Abstract
BACKGROUND. There is currently no clinical distinction between different TP53 mutations, despite increasing evidence that not all mutations have equally deleterious effects on the activity of the encoded tumor suppressor protein p53. The objective of this study was to determine whether these biological differences have clinical significance.
METHODS. This retrospective cohort analysis included 2,074 patients with sporadic TP53 mutations (403 unique mutations) and 1,049 germline TP53 mutation carriers (188 unique mutations). Survival was projected by stratifying patients according to their p53 mutant–specific residual transcriptional activity scores.
RESULTS. Pan-cancer survival analyses revealed a strong association between increased mutant p53 residual activity and improved survival in males with glioma and gastric adenocarcinoma (P = 0.002 and P = 0.02) that was not present in the female cohorts (P = 0.16 and P = 0.50). Male glioma and gastric cancer patients with TP53 mutations resulting in >5% transcriptional activity had 3.1-fold (95% CI, 2.4–3.8; P = 0.002; multivariate analysis hazard ratio [HR]) and 4.6-fold (95% CI, 3.7–5.6; P = 0.001; multivariate analysis HR) lower risk of death as compared with patients harboring inactive (0% activity) p53 mutants. The correlation between mutant p53 residual activity with survival was recapitulated in the dataset of germline TP53 mutation carriers (HR = 3.0, 95% CI, 2.7–3.4, P < 0.001 [females]; HR = 2.2, 95% CI, 1.8–2.6, P < 0.001 [males]), where brain and gastric tumors were more common among males (P < 0.001 and P = 0.001, respectively).
CONCLUSION. The retention of mutant p53 transcriptional activity prognosticates superior survival for men with glioma and gastric adenocarcinoma harboring sporadic TP53 mutations. Among germline TP53 mutation carriers, increased residual transcriptional activity is correlated with prolonged lifetime cancer survival and delayed tumor onset, and males are more prone to develop brain and gastric tumors.
FUNDING. Canadian Institutes of Health Research (no. 148556).
Keywords: Genetics, Oncology
Keywords: Brain cancer, Gastric cancer, Tumor suppressors
The residual transcriptional activity associated with individual TP53 mutations was used to stratify cancer patient survival and is potentially useful as a prognostic tool.
Introduction
The future of personalized cancer medicine will require an understanding of the biological function of biomarkers to improve their prognostic value in terms of risk assessment and selecting optimal treatment options for cancer patients. Mutations in the TP53 gene that encodes the tumor suppressor protein p53 occur in up to 60% of human cancers, with most alterations being missense substitutions (1). Despite the frequent occurrence of TP53 mutations in cancer, knowledge of such mutations in patients has had limited impact in terms of clinically useful biomarkers (2). Specifically, many clinical studies investigating the prognostic and predictive value of TP53 mutations in different cancer types have yielded variable results between studies, with their clinical utility being largely inconclusive, likely due to grouping all mutations as equal (2, 3).
IHC has been widely used to assess TP53 gene status, where the overexpression and accumulation of p53 protein is a surrogate marker for detecting TP53 mutations in tumors. The exact mutation within the TP53 gene can now be readily established by sequencing patient-derived tumor DNA. Although a handful of conventional hotspot mutations have been demonstrated to possess some predictive clinical value, most mutations that occur across the entire TP53 gene provide no clinical insight toward treating individual cancer patients (4, 5).
In addition to the frequent occurrence of sporadic TP53 mutations in cancer, germline TP53 mutations are the underlying cause of the rare hereditary cancer disorder Li-Fraumeni syndrome (LFS) (6). These individuals are predisposed to multiorgan tumorigenesis, with a substantially increased risk of cancer-related morbidity and mortality during childhood and adulthood (7). Nearly 100% of affected females and 75% of affected males develop cancer throughout their lifetimes (8). Unfortunately, the considerable phenotypic heterogeneity presents a major challenge for disease management of these patients (9). As a result, it is difficult to provide accurate risk assessments and treatment options for LFS patients who undergo rigorous cancer screening programs (10).
As a transcription factor, p53 regulates the expression of genes involved in cellular growth, repair, and damage in response to stress, acting as a crucial barrier to oncogenesis (11). This 393–amino acid protein has 4 main functional domains: an N-terminal transactivation domain, central DNA-binding domain, oligomerization domain, and C-terminal regulatory domain (12). The mutational landscape of p53 in cancer spans the entire protein, but alterations (~75% being missense mutations) are predominantly located in the DNA-binding domain (residues 94–292, exons 5–8). Sequencing bias limited to these exons can account, in part, for this disparity (13).
Increasing evidence suggests that not all mutations have equally deleterious effects on p53 activity, which has been previously described as the mutant p53 functional gradient effect (14–18). In vitro studies demonstrate a broad range of activities when comparing different p53 mutants, where some retain near WT transcriptional activity and others are inactive (15). This functional diversity between different TP53 mutations is overlooked when evaluating p53 as a biomarker. Here, we hypothesized that the functional heterogeneity of p53 mutants could, in part, explain the inconclusive results of studies investigating the prognostic value of TP53 mutations in the clinic (2). A meta-analysis was performed to integrate mutant p53 functional information with cancer survival data, where each patient was assigned a transcriptional activity score according to their specific TP53 mutation, relative to WT p53. This analysis was used to generate a prognostic guide (Supplemental Table 3; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.121364DS1) with the aim of improving risk assessment and choice of therapy.
Results
Stratification of cancer patients with TP53 mutations based on transcriptional activity scores.
