Abstract
MicroRNAs (miRs) posttranscriptionally regulate mRNA and its translation into protein, and are considered master controllers of genes modulating normal physiology and disease. There is growing interest in how miRs change with drug treatment, and leveraging this for precision guided therapy. Here we contrast 2 closely related therapies, inhibitors of phosphodiesterase type 5 or type 9 (PDE5-I, PDE9-I), given to mice subjected to sustained cardiac pressure overload (PO). Both inhibitors augment cyclic guanosine monophosphate (cGMP) to activate protein kinase G, with PDE5-I regulating nitric oxide (NO) and PDE9-I natriuretic peptide–dependent signaling. While both produced strong phenotypic improvement of PO pathobiology, they surprisingly showed binary differences in miR profiles; PDE5-I broadly reduces more than 120 miRs, including nearly half those increased by PO, whereas PDE9-I has minimal impact on any miR (P < 0.0001). The disparity evolves after pre-miR processing and is organ specific. Lastly, even enhancing NO-coupled cGMP by different methods leads to altered miR regulation. Thus, seemingly similar therapeutic interventions can be barcoded by profound differences in miR signatures, and reversing disease-associated miR changes is not required for therapy success.
Keywords: Cardiology, Cell Biology
Keywords: Heart failure, Noncoding RNAs, Phosphodiesterases
PDE5A and PDE9A inhibition regulate microRNAs in a highly disparate fashion in the failing mouse heart, despite a shared effector kinase and phenotypic improvement.
Introduction
MicroRNAs (miRs) are small ribonucleic acids that provide posttranscriptional control of mRNA translation and degradation (1). First discovered in 1993, there are now over 24,000 identified miRs. Their primary role is repressing gene expression by facilitating mRNA degradation, inhibiting protein translation, or degrading polypeptides through complementary binding to the 3′-UTR of target mRNAs. Each miR impacts around 1,000 individual mRNAs, generally clustering around a thematically coherent set of signaling pathways. Importantly, their expression changes with disease and therapeutic interventions, which has spawned interest in their use as both diagnostic and therapeutic agents (2, 3). The first reports of the role of miRs in heart disease came in 2006 when the Olson lab reported enhancement of miR-208 in pathological hypertrophy and therapeutic benefits from its reversal (4–6). Other similar examples followed, including suppressing miR-122/132 or increasing miR-1 or miR-133 (7–11), with now well over 2,000 studies on various miRs and heart disease.
From a systems biology perspective, the position of miRs is generally placed above gene translation. However, growing evidence demonstrating their modification by intracellular and environmental signaling have led to their exploration as markers of disease therapy, of particular interest to the efforts to better personalize medical treatment (12, 13). However, such data remain fairly limited, and the sensitivity of miR signatures to treatments that share common signaling little explored.
Here, we tested the hypothesis that miR profiling may even provide a useful tool to distinguish between mechanistically very similar therapeutic interventions. To do this, we contrasted the effects of pharmacologically inhibiting either cGMP-selective phosphodiesterase type 5 or type 9 (PDE5-I, PDE9-I). Both activate protein kinase G (PKG), though they do so by modulating different cGMP pools, the former coupled to nitric oxide (NO) stimulation of guanylyl cyclase-1 (GC-1) and the latter to natriuretic peptide (NP) stimulation of GC-2A (14). Importantly, both drugs potently and very similarly improve heart disease stimulated by sustained pathological hemodynamic stress (14). Despite this, the miR profiles were dramatically and surprisingly different, with PDE5-I reducing a very broad array of miRs associated with the disease state, whereas PDE9-I had virtually no impact.
Results
PDE5-I and PDE9-I applied to pressure-overloaded heart yield very disparate miR profiles.
Mice were subjected to sustained pressure overload (PO) for 5 weeks and further randomized to receive either PDE5-I (sildenafil) or PDE9-I (PF-04449613), each initiated 1 week after PO. Reported hemodynamic and morphological data from these mice documents similar reduction of hypertrophy and cardiac dilation, as well as reversal of abnormal molecular signaling and fibrosis, and improved heart function (14, 15). Salient data are provided in Supplemental Figure 1 (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.121739DS1).
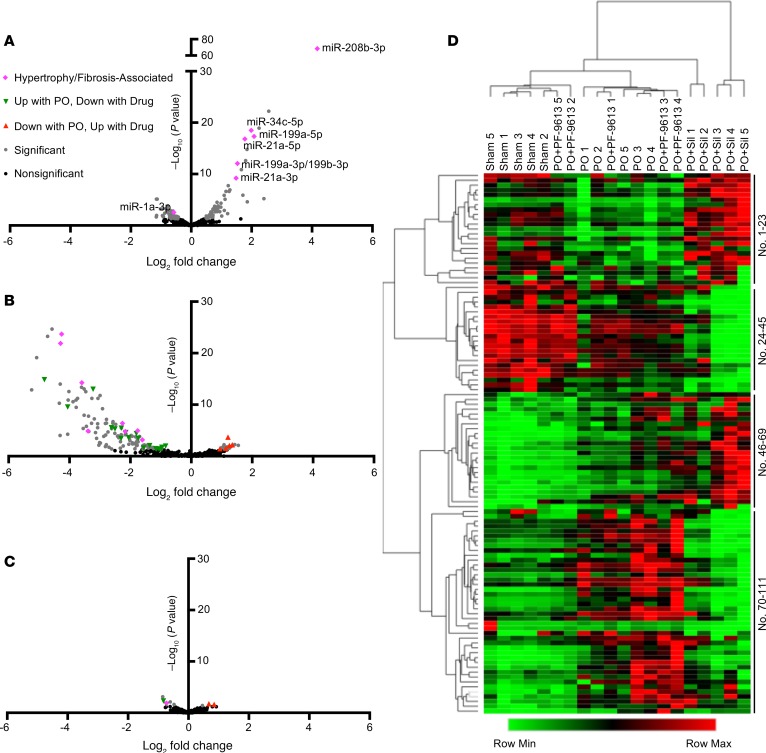
Figure 1 displays miR-seq results as volcano and heatmap plots for 3 group comparisons: PO+vehicle versus sham-control, PO+PDE5-I versus PO+vehicle, and PO+PDE9-I versus PO+vehicle. Each analysis was derived from whole myocardial tissue isolates obtained at terminal study (5 weeks after PO). With PO, 63 miRs significantly increased and 48 declined (Figure 1A and Supplemental Table 1). Many miRs reported to be either enhanced (miR-208b, 199, 199b, 21a, 34b, and 34c) or diminished (miR-133a, 133b, and 1a) by PO were observed in this analysis. PDE5-I reduced miR expression broadly (Figure 1B and Supplemental Table 2) as seen in the volcano plot with the vast majority of changes in the negative log-fold region. This either reversed or further diminished expression of many of the miRs altered by PO, and lowered others unchanged by PO. In stark contrast, PDE9-I minimally altered miRs associated with PO, impacting only 9, 5 of which reversed PO changes (Figure 1C and Supplemental Table 3). The magnitude of the change of even these 9 was modest. A heatmap using hierarchical clustering aggregated PO and PO+PDE9-I together, each being quite different from either sham or PO+PDE5-I groups (Figure 1D). The miR names for each row are provided in Supplemental Table 4. A heatmap clustered for miRs only is provided in Supplemental Figure 2.
