Abstract
Positron emission tomography (PET) imaging of functional metabolism has been widely used to investigate functional recovery and to evaluate therapeutic efficacy after stroke. The voxel intensity of a PET image is the most important indicator of cellular activity, but is affected by other factors such as the basal metabolic ratio of each subject. In order to locate dysfunctional regions accurately, intensity normalization by a scale factor is a prerequisite in the data analysis, for which the global mean value is most widely used. However, this is unsuitable for stroke studies. Alternatively, a specified scale factor calculated from a reference region is also used, comprising neither hyper- nor hypo-metabolic voxels. But there is no such recognized reference region for stroke studies. Therefore, we proposed a totally data-driven automatic method for unbiased scale factor generation. This factor was generated iteratively until the residual deviation of two adjacent scale factors was reduced by < 5%. Moreover, both simulated and real stroke data were used for evaluation, and these suggested that our proposed unbiased scale factor has better sensitivity and accuracy for stroke studies.
Keywords: Unbiased scale factor, Intensity normalization, Stroke, FDG-PET imaging, Voxel-wise analysis
Introduction
Positron emission tomography (PET) imaging of functional metabolism using fluorodeoxyglucose (18F-FDG) [1–5] has been widely used to investigate brain functions after stroke in both humans and animals. It images the distribution of the biological radiotracer at the molecular level with high sensitivity. The radiotracer FDG is directly correlated with energy consumption that supports synaptic activities and is presented as FDG-PET imaging intensity [6]. In each subject, the voxel intensity of hypo-metabolic regions is lower and that of hyper-metabolic regions is higher than normal. Therefore, the mission of FDG-PET data analysis is to automatically and accurately identify the dysfunctional regions.
However, in group analysis, the FDG-PET image intensity is not only determined by cellular activity, but is also affected by several other controllable factors, such as the injected dose of 18F-FDG, the total scan time, and the PET imaging count rate, as well as the uncontrollable factor of basal metabolic ratio in each subject [7, 8]. Therefore, despite the influence of brain diseases, the basic level of image intensity varies in each individual, and this should be eliminated prior to statistical analysis. Generally, a unified standardized operation is used to eliminate the controllable factors. To remove the internal influences, an intensity normalization algorithm is most commonly used.
The intensity normalization of FDG-PET images is frequently carried out by noise removal and subsequent global intensity scaling by a scale factor. The global mean value is one of the most popular scale factors, based on the assumption of a rather small area of dysfunction, but in stroke studies this does not perform well. In the acute stage after stroke, for example, there are large numbers of hypo-metabolic voxels because of the ischemia and edema [4, 5]. Including these voxels in scale factor calculations would lead to considerable false-positive results. Several studies have found that inhibition of the hyper-metabolic regions promotes functional recovery after stroke, so false-positive results must be avoided. Therefore, the global mean value is not a perfect scale factor for stroke studies.
Alternatively, in several studies, a specified scale factor has been used instead of the global mean value, in which the mean value of several reference regions is chosen according to a priori knowledge. These reference regions are always those that have been demonstrated to have the least correlation with the corresponding disease, such as cerebellar and pontine areas in Alzheimer disease studies [9]. However, in stroke studies, there are not only known hypo-metabolic regions related to ischemia and motor and cognitive impairment [10], but also unknown hyper-metabolic regions related to functional compensation [11]. So far, there are no commonly used reference regions in stroke studies [9]. Ideally, the reference region should be unbiased so that it contains neither hyper-metabolic nor hypo-metabolic voxels.
Given the above, we designed the current study to introduce totally data-driven and automatic method of unbiased scale factor generation for intensity normalization in FDG-PET imaging analysis. To evaluate the practicality and accuracy of this method, several sets of simulated stroke data were generated from a group of FDG-PET images of healthy rats. To further verify the proposed method of scale factor setting, a group of FDG-PET image data from groups of rats with and without left middle cerebral artery occlusion (MCAO) were compared.
Materials and Methods
The Unbiased Scale Factor Generation Method
The scale factor in FDG-PET image analysis is used to account for global confounds, so it should be uncorrelated with a specific brain disease, such as stroke. In this study, our concept for unbiased scale factor generation was to remove regions with abnormal values before calculation.
Therefore, the proposed unbiased scale factor was generated iteratively in a data-driven manner, in which the conventional global mean value was chosen as the initial factor. The generation procedure (Fig. 1) can be summarized as follows. (1) All the individual images were preprocessed and divided into several groups. (2) The global mean value was chosen as the initial scale factor (SF). (3) Statistical analysis, such as the two-sample t-test or one-way ANOVA, was performed to quantify the differences in FDG signals between groups, in which proportional scaling and intensity normalization based on SF were applied to account for global confounds. (4) Several regions with significant FDG changes were obtained based on a threshold of voxel-level height and cluster size. These regions were regarded as hyper/hypo-metabolic voxels and excluded from the new scale-factor calculation. The mean value of the remaining voxels was then calculated (SFn). (5) SFn was selected as a new SF and steps 3–5 were repeated until the residual deviation between the SFn and SF was reduced by < 5%. (6) The last SF was taken as the optimal unbiased scale factor.
