Abstract
Little is known about the biological function of histone deacetylase 11 (HDAC11), which is the lone class IV HDAC. Here, we demonstrate that deletion of HDAC11 in mice stimulates brown adipose tissue (BAT) formation and beiging of white adipose tissue (WAT). Consequently, HDAC11-deficient mice exhibit enhanced thermogenic potential and, in response to high-fat feeding, attenuated obesity, improved insulin sensitivity, and reduced hepatic steatosis. Ex vivo and cell-based assays revealed that HDAC11 catalytic activity suppresses the BAT transcriptional program, in both the basal state and in response to β-adrenergic receptor signaling, through a mechanism that is dependent on physical association with BRD2, a bromodomain and extraterminal (BET) acetyl-histone-binding protein. These findings define an epigenetic pathway for the regulation of energy homeostasis and suggest the potential for HDAC11-selective inhibitors for the treatment of obesity and diabetes.
Keywords: Metabolism
Keywords: Adipose tissue, Transcription
HDAC11 epigenetically regulates thermogenic gene expression, brown adipocyte differentiation, and beiging of white adiopose tissue.
Introduction
It is estimated that more than one-third of the US population is obese (1). Obesity is coupled to the development of many chronic diseases, including type 2 diabetes (T2D), which is projected to affect more than a half billion adults worldwide by 2040 (2). Obesity is a consequence of a disparity between energy intake and usage (3), and decreasing obesity requires a reduction in energy consumption and/or an increase in energy expenditure. Aside from diet and exercise, therapeutic interventions for obesity include bariatric surgery and the use of drugs that decrease energy intake (4).
There is intense interest in developing alternative pharmacotherapy for obesity based on increasing energy expenditure via brown adipose tissue (BAT) (5). In contrast to white adipose tissue (WAT), which functions mainly to store energy in the form of triglycerides in unilocular white adipocytes, brown adipocytes within BAT harbor small, multilocular lipid droplets and an abundance of mitochondria, which produce heat through nonshivering thermogenesis (6). Heat production by BAT is governed by uncoupling protein–1 (UCP1), which resides in the inner mitochondrial membrane in brown adipocytes and functions as a long-chain fatty acid/H+ symporter to catalyze mitochondrial proton leak and thereby uncouple electron transport from ATP synthesis (7–9). BAT is highly metabolically active and has been shown to contribute to energy expenditure in humans (10). Additional studies in humans have revealed that body mass index and percent body fat negatively correlate with BAT abundance (11), and a polymorphism in the gene encoding UCP1 is associated with fat gain and obesity (12). Together, these findings validate the potential of BAT-targeted therapies for the treatment of obesity.
Pharmacological approaches to promote BAT formation and function have included the use of β3-adrenergic receptor (β3-AR) agonists. β3-AR stimulation directly enhances lipolysis and energy expenditure, and also triggers downstream signaling events that lead to induction of a thermogenic gene program, which includes the gene encoding UCP1 (13). Furthermore, β3-AR stimulation of WAT can trigger the emergence of UCP1-expressing cells, termed beige or brite fat (14, 15), with morphological and functional characteristics of brown adipocytes (5, 16, 17). However, while β3-AR agonists were shown to acutely increase energy expenditure and insulin sensitivity in humans, they failed to promote weight loss upon chronic administration (18–21). Other approaches have targeted transcriptional regulators, primarily nuclear hormone receptors, which stimulate thermogenic gene expression. For example, PPARs and thyroid hormone receptors function downstream of the β3-AR to promote BAT gene expression and beiging of WAT, and PPAR agonists and thyromimetics have been shown to be efficacious in animal models of obesity (22–30). However, it is unclear whether pharmacological activation of these transcription factor pathways will be sufficiently tolerated in humans to provide a viable avenue for treatment of obesity (31).
Here, using multiple in vivo, ex vivo, and cell-based approaches, we demonstrate that histone deacetylase 11 (HDAC11) functions as a repressor of the thermogenic gene program in BAT and prevents beiging of WAT. Compared with WT controls, mice lacking HDAC11 are lean and harbor excess BAT. Accordingly, HDAC11 deficiency leads to enhanced cold-induced thermogenesis, reduced weight gain and lipid accumulation in response to high-fat feeding, and improved glucose tolerance. The global metabolic effects of HDAC11 deletion correlate with enhanced UCP1 expression in BAT, a profound increase in beiging of WAT, and augmented thermogenic gene expression in response to β3-AR signaling. Using cell-based models, we provide evidence for cell-autonomous roles for HDAC11 as a repressor of brown adipocyte differentiation and thermogenic gene expression, functions that are dependent on association of HDAC11 with the bromodomain and extraterminal (BET) family member BRD2. These data demonstrate a previously unrecognized role for HDAC11 as an epigenetic regulator of whole-body metabolism. Furthermore, since HDAC11-deficient mice are healthy (32), and HDAC11 has a unique catalytic domain compared with other HDAC isoforms (33), the findings suggest the possibility that selective HDAC11 inhibitors could be developed to increase energy expenditure for the treatment of obesity and metabolic disease.
Results
HDAC11 deficiency increases BAT abundance and function and triggers beiging of WAT.
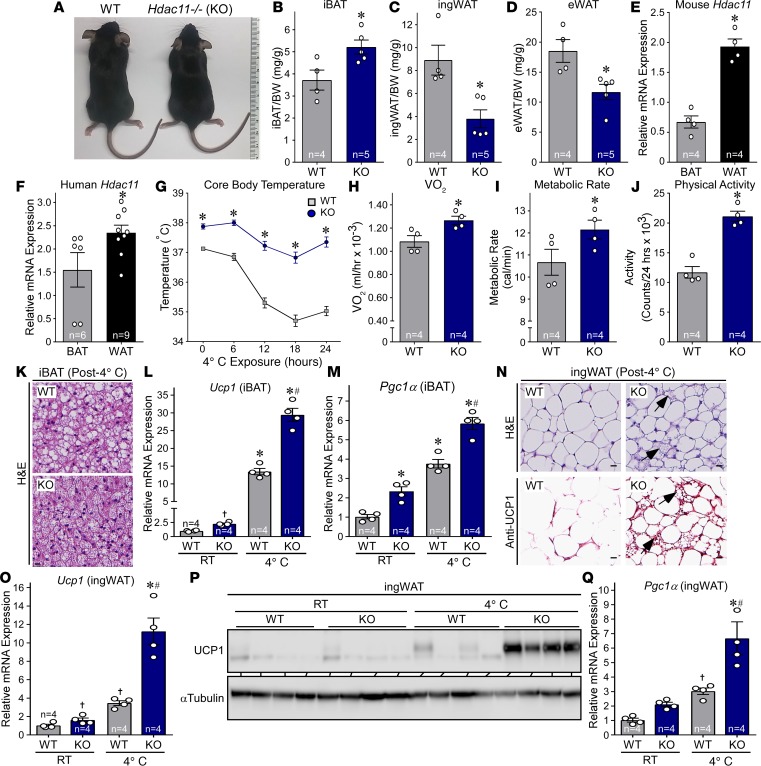
To further define the biological functions of HDAC11, we began investigating mice in which the Hdac11 gene was deleted globally (KO). KO mice were indistinguishable from WT controls with regard to overt morphological characteristics (Figure 1A). However, necropsy revealed increased interscapular BAT (iBAT) mass accompanied by a significant reduction in inguinal WAT (ingWAT) and epididymal WAT (eWAT) mass in KO mice compared with WT littermates (Figure 1, B–D). Consistent with a role for HDAC11 in the differential control of distinct fat depots, Hdac11 mRNA was more highly expressed in WAT compared with BAT in mouse and human samples (Figure 1, E and F). These findings suggest a previously unrecognized role for HDAC11 in the control of adipose tissue.
Figure 1. HDAC11 deficiency increases thermogenic potential in mice.
(A) Gross morphology of 10-week-old male WT and Hdac11–/– (KO) littermates fed standard rodent chow. (B–D) iBAT, ingWAT, and eWAT from 10-week-old male mice were dissected and weighed. n = 4 WT, n = 5 KO. (E and F) qPCR analysis of Hdac11 mRNA expression in mouse and human BAT and WAT. E: n = 4, BAT and WAT, ; F: n = 6 BAT, n = 9 WAT. (G) WT and KO mice were fed standard rodent chow and exposed to 4°C for 24 hours, and core body temperature was recorded at the indicated time points using a rectal thermal probe; n = 4 mice/group. (H and I) Oxygen consumption (VO2) and metabolic rate (MR) were measured using a CLAMS system for 24 hours. (J) Activity was calculated using laser beam breaks per 24 hours. H–J: n = 4 mice/group. (K) H&E staining of iBAT after 24 hours of 4°C exposure. Scale bar: 50 μm. (L and M) Quantitative PCR analysis of Ucp1 and Pgc1α mRNA expression in iBAT from mice housed at room temperature (RT) or exposed to 4°C for 24 hours; n = 4 mice/group. (N) H&E staining and UCP1 immunohistochemistry of ingWAT 24 hours after 4°C exposure. Arrows indicate evidence of beiging. Scale bars: 50 μm. (O) qPCR analysis of Ucp1 mRNA expression in ingWAT; n = 4 mice/group. (P) Immunoblot analysis of UCP1 protein expression in ingWAT. (Q) qPCR analysis of Pgc1α mRNA expression in ingWAT; n = 4 mice/group. Values for all graphs represent mean ± SEM; *P < 0.05 vs. WT (or BAT for E and F), #P < 0.05 vs. WT 4°C using ANOVA with Tukey’s multiple-comparisons test. Student’s t test was employed for 2 groups, and †P < 0.05 vs. WT RT; unpaired Student’s t test was employed for 2 groups, after ANOVA indicated significance.
