Abstract
Background
The consumption of omega 3 polyunsaturated fatty acids (PUFAs) is important for human health and is closely associated with cell proliferation and differentiation. This study aimed to investigate the influence of omega 3 PUFAs on embryonic stem cell (ESC) proliferation and explore potential mechanisms that mediate these effects.
Methods
In this study, we isolated ESCs from fad3b-expressing transgenic mice. We detected the fatty-acid composition of ESCs using gas chromatography-mass spectroscopy, analyzed cell-cycle phases using flow cytometry, and detected gene expression using real-time polymerase chain reaction (PCR) and western blots.
Results
The amount of omega 3 PUFAs significantly increased in fad3b versus control ESCs. However, the growth of fad3b ESCs was slower than that of control cells, and most fad3b ESCs were in a prolonged G0/G1 phase after being passaged for 18 h. Therefore, we hypothesized that fad3b expression inhibited the cell cycle in ESCs by increasing the expression of P21, which then decreased the expression of cyclin-dependent kinase 4 (Cdk4). We found that pretreatment of fad3b ESCs with PD0325901, a P21 inhibitor, clearly attenuated the inhibitory effects of P21 on Cdk4, and resumed the cell cycle.
Conclusions
Expression of the fad3b gene in ESCs increased the omega 3 PUFA content, which inhibited cell proliferation by prolonging the G1 phase but did not arrest the G0-to-G1 or G1-to-S transitions. The prolonged G1 phase in fad3b ESCs was probably induced by downregulation of Cdk4 expression via p21 upregulation. These results suggest that accumulation of omega 3 PUFAs in vivo may beneficially affect ESC differentiation and that fad3b ESCs may be a useful tool for investigating related mechanisms.
Electronic supplementary material
The online version of this article (10.1186/s12944-018-0862-x) contains supplementary material, which is available to authorized users.
Keywords: Embryonic stem cells, fad3b, Cell cycle, P21, Cdk4
Background
Omega 3 polyunsaturated fatty acids (PUFAs) are believed to be closely involved in immune regulation, aging, anti-inflammatory activity, cell proliferation, and cell differentiation [1]. In mammals, omega 3 PUFAs must be sourced from dietary components due to a lack of fatty acid desaturases, which catalyze omega 6 fatty acids into omega 3 PUFAs [2]. However, transgenic expression of fatty acid desaturase genes could provide an alternative pathway for mammals to transform omega 6 to omega 3 PUFAs themselves.
Previous studies primarily used the Caenorhabditis elegans fat1 gene as a transgenic fatty acid desaturase [3–6]. Fad3b is an endoplasmic reticulum transmembrane protein that functions similarly to Fat1 [7] and is relatively suitable for expression in mammalian cells [8]. The primary omega 3 PUFAs are docosahexanoic acid (DHA) and eicosahexanoic acid (EPA). The mechanism that controls the effect of omega 3 PUFAs on cell-cycle regulation and physiological activity is not well characterized [9]. It is possible that variations in the concentrations of omega 3 PUFAs and in treatment times of the exogenous fatty acids resulted in the inconsistent results observed by different research groups [10]. For example, the addition of DHA to tumor cells arrested in G1 phase increased expression of p21 and decreased expression of cyclin D1 and cyclin E in one study [11], but decreased expression of the Cdk2 and cyclin E proteins and induced apoptosis in another study [12]. In endothelial cells, the addition of 17,18-epoxy-EPA decreased cell proliferation by down-regulating the cyclin D1/cyclin-dependent kinase (Cdk)-4 complex [13]. By contrast, EPA addition to leukemic k-562 cells promoted accumulation of G0/G1 cells and down-regulated cyclin E expression [14]. Interestingly, addition of both DHA and EPA to myoblast cells decreased cell growth and cell accumulation at G1 by decreasing expression of Cdk2 and cyclin E expression [15]. However, DHA addition in neural stem cells promoted cell-cycle progression, inhibited apoptosis, and induced neurogenesis [16].
The cell cycle and proliferation of ESCs is different than that of somatic cells in that ES cells have a short G1 phase and devote about half of their entire cycle to S phase [17]. In most cases, a prolonged G1 phase is associated with differentiation, but artificially extending the G1 phase by knocking down Cdk4/6 or by overexpressing the Cdk inhibitor p21 does not significantly affect ESC pluripotency [18].
In this study, we used a transgenic mouse model expressing the fad3b gene from flax (Linum usitatissimum) to investigate the mechanism of how endogenously expressed omega 3 PUFAs affect cell-cycle regulation in ESCs. We show that fad3b expression in ESCs increased the omega 3 PUFA content, and then induced a prolonged G1 phase by down-regulating Cdk4 expression via p21 upregulation.
Methods
Animals
The fad3b mice aged 6–8 weeks were obtained from the Research Center for Laboratory Animal Science Inner Mongolia University. All experimental mice were maintained in conventional animal housing with a 12 h light/dark photoperiod and free access to food and water. This study was carried out in strict accordance with the guidelines of Experimental Animal Management and Operation Standard of Inner Mongolia University.
Isolation and culture of fad3b ESCs
The blastocysts were collected at 3.5 days post coitum from the uterus of fad3b mice and inoculated onto 24-well plates with mouse embryonic fibroblast feeder cells. After 4–6 d, we selected well-shaped clones, digested these with 0.05% trypsin, and then transferred cells onto a new feeder layer [19]. The cells were cultured over 5–30 generations for subsequent cell identification experiments. To verify the regulatory role of fad3b in the cell cycle, we added 1 μM PD0325901 (Sigma, USA) to the culture medium after passage and collected the cells 18 h later.