Fig. 1.
Schematic of unbiased, totally data-driven, scale factor generation.
Animals and Data Acquisition
Twenty-two Sprague-Dawley rats of either sex (11 males), 10 weeks–13 weeks old, weighing 350 g ± 20 g, were used for quantitative evaluation of the influence of the scale factor in voxel-wise analysis. Eleven of the rats (6 males) were randomly selected for simulated data sets and the other 11 served as healthy controls. Another group of 16 male rats, 9 weeks–11 weeks old, weighing 300 g ± 20 g, was also used, of which 8 underwent left MCAO and the other 8 were healthy controls. Intraluminal occlusion of the MCA was accomplished using a modification of the Longa technique [7, 12]. All the rats were deprived of food for 12 h–15 h before 18F-FDG injection, but had free access to drinking water [13].
Prior to PET scanning, 18F-FDG was prepared at the PET Center of the China PLA General Hospital. For each rat, 18F-FDG (18.5 MBq/100 g body weight) was administered via tail vein injection without anesthesia. Then the rats were kept in their cages in a room with minimal ambient noise for 18F-FDG uptake. The uptake period was 40 min for maximization of uptake in the brain [14]. Then the rats were anesthetized with isoflurane inhalation (2% in 100% oxygen; IsoFlo, Hebei Jiumu Pharma, Ltd, China) using a nose cone.
For the 22 rats used for simulated data set generation, FDG-PET imaging was performed at the PET Center of the China PLA General Hospital in a MicroPET/CT imaging system (eXplore Vista-CT, General Electric, USA), of which the radial spatial resolution was 1.0 mm full-width at half-maximum (FWHM) at the center of the field of view (FOV). During FDG-PET scans, all the rats were anesthetized with isoflurane (as above) and placed in the scanner in prone position with a plastic stereotactic head holder on the scanner bed. The brain was centered in the FOV for a static acquisition of 10 min. Images were subsequently reconstructed using the 3D Ordered Set Expectation Maximization algorithm (General Electric). Corrections for dead time, decay, attenuation, random coincidences, and scattering were applied. Images were reconstructed on a 175 × 175 × 61 matrix, where the voxel size was 0.39 mm × 0.39 mm × 0.77 mm. All scans were saved in Analyze format.
To demonstrate that our proposed method is not affected by different imaging techniques, FDG-PET images of the 8 rats with MCAO and 8 without were acquired using another imaging system. Twenty-four hours after operation, FDG-PET images were acquired on a Siemens Inveon PET (Siemens Medical Solutions, USA), of which the radial spatial resolution was 1.4 mm FWHM at the center of the FOV. During FDG-PET scans, all rats were anesthetized with isoflurane (as above) and placed in the scanner in prone position with a plastic stereotactic head holder on the scanner bed. The brain was centered in the FOV for a static acquisition for 20 min. The images were subsequently reconstructed using the Filtered Back Projection (Siemens Medical Solutions, USA) algorithm. Corrections for dead time, decay, attenuation, random coincidences, and scattering were applied. Images were reconstructed on a 128 × 128 × 159 matrix, where the voxel size was 1.4 mm × 1.4 mm × 0.79 mm. All scans were saved in Analyze format.
All experiments were performed with the approval of the Animal Care and Use Committee of the Chinese Academy of Sciences.
Simulated FDG-PET Data Sets
The simulated FDG-PET data sets were created by decreasing voxel values [15] and the details were as follows. First, the body tissues and background of all 22 images were manually removed using MRIcro (Chris Rorden’s Neuropsychology Lab) [16] and the origin of the image was repositioned at dorsal 3rd ventricle (D3V), which corresponded to the standard FDG-PET template in Paxinos & Watson space [17]. Then, individual images of the brain were spatially normalized into Paxinos & Watson space, comprising scaling up the voxel size in the Analyze header by a factor of 4 [18–21], registering to the FDG-PET template, subsequently removing extracranial tissues via the intracranial image, and shearing the matrix to cut off the background.