Given the function of BAT in mediating nonshivering thermogenesis, we subsequently placed mice in metabolic cages and measured core body temperature and metabolic parameters at ambient temperature (22°C) and during a 24-hour cold challenge at 4°C. Compared with WT controls, KO mice had elevated body temperature prior to and during the cold challenge, which correlated with increased oxygen consumption, metabolic rate, and physical activity (Figure 1, G–J). Consistent with this, histological analysis following cold exposure revealed smaller lipid droplets in iBAT from KO mice compared with WT controls, which is a characteristic of accelerated triglyceride liberation (Figure 1K). Furthermore, expression of BAT-selective thermogenic genes, including Ucp1 and Pgc1α, was also increased in KO iBAT compared with WT controls (Figure 1, L and M). Strikingly, subcutaneous ingWAT from KO mice subjected to cold challenge exhibited marked signs of beiging, including increased abundance of multilocular adipocytes, pronounced induction of UCP1 protein expression, and increased levels of Ucp1 and Pgc1α mRNA expression (Figure 1, N–Q); quantitative PCR (qPCR) analysis confirmed the absence of Hdac11 mRNA transcripts in KO adipose tissue (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.120159DS1). Together, these data suggest that BAT function is ameliorated by the absence of HDAC11 and that deletion of HDAC11 promotes beiging of WAT.
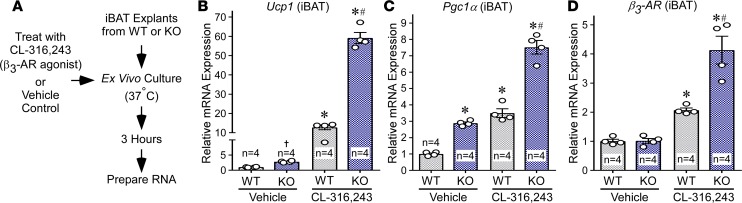
Induction of thermogenic gene expression upon cold challenge is governed by β3-AR signaling. To begin to address whether HDAC11 functions in an adipose tissue–autonomous manner to control thermogenesis, we performed acute ex vivo studies with iBAT explants from WT and KO mice (Figure 2A). Induction of Ucp1 and Pgc1α mRNA expression by the β3-AR–selective agonist CL-316,243 was significantly potentiated in KO iBAT compared with WT controls (Figure 2, B and C). Although expression of β3-AR mRNA expression was also elevated in KO iBAT following agonist treatment, baseline expression of the receptor was equivalent in WT and KO iBAT (Figure 2D). These data suggest that HDAC11 functions, at least in part, within adipose tissue to repress thermogenic β3-AR target gene expression. Nonetheless, we note that KO mice have elevated levels of circulating norepinephrine relative to WT controls, which could also contribute to the observed adipose phenotypes (Supplemental Figure 2).
Figure 2. HDAC11 deficiency augments β3-AR–induced target gene expression in BAT ex vivo.
(A) Schematic depiction of the iBAT ex vivo signaling assay. (B–D) Quantitative PCR analysis of Ucp1, Pgc1α, and β3-AR mRNA expression in iBAT treated ex vivo with CL-316,243 or vehicle control for 3 hours. n = 4 mice/group. Values for all graphs represent mean ± SEM; *P < 0.05 vs. WT vehicle, #P < 0.05 vs. WT CL-316,243 using ANOVA with Tukey’s multiple-comparisons test; †P < 0.05 vs. WT vehicle using unpaired t test following significant ANOVA.
HDAC11 deficiency protects against deleterious effects of high-fat feeding.
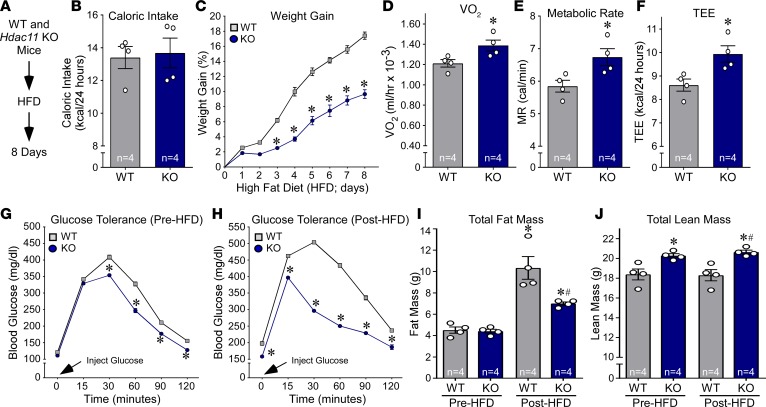
Activation of BAT and beiging of WAT can attenuate obesity and metabolic dysfunction. To initially assess the impact of deleting HDAC11 on metabolic homeostasis, we individually housed WT and KO mice in metabolic cages, fed them a high-fat, high-sucrose diet (HFD), and monitored them continuously over an 8-day period during which mice were given free access to food (Figure 3A). Remarkably, despite equivalent food intake between WT and KO mice, weight gain was reduced by approximately 50% in KO mice (Figure 3, B and C). Relative to WT controls, KO mice exhibited increased oxygen consumption as well as a higher metabolic rate and total energy expenditure, suggesting enhanced fatty acid oxidation in KO adipose tissue (Figure 3, D–F). Glucose tolerance was also enhanced in KO mice compared with WT controls at baseline and following high-fat feeding (Figure 3, G and H). Quantitative MRI (QMRI) revealed that the overall improvement in systemic metabolism in KO mice correlated with reduced fat mass after HFD feeding, and elevated lean mass before and after HFD compared with WT controls (Figure 3, I and J).
Figure 3. HDAC11 deficiency increases metabolism and glucose tolerance in response to acute high fat feeding.
(A) Schematic representation of the HFD experiment. (B) Daily food consumption was determined using an automated food hopper attached to CLAMS chambers. (C) Weight gain (%) was calculated each day for 8 days. n = 4 mice/group. *P < 0.05 by ANOVA. (D–F) VO2, metabolic rate (MR), and total energy expenditure (TEE) were measured using the CLAMS system; n = 4 mice/group. *P < 0.05 by Student’s t test. (G) Glucose tolerance was assessed in mice prior to high fat feeding; n = 4 mice/group. (H) Glucose tolerance was assessed in mice following 8 days of high-fat feeding. (I and J) Body composition (fat and lean mass) was determined using QMRI; n = 4 mice/group. Values for all graphs represent mean ± SEM. C–H: *P < 0.05 vs. WT; I and J: *P < 0.05 vs. WT before HFD and #P < 0.05 vs. WT after HFD. Two-way ANOVA with Šidák’s or Tukey’s multiple-comparisons test for 4 groups was employed for C, G, and H and for I and J, respectively; Student’s t test was used for B and D–F.
To determine whether HDAC11 deficiency confers chronic protection from deleterious effects of high-fat feeding, we fed WT and KO mice a HFD for 12 months (Figure 4A). Consistent with findings from the acute study, weight gain was blunted and glucose tolerance enhanced in KO mice compared with WT controls (Figure 4, B and C). These improvements in KO mice were associated with reduced circulating levels of insulin and leptin, and a concomitant increase in circulating adiponectin, a fat-derived hormone that is negatively correlated with metabolic dysfunction (Figure 4, D–F). QMRI demonstrated significantly lower fat mass in KO mice fed HFD compared with WT controls, with no change in lean mass between the groups (Figure 4, G and H). In line with the lower fat mass in KO mice, histological analyses revealed small lipid droplets in iBAT, reduced size of triglyceride-storing adipocytes in eWAT, and attenuated hepatic steatosis in KO mice compared with WT controls (Figure 4, I–L). Together, these findings demonstrate that HDAC11 deficiency has a general salutary impact on mouse metabolic health in the context of acute and chronic fat and caloric excess.
Figure 4. HDAC11 deficiency protects against deleterious effects of chronic high-fat feeding.
(A) Schematic representation of the experiment. (B) Weight gain (%) was calculated at 12 months; n = 4 WT, n = 5 KO. (C) Glucose tolerance was assessed in mice following 12 months of high-fat feeding; n = 4–5/group. (D and E) Plasma insulin and leptin concentrations were determined by ELISA; n = 4/group. (F) Plasma adiponectin concentrations were determined via immunoblotting and normalized to IgG levels; n = 4/group. (G and H) Body composition (total fat and lean mass) was determined using QMRI; n = 4 WT, n = 5 KO. (I) H&E staining of iBAT, eWAT and liver following 12 months of high-fat feeding. Scale bars: 10 μm. (J–L) Adipocyte area in iBAT and eWAT and lipid accumulation in liver were quantified as described in Methods.; n = 4 WT, n = 5 KO. Values for all graphs represent mean ± SEM; *P < 0.05 vs. WT. Two-way ANOVA with Šidák’s multiple-comparisons test for 4 groups was employed for C; Student’s t test was used for all other graphs.
