALBA) | An immune response is induced by this invented method against α-lactalbumin via leading a patient to an immunogenic composition, comprising an adjuvant; and a purified recombinant polypeptide including a human α-lactalbumin | Sep. 8, 2015 49 |
| US9327026 B2 | The Cleveland Clinic Foundation | α-Lactalbumin (LALBA) | The invented method is to prevent or treat breast tumors that express human αS1 casein, human α-lactalbumin, human κ-casein or human β-casein in a non-lactating human female of a non-child bearing age in where these genes are not expressed by normal breast tissue. | May. 3, 201650 |
| US5744144A | National Institutes Of Health | muc-1 | A synthetic peptide vaccine is invented for cancer. The peptide includes at least 2 20-amino acid tandem repeats of muc-1 and at least one foreign amino acid sequence corresponding to an epitope present on a cancer cell that does not express muc-1. | Apr. 28, 199851 |
| US20160101169 A1 | He Cleveland Clinic Foundation | α-Lactalbumin (LALBA) | A patient with non-lactating female human is immunized by this method against a human α-lactalbumin. The process of inducing an immune response against α-lactalbumin is comprised by the method by a step-by-step managing an immunogenic composition, the immunogenic composition comprising an adjuvant; and a polypeptide comprising a human α-lactalbumin sequence | Apr. 14, 201652 |
| US5660834A | The Biomembrane Institute | muc-1 | Monoclonal antibodies preparation and establishment are invented through the invented method to manage human cancer-associated mucin-type glycoprotein antigens. The method reduces the extra steps giving hybridomas with high quality. | Aug.26, 1997 53 |
| US6344203B1 | Austin Research Institute | muc-1 | A peptide is designed in this invention as a cancer vaccine including mimics MUC1 or other cancer peptides and one or more pharmaceutically acceptable carrier or diluent, | Feb.5, 2002 54 |
|
US5922836A |
Washington University |
Mamma globin |
The invention proposes a vaccine for treatment of breast cancers expressing mammaglobin. It consists of at least one B cell mammaglobin antigen, one TC mammaglobin antigen for inducing antibody and/or cell-mediated immune responses against mammaglobin-expressing tumors. | Jul.13, 1999 55 |
The aforementioned problems make the development of efficient vaccines very difficult. The essential problem is the self-antigenic action of tumor Ags, at which they may act moderately immunogenic. Several attempts have been done to overcome these difficulties and increase the response of the immune system by choosing proper Ags and effectively presenting them to the immune system. In fact, these Ags may be found in the regular cells, while they are mutated or overexpressed in the cancerous cells. The explored vaccine platforms used in the vaccination of breast cancer include (i) whole tumor cell vaccines (allogeneic, autologous), (ii) dendritic cell vaccines, (iii) recombinant protein vaccines, (iv) peptide vaccines, (v) DNA vaccines, and (vi) recombinant viral vectors. In the following part, we briefly review these platforms.
Autologous tumor cell-based vaccines
Autologous tumor cell-based vaccines (ATCVs) are based on the tumor cells lysate obtained from patients. Then, the vaccine is used to train the immune system to detect and kill the cancerous cells. The ATCVs consist of several known and unknown potential Ags, and therefore, they have a variety of epitopes giving the capability of replying to a highly different set of tumor cells.56 In addition, all the existing Ags within the ATCVs are extracted from the antigen repertoire of a patient. In summary, the major advantages of this kind of vaccines are their safety, multivalency, and patient specificity, at which it can be called as personalized vaccination. However, these vaccines have poor immunogenicity and production inconsistency.57
Allogeneic tumor cell-based vaccines
Allogeneic tumor cell-based vaccines are another type of vaccines that use the collected cell lines from a similar class of cells. This kind of vaccine is a cost-effective, reproducible and easily designed type of vaccines in comparison with ATCVs, in large part because of simplicity in mass production and storage. Furthermore, the allogeneic cell lines have the capability to contain one/two tumor-associated Ags for a specific tumor with a low complexity of tumor's Ag, inducing unfavorable selective pressure and promote tumor escape.57
Peptide-based vaccines
Peptide-based vaccines are another approach of the cancer immunotherapy, in which peptides are directly derived from the tumor-associated Ags (TAAs). These vaccines are specifically designed to associate with T cells in the presence of the MHC class I or II molecules. The most common TAA epitopes of the breast cancer are extracted from HER2/neu, MUC1, and CEA proteins. The TAA extracted peptides can be effectively utilized to stimulate the responses of CD4+ and CD8+ T cells. Hence, it is important to find appropriate peptides adapting to natural variations of human leukocyte antigens (HLAs). Comparing with other therapies and vaccines, the most attractive advantage of this approach is the specificity of the target response and its low expected toxicity. The peptide-based vaccines are easily manufactured, and can induce a fairly high-level of immunological response. The efficiency of peptide-based vaccines as neo-adjuvant immunotherapy has experimentally been proven in some experiments on NeuVax-E75 (epitope for HER2/neu and GM-CSF) and DPX-0907 (HLA-A2-TAAs) expressed in the breast, ovarian and prostate cancers.58 Peptides have important roles in the early diagnosis of breast cancer, and consequently, in decreased mortality.
Dendritic cell vaccines
Dendritic cells are principally the major regulators of the immune response. They are known as the major Ag-presenting cells for the preparation and activation of the immature T cells against tumor cells, at which many researchers have been motivated to develop several dendritic cell-based immunotherapies.59 The dendritic cell vaccines are constructed from the immune-stimulating white cells of patients – a type of personalized medicine. In other words, the cells are taken out of the patient's blood, activated in the lab away from the tumor influence, and then, injected back into the patient. It has previously proven that the immune cells have less ability to recognize and target HER2 as a most frequently overexpressed protein in the breast cancer (20-25%).29 Thus, the DC-based vaccines can be an effective solution to re-stimulate the immune system. This strategy has previously been developed by researchers in the form of HER2 targeted vaccine on the breast cancer cells. These kinds of vaccines seem to be well-tolerated, and by use of which patients might only experience trivial toxicity.