Figure 1. PDE5-I and PDE9-I applied to PO heart yield disparate miR profiles.
Left ventricular myocardium from mice subjected to sham or PO surgery and subsequently given either vehicle, PDE5-I (Sil), or PDE9-I (PF-9613) were subjected to miR sequencing and subsequent differential expression analysis (n = 5 per group). (A) Volcano plot of miRs altered in PO versus sham, with miRs relevant to cardiac hypertrophy/fibrosis labeled. (B) Volcano plot of miRs altered in PO+Sil versus PO. (C) Volcano plot of miRs altered in PO+PF-9613 versus PO. For all volcano plots, dark gray dots indicate differentially expressed miRs; green triangles indicate miRs increased with PO, and decreased with drug treatment; red triangles indicate miRs decreased with PO, and increased with drug treatment; and pink diamonds indicate miRs labeled in panel A that are associated with cardiac hypertrophy and fibrosis (legend can be found in panel C). (D) Heatmap of all miRs changed significantly with PO for all treatment groups, clustered by both rows (miRs) and columns (samples). Row labels (i.e., miR names) can be found in Supplemental Table 4.
Transcriptome for each treatment shows many changes but few are overlapping.
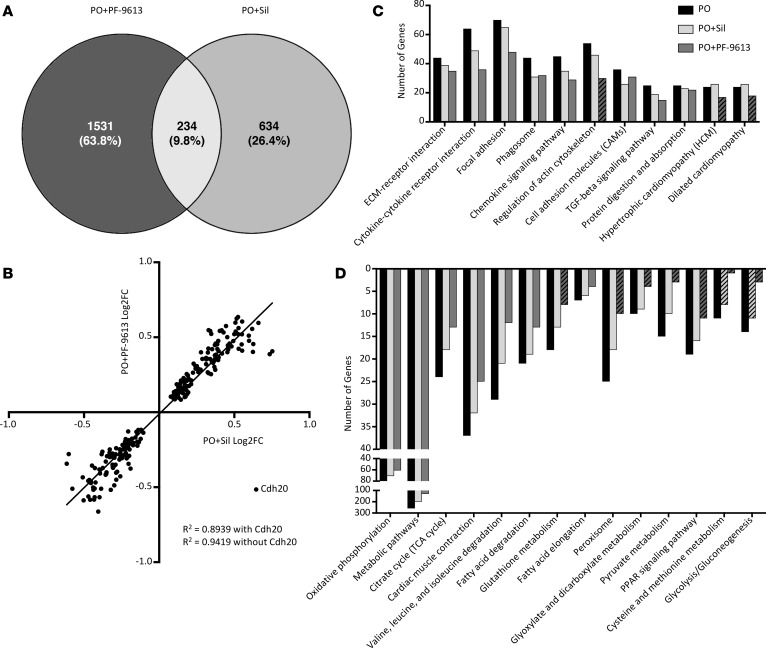
Given the role of miRs, these results might predict minimal transcriptome changes from PDE9-I, whereas PDE5-I treatment would be expected to more broadly alter mRNA expression. This was tested by RNA-seq on the same samples. To our surprise, more than twice as many genes were significantly altered by PDE9-I (1,756 genes) as compared with PDE5-I (868 genes) (Figure 2A), 87% and 73% of them being unique to PDE5-I or PDE9-I treatment, respectively. Among the shared genes, all but one changed in the same direction and magnitude (Figure 2B), the one exception being Cdh20 encoding cadherin-20 precursor.
Figure 2. Transcriptome for PDE5-I and PDE9-I shows many changes but few overlapping ones.
Samples from the same cohort of mice from Figure 1 were analyzed by RNA-seq. (A) RNA-seq analysis revealed 234 shared genes between PDE5-I (Sil) and PDE9-I (PF-9613), with more genes changed overall by PDE9-I (1,756) than PDE5-I (868). (B) Correlation analysis of fold changes of the genes shared between PDE5-I and PDE9-I. (C) Gene numbers in KEGG pathways identified to be upregulated in PO compared with sham for PO, PO+PDE5-I, and PO+PDE9-I. (D) Gene numbers in KEGG pathways identified to be downregulated in PO compared with sham for PO, PO+PDE5-I, and PO+PDE9-I. Striped bars in the KEGG pathway graphs indicate pathways that are not significantly different from sham.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis for the PO condition revealed typical changes, increasing extracellular matrix, cytoskeletal, and hypertrophy and heart failure–related genes, and decreasing metabolic pathway–related genes. While the specific genes altered by each treatment mostly differed, pathway analysis yielded similar functional clusters, with the number of genes altered declining relative to PO in some cases to levels similar to sham control (Figure 2, C and D). Thus, despite targeting a similar kinase pathway, PDE5-I and PDE9-I impacted genes very differently, while still converging on similar signaling pathways altered by PO stress.