HDAC11 knockdown promotes brown adipocyte differentiation.
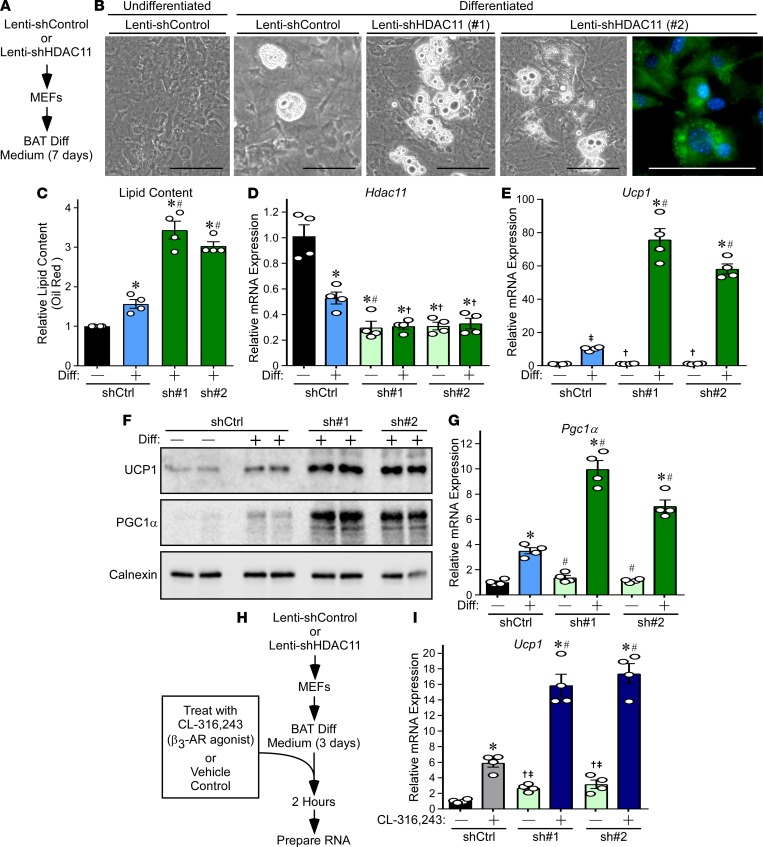
A loss-of-function approach was employed to address the possibility that HDAC11 functions in a cell-autonomous manner to suppress brown adipocyte differentiation. Initially, shRNA was employed to diminish expression of endogenous HDAC11 in mouse embryonic fibroblasts (MEFs), and the cells were subsequently exposed to BAT differentiation medium for 7 days (Figure 5A). Reduced HDAC11 expression correlated with enhanced differentiation of MEFs into adipocyte-like cells harboring multilocular lipid droplets that are characteristic of BAT (Figure 5, B–D). Furthermore, HDAC11 knockdown led to a dramatic enhancement of Ucp1 mRNA and protein expression, which correlated with augmented Pgc1α mRNA and protein levels (Figure 5, E–G).
Figure 5. HDAC11 knockdown enhances differentiation of MEFs into brown adipocyte–like cells.
(A) Schematic representation of the MEF differentiation (Diff) assay. (B) Phase-contrast images of MEFs differentiated into brown adipocyte–like cells. BODIPY staining shows multilocular lipid droplets characteristic of brown adipocytes. Scale bars: 100 μm. (C) Lipid content was quantified using colorimetric oil red O staining; n = 4 plates/condition. (D and E) qPCR analysis of Hdac11 and Ucp1 mRNA expression; n = 4 plates of cells/condition. (F) Immunoblot analysis of UCP1 and PGC1α protein expression; each lane represents protein from an independent plate of cells. Calnexin served as a loading control. (G) qPCR analysis of Pgc1α mRNA expression; n = 4 plates of cells/condition. (H) Schematic representation of the β3-AR signaling assay. (I) qPCR analysis of Ucp1 mRNA expression following 2 hours of treatment of cells with CL-316,243 or vehicle control; n = 4 plates of cells/condition. Values for all graphs represent mean ± SEM; *P < 0.05 vs. shControl undifferentiated or unstimulated; #P < 0.05 vs. shControl differentiated or stimulated using ANOVA with Tukey’s multiple-comparisons test. †P < 0.05 vs. shControl differentiated or stimulated; ‡P < 0.05 vs. shControl undifferentiated or unstimulated using Student’s t test following significant ANOVA.
Next, an acute study was performed to address whether HDAC11 regulates intrinsic responsiveness of cells to β3-AR signaling (Figure 5H). Consistent with the findings from the ex vivo study of BAT (Figure 2B), β3-AR agonist–mediated Ucp1 induction was substantially increased in partially differentiated MEFs in which HDAC11 expression was attenuated using shRNA (Figure 5I).
Repression of brown adipocyte differentiation by HDAC11 is dependent on association with BRD2.
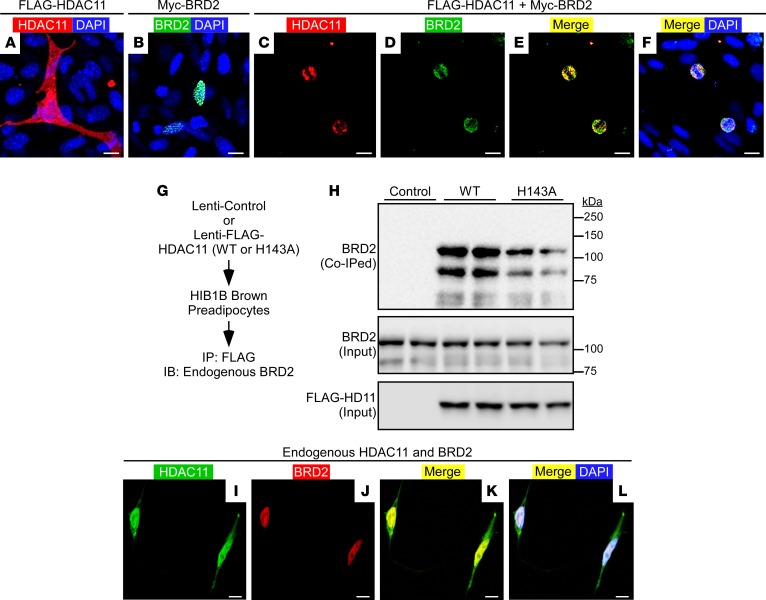
A prior proteomics study suggested that HDAC11 is capable of associating with BRD2, a member of the BET family of acetyl-histone-binding proteins that has been implicated as a negative regulator of brown adipocyte differentiation (34, 35). Thus, we hypothesized that HDAC11-mediated control of BAT differentiation is dependent on its interaction with BRD2. To initially address this hypothesis, we performed experiments to confirm the association between HDAC11 and BRD2. Confocal microscopy of HIB1B pre-brown adipocytes ectopically expressing tagged forms of HDAC11 and BRD2 revealed that, when expressed individually, the proteins were pan-cellular and nuclear localized, respectively (Figure 6, A and B). However, coexpression of the proteins led to redistribution of HDAC11 exclusively to the nuclear compartment, where it colocalized with BRD2 (Figure 6, C–F).
Figure 6. HDAC11 associates with BRD2.
(A–F) Confocal imaging of epitope-tagged HDAC11 and BRD2 in transiently transfected HIB1B preadipocytes. Scale bars: 10 μm. (G) Schematic representation of the HDAC11:BRD2 co-IP assay. (H) Immunoblot assessment of endogenous BRD2 coimmunoprecipitating (Co-IPed) with ectopic HDAC11 or H143A mutant. Total amounts of HDAC11 and BRD2 in protein homogenates were also determined (Input). (I–L) Confocal imaging of endogenous HDAC11 and BRD2 in untransfected HEK293 cells. Scale bars: 10 μm.
To further validate that HDAC11 and BRD2 associate within a common regulatory complex, we performed coimmunoprecipitation (co-IP) assays with cells ectopically expressing WT HDAC11 or a catalytically inactive version of HDAC11 harboring a histidine-to-alanine substitution at position 143 (H143A) within the deacetylase domain (Figure 6G). Endogenous BRD2 was effectively coimmunoprecipitated with HDAC11 (both WT and H143A) in HIB1B cells and HEK293 cells (Figure 6H and data not shown). Furthermore, confocal microscopy demonstrated nuclear colocalization of endogenous HDAC11 and endogenous BRD2 in untransfected HEK293 cells (Figure 6, I–L).