DNA vaccine
The DNA vaccines represent an attractive immunotherapeutic method for the cancer treatment regarding its simplicity, and stability. Norell et al60 developed a vaccine to target the HER2 Ag and to serve as an immunotherapy modality against breast cancer. In a primary clinical test, a DNA plasmid encoding full-length version of HER2 was used along with low doses of IL-2 and granulocyte-macrophage-colony-stimulating factor (GM-CSF) in an experimentally handled vaccine in the metastatic HER2-expressing breast cancer patients. In another preclinical attempt,61 the human mammaglobin-A (Mam-A/MGBA) cDNA encoding vaccine was engineered to activate the Mam-A-specific CD8 T-cell immune responses. Several clinical studies have proven that DNA vaccines can be safely used without concerning clinical toxicity or autoimmunity.62,63 DNA vaccines can also be designed for long-term cancer protection, in large part because of being very cost-effective. Although DNA vaccines display some advantages, the induction of powerful Ag-specific cellular immune responses against endogenous self-antigens of solid tumors remains as a major striking challenge.22
Viral vector-based vaccine
The viral vectors have also been used for the vaccination, in part due to their ability in an efficient delivery of genes. They show high in vitro and in vivo transgene expression and elicit low toxicity, yet are often immunogenic. They have been used to serve as Ag-specific vaccines for the activation of the immune response in the tumor microenvironment. Mam-A, as one of the most frequently overexpressed proteins in the primary and metastatic breast cancers in the human, has been exploited for the production of the viral vector-based vaccines. In a study, the Mam-A was inserted into a replication-deficient adenovirus vector, by which the dendritic cells prepared from the healthy female individuals were transduced resulting in the stimulation of CD8+ CTLs in vitro. The transfection with adenoviral vectors encoding Mam-A was shown to improve the maturation of DCs and secretion of IL-12.64
The impact of bioinformatics on vaccine design
The use of high-throughput methods in different experimental applications of biological studies such as genomics and proteomics have produced a variety of data that could not be handled and analyzed by the traditional methods. During the last years, techniques and applications of bioinformatics have played critical roles in the mining and understanding of large volumes of biological data. Furthermore, rapidly growing of computational methods along with the extensive volume of experimental data on the immunology has created a novel field of research entitled immunoinformatics. The field of immunoinformatics is a subdivision of bioinformatics focused on design and development of computational and mathematical models for the analysis of the immune systems by means of immunological data.65 The efficiency of developed vaccines against cancer can excitedly be increased through in-silico modeling and analysis of the cancer cells mechanisms by the immunoinformatics tools. We provide some key insights into the immunoinformatics applications in the following contexts.
Cancer vaccine modeling and design by computational approaches
The computational models can principally guide biologists from a quantitative data to a qualitative and thus predictive knowledge. In particular, the therapeutic effects at the organism level can be efficiently studied, predicted and optimized by the mathematical modeling of the immune system. These models can be used to simulate the behavior of the immune system in response to various pathogens and immunosurveillance of cancer. Furthermore, in-silico simulation of physiological and pathophysiological interactions at the cellular and molecular levels can be used to provide key information prior to laboratory-based experiments.
In computational biology, designing an effective vaccine for cancer therapy is one of the most exciting challenges regarding the highly complex system of tumors, where various interactions may occur, including events and conditions leading to the initiation, progression, invasion and metastasis of cancer cells. It seems that adaptive learning techniques might be useful for the development of the models at the hierarchy of organism, organ, cell, and molecule levels. The developed models simplify understanding of the general behavior of the system such as cellular and molecular interactions, the course of a disease and the effects of treatment through computational simulation of the system.
During the last couple of years, a number of models have been proposed for the computational modeling of the cancer vaccines. SimTriplex is an agent-based approach for the modeling of cancer vaccine, which has especially been designed for the simulation of triplex tumor-preventive cell vaccines and its effects on HER2 transgenic mice prone to the breast cancer development.66 MetastaSim is another model inspired by the SimTriplex model sharing a similar framework for modeling the biological mechanisms.67 Furthermore, several types of research have been done to investigate the response of the immune system against the cancer cells through computational modeling.68 To this end, a mathematical model 69 has been developed to simulate the dynamic growth of an immunogenic tumor in which an active immune response exists. The model especially focuses on the interactions between cancer cells and cytotoxic T cells and antigen presenting cells in small multicellular tumors. The above developed computational models provided the ability to design and establish efficient therapeutic strategies for the cancer treatment.
Besides the above tools for the computational modeling of cancer vaccines, a number of tools are available for the development of cancer vaccines. The tools can be used in a step-by-step in-silico procedure to design and construct a targeted vaccine. First, several analyses are done over the primary sequence of the target protein to investigate its structural and biochemical characteristics. Then, a set of tools are utilized to predict and select the appropriate B and T-cell epitopes using epitope databases. Finally, the selected epitopes are fused together via proper linkers to construct the final cancer vaccine sequence. The constructed vaccine sequence is submitted to a homology modeling software to build its 3D structure and validate its functional behavior.
B- and T-cell epitope prediction
An important research field in immunoinformatics is the development of algorithms for analyzing potential B- and T-cell epitopes. The algorithm has the ability to efficiently analyze the potential binding sites within the sequences of B- and T-cell epitopes instead of the laboratory analysis of pathogen gene products. The anti-tumor behavior of the immune system is potentially stimulated when a short deletion or insertion or substitution in DNA is discriminated as non-self. A mutation imposes novel peptide(s), which are presented by the MHC molecules on the surface of the cell and can be detected by T helper or cytotoxic T lymphocytes (CTL). Thus, it is required to look for a set of potentially immunogenic peptides, and then, analyze these sequences by the epitope prediction tools. Results of the analysis lead an immunologist to in silico design and development of new vaccines. This strategy is called reverse vaccinology, where the pathogen genome is analyzed to identify the potential antigenic proteins.70
Additionally, a major goal in the epitope prediction is to analyze the binding affinity of antigenic peptides to the MHC molecules. The most fruitful proposed methods for the prediction of T-cell epitopes have been designed based on data-driven approaches. The methods first look for the peptide-binding property of particular class I or class II MHC alleles, and then, in-silico prediction of epitopes is done. A variety of tools and techniques has been proposed for the epitope prediction and investigation, including (i) homology modeling, (ii) protein threading, (iii) docking techniques, and (iv) identification of structural binding motifs. Alternatively, the role of interactions between antigen and antibody in the human immune system can be studied by analyzing whole mutated antigen sequence. Conventional methods may face different issues and limitations, including the use of high cost and time-demanding procedures for the pathogen cultivation and subsequent protein extraction and testing.71 Taken all, computational tools provide the ability for the design of efficient vaccines by saving time and resources.