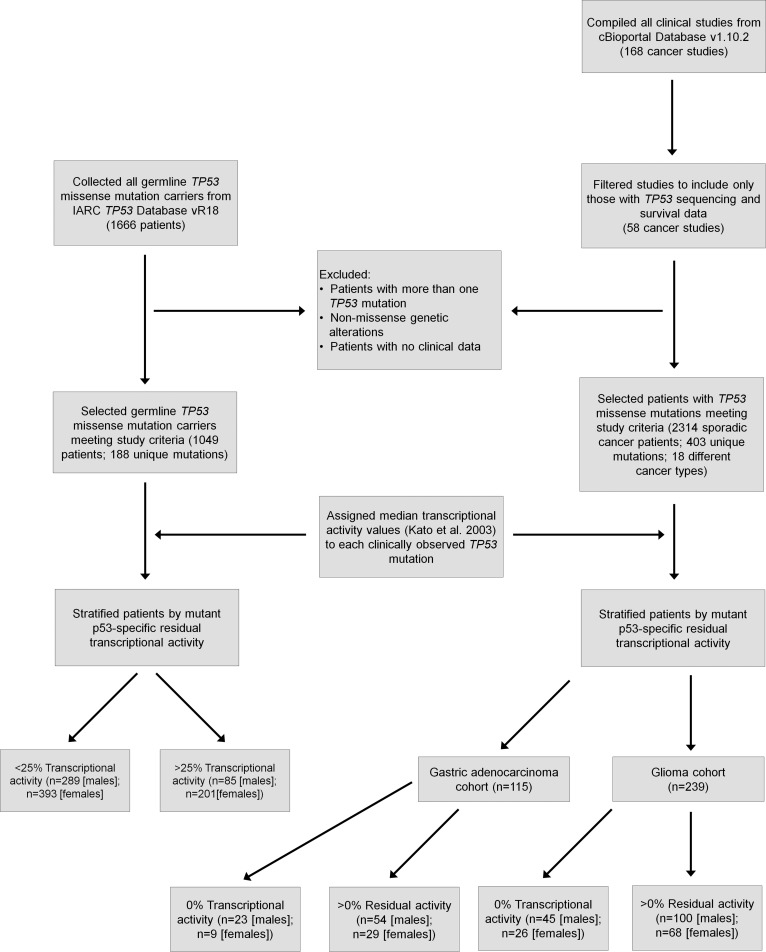
In order to establish a prognostic tool that would link mutant p53 activity to patient survival, an integrated functional genomics meta-analysis was carried out that included 2,314 cancer patients with sporadic TP53 missense mutations and 1,049 LFS patients with germline TP53 mutations (Figure 1). In total, 58 cancer studies (spanning at least 18 different cancer types; Supplemental Table 1) were compiled to generate a sporadic cancer patient dataset that included 403 unique p53 single amino acid substitutions (19, 20). Germline TP53 mutation carriers, including 188 unique p53 single amino acid substitutions, were collected from the International Agency for Research on Cancer (IARC) TP53 database (21). All patients with TP53 mutations were subsequently divided into subgroups along a gradient of residual transcriptional activity based on a comprehensive panel of p53 mutants originally generated by saturation mutagenesis throughout the p53 coding region that covers all clinically identified TP53 mutations (22). Specifically, we assigned a median transcriptional activity value to each clinically observed TP53 mutation relative to WT p53 (Supplemental Tables 2 and 3), as derived from monitoring 8 different p53 responsive promoter elements (p21, Mdm2, Bax, 14-3-3, p53AIP1, GADD45, Noxa, and p53R2) (22). Importantly, this analysis includes many low-abundance mutations that have been identified due to the increased use of next-generation sequencing for mutation testing in the clinic.
Figure 1. Study design and patient selection criteria for the survival analysis of cancer patients with sporadic or germline TP53 missense mutations.
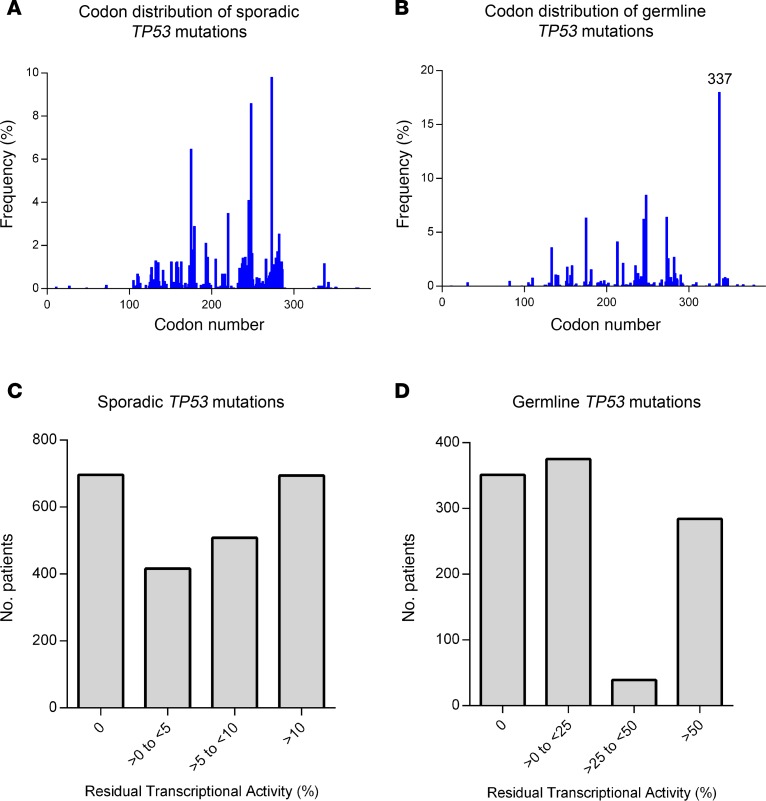
The codon distribution of TP53 missense mutations from patients in the sporadic and germline datasets both display a broad spectrum of mutations across the p53 protein (Figure 2, A and B). In addition, a large gradient of residual transcriptional activity is observed among the collection of TP53 mutations found in patients from both datasets (Figure 2, C and D). The functional landscape of the mutations observed in the 2 datasets are considerably different. Whereas only 4% of cancer patients with sporadic TP53 mutations result in mutants with >25% transcriptional activity, over 30% of germline carriers harbor p53 mutants with at least 25% activity. This finding reflects the fact that the broader range of occurring sporadic TP53 mutations (Supplemental Table 3) tend to display a lower level of transcriptional activity than the more tolerable p53 mutations observed among germline carriers.
Figure 2. Codon distribution, frequencies and transcriptional spectrum of sporadic and germline TP53 mutations.
(A and B) The codon distribution and frequencies of TP53 missense mutations in the sporadic (n = 2,314) (A) and germline (n = 1,049) (B) datasets from the current analysis. (C and D) Histogram displaying the range of residual transcriptional activity associated with TP53 mutations observed in patients from the sporadic (C) and germline (D) datasets.
Mutant p53 residual transcriptional activity stratifies survival outcomes of cancer patients with sporadic TP53 mutations.