Immunohistochemical analysis
The detected cells were washed three times with Dulbecco’s phosphate-buffered saline (DPBS), fixed for 30 min in 4% paraformaldehyde (PFA) at room temperature, then treated with 0.5% Triton X-100 for 10 min. Cells were incubated with primary antibodies against Oct4, Sox2, Nanog, and SSEA-1 (1:100, cell signaling technology, USA) at 4 °C overnight then incubated with fluorescence-tagged secondary antibodies (1:500, Santa Cruz, USA) at 37 °C for 2 h. After incubation, we stained with 4′,6-diamidino-2-phenylindole (DAPI) for 10 min after mounting and then observed and photographed samples under a confocal fluorescence microscope (A1R/A1, Nikon, Japan). For the alkaline phosphatase activity assay, we followed the instructions of the BCIP/NBT Alkaline Phosphatase Color Development Kit (Beyotime, China).
Teratoma preparation and analysis
The ESCs were digested, removed from feeder cells, and collected at 107 cells per 0.5 ml of phosphate-buffered saline (PBS) [19]. The resuspended ESCs were injected into the groins of nude mice. After 4–5 weeks, we removed the resultant tumors, performed paraffin sectioning and hematoxylin and eosin staining, and examined the sections under the microscope (A1, Carl Zeiss, German).
Fatty acid methyl ester analysis
The ESCs were collected in 15 ml tubes, washed twice with DPBS, and then 1 ml methanol solution containing 2.5% H2SO4 was added. Samples were then placed in an 80 °C water bath for 90 min. After cooling to room temperature, we added 1.5 ml of 0.9% NaCl to the tubes, mixed well, vortexed for 5 min, and centrifuged for 5 min at 2000 rpm/min to extract fatty acids into the organic phase. The supernatant was then transferred to new 1.5 ml tubes. We added 0.4 ml saturated KOH methanol solution, mixed well, vortexed for 5 min, and centrifuged for 10 min at 2000 rpm/min. The supernatant was collected in a sample bottle, and we analyzed the fatty-acid content using gas chromatography-mass spectroscopy (GC-MS) (GC-MS 2010 plus, Shimadzu, Japan). The fatty-acid levels are reported as percentage of total content.
Detection of the cell-cycle phase using flow cytometry
The ESCs were collected in centrifuge tubes, resuspended with propidium iodide (PI) staining solution (10 mg/ml RNase 10 μl, 1 mg/ml PI 50 μl, Triton X-100 2.5 μl, DPBS 940 μl), and mixed gently and evenly. After 15 min, we performed flow cytometry (FACSAria II, BD Company, USA) to detect the cell-cycle phase using a 488 nm laser. At least 1 × 104 cells were recorded. BD FACSDiva software was used to analyze the percentage of cells in G0/G1, S, and G2/M phases based on DNA content [20].
Calculations of cell growth rate and doubling time
We inoculated 1×105 ESCs into each well of a 6-well plate. After 12 h, we digested the ESCs with 0.05% trypsin and then counted cells. We repeated the digestion and cell count every 6 h until the 96-h endpoint. Then, we drew ESC growth curves and calculated the doubling time of the cells according to the formula: DT = t × [ln2/ (lnNt-lnNo)], where T is the culture time, No is the initial number of cells, and Nt is the number of cells at the time t.
Real-time PCR
The RNA from ESCs was isolated using the RNAiso Plus kit (Takara 9108) and reverse transcribed into cDNA using an Omniscript RT system (Takara RR047A) and oligo-dT primers. We amplified cDNA using an ABI7500 real-time PCR system (Applied Biosystems, America) and SYBR Green (Takara RR820A). The PCR primers are listed in Additional file 1: Table S1. The protocol for PCR amplifications was as follows: 95 °C for 30 s initially, followed by 40 cycles at 95 °C for 5 s, 60 °C for 34 s, and a final melt curve stage.The cycle threshold (Ct) values of targeted genes were normalized to the housekeeping gene Gapdh using the 2-ΔΔCt method [21].
Western blotting
The collected ESCs were lysed on ice using the Mammalian Protein Extraction Reagent (CWBiotech, China). The protein concentration was determined using Varioskan Flash (Thermo Fisher Scientific, USA). We boiled the lysate supernatant for 10 min, separated samples on a 12% sodium dodecyl sulfate (SDS)-polyacrylamide gel, and transferred proteins to a polyvinylidene fluoride (PVDF) membrane (Millipore, USA). The membrane was blocked in 5% skim milk in Tris-buffered saline with Tween (TBST) for 1 h and then incubated with antibodies against Fad3b (1:50000, ABclonal, China),P21, Cdk4, Cdk6, and α-tubulin (1:500, Abcam, USA) overnight at 4 °C. The membrane was then incubated with donkey peroxidase-conjugated goat anti-rabbit immunoglobulin G (IgG) antibody (diluted 1:5000; Jackson Immunoresearch, USA) for 3 h at room temperature before washing with TBST. Signal was detected using a Tanon-5200 chemiluminescence detector (YuanPingHao Biotech, China).
Calculations and statistical analyses
The data are expressed as means ± standard error of the mean of at least three independent experimental repeats. In graphs, all bars represent means and error bars represent one standard deviation. The statistical analyses were performed by Welch’s two-tailed t-test when comparing two groups with unequal standard deviations. When comparing multiple groups, a one-way analysis of variance test without replication was performed followed by post hoc analysis using Bonferroni correction to adjust for multiple comparisons. We considered *p < 0.05 and **p < 0.01 to be statistically significant.