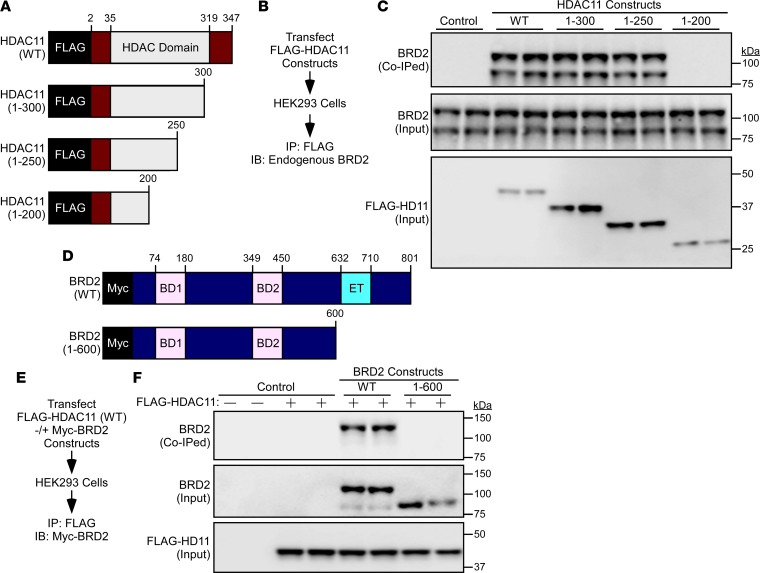
Employing a series of deletion constructs in transiently transfected HEK293 cells, the region between amino acids 200 and 250 within the catalytic domain of HDAC11 was determined to be critical for association with BRD2, while the extraterminal (ET) domain of BRD2 was found to be required for association with HDAC11 (Figure 7, A–F). Of note, the related BET family member BRD4 failed to associate with HDAC11, which suggests that HDAC11 selectively interacts with BRD2 as opposed to generally targeting BET proteins (Supplemental Figure 3).
Figure 7. Mapping of HDAC11:BRD2 association domains.
(A) Schematic representation of human HDAC11 and carboxy terminal deletion constructs. (B) Schematic representation of the HDAC11:BRD2 co-IP assay with HDAC11 deletion constructs. (C) Immunoblot assessment of endogenous BRD2 coimmunoprecipitating (co-IPed) with ectopic HDAC11. Total amounts of HDAC11 and BRD2 in protein homogenates were also determined (Input). (D) Schematic representation of human BRD2 and a carboxy terminal deletion construct. BD1, bromodomain 1; BD2, bromodomain 2; ET, extraterminal domain. (E) Schematic representation of the HDAC11:BRD2 co-IP assay with ectopic HDAC11 and ectopic BRD2. (F) Immunoblot assessment of ectopic BRD2 co-immunoprecipitating with ectopic HDAC11. Total amounts of epitope-tagged HDAC11 and BRD2 in protein homogenates were also determined (Input).
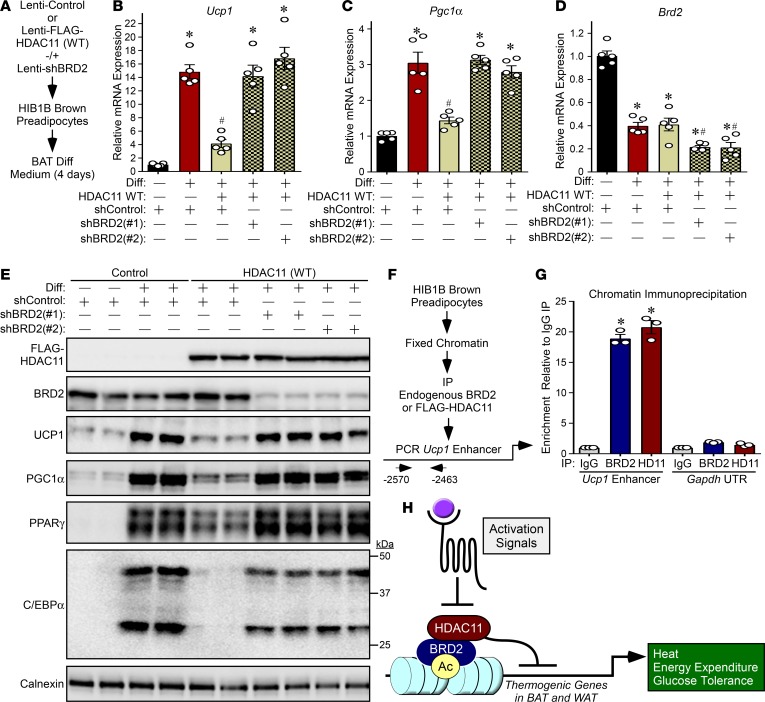
Having confirmed the association between HDAC11 and BRD2, we next sought to determine whether BRD2 contributes to HDAC11-mediated regulation of brown adipocyte differentiation. WT HDAC11 was ectopically overexpressed in HIB1B brown preadipocytes in the absence or presence of shRNA targeting BRD2, and the cells were subsequently exposed to BAT differentiation medium for 4 days (Figure 8A). Overexpression of HDAC11 WT significantly reduced Ucp1 and Pgc1α mRNA expression upon HIB1B differentiation (Figure 8, B and C). In contrast, catalytically inactive HDAC11 failed to repress expression of Ucp1 or Pgc1α (Supplemental Figure 4). Remarkably, BRD2 knockdown completely blocked the ability of HDAC11 to suppress expression of Ucp1 and Pgc1α mRNA and protein expression, and also rescued expression of PPARγ and C/EBPα (Figure 8, B–E).
Figure 8. HDAC11-dependent suppression of brown adipocyte differentiation requires BRD2.
(A) Schematic representation of the experiment. (B–D) qPCR analysis of Ucp1, Pgc1α, and Brd2 mRNA expression; n = 4 plates of cells/condition. (E) Immunoblot analysis of the indicated proteins; each lane represents protein from an independent plate of cells. Calnexin served as a loading control. Values for all graphs represent mean ± SEM; *P < 0.05 vs. shControl undifferentiated; #P < 0.05 vs. shControl differentiated. Two-way ANOVA with Tukey’s multiple-comparisons test was employed. (F) Schematic representation of the ChIP experiment. (G) ChIP-qPCR analysis of enrichment of BRD2 and HDAC11 at the Ucp1 enhancer and Gapdh UTR (negative control). Values for the graph represent mean ± SEM; *P < 0.05 vs. corresponding IgG IP. Two-way ANOVA with Tukey’s multiple-comparisons test was employed. (H) A model for HDAC11-mediated control of thermoregulation and energy homeostasis.
In a complementary experiment, the ability of ectopically expressed BRD2 to suppress Ucp1 and Pgc1α expression in differentiating HIB1B cells was impaired upon knockdown of endogenous HDAC11 and by deletion of the HDAC11-binding domain in BRD2 (Supplemental Figure 5). Together, these findings suggest that HDAC11 inhibits BAT differentiation and thermogenic gene expression, at least in part, through association with BRD2. Consistent with this notion, ChIP revealed the presence of HDAC11 and BRD2 on an enhancer that regulates Ucp1 expression (Figure 8, F and G). Furthermore, ectopically expressed HDAC11 WT, but not catalytically inactive HDAC11, blunted H3K27 acetylation at this enhancer in differentiating HIB1B cells (Supplemental Figure 6, A and B).
Discussion
Here, we describe a role for an obscure lysine deacetylase, HDAC11, in the transcriptional control of thermogenic gene expression in adipose tissue through association with BRD2, an acetyl-lysine-binding epigenetic “reader” protein. HDAC11-deficient mice are healthy and phenotypically indistinguishable from WT controls, yet are resistant to cold challenge and the adverse metabolic consequences of high-fat feeding due to enhanced metabolic activity in BAT, and increased beiging of WAT. Using cell-based models, we provide evidence to suggest that HDAC11 inhibits BAT differentiation, WAT beiging, and thermogenic gene expression through association with BRD2 on critical gene regulatory elements (Figure 8H). The findings reveal a druggable transcriptional pathway for the regulation of energy expenditure, and thus suggest novel approaches for combatting the world-wide pandemics of obesity and T2D via selective inhibition of HDAC11 or disruption of HDAC11:BRD2 association.
Increased thermogenesis and energy expenditure in HDAC11-deficient mice is associated with augmented BAT formation and function, and beiging of WAT. Although we cannot rule out the possibility that HDAC11 controls adipose tissue through indirect mechanisms, such as enhancing sympathetic drive, evidence is presented to suggest that HDAC11 functions in a cell-autonomous manner to control thermogenic gene expression in adipose tissue. For example, BAT explants from KO mice exhibited dramatically enhanced responsiveness to acute CL-316,243 stimulation compared with WT controls, despite equivalent expression of the β3-AR. Furthermore, knockdown of HDAC11 expression in fibroblasts was sufficient to promote Ucp1 and Pgc1α expression and amplify β3-AR–stimulated thermogenic gene expression, and ectopic expression of HDAC11 in preadipocytes had the converse actions. Future studies with mice in which HDAC11 is conditionally deleted in a tissue-specific manner will elucidate the relative contributions of HDAC11 in distinct cell types to the control of systemic metabolism.