Epitope databases
Several publicly available databases have been established to record computationally and experimentally extracted data on T-cell and B-cell epitopes and also binders to the major MHC molecules. Currently, the largest databases in use are SYFPEITHI,69 MHCBN,72 and IEDB.73 Of these, IEDB is a well annotated and frequently maintained repository that provides wide-ranging tumor-related information and their experimental details. The SYFPEITHI is a continuously maintained database that is used in epitope prediction and binding motifs analysis and consists of constitutive MHC binders and T-cell epitopes collected from different resources. A number of databases have been established for the discovery of cancer vaccine target, including DFRMLI,74 CIG-DB,75 TANTIGEN,76 and CTDatabase.77 The DFRMLI is a database of immunological datasets from important databanks that is used for the machine learning applications for training and testing purposes. The CIG-DB classifies T-cell receptors and immunoglobulins for the cancer treatment and hematological tumors groups by mining, training, and clustering of literatures (the server is currently not available). The Peptide Database includes a list of manually organized T-cell defined tumor antigens as well as an integrated unique categorized library of overexpressed tumor antigens. The TANTIGEN provides a scheme for the classification of Ags and their annotation. It also provides detailed information on the T-cell epitopes and experimentally confirmed HLA ligands. The CTDatabase is also called "Cancer-Testis" and consists of only Ags from the last category. Furthermore, Bcipep78 and CED79 are databases for the B-cell epitopes, providing a great possibility for an expressive measurement of epitope immunoproperty. Except for the MHCBN and EPIMHC, all the aforementioned databases can be integrated with experimentally derived information collected from the literature. Fig. 3 shows schematic representation for the publicly available databases for the epitope prediction that are grouped into three categories, including B-cell, T-cell and HLA databases.
Fig. 3.
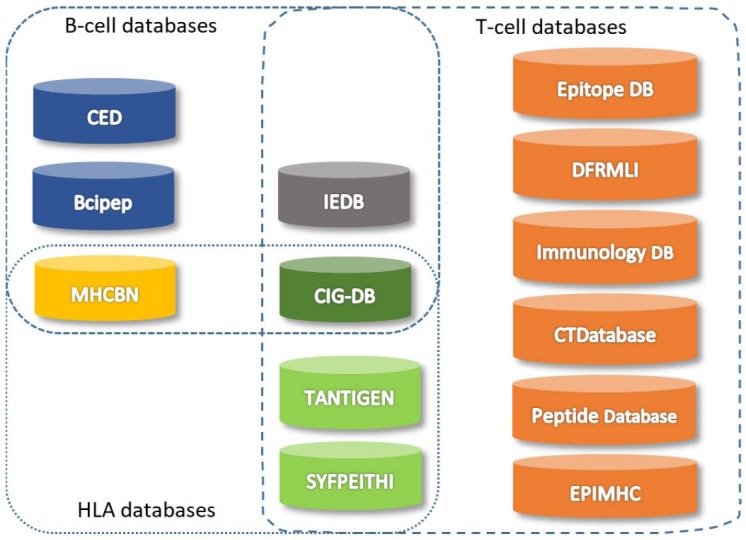
Databases for B-cell and T-cell epitope selection.
Molecular docking
Protein-protein interactions (PPIs) provide important underlying mechanisms of the biological systems.62,80 The structure and mechanism of protein-protein complexes provide enormously valuable knowledge for several biological applications especially for various groups of medicines, including chemotherapies, antimicrobial drugs and vaccine design. In silico molecular docking is known as a promising approach for the structural investigation and systematic analysis of PPI mechanism at the atomic level. A docking algorithm generally predicts the 3D-structure of PPIs through analyzing its molecular component structures. The algorithm uses various computational methods over the structural profile of proteins to decipher potentially suitable binders. Some methods have previously been developed to investigate the PPIs through molecular docking of proteins. The methods are available in the form of the standalone programs, including Autodock,81 Gold,82 ZDOCK,83 RosettaDock,84 and some others. Besides, some of the programs are also available via public Web servers that may display some limited options for the simulation of a single protein-protein docking, while they are not relevant for large-scale studies. Docking tools can be used to analyze the MHC molecules by investigating peptides. The EPIDOCK is a structure-based Web server that is broadly employed for the prediction of MHC binding peptides.85 The output of a docking tool is deemed to be very useful for the prediction and binding of a T-cell epitope. Consequently, for in-silico search for the possible antigenic determinant of the MHC molecule binding that can be used to successfully design and develop cancer vaccines.
Protein 3D structure prediction
Computational prediction of protein 3D structure is an essential and critical step in different bioinformatics studies. The methods generally work based on the fact that similar sequences are folded into similar 3D structures. Thus, they look at sequences with the known 3D structure for high identical sequences of the protein of interest. Then, a 3D model is constructed for the novel sequence using the structure of the most similar sequence as a template. Finally, the constructed model is refined to minimize its energy level and increase its quality.86
In the field of vaccine design and construction via multi-epitope peptides, it is required to predict an appropriate 3D structure for the designed vaccine sequence. To this end, the sequence can be submitted to online homology modeling web servers such as Phyre2,87 I-TASSER,88 and SWISS-MODEL,89 or use standalone programs such as MODELLER.90 The quality of the constructed model is evaluated using different online tools, including ERRAT,91 Verify 3D,92 and ProCheck.93
Molecular dynamics simulation
The computational process of molecular motions is called molecular dynamics. In molecular dynamics simulation, the physical movements of atoms and molecules are studied through a computational method. During the simulation, the interaction between atoms and molecules are modeled for a fixed period of time. The trajectories of atoms and molecules are simulated using Newton's equations for interacting particles motion within a system. Furthermore, the potential energies of particles and their interatomic forces are computed via molecular mechanics force fields. Such approaches were initially proposed by Fermi et al94 and Alder et al,95 and then, widely used in related fields of biomolecules, materials science and chemical physics.