Results
Establishment of fad3b ESCs
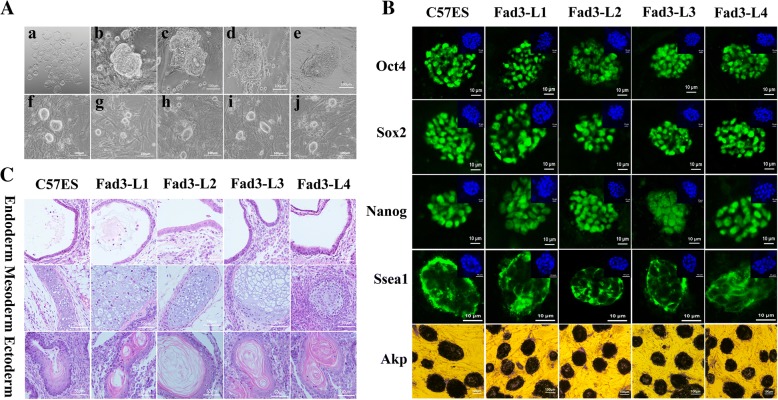
We collected a total of 29 blastocysts (Fig. 1aa) with normal morphology from five fad3b transgenic mice. From these blastocysts, we obtained four fad3b ESC lines and defined these as the Fad3-L1 to L4 lines (Fig. 1ab-e). The lab-kept C57 ESC line was used as a control for subsequent experiments (Fig. 1af). Compared with the control, the fad3b ESC lines had characteristics consistent with those of undifferentiated mouse ESCs and had no obvious morphological difference from control cells (Fig. 1ag-h). The fad3b ESCs both expressed the key markers of ESCs (Fig. 1b) and were able to differentiate into three germ-layer structures in vivo (Fig. 1c).
Fig. 1.
Establishment and identification of fad3b-expressing mouse embryonic stem cells (ESCs) lines. a. The establishment of fad3b mouse ESCs. a: fad3b blastocysts; b-e: primary cells grown from fad3b blastocysts; f: C57 ESCs; g-h: fad3b ESCs. b. Expression of Oct4, Sox2, Nanog, Ssea1, and AKP in fad3b-expressing mouse ESCs. c. Teratoma staining of in vivo germ-layer differentiation in fad3b mouse ESCs
Expression fad3b altered fatty-acid metabolism and the composition of ESCs
The expression of fad3b in the Fad3-L1 to L4 cell lines was relatively stable. However, the expression of fad3b mRNA was not consistent across the four cell lines, the fad3b mRNA expression in Fad3-L1 was less than that in L2, L3, and L4 (Fig. 2a). The fad3b protein expression was showed in Fig. 2b. The expression of lipid metabolism-related genes Lpl, Fabp4, Pnpla2, PPARα, and Cebpα at the 10th to 30th passages in the fad3b ESCs were upregulated (Fig. 3a).
Fig. 2.
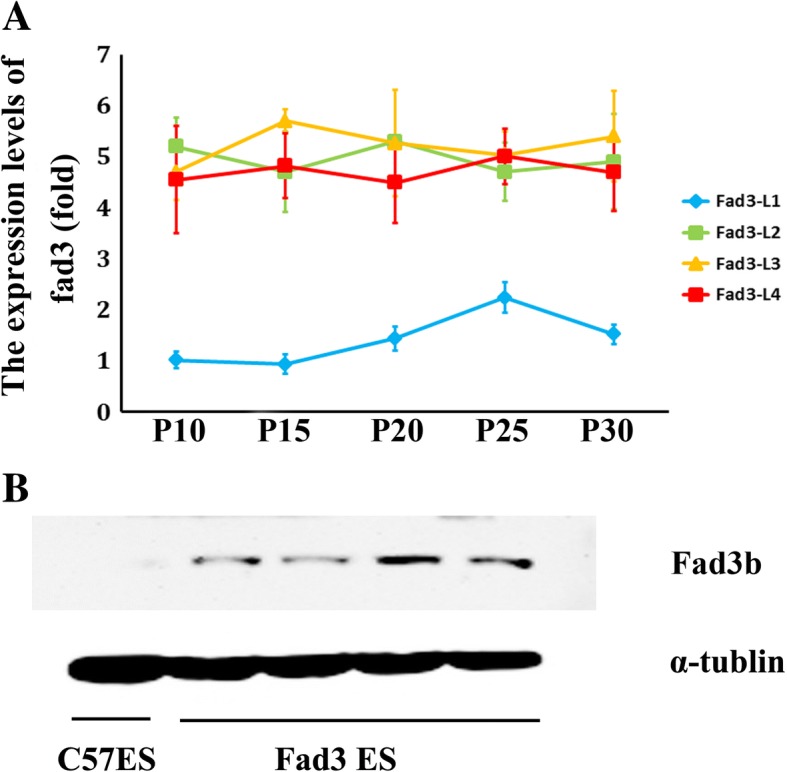
The fad3b expressing in ESCs. a. Stable fad3b mRNA expression in Fad3-L1 to L4 cell lines. b. Fad3b protein expression in ESCs
Fig. 3.