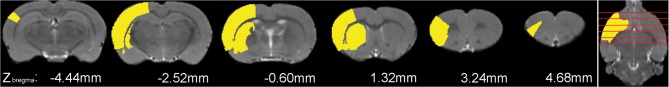
The MCAO data set simulation represented the actual situation of the acute stage after intraluminal occlusion. At 24 h after the operation, the hypo-metabolic regions in FDG-PET images not only contained the infarction area, but also the edematous and functionally impaired areas (e.g., motor impairment areas). Therefore, according to the FDG-PET images of the MCAO rat model, several unilateral regions were selected, comprising the corpus striatum and sensory cortex (Fig. 2). However, as the location and extent of hyper-metabolic regions were unclear, only clearly hypo-metabolic regions were simulated. Because of individual variations, the attenuation of FDG-PET signal amplitude differed within the range of 10%–70%. Hence, the voxel value was decreased by seven levels (10%, 20%, 30%, 40%, 50%, 60%, and 70%) to simulate cellular dysfunction after stroke. The other 11 rats served as the control group. Finally, all the simulated data sets of both groups were smoothed by a Gaussian kernel of 2 mm × 2 mm × 4 mm FWHM.
Fig. 2.
Superimposition of the simulated stroke region on the corresponding structural rat brain image in Paxinos & Watson space. The simulated region was extracted from an atlas image in Paxinos & Watson space, mainly comprising the caudate-putamen, somatosensory cortex, claustrum, and dorsal endopiriform nucleus. Six coronal planes were selected at coordinates Zbregma − 4.44 mm, − 2.52 mm, − 0.60 mm, 1.32 mm, 3.24 mm, and 4.68 mm. The simulated region is indicated in yellow, while the structural brain image is presented in gray-scale as background.
Voxel-Wise Analysis of Simulated FDG-PET Data Sets
Voxel-wise analyses based on different scale factors were performed in the spmratIHEP toolbox [18, 19] of SPM8 (Wellcome Department of Cognitive Neurology, London, UK). In each voxel-wise analysis, the simulated MCAO group was compared with the healthy control group.
In spmratIHEP, all the smoothed simulated data were analyzed voxel-wise, based on the framework of the general linear model (GLM). In order to identify differences in the FDG signals between the simulated data sets with MCAO and healthy controls, we used the two-sample t-test in SPM8. Proportional scaling and intensity normalization were applied to account for global confounds, based on three different scale factors: the truth value, the conventional global mean value, and our proposed unbiased value. In the simulated data sets, the truth value was calculated from the contralateral voxels and regarded as the ‘gold-standard’ reference. Our proposed unbiased value was generated based on the threshold of P < 0.001 (uncorrected) and a cluster size of no less than 50 voxels. As detailed above, this unbiased value was generated iteratively until the residual deviation between the nearest two values was reduced by < 5%. Finally, based on these three scale factors, brain regions with significant FDG changes were identified. The voxel-level height threshold was P < 0.05 (FWE corrected) and the cluster size was ≥ 50 voxels.
At this point, these results of the voxel-wise statistical analysis based on different scale factors were ready for further qualitative and quantitative evaluations.
Quantitative Evaluation of the Simulated FDG-PET Data Sets
The accuracy of the different scale factors was quantitatively evaluated from the voxel-wise analysis results. Using the statistical result generated by the truth mean value as the reference standard, the accuracy of the proposed unbiased value and the global mean value were evaluated with two volumetric and spatial indexes:
- The Dice similarity coefficient (Dc), which describes the similarity of the volume and position between the results generated by the truth mean value (Mth) and the values to be evaluated (Me) (Eq. 1). The derivation of the Dc has been detailed previously [22]. An excellent agreement value of Dc is > 80%.
1 - False-negative (FN), which describes the proportion of false-negatives (Eq. 2). The optimal value of FN is 0%.
2
Real FDG-PET Data Sets of the MCAO Rat Model
Images of the MCAO rat model were processed in the spmratIHEP toolbox [18, 19] similar to the simulated data set. In detail, the real MCAO data set was preprocessed by skull stripping, repositioning of the origin point, and spatial normalization and smoothing (as above). Then, based on the GLM framework, the two-sample t-test was used to assess differences between the MCAO rats and the healthy controls. Proportional scaling and intensity normalization were also performed based on either the global mean value or our proposed unbiased value. This unbiased value was generated iteratively based on P < 0.001 (uncorrected) and a cluster size of ≥ 50 voxels, as described above. Finally, based on these two scale factors, the brain regions with significant FDG changes in rats with MCAO were identified. The voxel-level height threshold was P < 0.05 (FWE corrected) and the cluster size was ≥ 50 voxels.