PDE5-I– and PDE9-I–mediated miR disparities occur at late-stage processing.
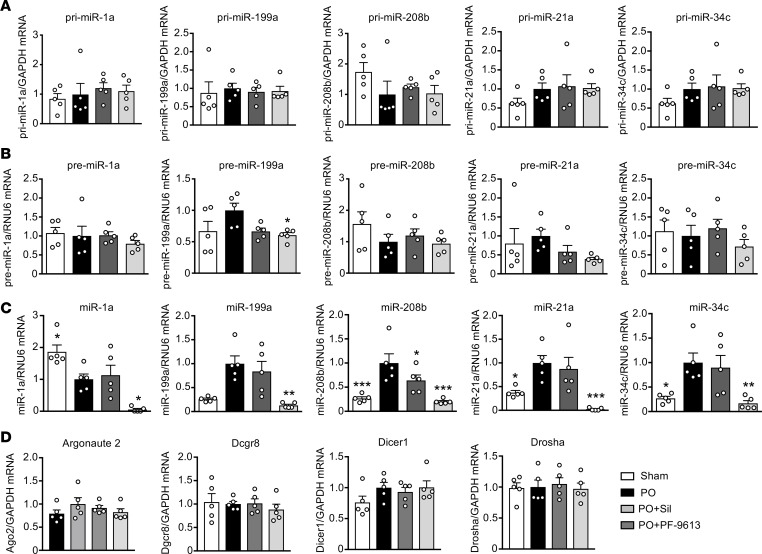
miRs are transcribed from the genome and processed from a pri to pre form in the nucleus by Drosha and DGCR8. The pre-miR is then exported to the cytosol, and converted to its mature form by Dicer and its partner TRBP, and growing evidence supports kinase signaling control over this process (16). No study has reported a specific influence of PKG, so we tested whether different miR profiles evolve from nuclear or cytosolic processing. We focused on a subset of relevant miRs (miR-1, 199, 208b, 21a, and 34c), each known to be involved with cardiac hypertrophy and/or fibrosis, and all expressed in cardiomyocytes (5, 17, 18). Pre- and pri-miR levels were similar between treatments (Figure 3, A and B), whereas differences in expression appeared in the mature miR as found by miR-seq (Figure 3C). Thus, the disparity in miR profiles from PDE5-I versus PDE9-I in the PO heart occurred at the level of cytosolic processing.
Figure 3. miR disparities from different PDE inhibitors occur at late-stage processing.
(A–C) qRT-PCR analysis for (A) pri-miRs, (B) pre-miRs, and (C) mature miRs for a panel of miRs selected from the larger sequencing data set that are associated with cardiac hypertrophy and fibrosis (pink diamond miRs from Figure 2, A–C). Analysis was performed on the same samples used for sequencing analysis (n = 5 per group). *P < 0.05, **P < 0.01, ***P < 0.001 as compared with PO. Data are presented as the mean ± SEM. Data were analyzed using 1-way ANOVA with Dunnett’s post hoc test, or Kruskal-Wallis test with Dunn’s post hoc test.
We further tested if either PDE5 or PDE9 inhibition modifies gene expression of the primary miR processing proteins, Argonaute (Ago2), microprocessor complex subunit DGCR8, Dicer1, and Drosha. There were no significant differences in expression for any of these genes between the groups (Figure 3D).
Effect on miRs from alternative stimulation of GC-1 or GC-2A signaling.
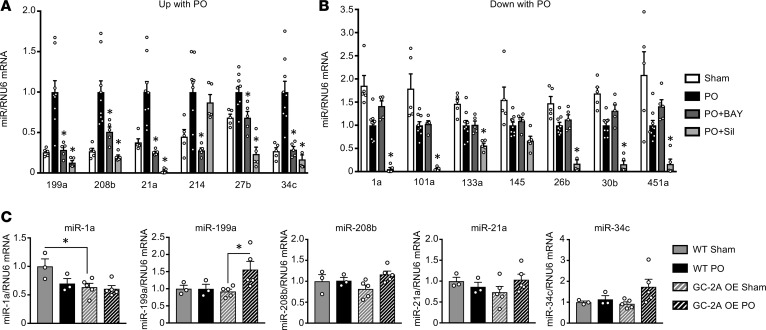
PDE5-I primarily regulates cGMP generated from GC-1, which is activated by NO. To further test if this pathway is important for broad suppression of miRs by PDE5-I, we studied mice exposed to PO and treated with a direct GC-1 activator, BAY 60-2770. BAY 60-2770 treatment also reduces cardiac hypertrophy, fibrosis, and improves function similar to PDE5-I (Supplemental Figure 3) (19). Thirteen miRs were assayed, each known to be relevant to pro- or antihypertrophic signaling (6–8, 20–31) and divided into those significantly increased or decreased by PO. Both PDE5-I and GC-1 activation displayed similar reductions of miRs that were increased by PO, (P = not significant, Figure 4A). However, among those that declined with PO (antihypertrophic miRs), PDE5-I further reduced 6 out of 7, whereas GC-1 activation impacted none (P < 0.05 by Fisher’s exact test, Figure 4B). Thus, triggering PKG by enhancing cGMP synthesis via GC-1 activation or by impeding its hydrolysis by PDE5-I had similar effects on miRs positively associated with hypertrophy, but only PDE5-I impacted miRs that were antihypertrophic. Thus, even different ways of stimulating the same pathway do not guarantee identical miR modulation.