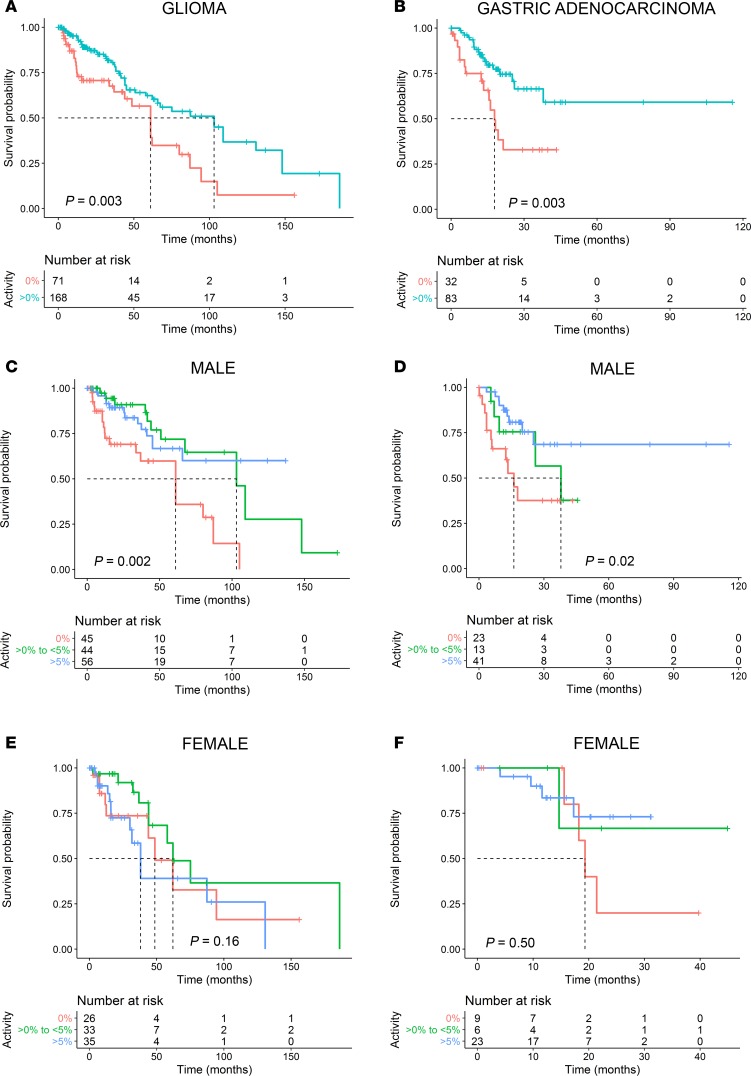
An initial pan-cancer analysis, including all cancer types pooled together, indicated that patients harboring sporadic TP53 mutations that retain any residual transcriptional activity (>0% activity) had improved survival compared with those with inactive (0% activity) p53 mutants (P = 0.01) (Supplemental Figure 1). Subsequently, individual cancer-specific survival analyses were performed. A significant difference between survival outcomes was observed for patients with glioma (low- and high-grade glioma merged cohort), as well as gastric adenocarcinoma. Specifically, patients with sporadic TP53 mutations leading to inactive p53 had a considerably worse survival outcome for both glioma and gastric adenocarcinoma (P = 0.003 and P = 0.003, respectively; Figure 3, A and B). No statistically significant difference was observed between the survival outcomes of the other cancer types harboring TP53 mutations (Supplemental Figure 1). For example, there was no survival difference when comparing TP53 mutations based on residual transcriptional activity in breast cancer, although it is well established that breast cancer patients with TP53 mutations have poorer outcomes compared with WT carriers (23, 24). These findings might indicate that a TP53 mutation represents only 1 key factor in projecting survival in certain cancer types, which may also be subtype specific (25).
Figure 3. Sex-specific associations between mutant p53 residual transcriptional activity and survival time after diagnosis of glioma and gastric adenocarcinoma.
(A) Kaplan-Meier survival curves of glioma patients and (B) gastric adenocarcinoma patients harboring sporadic TP53 mutations resulting in inactive mutant p53 (0% activity) versus those having mutations that retain transactivation function (>0% activity). (C and D) Kaplan-Meier survival curves of male patients stratified by a gradation of mutant p53 residual transcriptional activity (0% activity, >0%-to-<5% activity, and >5% activity) with glioma (C) and gastric cancer (D). (E and F) Kaplan-Meier survival curves of female patients stratified by a gradation of mutant p53 residual transcriptional activity (0% activity, >0%-to-<5% activity, and >5% activity) with glioma (E) and gastric cancer (F). Dotted lines indicate median survival times, and vertical lines on the Kaplan-Meier curves indicate patients who were censored. Two-sided P values were calculated using the log-rank test.
The strongest correlation linking mutant p53 transcriptional activity to survival is observed in male patients with glioma and gastric adenocarcinoma.
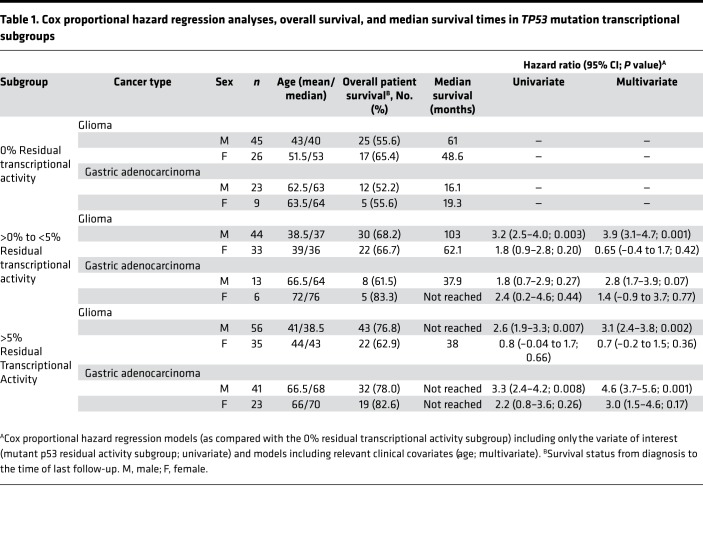
A further analysis of patient cohorts of both the glioma and gastric cancer datasets uncovered a strong association between mutant p53 residual activity and survival in males (P = 0.002 and P = 0.02, respectively) that was not present in the female cohorts (P = 0.16 and P = 0.50, respectively) (Figure 3, C–F). Specifically, the subgroups with residual mutant p53 activity (>0%-to-<5% and >5%) in the male glioma cohort had a 3.9-fold (95% CI 3.1–4.7; P = 0.001; based on multivariate analysis hazard ratio [HR]) and 3.1-fold (95% CI, 2.4–3.8; P = 0.002; based on multivariate analysis HR) lower risk of death as compared with the inactive (0% activity) p53 subgroup, respectively (Figure 3C and Table 1). The median survival time of the >0%-to-<5% activity subgroup was prolonged by 42 months as compared with the 0% activity subgroup (103 versus 61 months, respectively; P = 0.002). Survival was greatest in the >5% subgroup (median survival time not reached) and significantly extended as compared with the 0% activity subgroup (P = 0.006). In the male gastric cancer cohort, the >5% activity subgroup had a 4.6-fold (95% CI, 3.7–5.6; P = 0.001; based on multivariate analysis HR) lower risk of death with significantly improved survival (median survival time not reached) as compared with the 0% activity subgroup (16.1 months; P = 0.005; Figure 3D and Table 1). There was no statistical difference between the >0%-to-<5% activity and >5% activity subgroups of the male glioma or gastric cancer cohorts, although the shift between survival curves corresponds with increased mutant p53 residual transcriptional activity. There were no statistical differences between the ages of the male subgroups in glioma or gastric cancer (Table 1). In the female glioma cohort, the 0% activity subgroup had a significantly higher age as compared with the >0%-to-<5% activity subgroup (P = 0.001), although no other statistical differences between ages were observed among any other female subgroups. There were no differences in survival between female subgroups in either glioma or gastric cancer.