Suppression of thermogenic gene expression by HDAC11 is due, at least in part, to its association with BRD2. However, HDAC11 deletion does not completely phenocopy BRD2 hypomorphic mice engineered using gene trap mutagenesis (35). In these animals, diminishing global BRD2 expression by approximately 50% resulted in increased BAT formation and thermogenesis and reduced blood glucose in the fasting and fed states, consistent with what was observed in HDAC11-KO mice. However, disruption of BRD2 expression in mice also led to extreme obesity, which contrasts with the lean phenotype of HDAC11-deficient animals. These data suggest that HDAC11 selectively regulates only a subset of BRD2 target genes, and that inhibition of HDAC11 catalytic activity or interference of HDAC11 association with BRD2 may be better tolerated than general inhibition of BRD2 with compounds such as bromodomain inhibitors. Given that HDAC11 can also localize to the cytoplasmic compartment, and has been shown to associate with a multitude of extranuclear proteins, including splicing factors (36), it remains possible the HDAC11 also governs thermogenic gene expression through BRD2-independent and non-genomic actions.
The 18 mammalian HDACs are clustered into 4 classes: class I HDACs (HDAC1, -2, -3, and -8), class II HDACs (HDAC4, -5, -6, -7, -9, and -10), class III HDACs (SirT 1–7), and class IV (HDAC11) (33). Class I, II, and IV HDACs are zinc-dependent enzymes, while class III HDACs, which are also known as sirtuins, require NAD+ as a cofactor for catalytic activity. Sirtuins are most commonly associated with regulation of metabolism (37), although zinc-dependent HDACs have recently emerged as regulators of thermogenic gene expression. For example, HDAC1 has been shown to associate with regulatory elements of BAT-specific genes, resulting in reduced histone H3K27 acetylation and consequent transcriptional repression (38). Another class I HDAC, HDAC3, has also been linked to the regulation of BAT gene expression and thermogenesis in mice (39–41). The class II HDAC HDAC9 has been demonstrated to repress adipogenesis, and deletion of HDAC9 in mice is sufficient to promote beiging and increase energy expenditure and adaptive thermogenesis in the context of high-fat feeding (42–44). However, HDAC9-mediated, unlike HDAC1- and HDAC3-mediated, regulation of adipogenic gene expression is not dependent on its deacetylase domain and instead is controlled by the amino-terminal cofactor interaction module of the protein (42).
HDAC11 enzymatic activity appears to be required for inhibition of adipocyte gene expression, as evidenced by the failure of catalytically inactive HDAC11 (H143A) to effectively suppress expression of Ucp1 and Pgc1α in HIB1B cells. This finding supports the concept of employing selective small molecule inhibitors of HDAC11 to increase energy expenditure, and the feasibility of this therapeutic approach is bolstered by the fact that 4 HDAC inhibitors are FDA-approved drugs (45). Nonetheless, pan-HDAC inhibition is accompanied by untoward toxicities, such as thrombocytopenia, nausea, and fatigue (46). We posit that selective inhibition of HDAC11 would provide a safer and more effective alternative to pan-HDAC blockade. This notion is supported by the observation that global deletion of HDAC11 in mice is well tolerated, while whole body removal of the previously mentioned HDACs that are implicated in the control of obesity and metabolism results in embryonic lethality or heart failure (47–50).
A recent discovery highlights the attainability of selective HDAC11 inhibitors. Unlike other HDACs, HDAC11 is a weak histone deacetylase and instead is a highly effective lysine defatty-acylase; HDAC11 defatty-acylates substrates with an efficiency that is more than 10,000-fold greater than its deacetylase activity (51–53). This finding suggests that HDAC11 functions, at least in part, in a manner analogous to sirtuins, which have the ability to proficiently catalyze removal of acyl groups in addition to acetyl, as opposed to other zinc-dependent HDACs, which primarily target acetyl moieties on lysine (54). The combination of this unique catalytic activity among the zinc-dependent HDACs and the divergent phylogeny of HDAC11 underscore the potential to selectively target HDAC11 with small molecule inhibitors.
An alternative approach to alleviate HDAC11-mediated suppression of thermogenesis would be to inhibit its interaction with BRD2. In this regard, a region of BRD2 that encompasses the ET domain was found to be required for association with HDAC11. ET domains of BET family members control gene transcription by functioning as interaction modules for cofactors such as the NSD3 histone methyltransferase (55). Recent NMR-derived structural analyses revealed compact motifs governing ET domain binding with partner proteins, suggesting the feasibility of employing small molecule ligands to disrupt ET domain–dependent functions to achieve therapeutic benefit (56, 57).
In conclusion, the present work uncovered insights into the molecular underpinnings of thermogenic gene regulation and the control of metabolism via BAT and beiging of WAT. HDAC11-deficient mice are healthy and exhibit amelioration of multiple metabolic parameters in mouse models of obesity. Given the unique features of HDAC11 highlighted above, the current findings suggest innovative avenues for the development of epigenetic therapies for obesity and obesity-related metabolic disorders by targeting the HDAC11/BRD2 axis.
Methods
Animals.
Global HDAC11-KO mice on a C57BL/6J background were provided by Merck Research Laboratories and generated by a targeted deletion of floxed exon 3 of the HDAC11 gene utilizing Rosa26 promoter–driven Cre recombinase expression. The Cre transgene was removed by segregation. These mice are now available from Taconic (model 6978). HDAC11 heterozygous (+/–) mice were bred to obtain HDAC11-KO mice (–/–) and their WT (+/+) littermate controls. Genotypes were confirmed via PCR using specific primers (Supplemental Table 3). Mice were maintained in a temperature- (22°C) and light-controlled vivarium with free access to water and standard rodent chow. All animal studies were initiated with 10-week-old male mice.
Human tissue biopsies.
BATs were collected from deep neck regions of patients undergoing thyroid surgeries at Florida Hospital. Subcutaneous abdominal WAT samples were obtained from participants of a cross-sectional study conducted at the Florida Hospital Translational Research Institute for Metabolism and Diabetes. Tissues were immediately snap-frozen and stored in liquid nitrogen. Tissue samples used in this study are a subset of those described in a prior report (Supplemental Table 4 and ref. 58).
Cold challenge and high-fat feeding.
For cold exposure experiment, WT and HDAC11-KO mice were kept at 4°C for 24 hours in indirect calorimetry metabolic cages (Oxymax/Comprehensive Lab Animal Monitoring System [CLAMS]; Columbus Instruments). Core body temperature was monitored every 2 hours for the duration of the study using a rectal probe (Physitemp Instruments Inc.) connected to a physiological monitoring unit (THM150; VisualSonics). Mice were fed an HFD (Research Diets Inc., D09071702; 58% calories derived from fat) for 8 days or 12 months. Metabolic monitoring of mice in response to acute HFD was performed using CLAMS. For glucose tolerance tests, mice were fasted for 6 hours prior to administration of glucose (2 mg/g body weight) via intraperitoneal injection. Tail vein blood was collected and blood glucose measured using a commercial glucometer (Bayer Contour) at the indicated time points. Body composition was determined using MRI (EchoMRI) to assess fat and lean mass. Mice were sacrificed by isoflurane inhalation followed by cervical dislocation, blood was collected via cardiocentesis, and adipose tissues and liver were harvested following tissue perfusion with saline.
Histological analyses.
Adipose and liver tissues were formalin fixed and embedded in paraffin. Tissue sections (5-μm-thick) were deparaffinized, hydrated, and processed for H&E staining using conventional methods. Sections were then dehydrated and mounted using Organo/Limonene Mount (Sigma-Aldrich). Tissue sections (5-μm-thick) of ingWAT from HDAC11-KO and WT mice exposed to cold challenge were deparaffinized, hydrated, and subjected to antigen retrieval using citrate buffer (Vector Laboratories). Endogenous peroxidase activity was inhibited using BLOXALL blocking solution (Vector Laboratories) for 10 minutes, and sections were incubated in 5% BSA for 1 hour at room temperature. Tissue sections were incubated overnight with anti-UCP1 antibody (Abcam), followed by incubation with biotinylated secondary antibody. Immunoperoxidase detection was performed using commercially available reagents as per the manufacturer’s instructions (Vector Laboratories). Images were acquired using an Olympus microscope (BX51) equipped with a multicolor camera (DP72). To quantify adipocyte size or hepatic lipid droplet accumulation, 5-μm-thick sections per paraffin-embedded tissue block were stained with H&E and imaged at ×20 magnification. High-resolution images (4–5 fields per section per animal) were acquired for adipocyte morphometry analysis. Automated analysis of adipose tissue cellularity and liver lipid content area was performed using Adiposoft open-source software (59). Average adipocyte areas were calculated for each animal, grouped, and plotted as mean adipocyte area for each genotype. For hepatic lipid accumulation quantification, total percent area was calculated for 3 random fields from each tissue section and plotted individually for every field for each genotype.
Quantification of circulating insulin, leptin, adiponectin, and catecholamine levels.
Blood was collected in 1.5 ml microfuge tubes containing EDTA and stored on ice. Tubes were centrifuged at 20,817 g for 5 minutes at 4°C, and supernatants were transferred to new microfuge tubes and stored at –80°C. Samples were used to determine levels of plasma insulin and leptin by ELISA according to the manufacturer’s instructions (Crystal Chem; 90080, 90030). Plasma adiponectin estimation was performed by immunoblotting with adiponectin-specific antibody (Acrp30), followed by quantification of adiponectin-positive bands by densitometry, as previously described (60). Levels of epinephrine and norepinephrine in plasma were determined by ELISA according to the manufacturer’s instructions (Abnova; KA1877).