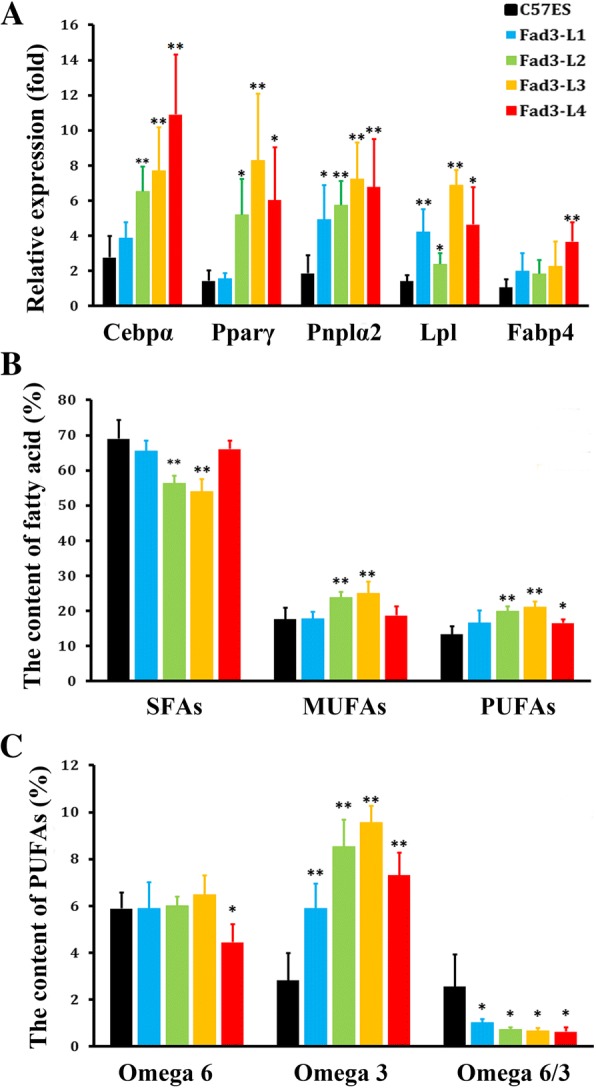
The ESCs expressing fad3b had fatty acid desaturase activity. a. The expression of lipid metabolism genes in fad3b ESCs. b. Saturated fatty acid (SFA), monounsaturated fatty acid (MUFA), and polyunsaturated fatty acid (PUFA) content in fad3b ESCs and in control cells. c. Levels of omega 6 PUFAs and omega 3 PUFAs in fad3b ESCs and control cells
The fatty-acid content in the fad3b ESCs was changed (Tab.S2), along with the average levels of saturated fatty acids (SFAs), monounsaturated fatty acids (MUFAs), and PUFAs at 10th to 30th passages, as shown in Fig. 3b and c. The SFA content was lower in fad3b ESCs, especially in L2 and L3 (56.39 ± 2.03% and 53.93 ± 3.48%, respectively, p < 0.01). The MUFAs content was higher in the fad3b ESCs, especially L2 and L3 (23.77 ± 1.64% and 25.02 ± 3.30%, respectively, p < 0.01) than in control cells (17.61 ± 3.30%). The PUFA content was higher in the fad3b ESCs, especially in L2, L3, and L4 (19.22 ± 1.31%, 21.06 ± 1.69%, and 16.61 ± 1.08%, respectively, p < 0.01or 0.05) than in control cells (13.34 ± 2.21%). The specific analysis of PUFA composition found that the omega 6 PUFAs in the fad3b ESCs were almost the same as in controls, except for the L4 line. However, the omega 3 PUFAs content was significantly higher in L1 to L4 cell lines (5.90 ± 1.04%, 8.52 ± 1.16%, 9.55 ± 0.71%, and 7.30 ± 0.97%, respectively, p < 0.01) than in the controls (2.83 ± 1.17%). The ratio of omega 6 to omega 3 fatty acids was significantly lower in L1 to L4 cell lines (1.01 ± 0.16%, 0.72 ± 0.08%, 0.68 ± 0.11%, and 0.64 ± 0.18%, respectively, p < 0.05) than in controls (2.56 ± 1.35%). Thus, expression of fad3b altered the fatty-acid composition in the ESCs by increasing the proportion of MUFAs and PUFAs, particularly that of omega 3 PUFAs.
Fad3b expression prolonged ESCs in G0/G1 phase by regulating p21 and Cdk4
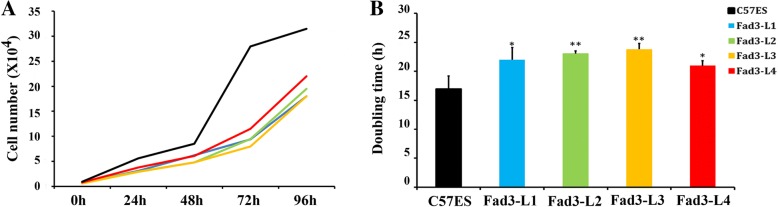
The cell-cycle transition of undifferentiated mouse ES cells is dependent on the cell line and cultivation conditions [22]. As showed in Fig. 4, the growth rate of fad3b ESCs was slower down than that of the control cells (Fig. 4a). Our calculation of cell doubling times showed that the control cell (C57 ESCs) doubling time was 16.99 ± 2.22 h, whereas the doubling times of Fad3-L1 to L4 cells were 21.98 ± 2.13 h, 23.07 ± 0.46 h, 23.80 ± 0.98 h, and 20.97 ± 0.87 h, respectively (Fig. 4b). The doubling times of the fad3b ESCs were significantly longer than that of the controls, suggesting that fad3b expression affected cell growth and prolonged the cell cycle.
Fig. 4.
The growth rate of fad3b ESCs decreased significantly. a. The growth curve of fad3b ESCs. b. The doubling time of fad3b ESCs
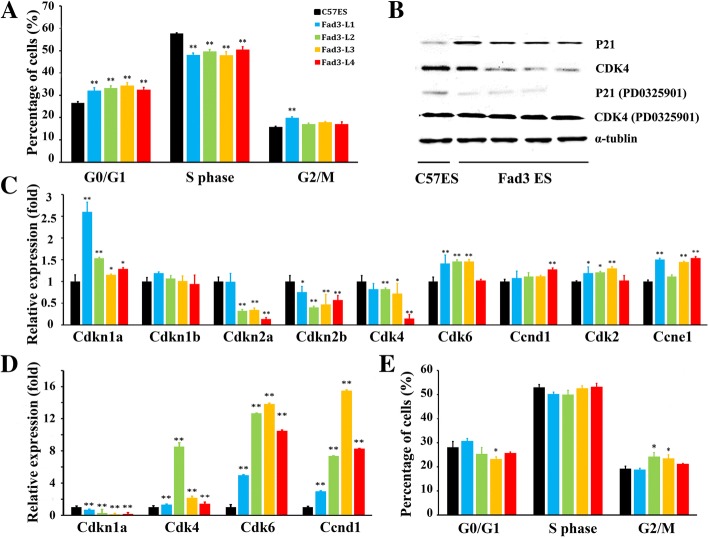
To further study the prolonged cell cycle induced by fad3b expression, we next detected the cell-cycle distributions of fad3b ESCs. The results showed that 18 h after the 15th passage, only 26.57 ± 0.56% of the control cells were in the G0/G1 phase, whereas significantly more fad3b ESCs (L1: 32.05 ± 0.56%, L2: 33.27 ± 1.33%, L3: 34.24 ± 0.95%, and L4: 32.47 ± 1.05%) were in the G0-G1 phase (all p < 0.01) (Fig. 5a). At the same time, 57.67 ± 0.29% of the C57 ESCs were in S phase, which was significantly higher than that of the fad3b ESCs (L1: 48.06 ± 0.85%, L2: 49.60 ± 0.93%, L3: 47.89 ± 1.55%, and L4: 53.50 ± 1.29%; all p < 0.01). At 18 h after passaging, only the Fad3-L1 had more M phase cells than control cells (19.89 ± 0.48% vs. 15.76 ± 0.35%, p < 0.01), and there was no significant difference for the other fad3b ESC lines. From the cell-cycle analysis, we observed that the fad3b ESCs were arrested or prolonged in the G0/G1 phase, and this is perhaps the main reason for their growth slowdown.