Table 1. Cox proportional hazard regression analyses, overall survival, and median survival times in TP53 mutation transcriptional subgroups.
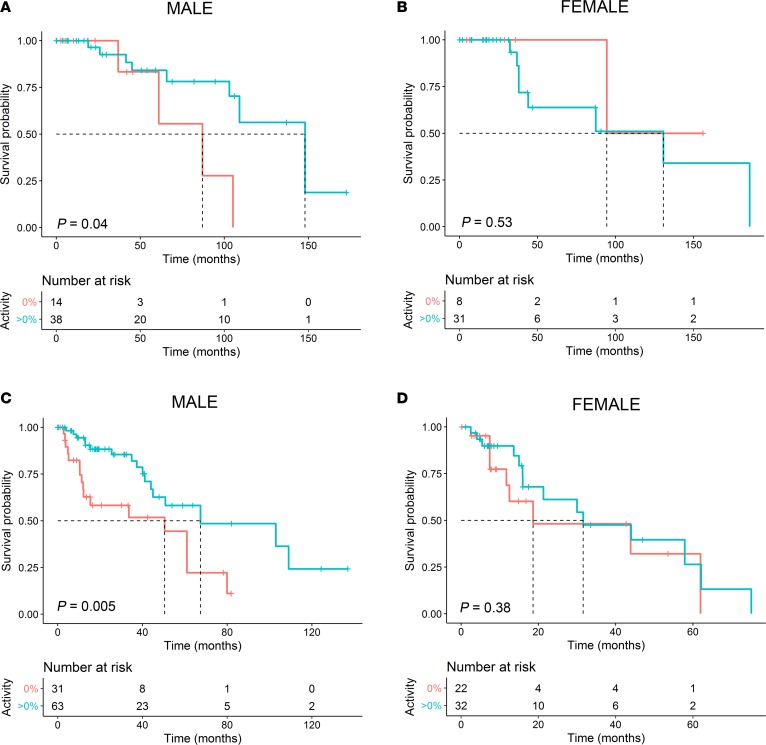
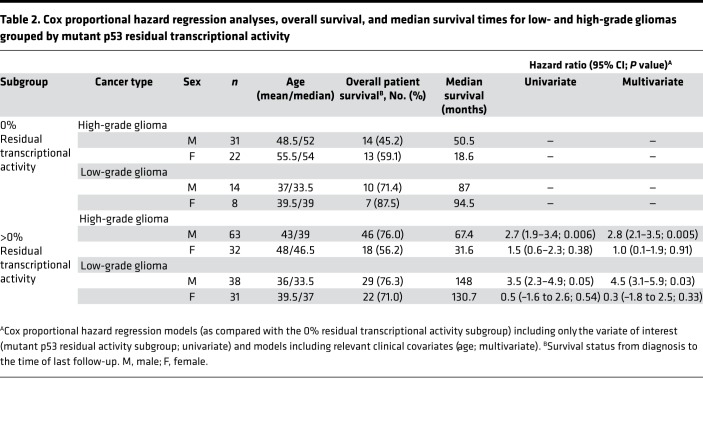
Next, the glioma cohorts were separated into low-grade (grades 1 and 2) and high-grade (grades 3 and 4). Male low- and high-grade glioma patients with mutant p53 having residual transcriptional activity had 4.5-fold (95% CI, 3.1–5.9; P = 0.03; based on multivariate analysis HR) and 2.8-fold (95% CI, 2.1–3.5; P = 0.005; based on multivariate analysis HR) lower risk of death as compared with the inactive (0% activity) subgroup, respectively (Figure 4, A and C, and Table 2). The median survival times for men with low- and high-grade gliomas having a mutant p53 with residual activity was significantly extended as compared with those harboring an inactive mutant p53 (148 versus 87 months, P = 0.04, and 67.4 versus 50.5, P = 0.005, respectively). No statistical differences were observed in the female low- or high-grade glioma cohorts (P = 0.53 and P = 0.38, respectively; Figure 4, B and D, and Table 2). There were no statistical differences between the ages of the male or female subgroups in the glioma subgroups.
Figure 4. A male-specific association between increased mutant p53 residual transcriptional activity and improved survival for low- and high-grade gliomas.
(A and B) Kaplan-Meier survival curves of male (A) and female (B) cohorts of low-grade gliomas. (C and D) Kaplan-Meier survival curves of male (C) and female (D) cohorts of high-grade gliomas. Two-sided P values were calculated using the log-rank test.
Table 2. Cox proportional hazard regression analyses, overall survival, and median survival times for low- and high-grade gliomas grouped by mutant p53 residual transcriptional activity.
The lifetime cancer survival of germline TP53 mutations carriers is associated with p53 mutant–specific transcriptional activity.