Ex vivo BAT assay.
iBAT was collected from 10-week-old male mice. Each BAT explant was dissected in half and placed in medium (5 ml DMEM supplemented with 10% FBS) in sterile 14-ml round-bottom polystyrene tubes. Immediately thereafter, the explants were treated with CL-316243 (10 μM, Cayman Chemical) or vehicle control (DMSO, 0.1% final concentration) and incubated at 37°C for 3 hours in a cell culture incubator. The tissues were flash frozen in liquid nitrogen prior to RNA extraction.
qPCR.
RNA was prepared from cells and adipose tissues using QIAzol Lysis Reagent (QIAGEN). 500 ng total RNA was reverse transcribed and cDNA synthesized using a Verso cDNA Synthesis Kit (Thermo Fisher Scientific). qPCR was performed on a StepOnePlus Real-Time PCR System (Applied Biosystems) using PowerUp SYBR Green Master Mix (Thermo Fisher Scientific) and specific primers (Supplemental Table 1). Amplicon abundance was calculated using the 2–ΔΔCT method and normalized to 18S RNA or Hprt1. For analysis of mRNA expression in human tissue biopsies, RNA was extracted using commercial reagents (RNeasy Lipid Tissue Kit; QIAGEN), and qPCR was performed using TaqMan Fast Virus 1-Step Master reaction mix (Life Technologies) on a ViiAR 7 Real-Time PCR system (Applied Biosystems). Predesigned primer probe sets were used (Hs00978041_m1 for Hdac11, Hs02800695_m1 for Hprt1; Life Technologies). Relative gene expression data were calculated using the ΔCt method.
Immunoblotting.
Total protein was isolated from undifferentiated or brown adipocyte–differentiated cells (HIB1B cells or MEFs) using lysis buffer comprising NaCl (300 mM) and Triton X-100 (0.5% in PBS pH 7.4) supplemented with protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific). Proteins were separated using SDS-PAGE and transferred onto nitrocellulose membranes (0.45 μm; Life Science Products). Blots were probed with primary antibodies specific for UCP1 (Abcam; ab23841), PGC-1α (Calbiochem; ST1202), C/EBPα, PPARγ, BRD2 (Cell Signaling Technology; 8178, 2435, 5848), FLAG M2-HRP (Sigma-Aldrich; A8592), calnexin, or α-tubulin (Santa Cruz Biotechnology Inc.; sc11397, sc23948) overnight at 4°C. Following incubation with primary antibodies, blots were then probed with appropriate HRP-conjugated mouse or rabbit secondary antibodies (Southern Biotech). Protein bands were visualized using enhanced chemiluminescent HRP substrate (SuperSignal West Pico Chemiluminescent Substrate; Thermo Fisher Scientific) on a FluorChem HD2 Imager (Alpha Innotech). Full, uncut gels are presented in Supplemental Methods.
Plasmids.
pcDNA3.1(+)-based plasmids encoding human HDAC11, human BRD2, and deletion constructs fused in-frame to amino-terminal epitope tags were generated using PCR with Pfu Turbo polymerase (Agilent Technologies) and specific primers (Supplemental Table 2). Site-directed mutagenesis to create pcDNA3.1 encoding human HDAC11 (H143A) was performed using the QuickChange method (Agilent Technologies). For lentivirus production, complementary DNAs encoding human HDAC11, H143A mutant, BRD2, and BRD2 1-600 were cloned into pLenti CMV Hygro DEST. pLKO.1 plasmids (MilliporeSigma) encoding shRNA for murine HDAC11 (TRCN0000339501 and TRCN0000377043), BRD2 (TRCN0000023960 and TRCN0000023962), and a negative control (SHC002) were obtained through the Functional Genomics Facility at the University of Colorado Cancer Center.
Lentivirus production.
Lentiviruses were generated by cotransfecting L293 cells with pLenti or pLKO.1 vectors in combination with packaging plasmid (psPAX2) and envelope plasmid (pMD2.G). Virus-containing cell culture supernatants were collected, filtered, and used for experiments. pLenti-CMV-Hygro DEST (w117-1) was a gift from Eric Campeau and Paul Kaufman (Addgene plasmid 17454). psPAX2 and pMD2.G were gifts from Didier Trono (Addgene plasmids 12260, 12259) Virus-containing cell culture supernatants were collected, filtered (0.45 μm; Life Science Products), and frozen at –800C until needed.
Cell culture and differentiation.
MEFs were isolated from C57BL/6 E14.5 embryos using enzymatic digestion and maintained in DMEM/Hi glucose (HyClone) supplemented with 10% FBS (Gemini), 1% penicillin-streptomycin (Gibco), and 1.1% GlutaMAX (Gibco), as previously described (61). For differentiation into brown adipocytes, 4 × 105 cells were seeded per 60-mm dish and cultured in DMEM medium containing dexamethasone (1 μM), isobutylmethylxanthine (0.5 mM), T3 (50 nM), insulin (5 μg/ml), and rosiglitazone (2.5 μM) for 48 hours after lentivirus-mediated knockdown of HDAC11 (shHDAC11, or control shRNA). Differentiated cells were then maintained in media supplemented with insulin (5 μg/ml) and rosiglitazone (0.5 μM). Cells were treated with a β3-AR agonist, CL-316,243 (1 μM; Cayman Chemical), for 3 hours. Differentiated MEF cultures were stained with 0.5% Oil Red O to determine lipid content. Lipid droplets were visualized using BODIPY 493/503 (2 μM; 4,4-Difluoro-1,3,5,7,8-Pentamethyl-4-Bora-3a,4a-Diaza-s-Indacene; Molecular Probes). HIB1B brown preadipocytes were maintained in DMEM with 10% FBS and differentiated into adipocytes by the addition of rosiglitazone (1 μM) for 4 days (62). For experiments with lentiviruses, 3 ml filtered viral supernatant supplemented with 3 μl polybrene (Sigma-Aldrich) was added to cells, and the medium was changed 24 hours after infection. For MEFs, cells were grown to confluency (48 hours), the medium was replenished, and cells were maintained in a growth-arrested state for an additional 48 hours prior to induction of differentiation. For HIB1B adipocytes, cells were infected at approximately 60% confluency (24 hours), the medium was subsequently replenished, and cells recovered for 24 hours prior induction of differentiation.
co-IP.
HIB1B brown preadipocytes were infected with lentiviruses encoding FLAG-tagged WT human HDAC11 or catalytically inactive mutant form (H143A). HEK293 cells were transfected with tagged cDNA expression vectors encoding human HDAC11 (FLAG-tagged), deletion constructs of HDAC11 (FLAG-tagged), human WT BRD2 (Myc-tagged) or ET domain–deleted BRD2 (Myc-tagged) using polyethylenimine (PEI) for 48 hours. Cells were lysed in buffer containing Tris (50 mM, pH 7.5), 150 mM NaCl, and Triton X-100 (0.5 %) using a syringe with a 25-gauge needle. Total protein was collected after centrifugation of lysates at 20,817 g at 4°C for 20 minutes. Protein homogenates (0.5 mg) were diluted in equilibration buffer (50 mM Tris, 150 mM NaCl, pH 7.4; 500 μl total volume) and were immunoprecipitated using anti-FLAG IP resin (25 μl packed resin, GenScript), overnight at 4°C on a rotator, washed (3 times with equilibration buffer; 0.5 ml per wash), denatured in sample loading buffer, and resolved through 10% SDS polyacrylamide gels. Proteins were transferred to nitrocellulose membranes (0.45 μm; Life Science Products) and immunoblotted as described above using anti-BRD2 (Cell Signaling Technology; 5848), anti-Myc (Santa Cruz Biotechnology; sc40) or anti-BRD4 (Bethyl Laboratories; A301-985A) antibodies. Whole-cell lysates were used as input controls.
ChIP.
Treated or untreated HIB1B brown preadipocytes were processed for ChIP assay as described previously using commercially available reagents as per the manufacturer’s instructions (Millipore; 17-10085) (51–53, 62). Briefly, HIB1B brown preadipocytes were infected with lentiviruses encoding FLAG-tagged WT human HDAC11 for 48 hours. Crosslinking was performed using formaldehyde (1% final volume), followed by cell lysis and sonication for shearing of chromatin. Sheared chromatin was incubated with anti-FLAG (Sigma-Aldrich; F1804) or BRD2 (Cell Signaling Technology; 5848) antibodies overnight at 4°C with rotation. In a parallel experiment, HIB1B preadipocytes were allowed to undergo differentiation, and the resulting brown adipocytes were infected with lentiviral constructs encoding FLAG-tagged HDAC11 or H143A mutant for 48 hours. Cells were processed as mentioned above, and chromatin was subjected to incubation with anti-H3K27ac antibody (Abcam; ab4729). Genomic DNA was purified after reverse-crosslinking, and subjected to qPCR using primers specific to an approximately 108-bp region in the Ucp1 enhancer (Supplemental Table 1). Fold enrichment over IgG was calculated using the ΔCt method.
Indirect immunofluorescence.