Figure 4. Effect on miRs from alternative stimulation of GC-1 or GC-2A signaling.
(A and B) Left ventricular myocardium from mice subjected to sham or PO surgery and subsequently given either vehicle, PDE5-I (Sil), or an sGC activator (BAY 60-2770) were analyzed by qRT-PCR for (A) prohypertrophic miRs and (B) antihypertrophic miRs. Sil downregulates both categories of miRs, whereas BAY downregulates only prohypertrophic miRs. n = 4–11 per group. *P < 0.05 compared with PO for Sil and BAY analysis (smaller P values are not differentiated by different symbols, though some comparisons were very significant). (C) Mice overexpressing GC-2A and WT littermate controls were subjected to sham or PO surgery (n = 3–5 per group). Myocardial tissue was used for qRT-PCR analysis of miRs. *P < 0.05 as compared with the indicated group for GC-2A analysis. All data are presented as the mean ± SEM. Data were analyzed using 1-way ANOVA with Dunnett’s post hoc test, or Kruskal-Wallis test with Dunn’s post hoc test.
As myocardial PDE9 primarily hydrolyzes cGMP coupled to GC-2A (14), an alternative to PDE9-I is to sustain stimulation of this NP receptor–coupled cyclase. We used a genetic model with cardiomyocyte-targeted overexpression of GC-2A, which is also protective against PO-induced cardiac hypertrophy (32). As with PDE9-I, there were minimal changes in pro- or antihypertrophic miRs in PO myocardium from GC-2A overexpressors as compared with littermate controls (Figure 4C).
PDE5 inhibition in the normal heart and lung.
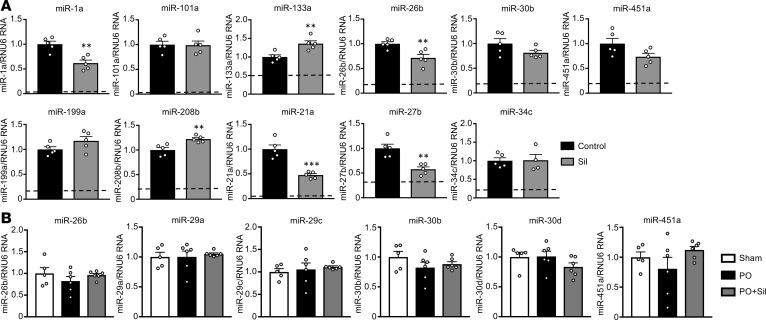
Given the broad impact of PDE5-I on miR expression, we wondered if similar effects would be observed under normal conditions or if a stressed or disease state was required. To test this, we exposed C57BL/6J mice to a PDE5-I (sildenafil) for 5 weeks and then assessed a representative panel of 11 miRs that all declined with this therapy in the PO heart. Of these, 6 showed significant changes (miR-1a, miR-21a, miR-26b, and miR-27b declined; miR-133a and miR-208b increased), so a majority (64%) were discordant with PO-induced changes (P = 0.0039 by Fisher’s exact test, Figure 5).
Figure 5. PDE5 inhibition in the normal heart and stressed lung.
(A) qRT-PCR was performed for various miRs in tissue from normal, nonstressed C57BL/6J mice treated with PDE5-I (Sil) for 5 weeks (n = 5 per group). Cardiac miRs that normally decrease with PO (top row) and increase with PO (bottom row) assessed from left ventricular tissue. Only miRs that were significantly decreased by Sil in Figure 4 were assayed. The dashed line in each graph represents the level to which Sil reduced miR expression in the PO model. (B) Lung tissue from C57BL/6J mice subjected to PO, PO+PDE5-I (Sil), or sham surgeries was analyzed by qRT-PCR for lung miRs (n = 5–6 per group). **P < 0.01, ***P < 0.001, ****P < 0.0001 as compared with control for sham Sil heart analysis. Data are presented as the mean ± SEM. For A, data were analyzed using 2-tailed unpaired t tests. For B, data were analyzed using 1-way ANOVA with Dunnett’s post hoc test, or Kruskal-Wallis test with Dunn’s post hoc test.
PDE5-I impacts the enzyme globally, including in the lung where PDE5 expression is prominent (33). To test if its effects on miR expression were organ independent, we examined lungs from normal and PO mice, assaying those known to be highly expressed in lung (34, 35). All were confirmed as being reduced in PO+PDE5-I myocardium by qPCR and/or miR-seq, yet none were altered in lungs by PDE5-I under either condition (Figure 5B and Supplemental Figure 4). This indicates that PDE5-I does not trigger a pathway broadly suppressing miR maturation machinery, as expected if Dicer or associated proteins were inhibited, and its effects are more prominent in the heart and largely depend on the existence of underlying disease.
Discussion
To our knowledge, the present study is the first to directly compare miR signatures from very closely related and phenotypically similar therapies (PDE5 and PDE9 inhibition) that both effectively counter the same pathological hypertrophic heart disease by ultimately activating the same protein kinase (i.e., PKG). The difference between them lies primarily in the subcellular localization of the 2 PDEs, and this ultimately defines where the effect is actuated. Despite strikingly similar improvement in PO-induced heart disease and many common features of both PDE5-I and PDE9-I, we found nearly opposite effects on miR expression, the former suppressing most of them while the latter had virtually no impact.
This study reveals several surprising aspects of miRs. First, it shows that their expression can be broadly and potently modified by a clinically widely used pharmaceutical, simultaneously reversing many miR changes pathophysiologically relevant to PO-induced heart disease. Importantly, this effect requires pharmaceutical biology in the appropriate intracellular compartment, and cannot be predicted purely based on the protein it affects. Secondly, it dramatically demonstrates that biological conditions that appear phenotypically very similar can have markedly disparate miR expression profiles. Lastly, it shows that miRs altered by disease do not need to be reversed for a treatment to effectively counter tissue- and organ-level pathology and pathophysiology. This poses some quandaries for their use as biomarkers for therapeutic responses.