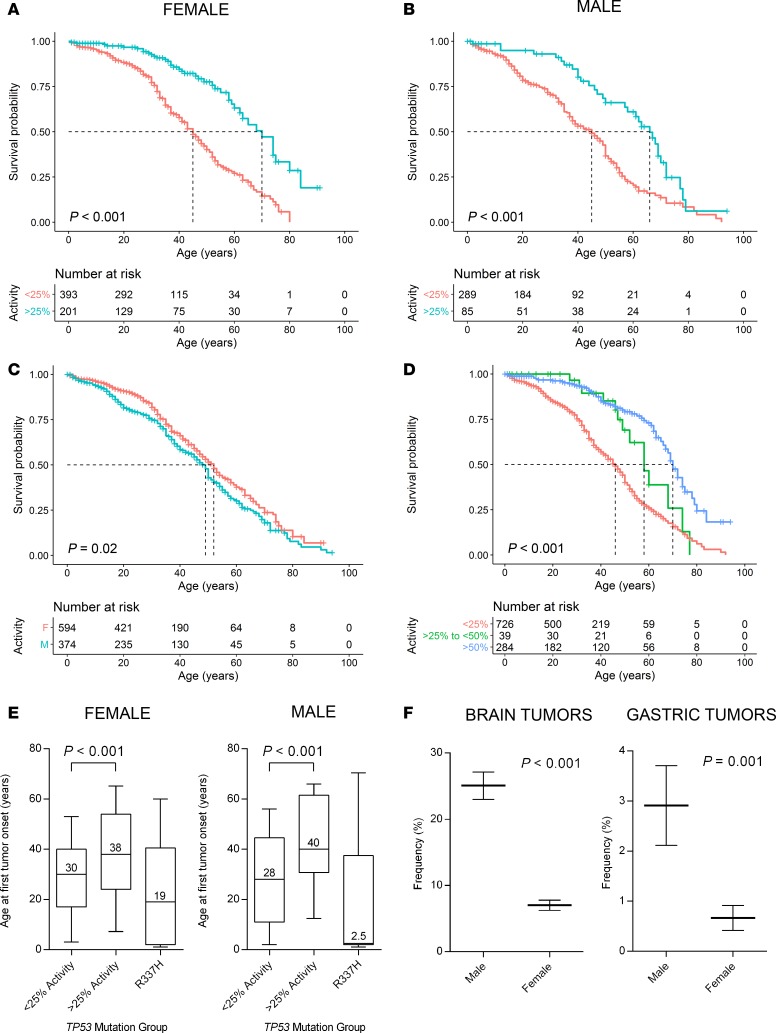
Survival data filtered through the germline cohort immediately revealed that patient survival outcomes can be projected from individual TP53 mutations. We determined that the threshold of transcriptional activity resulting in the greatest survival differences occurs at 25% residual activity for germline carriers of TP53 mutations. Remarkably, female germline TP53 mutation carriers with residual activity of at least 25% had a 3.0-fold lower risk of cancer-related death (95% CI, 2.7–3.4; P < 0.001) and median estimated lifespans extended by 25 years when compared with patients with TP53 mutations that result in less than 25% activity (70 versus 45 years, respectively; P < 0.001; Figure 5A). Among males, those carrying mutant p53 with residual activity of at least 25% had a 2.2-fold lower risk of cancer-related death (95% CI, 1.8–2.6; P < 0.001) and median estimated lifespans extended by 21 years when compared with patients with TP53 mutations that result in less than 25% activity (66 versus 45 years, respectively; P < 0.001; Figure 5B). Male TP53 mutation carriers had a shorter median estimated lifespan in comparison with female carriers (49 versus 52 years, respectively; P = 0.02; Figure 5C). Three subgroups covering 3 ranges of residual transcriptional activity (<25%, 25%-to-<50%, and >50%) indicated a strong association between prolonged lifetime survival with increased mutant p53 transactivation function in LFS patients (P < 0.001; Figure 5D).
Figure 5. Lifetime cancer survival of germline TP53 mutation carriers stratified by their mutant p53 residual transcriptional activity scores.
(A and B) Kaplan-Meier survival curves of female (A) and male (B) patients grouped by those carrying p53 mutants with <25% versus >25% transcriptional activity relative to WT p53. (C) Kaplan-Meier survival curves of the male and female cohorts of germline TP53 mutation carriers. (D) Kaplan-Meier survival curves of patients (pooled male and female) subdivided by carriers of mutant p53 with <25% activity, >25%-to-<50% activity, and >50% activity. Dotted lines indicate median survival times, and vertical lines on the Kaplan-Meier curves indicate patients who were censored. Two-sided P values were calculated using the log-rank test. (E) Box and whiskers plots (10th–90th percentile) representing the age at first tumor onset according to the TP53 mutation activity group or R337H carriers (P value calculated using proportion test). (F) The frequency of brain and gastric tumors that occur in male versus female germline TP53 mutation carriers (data represents mean ± SEM; P value calculated using 2-tailed Student’s t test).
The age at first tumor onset of germline TP53 mutation carriers is associated with p53 mutant–specific transcriptional activity.
The age at first tumor development for germline TP53 mutation carriers is highly variable (8). This classification of TP53 mutations reveals that the age at tumor onset is associated with p53 mutant–specific activity (Figure 5E). Specifically, carriers of p53 mutants that retain >25% activity had significantly delayed tumor manifestation as compared with those harboring mutants with <25% activity (median ages of 40 versus 28, P < 0.001 [males], respectively; 38 versus 30, P < 0.001 [females], respectively). Of note, R337H is a founder mutation that occurs at a high frequency in southern Brazil and accounts for 16% of the mutations in the germline dataset (26). This mutation has been associated with early childhood onset and a high prevalence of adrenocortical carcinomas (Figure 5E) (16, 27). The R337H allele exhibits a unique pH-dependent loss of function, although it retains 69% WT activity (median activity across 8 p53-responsive promoter elements) under normal physiological conditions (16, 28). However, the exclusion of R337H carriers from the germline dataset does not considerably affect the results of the analysis on lifetime cancer survival (Supplemental Figure 2).
The lifetime cancer risk is 96% for females carrying mutant p53 with <25% residual activity as compared with 87% in carriers of mutants with >25% activity (P < 0.001), based on the number of individuals affected by cancer in the germline dataset. In males, there is no difference between the 2 groups (92% versus 91%, respectively; P = 0.93). In terms of the tumor types manifested by germline TP53 mutation carriers, brain tumors are the most common tumors developed in male carriers (25%) and occur at a significantly higher frequency in contrast to female carriers, where only 7% develop brain tumors (P < 0.001; Figure 5F). In addition, gastric cancer has been reported in 3% of male carriers and occur at a significantly higher frequency when compared with female carriers (0.7%; P = 0.001). These observations provide additional support for a sex-specific link between mutant p53 and glioma or gastric cancer.
Discussion
Most studies investigating the prognostic or predictive value of TP53 mutations in cancer have not considered the diverse functional impact of different TP53 mutations on clinical outcomes. However, a large functional gradient exists among hundreds of unique mutations that occur along the entire TP53 gene in patient tumors, from transcriptionally inactive mutants to near WT activity levels. Here, we delineated this functional spectrum using a cancer-wide meta-analysis that integrated mutation-specific functional information with clinical data. This analysis revealed a male-specific association between increased mutant p53 residual transcriptional activity and improved survival for glioma and gastric adenocarcinoma patients with sporadic TP53 mutations (P = 0.002 and P = 0.02, respectively). A strong association was also evident in the lifetime survival and age at tumor onset of LFS patients (P < 0.001). Remarkably, the male-specific association was recapitulated, where both brain and gastric tumors were significantly more frequent among males (P < 0.001 and P < 0.001, respectively).