HIB1B brown preadipocytes grown on coverslips were transiently transfected with pcDNA3.1-based plasmids encoding human HDAC11 (FLAG-tagged) and human BRD2 (Myc-tagged) for 48 hours using Lipofectamine 3000 (Life Technologies). Cells were fixed with 4% paraformaldehyde at room temperature for 10 minutes, followed by permeabilization using PBS-T (0.2% Triton X-100) for 15 minutes with gentle rocking. Cells on coverslips were blocked with PBS containing 5% BSA for 30 minutes at room temperature and incubated with primary antibodies (FLAG, Sigma-Aldrich, F1804, 1:200; c-Myc, Cell Signaling Technology, 2272; 1:200) for 2 hours. Secondary antibodies (anti-rabbit Alexa Fluor 488, 1:800; anti-mouse Alexa Fluor 555, 1:800; Molecular Probes) were applied for 30 minutes at room temperature. Coverslips were mounted on glass slides using anti-fade mounting reagent with DAPI (Vector Laboratories, H-1200). Images were obtained on an Olympus FluoView FV1000 confocal laser scanning microscope (University of Colorado Denver, Advanced Light Microscopy Core) and processed through FV-Viewer software (Olympus). In an alternate experiment, HEK293 cells plated on glass coverslips were processed similarly. Endogenous HDAC11 and BRD2 were detected using antibodies specific to these targets (HDAC11, Abcam, ab18973, 1:500; Alexa Fluor 555 BRD2, Abcam,ab213347, 1:500). Images were acquired on a Zeiss LSM780 confocal microscope and processed through ZEN Black software (Zeiss).
Statistics.
Statistical significance (P < 0.05) was determined using unpaired t test (2 groups; 2-tailed) or 2-way ANOVA with correction for multiple-comparisons via Tukey’s post hoc test (GraphPad Prism 7.02).
Study approval.
Animal studies were conducted using a protocol approved by the Institutional Animal Care and Use Committee of the University of Colorado Anschutz Medical Campus, following appropriate guidelines. Studies involving human subjects were approved by the Florida Hospital Institutional Review Board, and participants gave written informed consent.
Author contributions
RAB, BSF, MSS, TH, MAC, YHL, and MFP performed the research and analyzed data. LS, DL, PL, KS, PES, SC, and ES provided reagents, designed experiments, and analyzed data. LMS, SRS, and TAM designed and supervised the research and analyzed data. RAB and TAM wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Matthew Jackman for assistance with metabolic phenotyping studies at the Colorado Nutrition Obesity Research Center (NORC) Animal Satellite Facility; Kelley Brodsky for assistance with microscopy; Sara Wennersten, Katharina Lutter, and Korey Haefner for help with in vivo studies; and Andrew Riching for technical advice on ChIP assay. We are grateful to Dwight Klemm and Keith Koch for critical input. This work was supported by NIH (HL116848, HL127240, and AG043822) and American Heart Association (AHA; 16SFRN31400013) grants to TAM. RAB received funding from the Canadian Institutes of Health Research (FRN-216927). MSS was funded by a T32 training grant and an F32 fellowship from the NIH (T32HL007822 and F32HL126354). YHL was supported by an AHA postdoctoral fellowship (16POST30960017). KS was supported by funds from the Boettcher Foundation, AHA (13SDG17400031), University of Colorado Department of Medicine Outstanding Early Career Scholar Program, and NIH R01HL133230. SC was supported by NIH R01 DK103056.
Version 1. 08/09/2018
Electronic publication
Footnotes
BSF’s present address is Department of Agriculture, Nutrition, & Veterinary Sciences, College of Agriculture, Biotechnology, & Natural Resources, University of Nevada, Reno, Nevada, USA.
MSS’s present address is: Davis Heart and Lung Research Institute, and Department of Physiology and Cell Biology, The Ohio State University, Columbus, Ohio, USA.
DL and SC’s present address is: Division of Cardiovascular Medicine, Department of Medicine, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(15):e120159. https://doi.org/10.1172/jci.insight.120159.
Contributor Information
Rushita A. Bagchi, Email: RUSHITA.BAGCHI@UCDENVER.EDU.
Bradley S. Ferguson, Email: bferguson@unr.edu.
Matthew S. Stratton, Email: Matthew.Stratton@osumc.edu.
Tianjing Hu, Email: TIANJING.HU@UCDENVER.EDU.
Lei Sun, Email: leisun@gwu.edu.
Ying-Hsi Lin, Email: YING.LIN@UCDENVER.EDU.
Dianxin Liu, Email: Dianxin.liu@vanderbilt.edu.
Pilar Londono, Email: pilar.londono@ucdenver.edu.
Kunhua Song, Email: kunhua.song@ucdenver.edu.
Maria F. Pino, Email: Maria.Pino@flhosp.org.
Steven R. Smith, Email: StevenMD.Smith@flhosp.org.
Philipp E. Scherer, Email: philipp.scherer@utsouthwestern.edu.
Sheila Collins, Email: Sheila.collins@vanderbilt.edu.
Edward Seto, Email: seto@email.gwu.edu.
References
- 1.Ogden CL, Carroll MD, Fryar CD, Flegal KM. Prevalence of obesity among adults and youth: United States, 2011–2014. NCHS Data Brief. 2015;(219):1–8. [PubMed] [Google Scholar]
- 2.Ogurtsova K, et al. IDF Diabetes Atlas: global estimates for the prevalence of diabetes for 2015 and 2040. Diabetes Res Clin Pract. 2017;128:40–50. doi: 10.1016/j.diabres.2017.03.024. [DOI] [PubMed] [Google Scholar]
- 3.Hill JO, Wyatt HR, Peters JC. Energy balance and obesity. Circulation. 2012;126(1):126–132. doi: 10.1161/CIRCULATIONAHA.111.087213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heymsfield SB, Wadden TA. Mechanisms, pathophysiology, and management of obesity. N Engl J Med. 2017;376(3):254–266. doi: 10.1056/NEJMra1514009. [DOI] [PubMed] [Google Scholar]
- 5.Pfeifer A, Hoffmann LS. Brown, beige, and white: the new color code of fat and its pharmacological implications. Annu Rev Pharmacol Toxicol. 2015;55:207–227. doi: 10.1146/annurev-pharmtox-010814-124346. [DOI] [PubMed] [Google Scholar]
- 6.Tamucci KA, Namwanje M, Fan L, Qiang L. The dark side of browning. Protein Cell. 2018;9(2):152–163. doi: 10.1007/s13238-017-0434-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fedorenko A, Lishko PV, Kirichok Y. Mechanism of fatty-acid-dependent UCP1 uncoupling in brown fat mitochondria. Cell. 2012;151(2):400–413. doi: 10.1016/j.cell.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicholls DG, Bernson VS, Heaton GM. The identification of the component in the inner membrane of brown adipose tissue mitochondria responsible for regulating energy dissipation. Experientia Suppl. 1978;32:89–93. doi: 10.1007/978-3-0348-5559-4_9. [DOI] [PubMed] [Google Scholar]
- 9.Rosen ED, Spiegelman BM. What we talk about when we talk about fat. Cell. 2014;156(1–2):20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Virtanen KA, et al. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360(15):1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 11.van Marken Lichtenbelt WD, et al. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009;360(15):1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 12.van Marken Lichtenbelt WD, Schrauwen P. Implications of nonshivering thermogenesis for energy balance regulation in humans. Am J Physiol Regul Integr Comp Physiol. 2011;301(2):R285–R296. doi: 10.1152/ajpregu.00652.2010. [DOI] [PubMed] [Google Scholar]
- 13.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84(1):277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 14.Petrovic N, Walden TB, Shabalina IG, Timmons JA, Cannon B, Nedergaard J. Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J Biol Chem. 2010;285(10):7153–7164. doi: 10.1074/jbc.M109.053942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu J, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150(2):366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Champigny O, Ricquier D, Blondel O, Mayers RM, Briscoe MG, Holloway BR. Beta 3-adrenergic receptor stimulation restores message and expression of brown-fat mitochondrial uncoupling protein in adult dogs. Proc Natl Acad Sci U S A. 1991;88(23):10774–10777. doi: 10.1073/pnas.88.23.10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishibashi J, Seale P. Medicine. Beige can be slimming. Science. 2010;328(5982):1113–1114. doi: 10.1126/science.1190816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arch JR. Challenges in β(3)-adrenoceptor agonist drug development. Ther Adv Endocrinol Metab. 2011;2(2):59–64. doi: 10.1177/2042018811398517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larsen TM, et al. Effect of a 28-d treatment with L-796568, a novel beta(3)-adrenergic receptor agonist, on energy expenditure and body composition in obese men. Am J Clin Nutr. 2002;76(4):780–788. doi: 10.1093/ajcn/76.4.780. [DOI] [PubMed] [Google Scholar]
- 20.van Baak MA, et al. Acute effect of L-796568, a novel beta 3-adrenergic receptor agonist, on energy expenditure in obese men. Clin Pharmacol Ther. 2002;71(4):272–279. doi: 10.1067/mcp.2002.122527. [DOI] [PubMed] [Google Scholar]
- 21.Weyer C, Tataranni PA, Snitker S, Danforth E, Ravussin E. Increase in insulin action and fat oxidation after treatment with CL 316,243, a highly selective β3-adrenoceptor agonist in humans. Diabetes. 1998;47(10):1555–1561. doi: 10.2337/diabetes.47.10.1555. [DOI] [PubMed] [Google Scholar]
- 22.Gross B, Pawlak M, Lefebvre P, Staels B. PPARs in obesity-induced T2DM, dyslipidaemia and NAFLD. Nat Rev Endocrinol. 2017;13(1):36–49. doi: 10.1038/nrendo.2016.135. [DOI] [PubMed] [Google Scholar]