Furthermore, in the field of vaccine design, it is highly important to minimize the energy level of the constructed vaccine and to exclude poor molecular contacts. In this line, the vaccine must be submitted to an MD simulation software such as GROningen MAchine for Chemical Simulations (GROMACS)96 or Chemistry at Harvard Macromolecular Mechanics (CHARMM)97 or Assisted Model Building with Energy Refinement (AMBER).98
Machine learning applications in immunoinformatics
The increasing growth and complexity of the generated biological data prove the importance of the design and development of intelligent computational methods for understanding and analyzing these data. Today, machine learning techniques play an important role in different applications of the computational biology. The techniques have been widely utilized in the development of different immunoinformatics tools for in-silico modeling, prediction, and simulation of the immune system mechanisms.99 Specifically, the machine learning methods can be efficiently used to simulate the classical experiments to select vaccine targets with a reduced time and cost.74 Accordingly, researchers have recently paid high attention to the value of machine learning approaches in immunology.
To date, machine learning techniques have been used in a variety of immunoinformatics applications including analysis of antibodies and their mechanisms,100 investigations of antigens,101 studies of allergenicity,102 classifications of immunological data103 and establishment of protocols for vaccine design.104 The design, development, and optimization of vaccines are facilitated and improved using these practical applications. The task is highly complex and involves with the selection of components for the formulation of vaccines through a significant number of experiments and in silico analysis. Machine learning methods have recently been given a privileged role in proper selection of targets and reduction in the number of required experiments over a large domain of possible combinations of components. Several machine learning techniques have been employed for development of immunoinformatics tools, including artificial neural networks (ANNs),105,106 support vector machines (SVMs),107 hidden Markov models (HMMs),108,109 and many others. The techniques give to computers the ability to learn without explicitly developing a computer program. They commonly construct a model based on a set of training samples as input observations in order to produce an output for data-driven prediction or decision making. These data-driven algorithms are relevant for a range of computing tasks which programming a set of strict instructions is infeasible or highly complex. The useful tools in immunoinformatics are summarized in Table 3.
Table 3. A summary of tools useful in immunoinformatics .
| Application | Tool | Method | Type | Description |
| Molecular docking | Autodock | Protein–ligand docking | Software | Predicts bound conformations and binding energies of ligands with macromolecular targets using a grid-based technique |
| Gold | Flexible ligand. Partial flexibility for protein | Software | Calculates the docking modes of small molecules in protein binding sites and docking of protein-ligand using an evolutionary-based algorithm for optimization of the result | |
| ZDOCK | Rigid-body protein-protein docking | Server, Software | Predicts structures of protein-protein complexes and symmetric multimers based on the rigid-body docking | |
| RosettaDock | Local protein–protein docking | Server | Analyses protein–protein interaction using a multi-scale and multi-start technique based on Monte Carlo algorithm | |
| EPIDOCK | A molecular docking-based tool | Server | Predicts MHC binding peptides | |
| Structure prediction | MODELLER | Comparative Protein Structure Modeling | Software | Models protein 3D structure through homology or comparative modeling |
| Phyre2 | Protein homology/analogy recognition engine | Server | Predicts and analyzes protein structure, function and mutations | |
| I-TASSER | Iterative Threading ASSEmbly Refinement | Server | Predicts protein structure and function based on a hierarchical approach from sequence to structure to function paradigm | |
| SWISS-MODEL | Protein structure homology modelling | Server | Builds a protein model based on homology models at different levels of complexity | |
| Robetta | Full-chain protein structure prediction server | Server | Generates structural models through either comparative modeling or de novo structure prediction techniques | |
| Structure Evaluation | ERRAT | Verifies protein structure | Server | Assesses quality of a structure for nonbonded atomic interactions |
| Verify 3D | A protein model assessment based on its 3D profile | Server | Assesses the compatibility of a 3D model with its primary sequence | |
| ProCheck | Protein structure quality assessment based on stereochemical characteristics | Server | Investigates the backbone conformation using a Psi/Phi Ramachandran plot | |
| Molecular Modeling | CHARMM | Chemistry at Harvard Macromolecular Mechanics | Software | Simulates and analysis the molecular dynamics associated with widely used set of force fields |
| GROMACS | GROningen MAchine for Chemical Simulations | Software | Simulates the molecular dynamics of proteins, lipids and nucleic acids | |
| Amber | Assisted Model Building with Energy Refinement | Software | Simulates the molecular dynamics of biomolecules via a family of force fields |
Conclusion
Computational and mathematical techniques have been widely used for modeling, designing, developing and analysis of the cancer vaccines. They are considered as important research tools in drug discovery for cancer immunotherapy. Today, genome databases increasingly accumulate a large volume of data, including critical data on the human immunologic mechanism. This has coined the field of immunoinformatics that deal with data collected from computational approaches and experimental immunology. The output of immunoinformatics studies provides foundations for the design and development of drugs and vaccines using in silico approaches.
The role of the immune system in controlling cancer cells has recently been annotated for the cancer treatment and prevention based on immunotherapy approaches. The developed computational models for simulation of the immune system processes give pivotal insights to understand the kinetics of cancerous cells, their biological pathways, and underlying mechanisms. Furthermore, the complex interactions between evolving pathogens and the host cellular immune system is characterized and investigated through the immunoinformatics tools. As a result, design and development of laboratory experiments is made highly easier without the uncertainty of systems.
Ethical approval
None to be declared.
Conflict of interest
Authors declare no conflict of interest.
Acknowledgment
The authors like to acknowledge the Research Center for Pharmaceutical Nanotechnology at Tabriz University of Medical Sciences and also the Department of Computer Science, Faculty of Mathematical Sciences, the University of Tabriz for their technical support. Also, the authors would like to thank the Iranian Elite National Foundation “Bonyad Melli Nokhbegan” and the Iranian National Science Foundation (INSF) for the financial support (Grant No: 95005404).
Review Highlights
What is current knowledge?
√ Immunotherapy is an effective strategy which has shown great promise to combat cancer malignancies.