Fig. 5.
The expression of fad3b in ESCs caused G0/G1 arrest by regulating P21-Cdk4. a. Distribution of cell-cycle phases in fad3b ESCs and control cells. b. Western blot showing protein expression of cell-cycle-related proteins. c. The mRNA expression of cell-cycle-related genes in fad3b ESCs and control cells. d. The mRNA expression of cell-cycle-related genes after treatment with PD0325901 in fad3b ESCs and control cells. e. Distribution of cell-cycle phases after PD0325901 treatment in fad3b ESCs
According to the mRNA expression data, the expression of cyclin D1 in the fad3b ESCs was not significantly different from that of the controls (Fig. 5c). However, the expression of Cdk6, which has an important role during the G0-to-G1 transition [23], was significantly upregulated and that of Cdk4 was significantly down-regulated (Fig. 5c). The reduction of Cdk4 expression might elongate the G1 phase [24, 25]. The expression of Cdk2 and cyclin E was significantly upregulated, which has been associated with G1-to-S transition [26] (Fig. 5c).
Further analysis of the expression of checkpoint inhibition factors revealed that the expression of p21 (Cdkn1a) was significantly upregulated, the expression of p15 (Cdkn2b) and p16 (Cdkn2a) were significantly down-regulated, and the expression of p27 (Cdkn1b) did not change significantly (Fig. 5c). Downregulation of p15 and p16 does not inhibit the cell cycle [27]. The western blot results indicated that the protein expression of p21 in the fad3b ESCs was higher than that of the control cells and that of Cdk4 was lower (Fig. 5b). After addition of 1 μM PD0325901, an inhibitor of p21 expression, to the cell culture medium [28], the expression of p21 reduced, whereas the expression of Cdk4 increased (Fig. 5d and b), and the cell-cycle distributions of fad3b ESCs became similar to that of controls (Fig. 5e).
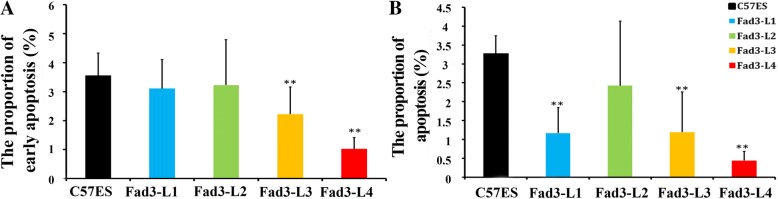
Because apoptosis is also an important factor that affects cell proliferation, we analyzed apoptosis using flow cytometry. The proportion of cells in early apoptosis and late apoptosis in fad3b ESCs was slightly lower than that of control cells (Fig. 6a, b). Therefore, apoptosis was not a factor that affected the proliferation of fad3b ESCs.
Fig. 6.
The expression of fad3b in ESCs had not effect on apoptosis. a. Early apoptosis frequency in fad3b ESCs and control cells. b. Apoptosis frequency in fad3b ESCs and control cells
These results show that the slowed cell proliferation in the fad3b ESCs was due to a prolonged G1 phase, which was probably due to p21-Cdk4, but not cyclin D-Cdk6 and cyclin E-Cdk2. Overall, our findings suggest that fad3b prolongs the cell cycle in the G1 phase but does not totally inhibit the G0-to-G1 or G1-to-S transition or apoptosis.
Discussion
In this study, we obtained fad3b-expressing ESCs from blastocysts of fad3b transgenic mice and found no obvious differences in morphology or pluripotency between the fad3b ESCs and control C57 ESCs [29]. The expression of fad3b was stable in fad3b ESCs but was different between fad3b ESC lines. This difference may be caused by the copy numbers of fad3b integrated into the genome; the transgenic mice used in the experiment had three to four copies of fad3b [30]. Our analysis showed that the omega 3 PUFA content of the fad3b ESCs was significantly higher than that in controls. In addition, with prolonged in vitro culture time, the omega 6 to omega 3 ratios of the control cells increased, but there was no such change in the fad3b ESCs, indicating that the fatty-acid desaturation activity of fad3b is very stable in the mice ESCs.
The expression of fad3b significantly affected the proliferation of fad3b ESCs. This change was likely mediated by the effect of omega 3 PUFAs on the cell cycle and the subsequent arrest of cells in the G0/G1 phases. ESC proliferation and cell-cycle properties are different from those of somatic cells, in that ES cells have short G1 phases and a very long S phase [31]. The G1 phase of the ESC cell cycle has been correlated with lineage specifications [18, 32, 33]. However, studies by Gonzales et al. have shown that upregulation of differentiation markers was inhibited upon knockdown of S and G2 phase progression genes but not upon prolonging of G1 phase [18].