Both PDE5 and PDE9 inhibition augment cGMP and activate PKG. This kinase activation confers antifibrotic and antihypertrophic effects by blocking Gq-protein receptor–coupled activation (36), and inhibiting transient receptor potential canonical ion channel (type 6) to blunt nuclear factor of activated T cells and serum response factor–regulated gene expression (14, 15, 37). The pools of PKG activated by either PDE5 or PDE9 inhibition share these properties, but also display downstream phosphorylation profiles unique to each intervention (14). We found only 2 prior studies reporting miRs altered by PDE5-I. Each involved very different models and did not assess miR expression broadly. For example, pig hearts subjected to cardiac arrest and resuscitation had reduced miR-1 and increased miR-133a with sildenafil treatment (38). Sildenafil reduced miR-1 in the current model as well. The second study examined adipose tissue miR expression in type 2 diabetic patients treated with chronic high-dose PDE5-I, and found PDE5-I decreased miR-22-3p expression (39). Again, the model and tissue examined are very different from the present work, and only selective analysis was provided. Our observation that PDE5-I does not simply suppress miR levels in normal mice or other noncardiac tissue from PO mice shows this is not a blanket effect, but more specific to the tissue and condition. We find no reports of miR changes with PDE9-I. Furthermore, the results with PDE9-I are unusual in that we also found no reports of a therapy conferring substantial benefits against disease yet with virtually no impact on miR signaling coupled with the disease. This does not mean gene transcription is unaltered, just that it is dissociated from miR regulation.
PDE5 and PDE9 each preferentially regulate different cGMP pools linked to either NO or NP signaling, respectively. However, we found even targeting presumably the same cGMP pool but by different strategies — e.g., GC-1 activation versus PDE5-I — that miR expression profiles consistently differed while organ- and tissue-level improvement of underlying heart disease was very similar (19). The specific differences were intriguing, as unlike PDE9-I, or upregulation of the NP-associated GC-2A cyclase, GC-1 activation and PDE5-I both consistently reduce expression of prohypertrophic miRs that are otherwise increased by PO. However, miRs that decline with PO are unaltered by GC-1 activation, but reduced further by PDE5-I. Statistically, this consistent pattern was unlikely due to chance, and shows that regional kinase signaling within the cytosol can yield directionally cohesive changes in miR expression. This too may relate to compartment differences between PDE5 and GC-1, with GC-1 residing in both the cytosol and at the plasma membrane (40), whereas PDE5 normally localizes to the sarcomere (14, 41). PDE5-I can only alter cGMP in the nanodomain in which it is expressed; while GC-1 stimulation generates cGMP that can also be regulated by other PDEs besides PDE5 (42), so their impact is not identical.
The mechanism by which PDE5-I broadly alters miR expression in the PO heart remains unknown. Given that pri- and pre-miRs are similar between PDE5-I and PDE9-I, the disparity seems likely to reside at the level of cytosolic Dicer/Argonaute miR processing and/or ribonucleases (16). While gene expression of the relevant proteins was unchanged, this does not preclude their posttranslational modification. Our phospho-kinome analysis of both PDE5-I and PDE9-I treatment in PO did not find changes in miR processing proteins from either (14), though lack of detection does not mean they do not exist. An example where kinase modulation of miR processing occurs is Argonaute 2 phosphorylation by casein kinase-1 α1 that broadly impacts miR-gene suppression (43). This study utilized a genomic CRISPR/Cas9 knockout strategy to identify proteins that blunt miR processing. Of note, PKG was not identified in this screen. Furthermore, if PKG had a general effect on Dicer/Argonaute processing, one would expect PDE5-I to suppress miR expression in other tissues and conditions (e.g., with or without disease), but this was not observed. Whatever changes PDE5 can induce also appear tissue and condition specific, as similar broad suppression was not observed in normal hearts or the lung. It is also unlikely that PDE5-I would necessarily ameliorate diseases generated by artificial overexpression of a pathogenic miR, or that its expression/activity is altered by such models. Unfortunately, the obvious reductionist model system — isolated cultured myocytes — proved inadequate for replicating miR signatures found in vivo. This is likely due to the absence of required environmental cues and stresses present in intact hearts. Mechanistic dissection of the current signaling would appear to require more complex in vivo genetic models where viability is often a limitation.
With respect to why PDE9 inhibitors do not affect miR expression in the PO model, this too may relate to where its cGMP modulation occurs. PDE9 functional interaction with NP receptor–coupled cGMP and colocalization with sarcoplasmic reticular membranes suggest compartments removed from cytosolic miR processing. It is not due to a failure to get the inhibitor into the myocyte, as we reported potent in vivo and in vitro effects on cGMP signaling from PDE9 inhibition, including effects on fibrotic gene expression and hypertrophy markers, that are absent when the gene for the protein is silenced (14). As shown here, PDE9-I also has marked effects on gene expression — it just does so without altering miRs, and like PDE5-I inhibition, impacts posttranslational protein phosphorylation.
In summary, we have shown how dramatically different miR expression profiles can be associated with similarly targeted kinase activation strategies that successfully treat the identical heart disease — one broadly suppressing expression and the other leaving it virtually unchanged. The results indicate that improving myocardial and chamber disease phenotype can be achieved without engaging miRs, and that with or without this engagement, gene expression impacting similar signaling pathways can still occur. Lastly, we reveal a barcode-like signature provided by miR profiles that can distinguish between treatments targeting the same overall pathway. This may provide a useful tool for drug development and precision therapy.
Methods
Cardiac PO model.