√ Computational tools and bioinformatics in the field of immunotherapy of malignancies have emerged the new dominion of immunoinformatics.
What is new here?
√ The impacts of bioinformatics on different stages of vaccine design for the breast cancer have been reviewed.
√ Immunoinformatics tools can be used to produce efficient vaccines through a rationalized time- and cost-effective in silico approaches prior to conducting costly experiments.
References
- 1.Shibuya K, Mathers CD, Boschi-Pinto C, Lopez AD, Murray CJ. Global and regional estimates of cancer mortality and incidence by site: II Results for the global burden of disease 2000. BMC Cancer. 2002;2:37. doi: 10.1186/1471-2407-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anand P, Kunnumakkara AB, Sundaram C, Harikumar KB, Tharakan ST, Lai OS. et al. Cancer is a preventable disease that requires major lifestyle changes. Pharm Res. 2008;25:2097–116. doi: 10.1007/s11095-008-9661-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhoo-Pathy N, Yip C-H, Hartman M, Uiterwaal CS, Devi BC, Peeters PH. et al. Breast cancer research in Asia: adopt or adapt Western knowledge? Eur J Cancer. 2013;49:703–9. doi: 10.1016/j.ejca.2012.09.014. [DOI] [PubMed] [Google Scholar]
- 4.Chitrala KN, Yeguvapalli S. Computational screening and molecular dynamic simulation of breast cancer associated deleterious non-synonymous single nucleotide polymorphisms in TP53 gene. PLoS One. 2014;9:e104242. doi: 10.1371/journal.pone.0104242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weir HK, Thompson TD, Soman A, Moller B, Leadbetter S. The past, present, and future of cancer incidence in the United States: 1975 through 2020. Cancer. 2015;121:1827–37. doi: 10.1002/cncr.29258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiang CL, Kandalaft LE, Tanyi J, Hagemann AR, Motz GT, Svoronos N. et al. A dendritic cell vaccine pulsed with autologous hypochlorous acid-oxidized ovarian cancer lysate primes effective broad antitumor immunity: from bench to bedside. Clin Cancer Res. 2013;19:4801–15. doi: 10.1158/1078-0432.CCR-13-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poland GA, Kennedy RB, Ovsyannikova IG. Vaccinomics and personalized vaccinology: is science leading us toward a new path of directed vaccine development and discovery? PLoS Pathog. 2011;7:e1002344. doi: 10.1371/journal.ppat.1002344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim SW, Goedegebuure P, Gillanders WE. Mammaglobin-A is a target for breast cancer vaccination. Oncoimmunology. 2016;5:e1069940. doi: 10.1080/2162402X.2015.1069940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 10.Rajendran BK, Deng C-X. Characterization of potential driver mutations involved in human breast cancer by computational approaches. Oncotarget. 2017;8:50252. doi: 10.18632/oncotarget.17225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma XJ, Wang Z, Ryan PD, Isakoff SJ, Barmettler A, Fuller A. et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–16. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 12.Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J. et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–72. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 13.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M. et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 14.Kantoff PW, Higano CS, Shore ND, Berger ER, Small EJ, Penson DF. et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N Engl J Med. 2010;363:411–22. doi: 10.1056/NEJMoa1001294. [DOI] [PubMed] [Google Scholar]
- 15.Sun Y-S, Zhao Z, Yang Z-N, Xu F, Lu H-J, Zhu Z-Y. et al. Risk Factors and Preventions of Breast Cancer. International journal of biological sciences. 2017;13:1387. doi: 10.7150/ijbs.21635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rogozińska-Szczepka J, Utracka-Hutka B, Grzybowska E, Mąka B, Nowicka E, Smok-Ragankiewicz A. et al. BRCA1 and BRCA2 mutations as prognostic factors in bilateral breast cancer patients. Annals of oncology. 2004;15:1373–6. doi: 10.1093/annonc/mdh352. [DOI] [PubMed] [Google Scholar]
- 17.Emens LA. Cancer vaccines: on the threshold of success. Expert Opin Emerg Drugs. 2008;13:295–308. doi: 10.1517/14728214.13.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Germain RN, Margulies DH. The biochemistry and cell biology of antigen processing and presentation. Annu Rev Immunol. 1993;11:403–50. doi: 10.1146/annurev.iy.11.040193.002155. [DOI] [PubMed] [Google Scholar]
- 19.Huang AY, Golumbek P, Ahmadzadeh M, Jaffee E, Pardoll D, Levitsky H. Role of bone marrow-derived cells in presenting MHC class I-restricted tumor antigens. Science. 1994;264:961–5. doi: 10.1126/science.7513904. [DOI] [PubMed] [Google Scholar]
- 20.Boon T, Cerottini JC, Van den Eynde B, van der Bruggen P, Van Pel A. Tumor antigens recognized by T lymphocytes. Annu Rev Immunol. 1994;12:337–65. doi: 10.1146/annurev.iy.12.040194.002005. [DOI] [PubMed] [Google Scholar]
- 21.Greten TF, Jaffee EM. Cancer vaccines. J Clin Oncol. 1999;17:1047–60. doi: 10.1200/JCO.1999.17.3.1047. [DOI] [PubMed] [Google Scholar]
- 22.Greenland JR, Letvin NL. Chemical adjuvants for plasmid DNA vaccines. Vaccine. 2007;25:3731–41. doi: 10.1016/j.vaccine.2007.01.120. [DOI] [PubMed] [Google Scholar]
- 23.Osada T, Clay TM, Woo CY, Morse MA, Lyerly HK. Dendritic cell-based immunotherapy. Int Rev Immunol. 2006;25:377–413. doi: 10.1080/08830180600992456. [DOI] [PubMed] [Google Scholar]
- 24.Copier J, Dalgleish A. Overview of tumor cell-based vaccines. Int Rev Immunol. 2006;25:297–319. doi: 10.1080/08830180600992472. [DOI] [PubMed] [Google Scholar]