The restriction (R) point divides the G1 phase of the cell cycle into the early phase and late phase. The transition from early to late G1 is primarily regulated by the cyclin D-Cdk4/Cdk6 complexes, and the late G1 phase is characterized by cyclin E-Cdk2 complex activity [26]. In most cases, a prolonged G1 phase is associated with differentiation, but artificially extending the G1 phase by knocking down Cdk4/6 or overexpressing p21 did not significantly affect ESC pluripotency [18]. Mouse ESCs have constitutive cyclin E-Cdk2 activity [26, 34] and low level cyclin D-Cdk4/Cdk6 activity [17]. Cdk4 and Cdk6 have similar structure and functions in the G1 transition [35]. Cdk6 has an important role during the G0-to-G1 transition [23], and overexpression of cyclin D1 and Cdk4 shortens the G1 phase [24, 25]. A shorter G1 phase and increased G0-to-G1 transition is the functional secession between Cdk4 and Cdk6. We found that Cdk4 not Cdk6 is responsible for the elongation of G1 phase in fad3b ESCs.
Cdk inhibitors, including INK (which inhibits p15, p16, and p18) and CIP (which inhibits p21, p27, and p57), modulate the activity of Cdk complexes. We found that the increased p21 expression in our fad3b ESC lines was key for G0/G1 arrest. Notably, p21 inhibits cyclin D-Cdk4 activity via c-Jun N-terminal kinases (JNKs) [36–38]. Cdk4 activation is required for phosphorylation of the T172 of Cdk4 [39], a residue that is not present in Cdk6 and Cdk1/2. Thus, the binding of Cyclin D1-Cdk4 to p21could be activated by JNKs via S130 phosphorylation of p21 and T172 phosphorylation of Cdk4 [38]. In cancer cells and tissue stem cells, omega 3 PUFAs increased p21 expression to induce cell-cycle arrest and apoptosis or differentiation [11, 40]. However, we observed that fad3b ESCs have reduced apoptosis and remain in an undifferentiated state, indicating that the role of omega 3 PUFAs is different in ESCs. p21 is an important downstream target of the Akt2 [41], p53 [42], and Erk [43], and the specific pathway that regulates omega 3 and p21 is still debated.
In summary, we found that fad3b expression in mouse ESCs increased the level of omega 3 PUFAs without affecting ESC morphology and pluripotency. However, fad3b expression inhibited cell proliferation such that cells were arrested in the G0/G1 phase due to p21-induced reduction of Cdk4. Understanding the mechanisms of this effect could be helpful for developing new methods of creating pluripotent cells or for differentiating cells that are suitable for clinical applications.
Conclusions
We found that expression of the fad3b gene in ESCs increased omega 3 PUFA content. Additionally, the proliferation of fad3b-expressing ESCs was inhibited, and a significantly higher proportion of fad3b ESCs were arrested in G0/G1 than control cells. The inhibition of cell proliferation was due to a prolonged G1 phase, but there was no inhibition of the G0-to-G1 transition or G1-to-S transition. The prolonged G1 phase in fad3b cells was probably induced by downregulation of Cdk4 expression via p21 upregulation. Therefore, omega 3 PUFAs may have unusual proliferative properties in undifferentiated ESCs.
Additional file
Table S1. The real time PCR primers. Table S2. The content of fatty acids in the cells. (DOCX 22 kb)
Acknowledgements
This work was supported by the National Major Project for Production of Transgenic Breeding (No. 2016ZX08007-002).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- Ccnd1
G1/S-specific cyclin-D1
- Ccne1
G1/S-specific cyclin-E1
- Cdk2
Cyclin-dependent kinase 2
- Cdk4
Cyclin-dependent kinase 4
- Cdk6
Cyclin-dependent kinase 6
- Cdkn1a
Cyclin-dependent kinase inhibitor 1A
- Cdkn1b
Cyclin-dependent kinase inhibitor 1B
- Cdkn2a
Cyclin-dependent kinase inhibitor 2A
- Cdkn2b
Cyclin-dependent kinase inhibitor 2B
- Cebpα
CCAAT/enhancer-binding protein alpha
- DAPI
4′,6-diamidino-2-phenylindole
- DHA
Docosahexanoic acid
- DPBS
Dulbecco’s phosphate-buffered saline
- EPA
Eicosahexanoic acid
- ESCs
Embryonic stem cells
- Fabp4
Fatty acid-binding protein 4
- JNKs
c-Jun N-terminal kinases
- Lpl
Lipoprotein lipase
- MUFAs
Monounsaturated fatty acids
- PFA
Paraformaldehyde
- Pnpla2
Patatin-like phospholipase domain-containing protein 2
- Pparγ
Peroxisome proliferator-activated receptor γ
- PUFAs
Polyunsaturated fatty acids
- PVDF
Polyvinylidene fluoride
- SDS
Sodium dodecyl sulfate
- SFAs
Saturated fatty acids
- TBST
Tris buffered saline with Tween
Authors’ contributions
ZW and GL conceived and designed the experiments; DL and LZ performed the experiments; ZW and CC analyzed the data, CB contributed reagents/materials; LY provide technical support for transgenic mouse experiment. ZW and GL drafted the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhuying Wei, Email: weizhuying2008@126.com.
Dongfang Li, Email: ndlidongfang@163.com.
Lin Zhu, Email: 1654012459@qq.com.
Lei Yang, Email: mrknowall@126.com.
Chen Chen, Email: chencheness@163.com.
Chunling Bai, Email: chunling1980_0@163.com.
Guangpeng Li, Phone: 86-10-471-5298583, Email: gpengli@imu.edu.cn.