Sex disparities in cancer are well documented, although the molecular basis for these differences remains enigmatic (29, 30). In particular, males are more susceptible to develop tumors and frequently respond poorer to treatment as compared with females (29–32). Some sex-specific effects of p53 loss of function have been previously described. The first evidence of sexual dimorphism in the p53 pathway was in mice with complete loss of p53 function that resulted in a deficiency of female progeny from neural tube defects later attributed to X chromosome dosage (33, 34). In cancer, such effects have been demonstrated in murine astrocytes (glial cells) leading to enhanced transformation and in vivo tumorigenesis in males compared with females (35). A network analysis of gene expression profiles in patients with glioblastoma identified sex-specific molecular mechanisms of cell death: TP53 in males and MYC in females (36). Recently, glycolytic gene overexpression has been identified as a factor in the decreased survival of male patients with glioma. The authors suggested TP53 status as 1 potential underlying contributor (37). Thus, sex differences in transformation and tumorigenesis have been demonstrated, and the loss of p53 function has been implicated in the molecular mechanism.
Gastric cancer is the third leading cause of cancer death and fifth most common cancer worldwide, with approximately 952,000 new cases and 723,000 deaths annually (38). TP53 is the most frequently mutated gene in gastric cancer (39). In a recent study of gastric cancers, TP53 mutations were identified in 48% of cases (40). In a current report of gliomas, TP53 was identified in 40% of cases as the second most frequently mutated gene next to isocitrate dehydrogenase 1 (IDH1; 51% of cases) (41). The high prevalence of TP53 mutations in gastric cancers and gliomas underscores their biological importance as a crucial barrier to oncogenesis. The association between residual transcriptional activity and survival suggests that these specific cancer types are particularly influenced by the level of p53 inactivation. In the cancer types that did not display this trend, there may exist other major driving factors (i.e., molecular, genetic, and epigenetic changes) that take place to influence tumor phenotype. In breast cancer, TP53 mutations are generally associated with poor prognosis, although the association between mutant p53 residual transcriptional activity and survival was not observed here. In this case, a higher sensitivity for p53 inactivation could lead to worse outcomes, regardless of the TP53 mutation and residual transcriptional activity.
Due to the phenotypic heterogeneity among LFS patients carrying germline TP53 mutations, it is presently difficult to provide accurate risk assessments and to optimally treat these patients based on this molecular determinant once their tumors have occurred (9, 42). As a result, these patients undergo rigorous screening programs consisting of physical examinations and frequent biochemical and imaging studies, including whole-body MRI, brain MRI, breast MRI, mammography, abdominal and pelvic ultrasound, and colonoscopy (10). The present analysis of 1,049 LFS patients showed that carriers of TP53 mutations resulting with at least 25% residual transcriptional activity had considerably longer lifespans (25 years, P < 0.001 [females]; 21 years, P < 0.001 [males]) with a significantly lower risk of cancer-related death (3.0-fold, 95% CI, 2.7–3.4, P < 0.001 [females]; 2.2-fold, 95% CI, 1.8–2.6, P < 0.001 [males]). Knowledge of this genotype/phenotype correlation could be considered for the counseling and clinical decision-making for LFS patients.
Previous attempts to classify TP53 mutations have been limited to a few hotspot mutations that occur at higher frequency or clustering based on structural domains (4, 5, 43–47). One previous attempt to classify TP53 mutations based on transcriptional activity in colorectal cancer reported prognostic significance only in patients with late-stage (Dukes’ D) tumors (48). No other studies investigating different cancer types have been conducted in this regard. Future investigations should recruit more patients with TP53 mutations to identify correlations between residual transcriptional activity and clinicopathologic markers such as tumor stage and grade at presentation. The current analysis did not account for variance between therapies administered to patients. Thus, further work is needed to determine the predictive value for the effect of mutant p53 residual transcriptional activity on therapeutic response.
The transactivation capacity of p53 is critical for tumor suppression; however, transcription-independent mechanisms of p53 may also be important for tumor suppression and tumorigenesis (49, 50). In addition to loss of function, missense mutations in p53 can promote cancer development through the gain of aberrant function (51). The present classification system did not specifically account for the impact of this effect. TP53 mutations that lead to the gain of oncogenic function (G245S, R248W, R248Q, R273H, R175H, and R282W) are often associated with significant loss of transactivation. As a result, these transcriptionally inactive mutants were found in the high-risk group, except for sporadic R175H mutants that categorized into the lower-risk group. This observation is consistent with reports of the aggressive nature of these p53 mutants in cells and murine models (52–54).
There is currently no clinical distinction between different TP53 mutations, which are considered equivalent in terms of cancer prognosis. Importantly, this meta-analysis has exposed the unequal prognostic value between individual TP53 mutations in terms of cancer patient survival and has uncovered potential sex-specific effects of p53 functional loss in glioma and gastric cancer patients. The clinical impact of the mutant p53 functional gradient effect, a term used to describe the functional diversity observed among different mutants, is clearly demonstrated by our stratification of cancer patients, where greater residual transcriptional activity prognosticates better outcome. This correlation leads to a deeper understanding of TP53 mutational status as a biomarker in cancer and supports the use of TP53 mutations as independent prognostic markers for males with glioma and gastric adenocarcinoma, in the context of mutation-specific functionality. Future work should consider the effect of diverse functional consequences resulting from different mutations in TP53 or other highly mutated genes in cancer on tumor phenotype and clinical outcomes. In addition, this work warrants further investigation into the sex-specific effects of p53 functional loss in cancer, particularly within brain and stomach tissues. Prospective studies are required to determine whether mutant p53 transcriptional activity status can be used for personalized treatment decisions.
Methods
Patient data collection.
This study was a retrospective cohort analysis, including datasets from patients with TP53 mutations obtained from the latest versions of cBioPortal (v1.10.2) and the IARC TP53 database (vR18) (19–21). All clinical studies that included TP53 sequence assessment of patient tumors were collected from the cBioPortal repository. Studies without survival data were excluded, and only patients with missense mutations resulting in an amino acid substitution were used for survival analyses. Patients with more than 1 TP53 missense mutation were excluded. In addition, tumors with both a TP53 missense mutation and TP53 frameshift mutation or any other type of mutation in the same allele were not included in the study. Data processing and figure generation was performed in the R statistical environment v3.4.3 and GraphPad Prism v6.01.