PO was induced by transaortic constriction, performed as previously described (14). Size-, age-, and sex-matched (male) C57BL/6J mice (Jackson Labs) were used for all drug intervention studies. Sham controls underwent similar surgery without ligature placement. Mice were followed for up to 6 weeks after PO, and were cotreated with PDE5-I (sildenafil, Pfizer, 200 mg/kg/day in Bioserv soft diet), PDE9-I (PF-04449613, Pfizer, 30 mg/kg twice daily by oral gavage), BAY 60-2770 (Bayer, 0.3 mg/kg/day once daily by oral gavage), or appropriate matched vehicle. Treatment started 1 week after PO. For the drug intervention studies, animals dying prior to drug assignment or that failed to develop disease after PO were excluded from analysis. The GC-2A overexpressor mice were generated as previously described (32), and subjected to PO or sham surgery. Animals were followed for 3 weeks after PO.
Conscious mouse echocardiography.
Intact heart morphology and function was determined in conscious mice by serial M-mode transthoracic echocardiography (VisualSonics Vevo 2100, 18–38 MHz linear array transducer; SanoSite Incorporated). Images were obtained and analyzed by an individual blinded to the animal condition.
miR-seq.
Total RNA was isolated from left ventricular myocardium using Qiazol Reagent and the miRNeasy kit (Qiagen) according to the manufacturer’s protocol. RNA was analyzed on an Agilent Bioanalyzer for RNA integrity number (RIN) values and concentrations. Only samples with RIN greater than 7 were used for sequencing. Libraries were prepared using the Illumina TruSeq Small RNA Sample Prep Kit. Sequencing was performed on an Illumina HiSeq 2500. Illumina’s CASAVA 1.8.4 was used to convert BCL files to FASTQ files. Sequencing read filtering and adapter trimming, read alignment, and generation of count data and RPM data were done using the miRge program v1.0 (44), which incorporates Cutadapt v1.18 and Bowtie v1.1.1. Differential expression analysis was performed using Bioconductor’s DESeq package (v1.26.0) (45). miRs were filtered according to the following: more than 50% of mice had reads for a given miR, and the miR was present in the Mus musculus miRgeneDB database (46). Heatmaps were generated using the Morpheus program (Broad Institute). Clustering was performed by the one minus Spearman rank correlation. The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (47), and are accessible through GEO Series accession number GSE112056 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE112056).
RNA-seq.
RNA samples were prepared and analyzed as described for miR-seq. Libraries were generated using the TruSeq Stranded Total RNA sample preparation kit (Illumina), and sequenced on an Illumina HiSeq 2500. Illumina’s CASAVA was used to convert BCL files to FASTQ files. RNA-seq reads were trimmed and mapped to the Ensembl mouse reference genome (mm10) using HISAT2 version 2.0.5 (48). Transcript counts were quantified using RSEM v1.3.0 with default parameters (49). Differential expression analysis of genes between different treatments was performed using R package DESeq2 v1.18.1 (50). Pathway enrichment analysis was done using the KEGG database and R package KEGG (db v3.2.3) (51). The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (47), and are accessible through GEO Series accession number GSE112056 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE112056).
Gene expression — qRT-PCR.
Total RNA was isolated from left ventricular myocardium, lung, or cultured cells using Qiazol Reagent and the miRNeasy kit (Qiagen) according to the manufacturer’s protocol. Reverse transcription (RT) to cDNA was performed using the miScript II RT kit (Qiagen). cDNA underwent PCR amplification for mRNA expression using TaqMan probes for atrial NP (ANP) (mouse Mm01255747_g1), brain or B-type NP (BNP) (mouse Mm01255770_g1), Argonaute 2 (Ago2) (mouse Mm00838341_m1), DGCR8 (mouse Mm01146851_m1), Dicer1 (mouse Mm00521722_m1), Drosha (mouse Mm01310009_m1), or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (mouse 99999915_g1) (Applied Biosystems). For mature miR expression, Qiagen miScript probes were used for miR-1a (mouse MS00011004), miR-101a (mouse MS00011011), miR-133a (mouse MS00032305), miR-145 (mouse MS00001631), miR-195 (mouse MS00001792), miR-199 (mouse MS00032529), miR-208b (mouse MS00011466), miR-21 (mouse MS00011487), mir-214 (mouse MS00032571), miR-26b (mouse MS00001344), mir-27b (mouse MS00001358), miR-29 (mouse MS00001372), miR-29c (mouse MS00001379), miR-30b (mouse MS00001386), miR-30d (mouse MS00011746), miR-34c (mouse MS00001422), miR-451a (mouse MS00002408), and RNU6-2 (human, MS00033740). For pre-miR expression, Qiagen probes were used for pre-miR-1a (mouse MP00003990), pre-miR-199 (mouse MP00004970), pre-miR-208b (mouse MP00005082), pre-miR-21a (mouse MP00005103), and pre-miR-34c (mouse MP00005628). For pri-miR expression, Taqman probes were used for pri-miR-1a (mouse Mm03306163_pri), pri-miR-199 (mouse 03306505_pri), pri-miR-208b (mouse Mm03308667_pri), pri-miR-21a (mouse Mm03306822_pri), and pri-miR-34c (mouse Mm03306660_pri). The threshold cycle value was determined using the crossing point method. Samples were normalized to the GAPDH value for each Taqman run, and RNU6-2 for each miScript run.
Statistics.
All values are presented as mean ± SEM. For analysis of multiple independent groups, a 1-way ANOVA or Kruskal-Wallis test was used with a post hoc Tukey’s (or Dunn’s) multiple-comparisons test to assess group differences. Post hoc testing was only performed if the 1-way ANOVA was significant (P < 0.001 in all cases). For analysis of only 2 independent groups, an unpaired 2-tailed t test was performed. Analysis was performed using GraphPad Prism software (ver. 7a, 2016).
Study approval.
All animal studies were approved by either the Johns Hopkins Medical Institutions Animal Care and Use Committee (Baltimore, MD, USA) or the University of Würzburg animal care committee (Würzburg, Germany).