- 25.Binder RJ. Heat shock protein vaccines: from bench to bedside. Int Rev Immunol. 2006;25:353–75. doi: 10.1080/08830180600992480. [DOI] [PubMed] [Google Scholar]
- 26.Mignot G, Roux S, Thery C, Segura E, Zitvogel L. Prospects for exosomes in immunotherapy of cancer. J Cell Mol Med. 2006;10:376–88. doi: 10.1111/j.1582-4934.2006.tb00406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dranoff G, Jaffee E, Lazenby A, Golumbek P, Levitsky H, Brose K. et al. Vaccination with irradiated tumor cells engineered to secrete murine granulocyte-macrophage colony-stimulating factor stimulates potent, specific, and long-lasting anti-tumor immunity. Proc Natl Acad Sci U S A. 1993;90:3539–43. doi: 10.1073/pnas.90.8.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campoli M, Ferrone S, Zea AH, Rodriguez PC, Ochoa AC. Mechanisms of tumor evasion. Cancer Treat Res. 2005;123:61–88. doi: 10.1007/0-387-27545-2_3. [DOI] [PubMed] [Google Scholar]
- 29.Mittendorf EA, Peoples GE, Singletary SE. Breast cancer vaccines: promise for the future or pipe dream? Cancer. 2007;110:1677–86. doi: 10.1002/cncr.22978. [DOI] [PubMed] [Google Scholar]
- 30.Schneble EJ, Berry JS, Trappey FA, Clifton GT, Ponniah S, Mittendorf E. et al. The HER2 peptide nelipepimut-S (E75) vaccine (NeuVax) in breast cancer patients at risk for recurrence: correlation of immunologic data with clinical response. Immunotherapy. 2014;6:519–31. doi: 10.2217/imt.14.22. [DOI] [PubMed] [Google Scholar]
- 31.Clive KS, Tyler JA, Clifton GT, Holmes JP, Ponniah S, Peoples GE. et al. The GP2 peptide: a HER2/neu-based breast cancer vaccine. J Surg Oncol. 2012;105:452–8. doi: 10.1002/jso.21723. [DOI] [PubMed] [Google Scholar]
- 32. Peace KM, Mittendorf EA, Perez SA, Tzonis P, Pistamaltzian NF, Anastasopoulou EA, et al. Subgroup efficacy evaluation of the AE37 HER2 vaccine in breast cancer patients in the adjuvant setting. American Society of Clinical Oncology; 2017.
- 33.Karkada M, Berinstein NL, Mansour M. Therapeutic vaccines and cancer: focus on DPX-0907. Biologics. 2014;8:27–38. doi: 10.2147/BTT.S55196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gilewski T, Adluri S, Ragupathi G, Zhang S, Yao TJ, Panageas K. et al. Vaccination of high-risk breast cancer patients with mucin-1 (MUC1) keyhole limpet hemocyanin conjugate plus QS-21. Clin Cancer Res. 2000;6:1693–701. [PubMed] [Google Scholar]
- 35.Goydos JS, Elder E, Whiteside TL, Finn OJ, Lotze MT. A phase I trial of a synthetic mucin peptide vaccine Induction of specific immune reactivity in patients with adenocarcinoma. J Surg Res. 1996;63:298–304. doi: 10.1006/jsre.1996.0264. [DOI] [PubMed] [Google Scholar]
- 36.Mohebtash M, Tsang KY, Madan RA, Huen NY, Poole DJ, Jochems C. et al. A pilot study of MUC-1/CEA/TRICOM poxviral-based vaccine in patients with metastatic breast and ovarian cancer. Clin Cancer Res. 2011;17:7164–73. doi: 10.1158/1078-0432.CCR-11-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang XP, Yang DC, Elliott RL, Head JF. Vaccination with a mixed vaccine of autogenous and allogeneic breast cancer cells and tumor associated antigens CA15-3, CEA and CA125--results in immune and clinical responses in breast cancer patients. Cancer Biother Radiopharm. 2000;15:495–505. doi: 10.1089/cbr.2000.15.495. [DOI] [PubMed] [Google Scholar]
- 38.Domchek SM, Recio A, Mick R, Clark CE, Carpenter EL, Fox KR. et al. Telomerase-specific T-cell immunity in breast cancer: effect of vaccination on tumor immunosurveillance. Cancer Res. 2007;67:10546–55. doi: 10.1158/0008-5472.CAN-07-2765. [DOI] [PubMed] [Google Scholar]
- 39.Vonderheide RH, Domchek SM, Schultze JL, George DJ, Hoar KM, Chen DY. et al. Vaccination of cancer patients against telomerase induces functional antitumor CD8+ T lymphocytes. Clin Cancer Res. 2004;10:828–39. doi: 10.1158/1078-0432.ccr-0620-3. [DOI] [PubMed] [Google Scholar]
- 40.Miles D, Roche H, Martin M, Perren TJ, Cameron DA, Glaspy J. et al. Phase III multicenter clinical trial of the sialyl-TN (STn)-keyhole limpet hemocyanin (KLH) vaccine for metastatic breast cancer. Oncologist. 2011;16:1092–100. doi: 10.1634/theoncologist.2010-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Svane IM, Pedersen AE, Johnsen HE, Nielsen D, Kamby C, Gaarsdal E. et al. Vaccination with p53-peptide-pulsed dendritic cells, of patients with advanced breast cancer: report from a phase I study. Cancer Immunol Immunother. 2004;53:633–41. doi: 10.1007/s00262-003-0493-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tiriveedhi V, Tucker N, Herndon J, Li L, Sturmoski M, Ellis M. et al. Safety and preliminary evidence of biologic efficacy of a mammaglobin-a DNA vaccine in patients with stable metastatic breast cancer. Clin Cancer Res. 2014;20:5964–75. doi: 10.1158/1078-0432.CCR-14-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Makhoul I, Hutchins L, Emanuel PD, Pennisi A, Siegel E, Jousheghany F. et al. Moving a Carbohydrate Mimetic Peptide into the clinic. Hum Vaccin Immunother. 2015;11:37–44. doi: 10.4161/hv.34300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dillon PM, Petroni GR, Smolkin ME, Brenin DR, Chianese-Bullock KA, Smith KT. et al. A pilot study of the immunogenicity of a 9-peptide breast cancer vaccine plus poly-ICLC in early stage breast cancer. J Immunother Cancer. 2017;5:92. doi: 10.1186/s40425-017-0295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Peoples GE, Sathibalan P. Vaccine for the prevention of breast cancer relapse. Google Patents; 2012.