- 23.Hondares E, et al. Peroxisome proliferator-activated receptor α (PPARα) induces PPARγ coactivator 1α (PGC-1α) gene expression and contributes to thermogenic activation of brown fat: involvement of PRDM16. J Biol Chem. 2011;286(50):43112–43122. doi: 10.1074/jbc.M111.252775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JY, et al. Triiodothyronine induces UCP-1 expression and mitochondrial biogenesis in human adipocytes. Am J Physiol Cell Physiol. 2012;302(2):C463–C472. doi: 10.1152/ajpcell.00010.2011. [DOI] [PubMed] [Google Scholar]
- 25.Lin JZ, et al. Pharmacological activation of thyroid hormone receptors elicits a functional conversion of white to brown fat. Cell Rep. 2015;13(8):1528–1537. doi: 10.1016/j.celrep.2015.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rachid TL, Penna-de-Carvalho A, Bringhenti I, Aguila MB, Mandarim-de-Lacerda CA, Souza-Mello V. Fenofibrate (PPARα agonist) induces beige cell formation in subcutaneous white adipose tissue from diet-induced male obese mice. Mol Cell Endocrinol. 2015;402:86–94. doi: 10.1016/j.mce.2014.12.027. [DOI] [PubMed] [Google Scholar]
- 27.Roberts LD, et al. β-Aminoisobutyric acid induces browning of white fat and hepatic β-oxidation and is inversely correlated with cardiometabolic risk factors. Cell Metab. 2014;19(1):96–108. doi: 10.1016/j.cmet.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Villicev CM, et al. Thyroid hormone receptor beta-specific agonist GC-1 increases energy expenditure and prevents fat-mass accumulation in rats. J Endocrinol. 2007;193(1):21–29. doi: 10.1677/joe.1.07066. [DOI] [PubMed] [Google Scholar]
- 29.Wang L, et al. PPARα and Sirt1 mediate erythropoietin action in increasing metabolic activity and browning of white adipocytes to protect against obesity and metabolic disorders. Diabetes. 2013;62(12):4122–4131. doi: 10.2337/db13-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiner J, et al. Thyroid hormone status defines brown adipose tissue activity and browning of white adipose tissues in mice. Sci Rep. 2016;6:38124. doi: 10.1038/srep38124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tseng YH, Cypess AM, Kahn CR. Cellular bioenergetics as a target for obesity therapy. Nat Rev Drug Discov. 2010;9(6):465–482. doi: 10.1038/nrd3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang J, et al. Histone/protein deacetylase 11 targeting promotes Foxp3+ Treg function. Sci Rep. 2017;7(1):8626. doi: 10.1038/s41598-017-09211-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gregoretti, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338(1):17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 34.Denis GV, McComb ME, Faller DV, Sinha A, Romesser PB, Costello CE. Identification of transcription complexes that contain the double bromodomain protein Brd2 and chromatin remodeling machines. J Proteome Res. 2006;5(3):502–511. doi: 10.1021/pr050430u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang F, Liu H, Blanton WP, Belkina A, Lebrasseur NK, Denis GV. Brd2 disruption in mice causes severe obesity without Type 2 diabetes. Biochem J. 2009;425(1):71–83. doi: 10.1042/BJ20090928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joshi P, et al. The functional interactome landscape of the human histone deacetylase family. Mol Syst Biol. 2013;9:672. doi: 10.1038/msb.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verdin E. NAD+ in aging, metabolism, and neurodegeneration. Science. 2015;350(6265):1208–1213. doi: 10.1126/science.aac4854. [DOI] [PubMed] [Google Scholar]
- 38.Li F, et al. Histone deacetylase 1 (HDAC1) negatively regulates thermogenic program in brown adipocytes via coordinated regulation of histone H3 Lysine 27 (H3K27) deacetylation and methylation. J Biol Chem. 2016;291(9):4523–4536. doi: 10.1074/jbc.M115.677930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emmett MJ, et al. Histone deacetylase 3 prepares brown adipose tissue for acute thermogenic challenge. Nature. 2017;546(7659):544–548. doi: 10.1038/nature22819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferrari A, et al. HDAC3 is a molecular brake of the metabolic switch supporting white adipose tissue browning. Nat Commun. 2017;8(1):93. doi: 10.1038/s41467-017-00182-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galmozzi A, et al. Inhibition of class I histone deacetylases unveils a mitochondrial signature and enhances oxidative metabolism in skeletal muscle and adipose tissue. Diabetes. 2013;62(3):732–742. doi: 10.2337/db12-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chatterjee TK, et al. Histone deacetylase 9 is a negative regulator of adipogenic differentiation. J Biol Chem. 2011;286(31):27836–27847. doi: 10.1074/jbc.M111.262964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chatterjee TK, Basford JE, Yiew KH, Stepp DW, Hui DY, Weintraub NL. Role of histone deacetylase 9 in regulating adipogenic differentiation and high fat diet-induced metabolic disease. Adipocyte. 2014;3(4):333–338. doi: 10.4161/adip.28814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chatterjee TK, et al. HDAC9 knockout mice are protected from adipose tissue dysfunction and systemic metabolic disease during high-fat feeding. Diabetes. 2014;63(1):176–187. doi: 10.2337/db13-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones PA, Issa JP, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17(10):630–641. doi: 10.1038/nrg.2016.93. [DOI] [PubMed] [Google Scholar]
- 46.Mottamal M, Zheng S, Huang TL, Wang G. Histone deacetylase inhibitors in clinical studies as templates for new anticancer agents. Molecules. 2015;20(3):3898–3941. doi: 10.3390/molecules20033898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang S, McKinsey TA, Zhang CL, Richardson JA, Hill JA, Olson EN. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol Cell Biol. 2004;24(19):8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lagger G, et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21(11):2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Montgomery RL, et al. Maintenance of cardiac energy metabolism by histone deacetylase 3 in mice. J Clin Invest. 2008;118(11):3588–3597. doi: 10.1172/JCI35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang CL, McKinsey TA, Chang S, Antos CL, Hill JA, Olson EN. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110(4):479–488. doi: 10.1016/S0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. doi: 10.1073/pnas.1815365116. Cao J, et al. HDAC11 regulates type I interferon signaling through defatty-acylation of SHMT2. bioRxiv. doi: 10.1101/211706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kutil Z, Novakova Z, Meleshin M, Mikesova J, Schutkowski M, Barinka C. Histone deacetylase 11 is a fatty-acid deacylase. ACS Chem Biol. 2018;13(3):685–693. doi: 10.1021/acschembio.7b00942. [DOI] [PubMed] [Google Scholar]
- 53. doi: 10.1016/j.chembiol.2018.04.007. Moreno-Yruela C, Galleano I, Madsen AS, Olsen CA. Histone deacetylase 11 is an ε-N-myristoyllysine hydrolase. Cell Chem Biol. doi: 10.1016/j.chembiol.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 54.Jing H, Lin H. Sirtuins in epigenetic regulation. Chem Rev. 2015;115(6):2350–2375. doi: 10.1021/cr500457h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rahman S, et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol. 2011;31(13):2641–2652. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crowe BL, Larue RC, Yuan C, Hess S, Kvaratskhelia M, Foster MP. Structure of the Brd4 ET domain bound to a C-terminal motif from γ-retroviral integrases reveals a conserved mechanism of interaction. Proc Natl Acad Sci U S A. 2016;113(8):2086–2091. doi: 10.1073/pnas.1516813113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Q, et al. Structural mechanism of transcriptional regulator NSD3 recognition by the ET domain of BRD4. Structure. 2016;24(7):1201–1208. doi: 10.1016/j.str.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pino MF, Divoux A, Simmonds AV, Smith SR, Sparks LM. Investigating the effects of Orexin-A on thermogenesis in human deep neck brown adipose tissue. Int J Obes (Lond) 2017;41(11):1646–1653. doi: 10.1038/ijo.2017.155. [DOI] [PubMed] [Google Scholar]
- 59.Galarraga M, et al. Adiposoft: automated software for the analysis of white adipose tissue cellularity in histological sections. J Lipid Res. 2012;53(12):2791–2796. doi: 10.1194/jlr.D023788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scherer PE, Williams S, Fogliano M, Baldini G, Lodish HF. A novel serum protein similar to C1q, produced exclusively in adipocytes. J Biol Chem. 1995;270(45):26746–26749. doi: 10.1074/jbc.270.45.26746. [DOI] [PubMed] [Google Scholar]
- 61.Zhao Y, et al. High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nat Commun. 2015;6:8243. doi: 10.1038/ncomms9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang H, et al. Liver X receptor alpha is a transcriptional repressor of the uncoupling protein 1 gene and the brown fat phenotype. Mol Cell Biol. 2008;28(7):2187–2200. doi: 10.1128/MCB.01479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.