Author contributions
KMKS designed and performed most of the experiments and analyzed data, as well as wrote the manuscript. AS analyzed RNA-seq data. TN and MJR designed and performed experiments. GZ performed PO surgery and DB performed echocardiography analysis for all mice subjected to PO at Johns Hopkins. MK designed and performed PO experiments with GC-2A–overexpressing mice. MKH analyzed miR-seq data. DIL designed and performed experiments. DAK conceived the study and assisted in experimental design and interpretation, and edited the manuscript.
Supplementary Material
Acknowledgments
This work was supported by NIH — National Heart Lung and Blood Institute grants HL-135827, HL-119012, HL-089297, HL-07227 (to D.A. Kass), and F31HL134196 (to K.M. Kokkonen-Simon). This work was also supported by the American Heart Association Strategically Focused Research Network 16SFRN28620000 grant (to D.A. Kass) and Post-Doctoral Fellowships (to M.J. Ranek, D.I. Lee, and T. Nakamura), as well as the Abraham and Virginia Weiss Professorship (to D.A. Kass).
Version 1. 08/09/2018
Electronic publication
Footnotes
TN’s present address is: Department of Medical Information Science and Administration Planning, Kumamoto University Hospital, Kumamoto, Japan.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(15):e121739. https://doi.org/10.1172/jci.insight.121739.
Contributor Information
Amir Saberi, Email: asaberi1@jhu.edu.
Taishi Nakamura, Email: taishin@kumamoto-u.ac.jp.
Mark J. Ranek, Email: mranek1@jhmi.edu.
Guangshuo Zhu, Email: gzhu8@jhmi.edu.
Djahida Bedja, Email: dbedja1@jhmi.edu.
Michaela Kuhn, Email: michaela.kuhn@mail.uni-wuerzburg.de.
David A. Kass, Email: dkass@jhmi.edu.
References
- 1.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 2.Olson EN. MicroRNAs as therapeutic targets and biomarkers of cardiovascular disease. Sci Transl Med. 2014;6(239):239ps3. doi: 10.1126/scitranslmed.3009008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shah R, et al. MicroRNAs associated with reverse left ventricular remodeling in humans identify pathways of heart failure progression. Circ Heart Fail. 2018;11(2):e004278. doi: 10.1161/CIRCHEARTFAILURE.117.004278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dangwal S, Schimmel K, Foinquinos A, Xiao K, Thum T. Noncoding RNAs in heart failure. Handb Exp Pharmacol. 2017;243:423–445. doi: 10.1007/164_2016_99. [DOI] [PubMed] [Google Scholar]
- 5.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316(5824):575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 6.van Rooij E, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA. 2006;103(48):18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carè A, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13(5):613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 8.Karakikes I, et al. Therapeutic cardiac-targeted delivery of miR-1 reverses pressure overload-induced cardiac hypertrophy and attenuates pathological remodeling. J Am Heart Assoc. 2013;2(2):e000078. doi: 10.1161/JAHA.113.000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ucar A, et al. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat Commun. 2012;3:1078. doi: 10.1038/ncomms2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Montgomery RL, et al. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124(14):1537–1547. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vegter EL, van der Meer P, de Windt LJ, Pinto YM, Voors AA. MicroRNAs in heart failure: from biomarker to target for therapy. Eur J Heart Fail. 2016;18(5):457–468. doi: 10.1002/ejhf.495. [DOI] [PubMed] [Google Scholar]
- 12.Castro-Villegas C, et al. Circulating miRNAs as potential biomarkers of therapy effectiveness in rheumatoid arthritis patients treated with anti-TNFα. Arthritis Res Ther. 2015;17:49. doi: 10.1186/s13075-015-0555-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez JP, Kos A, Turecki G. Major depression and its treatment: microRNAs as peripheral biomarkers of diagnosis and treatment response. Curr Opin Psychiatry. 2018;31(1):7–16. doi: 10.1097/YCO.0000000000000379. [DOI] [PubMed] [Google Scholar]
- 14.Lee DI, et al. Phosphodiesterase 9A controls nitric-oxide-independent cGMP and hypertrophic heart disease. Nature. 2015;519(7544):472–476. doi: 10.1038/nature14332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takimoto E, et al. Chronic inhibition of cyclic GMP phosphodiesterase 5A prevents and reverses cardiac hypertrophy. Nat Med. 2005;11(2):214–222. doi: 10.1038/nm1175. [DOI] [PubMed] [Google Scholar]
- 16.Shen J, Hung MC. Signaling-mediated regulation of microRNA processing. Cancer Res. 2015;75(5):783–791. doi: 10.1158/0008-5472.CAN-14-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5(3):R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laine SK, Alm JJ, Virtanen SP, Aro HT, Laitala-Leinonen TK. MicroRNAs miR-96, miR-124, and miR-199a regulate gene expression in human bone marrow-derived mesenchymal stem cells. J Cell Biochem. 2012;113(8):2687–2695. doi: 10.1002/jcb.24144. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura T, et al. Prevention of PKG-1α oxidation suppresses antihypertrophic/antifibrotic effects from PDE5 inhibition but not sGC stimulation. Circ Heart Fail. 2018;11(3):e004740. doi: 10.1161/CIRCHEARTFAILURE.117.004740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Q, et al. Attenuation of microRNA-1 derepresses the cytoskeleton regulatory protein twinfilin-1 to provoke cardiac hypertrophy. J Cell Sci. 2010;123(Pt 14):2444–2452. doi: 10.1242/jcs.067165. [DOI] [PubMed] [Google Scholar]