Transcriptional activity scores.
Mutant p53 transcriptional activity data was downloaded from the IARC TP53 database (vR18) (21, 22). A median value from the transcriptional activity of p53 mutants monitored at 8 different p53-responsive promoter elements (p21, Mdm2, Bax, 14-3-3, p53AIP1, GADD45, Noxa, and p53R2), relative to WT p53, was assigned to each patient in a mutation-specific manner (Supplemental Table 2). Patients were then classified into subgroups according to the transcriptional activity level associated with their mutant p53. Cut-offs of ≥ 0% (sporadic dataset) and >25% or <25% (germline dataset) were used to define low versus high mutant p53 residual transcriptional activity in the sporadic and germline datasets, respectively. These cut-offs were determined based on the functional landscape of mutations, the number of patients in each grouping (Figure 2), and the observed effect sizes. Thresholds for patient substratification were chosen where the largest effect sizes were identified with suitable sample sizes in each subgroup (0%, >0%-to-<5%, and >5% for patients with sporadic TP53 mutations, and <25%, >25%-to-<50%, and >50% for germline carriers; Figure 2, C and D).
Statistics.
The primary outcome was overall survival from diagnosis to the time of death for patients in the sporadic cohort. The primary outcome for the germline cohort was lifetime survival. Patients who did not have an event were censored at the time of their last follow-up. Survival estimates were generated using the Kaplan-Meier method, and curves were compared with log-rank tests. We assessed prognosis using univariate and multivariate Cox proportional hazard regression models, with multivariate analyses correcting for age. Two-tailed Student’s t tests were used to compare the age at first tumor onset. Proportion tests were used to compare the frequency of patients with brain or gastric tumors, as well as the frequency of patients affected versus unaffected by cancer (estimate for the probability of getting cancer) in germline TP53 mutation carriers. All P values were based on a 2-sided hypothesis, and statistical significance was set at P < 0.05. All data processing and statistical analyses were performed in the R statistical environment (v3.4.3), using R packages survival (v2.41-3), survminer (v0.4.1), cgdsr (v1.2.6), and miscTools (v0.6-22).
Author contributions
NWF and JG designed the study, interpreted the data, and wrote the article. NWF and AP collected data and wrote the R code for data analysis.
Supplementary Material
Version 1. 08/09/2018
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(15):e121364. https://doi.org/10.1172/jci.insight.121364.
References
- 1.Leroy B, et al. Recommended Guidelines for Validation, Quality Control, and Reporting of TP53 Variants in Clinical Practice. Cancer Res. 2017;77(6):1250–1260. doi: 10.1158/0008-5472.CAN-16-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robles AI, Harris CC. Clinical outcomes and correlates of TP53 mutations and cancer. Cold Spring Harb Perspect Biol. 2010;2(3):a001016. doi: 10.1101/cshperspect.a001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2(1):a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu J, et al. Heterogeneity of Li-Fraumeni syndrome links to unequal gain-of-function effects of p53 mutations. Sci Rep. 2014;4:4223. doi: 10.1038/srep04223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu J, et al. Unequal prognostic potentials of p53 gain-of-function mutations in human cancers associate with drug-metabolizing activity. Cell Death Dis. 2014;5:e1108. doi: 10.1038/cddis.2014.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malkin D, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250(4985):1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 7.Kratz CP, et al. Cancer Screening Recommendations for Individuals with Li-Fraumeni Syndrome. Clin Cancer Res. 2017;23(11):e38–e45. doi: 10.1158/1078-0432.CCR-17-0408. [DOI] [PubMed] [Google Scholar]
- 8.Malkin D. Li-fraumeni syndrome. Genes Cancer. 2011;2(4):475–484. doi: 10.1177/1947601911413466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malkin D, Garber JE, Strong LC, Friend SH. CANCER. The cancer predisposition revolution. Science. 2016;352(6289):1052–1053. doi: 10.1126/science.aag0832. [DOI] [PubMed] [Google Scholar]
- 10.Villani A, et al. Biochemical and imaging surveillance in germline TP53 mutation carriers with Li-Fraumeni syndrome: 11 year follow-up of a prospective observational study. Lancet Oncol. 2016;17(9):1295–1305. doi: 10.1016/S1470-2045(16)30249-2. [DOI] [PubMed] [Google Scholar]
- 11.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14(5):359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joerger AC, Fersht AR. The tumor suppressor p53: from structures to drug discovery. Cold Spring Harb Perspect Biol. 2010;2(6):a000919. doi: 10.1101/cshperspect.a000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hainaut P, Pfeifer GP. Somatic TP53 Mutations in the Era of Genome Sequencing. Cold Spring Harb Perspect Med. 2016;6(11):a026179. doi: 10.1101/cshperspect.a026179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zambetti GP. The p53 mutation “gradient effect” and its clinical implications. J Cell Physiol. 2007;213(2):370–373. doi: 10.1002/jcp.21217. [DOI] [PubMed] [Google Scholar]
- 15.Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26(12):1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wasserman JD, et al. Prevalence and functional consequence of TP53 mutations in pediatric adrenocortical carcinoma: a children’s oncology group study. J Clin Oncol. 2015;33(6):602–609. doi: 10.1200/JCO.2013.52.6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer NW, Prodeus A, Malkin D, Gariépy J. p53 oligomerization status modulates cell fate decisions between growth, arrest and apoptosis. Cell Cycle. 2016;15(23):3210–3219. doi: 10.1080/15384101.2016.1241917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer NW, Prodeus A, Tran J, Malkin D, Gariépy J. Association Between the Oligomeric Status of p53 and Clinical Outcomes in Li-Fraumeni Syndrome. J Natl Cancer Inst. doi: 10.1093/jnci/djy114. [published online ahead of print June 27, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bouaoun L, et al. TP53 Variations in Human Cancers: New Lessons from the IARC TP53 Database and Genomics Data. Hum Mutat. 2016;37(9):865–876. doi: 10.1002/humu.23035. [DOI] [PubMed] [Google Scholar]