- 46. Lee JY, Kim D-H, Chung Y, Chang S-Y, Lee K-C, Kang C-Y. Her-2/neu DNA vaccine having anti-cancer activity. Google Patents; 2013.
- 47. Peoples G, Ponniah S. Vaccine for the prevention of breast cancer recurrence. Google Patents; 2017.
- 48. Wiseman C, Kharazi A. Breast cancer cell lines and uses thereof. Google Patents; 2010.
- 49.Tuohy VK, Jaini R, Johnson JM, Loya MG, Wilk D, Downs-Kelly E. et al. Targeted Vaccination against Human alpha-Lactalbumin for Immunotherapy and Primary Immunoprevention of Triple Negative Breast Cancer. Cancers (Basel) 2016;?:8. doi: 10.3390/cancers8060056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tuohy VK. Multivalent breast cancer vaccine. Google Patents; 2016.
- 51. Finn OJ, Fontenot JD, Montelaro RC. Synthetic multiple tandem repeat mucin and mucin-like peptides, and uses thereof. Google Patents; 1998.
- 52. Tuohy VK, Johnson JM, Jaini R. Breast Cancer Vaccine. Google Patents; 2016.
- 53. Kjeldsen TJ, Clausen H, Singhal A, Toyokuni T, Takahashi HK, Hakomori S-i. Monoclonal antibodies and vaccine development directed to human cancer-associated antigens by immunization with carbohydrate-carrier conjugates. Google Patents; 1997.
- 54. Sandrin MS, McKenzie IFC, Apostolopoulos V. Mimicking peptides in cancer therapy. Google Patents; 2002.
- 55. Watson MA, Fleming TP. Mammaglobin antigens. Google Patents; 1999.
- 56.Keenan BP, Jaffee EM. Whole cell vaccines--past progress and future strategies. Semin Oncol. 2012;39:276–86. doi: 10.1053/j.seminoncol.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kurtz SL, Ravindranathan S, Zaharoff DA. Current status of autologous breast tumor cell-based vaccines. Expert Rev Vaccines. 2014;13:1439–45. doi: 10.1586/14760584.2014.969714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peres Lde P, da Luz FA, Pultz Bdos A, Brigido PC, de Araujo RA, Goulart LR. et al. Peptide vaccines in breast cancer: The immunological basis for clinical response. Biotechnol Adv. 2015;33:1868–77. doi: 10.1016/j.biotechadv.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 59.Chiang CL, Coukos G, Kandalaft LE. Whole Tumor Antigen Vaccines: Where Are We? Vaccines (Basel) 2015;3:344–72. doi: 10.3390/vaccines3020344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Norell H, Poschke I, Charo J, Wei WZ, Erskine C, Piechocki MP. et al. Vaccination with a plasmid DNA encoding HER-2/neu together with low doses of GM-CSF and IL-2 in patients with metastatic breast carcinoma: a pilot clinical trial. J Transl Med. 2010;8:53. doi: 10.1186/1479-5876-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tiriveedhi V, Fleming TP, Goedegebuure PS, Naughton M, Ma C, Lockhart C. et al. Mammaglobin-A cDNA vaccination of breast cancer patients induces antigen-specific cytotoxic CD4+ICOShi T cells. Breast Cancer Res Treat. 2013;138:109–18. doi: 10.1007/s10549-012-2110-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kutzler MA, Weiner DB. DNA vaccines: ready for prime time? Nature Reviews Genetics. 2008;9:776. doi: 10.1038/nrg2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klinman DM, Klaschik S, Tross D, Shirota H, Steinhagen F. FDA guidance on prophylactic DNA vaccines: analysis and recommendations. Vaccine. 2010;28:2801–5. doi: 10.1016/j.vaccine.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Larocca C, Schlom J. Viral vector-based therapeutic cancer vaccines. Cancer J. 2011;17:359–71. doi: 10.1097/PPO.0b013e3182325e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brusic V, Petrovsky N. Immunoinformatics and its relevance to understanding human immune disease. Expert Rev Clin Immunol. 2005;1:145–57. doi: 10.1586/1744666X.1.1.145. [DOI] [PubMed] [Google Scholar]
- 66.Celada F, Seiden PE. A computer model of cellular interactions in the immune system. Immunol Today. 1992;13:56–62. doi: 10.1016/0167-5699(92)90135-T. [DOI] [PubMed] [Google Scholar]
- 67.Pappalardo F, Pennisi M, Castiglione F, Motta S. Vaccine protocols optimization: in silico experiences. Biotechnol Adv. 2010;28:82–93. doi: 10.1016/j.biotechadv.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 68.Pappalardo F, Palladini A, Pennisi M, Castiglione F, Motta S. Mathematical and computational models in tumor immunology. Mathematical Modelling of Natural Phenomena. 2012;7:186–203. [Google Scholar]
- 69.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–9. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 70.Tomar N, De RK. Immunoinformatics: an integrated scenario. Immunology. 2010;131:153–68. doi: 10.1111/j.1365-2567.2010.03330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sobolev BN, Olenina LV, Kolesanova EF, Poroikov VV, Archakov AI. Computer design of vaccines: approaches, software tools and informational resources. Current Computer-Aided Drug Design. 2005;1:207–22. [Google Scholar]
- 72.Lata S, Bhasin M, Raghava GP. MHCBN 40: A database of MHC/TAP binding peptides and T-cell epitopes. BMC Res Notes. 2009;2:61. doi: 10.1186/1756-0500-2-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Salimi N, Fleri W, Peters B, Sette A. The immune epitope database: a historical retrospective of the first decade. Immunology. 2012;137:117–23. doi: 10.1111/j.1365-2567.2012.03611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang GL, Lin HH, Keskin DB, Reinherz EL, Brusic V. Dana-Farber repository for machine learning in immunology. J Immunol Methods. 2011;374:18–25. doi: 10.1016/j.jim.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nakamura Y, Komiyama T, Furue M, Gojobori T, Akiyama Y. CIG-DB: the database for human or mouse immunoglobulin and T cell receptor genes available for cancer studies. BMC Bioinformatics. 2010;11:398. doi: 10.1186/1471-2105-11-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Olsen LR, Tongchusak S, Lin H, Reinherz EL, Brusic V, Zhang GL. TANTIGEN: a comprehensive database of tumor T cell antigens. Cancer Immunol Immunother. 2017;66:731–5. doi: 10.1007/s00262-017-1978-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Almeida LG, Sakabe NJ, deOliveira AR, Silva MC, Mundstein AS, Cohen T. et al. CTdatabase: a knowledge-base of high-throughput and curated data on cancer-testis antigens. Nucleic Acids Res. 2009;37:D816–9. doi: 10.1093/nar/gkn673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saha S, Bhasin M, Raghava GP. Bcipep: a database of B-cell epitopes. BMC Genomics. 2005;6:79. doi: 10.1186/1471-2164-6-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang J, Honda W. CED: a conformational epitope database. BMC Immunol. 2006;7:7. doi: 10.1186/1471-2172-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Jafar Razmara SD, Sepideh Parvizpour. A rapid protein structure alignment algorithm based on a text modeling technique. Bioinformation 2011; 6. [DOI] [PMC free article] [PubMed]