References
- 1.Das UN. Essential fatty acids and their metabolites as modulators of stem cell biology with reference to inflammation, cancer, and metastasis. Cancer Metastasis Rev. 2011;30:311–324. doi: 10.1007/s10555-011-9316-x. [DOI] [PubMed] [Google Scholar]
- 2.Wallis JG, Watts JL, Browse J. Polyunsaturated fatty acid synthesis: what will they think of next? Trends Biochem Sci. 2002;27:467. doi: 10.1016/S0968-0004(02)02168-0. [DOI] [PubMed] [Google Scholar]
- 3.Kang JX, Wang J, Wu L, Kang ZB. Transgenic mice: fat-1 mice convert n-6 to n-3 fatty acids. Nature. 2004;427:504. doi: 10.1038/427504a. [DOI] [PubMed] [Google Scholar]
- 4.Lai L, Kang JX, Li R, Wang J, Witt WT, Yong HY, Hao Y, Wax DM, Murphy CN, Rieke A, et al. Generation of cloned transgenic pigs rich in omega-3 fatty acids. Nat Biotechnol. 2006;24:435–436. doi: 10.1038/nbt1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu X, Ouyang H, Duan B, Pang D, Zhang L, Yuan T, Xue L, Ni D, Cheng L, Dong S, et al. Production of cloned transgenic cow expressing omega-3 fatty acids. Transgenic Res. 2012;21:537–543. doi: 10.1007/s11248-011-9554-2. [DOI] [PubMed] [Google Scholar]
- 6.Saeki K, Matsumoto K, Kinoshita M, Suzuki I, Tasaka Y, Kano K, Taguchi Y, Mikami K, Hirabayashi M, Kashiwazaki N. Functional expression of a 螖12 fatty acid desaturase gene from spinach in transgenic pigs. Proc Natl Acad Sci U S A. 2004;101:6361–6366. doi: 10.1073/pnas.0308111101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lou Y, Schwender J, Shanklin J. FAD2 and FAD3 desaturases form heterodimers that facilitate metabolic channeling in vivo. J Biol Chem. 2014;289:17996–18007. doi: 10.1074/jbc.M114.572883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Indo Y, Tatemizo A, Abe Y, Suzuki I, Matsumoto K, Hosoi Y, Kinoshita M, Mikami K, Murata N, Iritani A, Saeki K. Functional expression of a humanized gene for an omega-3 fatty acid desaturase from scarlet flax in transfected bovine adipocytes and bovine embryos cloned from the cells. Biochim Biophys Acta. 2009;1791:183–190. doi: 10.1016/j.bbalip.2008.12.019. [DOI] [PubMed] [Google Scholar]
- 9.Newell M, Baker K, Postovit LM, Field CJ. A critical review on the effect of docosahexaenoic acid (DHA) on Cancer cell cycle progression. Int J Mol Sci. 2017;18:1784. doi: 10.3390/ijms18081784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang JX, Wan JB, He C. Concise review: regulation of stem cell proliferation and differentiation by essential fatty acids and their metabolites. Stem Cells. 2014;32:1092–1098. doi: 10.1002/stem.1620. [DOI] [PubMed] [Google Scholar]
- 11.Danbara N, Yuri T, Tsujita-Kyutoku M, Sato M, Senzaki H, Takada H, Hada T, Miyazawa T, Okazaki K, Tsubura A. Conjugated docosahexaenoic acid is a potent inducer of cell cycle arrest and apoptosis and inhibits growth of Colo 201 human colon cancer cells. Nutr Cancer. 2004;50:71. doi: 10.1207/s15327914nc5001_10. [DOI] [PubMed] [Google Scholar]
- 12.So WW, Liu WN, Leung KN. Omega-3 polyunsaturated fatty acids trigger cell cycle arrest and induce apoptosis in human neuroblastoma LA-N-1 cells. Nutrients. 2015;7:6956–6973. doi: 10.3390/nu7085319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui PH, Petrovic N, Murray M. The ω-3 epoxide of eicosapentaenoic acid inhibits endothelial cell proliferation by p38 MAP kinase activation and cyclin D1/CDK4 down-regulation. Br J Pharmacol. 2011;162:1143–1155. doi: 10.1111/j.1476-5381.2010.01113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiu LC, Ooi VE, Wan JM. Eicosapentaenoic acid modulates cyclin expression and arrests cell cycle progression in human leukemic K-562 cells. Int J Oncol. 2001;19:845. doi: 10.3892/ijo.19.4.845. [DOI] [PubMed] [Google Scholar]
- 15.Peng Y, Zheng Y, Zhang Y, Zhao J, Chang F, Lu T, Zhang R, Li Q, Hu X, Li N. Different effects of omega-3 fatty acids on the cell cycle in C2C12 myoblast proliferation. Mol Cell Biochem. 2012;367:165–173. doi: 10.1007/s11010-012-1329-4. [DOI] [PubMed] [Google Scholar]
- 16.Kawakita E, Hashimoto M, Shido O. Docosahexaenoic acid promotes neurogenesis in vitro and in vivo. Neuroscience. 2006;139:991–997. doi: 10.1016/j.neuroscience.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 17.Fujii-Yamamoto H, Kim JM, Arai K, Masai H. Cell cycle and developmental regulations of replication factors in mouse embryonic stem cells. J Biol Chem. 2005;280:12976–12987. doi: 10.1074/jbc.M412224200. [DOI] [PubMed] [Google Scholar]
- 18.Gonzales KA, Liang H, Lim YS, Chan YS, Yeo JC, Tan CP, Gao B, Le B, Tan ZY, Low KY, et al. Deterministic restriction on pluripotent state dissolution by cell-cycle pathways. Cell. 2015;162:564–579. doi: 10.1016/j.cell.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 19.Shao H, Wei Z, Wang L, Wen L, Duan B, Mang L, Bou S. Generation and characterization of mouse parthenogenetic embryonic stem cells containing genomes from non-growing and fully grown oocytes. Cell Biol Int. 2007;31:1336–1344. doi: 10.1016/j.cellbi.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 20.Zang QQ, Lu Z, Ning G, Cheng H. Ophiopogonin D inhibits cell proliferation, causes cell cycle arrest at G2/M, and induces apoptosis in human breast carcinoma MCF-7 cells. J Integr Med. 2016;14:51–59. doi: 10.1016/S2095-4964(16)60238-8. [DOI] [PubMed] [Google Scholar]
- 21.Shen SJ, Zhang YH, Xx G, Jiang SJ, LJ X. Yangfei Kongliu formula, a compound Chinese herbal medicine, combined with cisplatin, inhibits growth of lung cancer cells through transforming growth factor-β1 signaling pathway. J Integr Med. 2017;15:242–251. doi: 10.1016/S2095-4964(17)60330-3. [DOI] [PubMed] [Google Scholar]