- 22.Kato S, et al. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci USA. 2003;100(14):8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olivier M, et al. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res. 2006;12(4):1157–1167. doi: 10.1158/1078-0432.CCR-05-1029. [DOI] [PubMed] [Google Scholar]
- 24.Silwal-Pandit L, Langerød A, Børresen-Dale AL. TP53 Mutations in Breast and Ovarian Cancer. Cold Spring Harb Perspect Med. 2017;7(1):a026252. doi: 10.1101/cshperspect.a026252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silwal-Pandit L, et al. TP53 mutation spectrum in breast cancer is subtype specific and has distinct prognostic relevance. Clin Cancer Res. 2014;20(13):3569–3580. doi: 10.1158/1078-0432.CCR-13-2943. [DOI] [PubMed] [Google Scholar]
- 26.Achatz MI, et al. The TP53 mutation, R337H, is associated with Li-Fraumeni and Li-Fraumeni-like syndromes in Brazilian families. Cancer Lett. 2007;245(1-2):96–102. doi: 10.1016/j.canlet.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 27.Ribeiro RC, et al. An inherited p53 mutation that contributes in a tissue-specific manner to pediatric adrenal cortical carcinoma. Proc Natl Acad Sci USA. 2001;98(16):9330–9335. doi: 10.1073/pnas.161479898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DiGiammarino EL, et al. A novel mechanism of tumorigenesis involving pH-dependent destabilization of a mutant p53 tetramer. Nat Struct Biol. 2002;9(1):12–16. doi: 10.1038/nsb730. [DOI] [PubMed] [Google Scholar]
- 29.Ashley DJ. A male-female differential in tumour incidence. Br J Cancer. 1969;23(1):21–25. doi: 10.1038/bjc.1969.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun T, Plutynski A, Ward S, Rubin JB. An integrative view on sex differences in brain tumors. Cell Mol Life Sci. 2015;72(17):3323–3342. doi: 10.1007/s00018-015-1930-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siegel R, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62(4):220–241. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- 32.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 33.Armstrong JF, Kaufman MH, Harrison DJ, Clarke AR. High-frequency developmental abnormalities in p53-deficient mice. Curr Biol. 1995;5(8):931–936. doi: 10.1016/S0960-9822(95)00183-7. [DOI] [PubMed] [Google Scholar]
- 34.Chen X, et al. Sex difference in neural tube defects in p53-null mice is caused by differences in the complement of X not Y genes. Dev Neurobiol. 2008;68(2):265–273. doi: 10.1002/dneu.20581. [DOI] [PubMed] [Google Scholar]
- 35.Sun T, et al. Sexually dimorphic RB inactivation underlies mesenchymal glioblastoma prevalence in males. J Clin Invest. 2014;124(9):4123–4133. doi: 10.1172/JCI71048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colen RR, Wang J, Singh SK, Gutman DA, Zinn PO. Glioblastoma: imaging genomic mapping reveals sex-specific oncogenic associations of cell death. Radiology. 2015;275(1):215–227. doi: 10.1148/radiol.14141800. [DOI] [PubMed] [Google Scholar]
- 37.Ippolito JE, Yim AK, Luo J, Chinnaiyan P, Rubin JB. Sexual dimorphism in glioma glycolysis underlies sex differences in survival. JCI Insight. 2017;2(15):e92142. doi: 10.1172/jci.insight.92142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferlay J, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 39.Tan P, Yeoh KG. Genetics and Molecular Pathogenesis of Gastric Adenocarcinoma. Gastroenterology. 2015;149(5):1153–1162.e3. doi: 10.1053/j.gastro.2015.05.059. [DOI] [PubMed] [Google Scholar]
- 40.Cancer Genome Atlas Research Network, et al. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541(7636):169–175. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ceccarelli M, et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell. 2016;164(3):550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McBride KA, et al. Li-Fraumeni syndrome: cancer risk assessment and clinical management. Nat Rev Clin Oncol. 2014;11(5):260–271. doi: 10.1038/nrclinonc.2014.41. [DOI] [PubMed] [Google Scholar]
- 43.Børresen AL, et al. TP53 mutations and breast cancer prognosis: particularly poor survival rates for cases with mutations in the zinc-binding domains. Genes Chromosomes Cancer. 1995;14(1):71–75. doi: 10.1002/gcc.2870140113. [DOI] [PubMed] [Google Scholar]
- 44.Russo A, et al. The TP53 colorectal cancer international collaborative study on the prognostic and predictive significance of p53 mutation: influence of tumor site, type of mutation, and adjuvant treatment. J Clin Oncol. 2005;23(30):7518–7528. doi: 10.1200/JCO.2005.00.471. [DOI] [PubMed] [Google Scholar]
- 45.Berns EM, van Staveren IL, Look MP, Smid M, Klijn JG, Foekens JA. Mutations in residues of TP53 that directly contact DNA predict poor outcome in human primary breast cancer. Br J Cancer. 1998;77(7):1130–1136. doi: 10.1038/bjc.1998.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Migliavacca M, et al. TP53 in gastric cancer: mutations in the l3 loop and LSH motif DNA-binding domains of TP53 predict poor outcome. J Cell Physiol. 2004;200(3):476–485. doi: 10.1002/jcp.20053. [DOI] [PubMed] [Google Scholar]
- 47.Trbusek M, et al. Missense mutations located in structural p53 DNA-binding motifs are associated with extremely poor survival in chronic lymphocytic leukemia. J Clin Oncol. 2011;29(19):2703–2708. doi: 10.1200/JCO.2011.34.7872. [DOI] [PubMed] [Google Scholar]
- 48.Iacopetta B, et al. Functional categories of TP53 mutation in colorectal cancer: results of an International Collaborative Study. Ann Oncol. 2006;17(5):842–847. doi: 10.1093/annonc/mdl035. [DOI] [PubMed] [Google Scholar]
- 49.Brady CA, et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 2011;145(4):571–583. doi: 10.1016/j.cell.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Speidel D. Transcription-independent p53 apoptosis: an alternative route to death. Trends Cell Biol. 2010;20(1):14–24. doi: 10.1016/j.tcb.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 51.Yue X, Zhao Y, Xu Y, Zheng M, Feng Z, Hu W. Mutant p53 in Cancer: Accumulation, Gain-of-Function, and Therapy. J Mol Biol. 2017;429(11):1595–1606. doi: 10.1016/j.jmb.2017.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mello SS, Attardi LD. Not all p53 gain-of-function mutants are created equal. Cell Death Differ. 2013;20(7):855–857. doi: 10.1038/cdd.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, Coillie SV, Fang JY, Xu J. Gain of function of mutant p53: R282W on the peak? Oncogenesis. 2016;5:e196. doi: 10.1038/oncsis.2016.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hanel W, Marchenko N, Xu S, Yu SX, Weng W, Moll U. Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis. Cell Death Differ. 2013;20(7):898–909. doi: 10.1038/cdd.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.