- 81.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS. et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–91. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved protein-ligand docking using GOLD. Proteins. 2003;52:609–23. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 83.Chen R, Li L, Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins. 2003;52:80–7. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- 84.Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA. et al. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol. 2003;331:281–99. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- 85.Sankar S, Nayanar SK, Balasubramanian S. Current trends in cancer vaccines--a bioinformatics perspective. Asian Pac J Cancer Prev. 2013;14:4041–7. doi: 10.7314/apjcp.2013.14.7.4041. [DOI] [PubMed] [Google Scholar]
- 86.Parvizpour S, Razmara J, Jomah AF, Shamsir MS, Illias RM. Structural prediction of a novel laminarinase from the psychrophilic Glaciozyma antarctica PI12 and its temperature adaptation analysis. J Mol Model. 2015;21:63. doi: 10.1007/s00894-015-2617-1. [DOI] [PubMed] [Google Scholar]
- 87.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–38. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–58. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc. 2009;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- 90.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY. et al. Comparative protein structure modeling using MODELLER. Curr Protoc Protein Sci. 2007;Chapter 2:Unit 2 9. doi: 10.1002/0471140864.ps0209s50. [DOI] [PubMed] [Google Scholar]
- 91.Colovos C, Yeates TO. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2:1511–9. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Luthy R, Bowie JU, Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–5. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 93.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–86. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 94. Fermi I, Pasta P, Ulam S, Tsingou M. Studies of the nonlinear problems: Los Alamos Scientific Lab., N. Mex.1955.
- 95.Alder BJ, Wainwright TE. Studies in molecular dynamics I General method. The Journal of Chemical Physics. 1959;31:459–66. [Google Scholar]
- 96.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–18. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 97.Brooks BR, Brooks CL, 3rd 3rd, Mackerell AD, Jr Jr, Nilsson L, Petrella RJ, Roux B. et al. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Shave SR. Development of high performance structure and ligand based virtual screening techniques. 2010.
- 99. Zhang Y, Rajapakse JC. Machine learning in bioinformatics: John Wiley & Sons; 2009.
- 100.David MP, Concepcion GP, Padlan EA. Using simple artificial intelligence methods for predicting amyloidogenesis in antibodies. BMC Bioinformatics. 2010;11:79. doi: 10.1186/1471-2105-11-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lafuente EM, Reche PA. Prediction of MHC-peptide binding: a systematic and comprehensive overview. Curr Pharm Des. 2009;15:3209–20. doi: 10.2174/138161209789105162. [DOI] [PubMed] [Google Scholar]
- 102.Muh HC, Tong JC, Tammi MT. AllerHunter: a SVM-pairwise system for assessment of allergenicity and allergic cross-reactivity in proteins. PLoS One. 2009;4:e5861. doi: 10.1371/journal.pone.0005861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hertz T, Yanover C. Identifying HLA supertypes by learning distance functions. Bioinformatics. 2007;23:e148–55. doi: 10.1093/Bioinformatics/btl324. [DOI] [PubMed] [Google Scholar]
- 104.Palladini A, Nicoletti G, Pappalardo F, Murgo A, Grosso V, Stivani V. et al. In silico modeling and in vivo efficacy of cancer-preventive vaccinations. Cancer Res. 2010;70:7755–63. doi: 10.1158/0008-5472.CAN-10-0701. [DOI] [PubMed] [Google Scholar]
- 105.Nielsen M, Lundegaard C, Worning P, Hvid CS, Lamberth K, Buus S. et al. Improved prediction of MHC class I and class II epitopes using a novel Gibbs sampling approach. Bioinformatics. 2004;20:1388–97. doi: 10.1093/bioinformatics/bth100. [DOI] [PubMed] [Google Scholar]
- 106.Larsen MV, Lundegaard C, Lamberth K, Buus S, Brunak S, Lund O. et al. An integrative approach to CTL epitope prediction: a combined algorithm integrating MHC class I binding, TAP transport efficiency, and proteasomal cleavage predictions. Eur J Immunol. 2005;35:2295–303. doi: 10.1002/eji.200425811. [DOI] [PubMed] [Google Scholar]
- 107.Wan J, Liu W, Xu Q, Ren Y, Flower DR, Li T. SVRMHC prediction server for MHC-binding peptides. BMC Bioinformatics. 2006;7:463. doi: 10.1186/1471-2105-7-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang C, Bickis MG, Wu FX, Kusalik AJ. Optimally-connected hidden markov models for predicting MHC-binding peptides. J Bioinform Comput Biol. 2006;4:959–80. doi: 10.1142/s0219720006002314. [DOI] [PubMed] [Google Scholar]
- 109.Noguchi H, Kato R, Hanai T, Matsubara Y, Honda H, Brusic V. et al. Hidden Markov model-based prediction of antigenic peptides that interact with MHC class II molecules. J Biosci Bioeng. 2002;94:264–70. doi: 10.1263/jbb.94.264. [DOI] [PubMed] [Google Scholar]