- 21.Li R, et al. miR-145 inhibits isoproterenol-induced cardiomyocyte hypertrophy by targeting the expression and localization of GATA6. FEBS Lett. 2013;587(12):1754–1761. doi: 10.1016/j.febslet.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, et al. miR-199a impairs autophagy and induces cardiac hypertrophy through mTOR activation. Cell Death Differ. 2017;24(7):1205–1213. doi: 10.1038/cdd.2015.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei L, et al. MicroRNA-101 inhibits rat cardiac hypertrophy by targeting Rab1a. J Cardiovasc Pharmacol. 2015;65(4):357–363. doi: 10.1097/FJC.0000000000000203. [DOI] [PubMed] [Google Scholar]
- 24.Callis TE, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest. 2009;119(9):2772–2786. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng Y, Zhang C. MicroRNA-21 in cardiovascular disease. J Cardiovasc Transl Res. 2010;3(3):251–255. doi: 10.1007/s12265-010-9169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han M, et al. GATA4 expression is primarily regulated via a miR-26b-dependent post-transcriptional mechanism during cardiac hypertrophy. Cardiovasc Res. 2012;93(4):645–654. doi: 10.1093/cvr/cvs001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, et al. Cardiomyocyte overexpression of miR-27b induces cardiac hypertrophy and dysfunction in mice. Cell Res. 2012;22(3):516–527. doi: 10.1038/cr.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang T, et al. MicroRNA-214 provokes cardiac hypertrophy via repression of EZH2. Biochem Biophys Res Commun. 2013;436(4):578–584. doi: 10.1016/j.bbrc.2013.05.079. [DOI] [PubMed] [Google Scholar]
- 29.Bernardo BC, et al. Therapeutic inhibition of the miR-34 family attenuates pathological cardiac remodeling and improves heart function. Proc Natl Acad Sci USA. 2012;109(43):17615–17620. doi: 10.1073/pnas.1206432109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He J, et al. MicroRNA-30b-5p is involved in the regulation of cardiac hypertrophy by targeting CaMKIIδ. J Investig Med. 2013;61(3):604–612. doi: 10.2310/JIM.0b013e3182819ac6. [DOI] [PubMed] [Google Scholar]
- 31.Song L, et al. MiR-451 is decreased in hypertrophic cardiomyopathy and regulates autophagy by targeting TSC1. J Cell Mol Med. 2014;18(11):2266–2274. doi: 10.1111/jcmm.12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kishimoto I, Rossi K, Garbers DL. A genetic model provides evidence that the receptor for atrial natriuretic peptide (guanylyl cyclase-A) inhibits cardiac ventricular myocyte hypertrophy. Proc Natl Acad Sci USA. 2001;98(5):2703–2706. doi: 10.1073/pnas.051625598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin CS, Lin G, Xin ZC, Lue TF. Expression, distribution and regulation of phosphodiesterase 5. Curr Pharm Des. 2006;12(27):3439–3457. doi: 10.2174/138161206778343064. [DOI] [PubMed] [Google Scholar]
- 34.Williams AE, Perry MM, Moschos SA, Lindsay MA. microRNA expression in the aging mouse lung. BMC Genomics. 2007;8:172. doi: 10.1186/1471-2164-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao F, et al. Changes in miRNA in the lung and whole blood after whole thorax irradiation in rats. Sci Rep. 2017;7:44132. doi: 10.1038/srep44132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takimoto E, et al. Regulator of G protein signaling 2 mediates cardiac compensation to pressure overload and antihypertrophic effects of PDE5 inhibition in mice. J Clin Invest. 2009;119(2):408–420. doi: 10.1172/JCI35620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis J, Burr AR, Davis GF, Birnbaumer L, Molkentin JD. A TRPC6-dependent pathway for myofibroblast transdifferentiation and wound healing in vivo. Dev Cell. 2012;23(4):705–715. doi: 10.1016/j.devcel.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Q, Wang G, Yuan W, Wu J, Wang M, Li C. The effects of phosphodiesterase-5 inhibitor sildenafil against post-resuscitation myocardial and intestinal microcirculatory dysfunction by attenuating apoptosis and regulating microRNAs expression: essential role of nitric oxide syntheses signaling. J Transl Med. 2015;13:177. doi: 10.1186/s12967-015-0550-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fiore D, et al. PDE5 inhibition ameliorates visceral adiposity targeting the miR-22/SIRT1 pathway: Evidence from the CECSID trial. J Clin Endocrinol Metab. 2016;101(4):1525–1534. doi: 10.1210/jc.2015-4252. [DOI] [PubMed] [Google Scholar]
- 40.Tsai EJ, et al. Pressure-overload-induced subcellular relocalization/oxidation of soluble guanylyl cyclase in the heart modulates enzyme stimulation. Circ Res. 2012;110(2):295–303. doi: 10.1161/CIRCRESAHA.111.259242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takimoto E, et al. cGMP catabolism by phosphodiesterase 5A regulates cardiac adrenergic stimulation by NOS3-dependent mechanism. Circ Res. 2005;96(1):100–109. doi: 10.1161/01.RES.0000152262.22968.72. [DOI] [PubMed] [Google Scholar]
- 42.Kokkonen K, Kass DA. Nanodomain regulation of cardiac cyclic nucleotide signaling by phosphodiesterases. Annu Rev Pharmacol Toxicol. 2017;57:455–479. doi: 10.1146/annurev-pharmtox-010716-104756. [DOI] [PubMed] [Google Scholar]
- 43.Golden RJ, et al. An Argonaute phosphorylation cycle promotes microRNA-mediated silencing. Nature. 2017;542(7640):197–202. doi: 10.1038/nature21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baras AS, et al. miRge - A multiplexed method of processing small RNA-Seq data to determine microRNA entropy. PLoS ONE. 2015;10(11):e0143066. doi: 10.1371/journal.pone.0143066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fromm B, et al. A uniform system for the annotation of vertebrate microRNA Genes and the evolution of the human microRNAome. Annu Rev Genet. 2015;49:213–242. doi: 10.1146/annurev-genet-120213-092023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Carlson M. KEGG.db: A set of annotation maps for KEGG. R package version 3.2.3. Bioconductor. https://bioconductor.org/packages/release/data/annotation/html/KEGG.db.html Accessed July 24, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.