- 22.Boheler KR. Stem cell pluripotency: a cellular trait that depends on transcription factors, chromatin state and a checkpoint deficient cell cycle. J Cell Physiol. 2009;221:10–17. doi: 10.1002/jcp.21866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheicher R, Hoelbl-Kovacic A, Bellutti F, Tigan AS, Prchal-Murphy M, Heller G, Schneckenleithner C, Salazar-Roa M, Zöchbauer-Müller S, Zuber J. CDK6 as a key regulator of hematopoietic and leukemic stem cell activation. Blood. 2015;125:90–101. doi: 10.1182/blood-2014-06-584417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mende N, Kuchen EE, Lesche M, Grinenko T, Kokkaliaris KD, Hanenberg H, Lindemann D, Dahl A, Platz A, Höfer T. CCND1-CDK4-mediated cell cycle progression provides a competitive advantage for human hematopoietic stem cells in vivo. J Exp Med. 2015;212:1171–1183. doi: 10.1084/jem.20150308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lange C, Huttner WB, Calegari F. Cdk4/CyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell. 2009;5:320–331. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 26.Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet. 2008;9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- 27.Faast R, White J, Cartwright P, Crocker L, Sarcevic B, Dalton S. Cdk6-cyclin D3 activity in murine ES cells is resistant to inhibition by p16(INK4a) Oncogene. 2004;23:491–502. doi: 10.1038/sj.onc.1207133. [DOI] [PubMed] [Google Scholar]
- 28.Karkhanis M, Park JI. Sp1 regulates Raf/MEK/ERK-induced p21(CIP1) transcription in TP53-mutated cancer cells. Cell Signal. 2015;27:479–486. doi: 10.1016/j.cellsig.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 30.Wei Z, Jian L, Yang L, Gao Y, Chen C, Zuo Y, Li G. Study of flax fatty acid desaturase fad3b in transgenic mice model Chinese. J Cell Biol. 2017:172–81.
- 31.White J, Dalton S. Cell cycle control of embryonic stem cells. Stem Cell Rev. 2005;1:131. doi: 10.1385/SCR:1:2:131. [DOI] [PubMed] [Google Scholar]
- 32.Pauklin S, Vallier L. The cell-cycle state of stem cells determines cell fate propensity. Cell. 2013;155:135–147. doi: 10.1016/j.cell.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh AM, Chappell J, Trost R, Lin L, Wang T. Cell-cycle control of developmentally regulated transcription factors accounts for heterogeneity in human pluripotent cells. Stem Cell Rep. 2013;1:532–544. doi: 10.1016/j.stemcr.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stead E, White J, Faast R, Conn S, Goldstone S, Rathjen J, Dhingra U, Rathjen P, Walker D, Dalton S. Pluripotent cell division cycles are driven by ectopic Cdk2, cyclin a/E and E2F activities. Oncogene. 2002;21:8320–8333. doi: 10.1038/sj.onc.1206015. [DOI] [PubMed] [Google Scholar]
- 35.Nakamuraishizu A, Takizawa H, Suda T. The analysis, roles and regulation of quiescence in hematopoietic stem cells. Development. 2014;141:4656–4666. doi: 10.1242/dev.106575. [DOI] [PubMed] [Google Scholar]
- 36.Leontieva OV, Blagosklonny MV. CDK4/6-inhibiting drug substitutes for p21 and p16 in senescence: duration of cell cycle arrest and MTOR activity determine geroconversion. Cell Cycle. 2013;12:3063. doi: 10.4161/cc.26130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amin M, Razi M, Sarrafzadehrezaei F, Shalizar AJ, Najafi G. Berberine inhibits experimental varicocele-induced cell cycle arrest via regulating cyclin D1, cdk4 and p21 proteins expression in rat testicles. Andrologia. 2018:e12984. [DOI] [PubMed]
- 38.Colleoni B, Paternot S, Pita JM, Bisteau X, Coulonval K, Davis RJ, Raspé E, Roger PP. JNKs function as CDK4-activating kinases by phosphorylating CDK4 and p21. Oncogene. 2017;36:4349–4361. doi: 10.1038/onc.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bisteau X, Paternot S, Colleoni B, Ecker K, Coulonval K, De GP, Declercq W, Hengst L, Roger PP. CDK4 T172 phosphorylation is central in a CDK7-dependent bidirectional CDK4/CDK2 interplay mediated by p21 phosphorylation at the restriction point. PLoS Genet. 2013;9:e1003546. doi: 10.1371/journal.pgen.1003546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Katakura M, Hashimoto M, Okui T, Shahdat HM, Matsuzaki K, Shido O. Omega-3 polyunsaturated fatty acids enhance neuronal differentiation in cultured rat neural stem cells. Stem Cells Int. 2013;2013:490476. doi: 10.1155/2013/490476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Héron-Milhavet L, Franckhauser C, Rana V, Berthenet C, Fisher D, Hemmings BA, Fernandez A, Lamb NJ. Only Akt1 is required for proliferation, while Akt2 promotes cell cycle exit through p21 binding. Mol Cell Biol. 2006;26:8267–8280. doi: 10.1128/MCB.00201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan HM, Narita M, Lowe SW, Livingston DM. The p400 E1A-associated protein is a novel component of the p53 → p21 senescence pathway. Genes Dev. 2005;19:196. doi: 10.1101/gad.1280205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beck SE, Jung BH, Rosario ED, Gomez J, Carethers JM. BMP-induced growth suppression in colon cancer cells is mediated by p21 WAF1 stabilization and modulated by RAS/ERK. Cell Signal. 2007;19:1465–1472. doi: 10.1016/j.cellsig.2007.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The real time PCR primers. Table S2. The content of fatty acids in the cells. (DOCX 22 kb)
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.