Results
Evaluation of the Simulated FDG-PET Data Sets
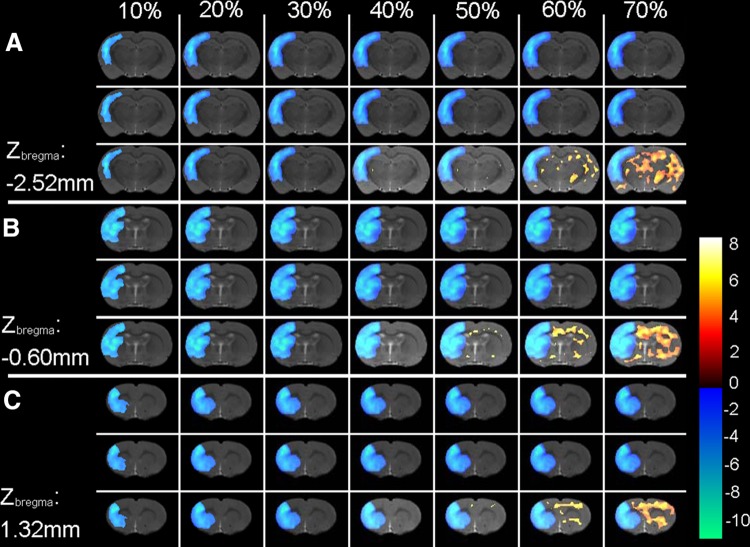
The accuracy of different scale factors was first inspected qualitatively by overlaying the voxel-wise analysis results onto an MRI T2WI single-brain image in Paxinos & Watson space (Fig. 2). Compared with the truth mean value, both the unbiased mean value and the global mean value were comparable in detecting hypo-metabolic voxels (Fig. 3). However, the unbiased mean value performed better than the global mean value in reducing the false-positives. When the voxel value decreased by > 30%, the global mean value produced increasing numbers of false-positive regions, while this did not occur for the unbiased mean value (Fig. 3).
Fig. 3.
Voxel-wise analysis results of simulated data. Three coronal planes were selected and are presented in (A), (B), and (C), whose coordinates were Zbregma − 2.52 mm, − 0.60 mm, and 1.32 mm. These statistical results were generated based on the truth mean value, the unbiased mean value, and the global mean value, as shown in the upper, middle, and lower rows in each panel. The regions with significant FDG changes of the simulated data sets were superimposed on a structural rat brain template image. The results of the simulated data set with different voxel values at decreasing levels are presented in different lines, from left to right representing voxel values decreasing from 10% to 70%. The results of the statistical analysis are presented as a color scale, while the structural brain image is presented in gray-scale as background. A warm pseudo-color indicates hyper-metabolic regions of the simulated data compared with the healthy controls, while a cold pseudo-color indicates hypo-metabolic regions. The color bar (right) indicates the t-value.
Furthermore, to quantitatively evaluate the accuracy of the unbiased scale factor, correspondence measures of three statistical results were calculated for the volumetric and spatial positions (Table 1). The statistical result generated by the truth mean value was selected as the reference, then the two correspondence measures were calculated between the reference and the result generated by our suggested mean value or the global mean value. The mean Dc of our suggested unbiased value was 89.03 ± 13.07, while it was 84.64 ± 11.84 for the global mean. The mean FN of our suggested unbiased value was 14.96 ± 21.09, while it was 20.68 ± 21.22 for the global mean (Table 1).
Table 1.
Volumetric and spatial correspondence measures between the true mean value and the unbiased/global mean value.
| Image intensity decrease rate | Dcoeff (%) | FN (%) | ||
|---|---|---|---|---|
| Unbiased | Global | Unbiased | Global | |
| 10% | 62.47 | 60.00 | 60.07 | 66.63 |
| 20% | 82.79 | 79.92 | 20.79 | 25.12 |
| 30% | 89.69 | 85.86 | 11.50 | 16.47 |
| 40% | 93.68 | 89.08 | 6.75 | 12.26 |
| 50% | 96.19 | 91.13 | 3.96 | 9.73 |
| 60% | 98.36 | 92.67 | 1.67 | 7.91 |
| 70% | 100.00 | 93.80 | 0.00 | 6.61 |
Dcoeff (%), Dice similarity coefficient (excellent agreement value > 80%); FN (%), proportions of false-negatives (optimal value, 0%); Unbiased, volumetric and spatial correspondence measure between the true mean value and the unbiased mean value; Global, volumetric and spatial correspondence measure between the true mean value and the global mean value.
Evaluation of Real MCAO Data Sets
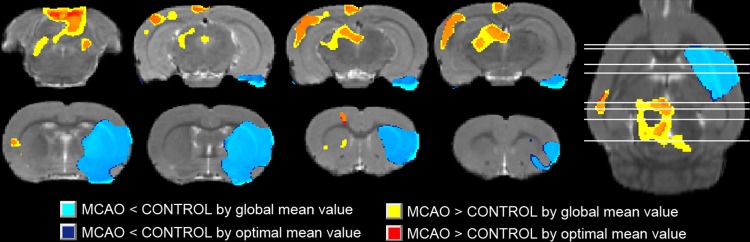
The brain regions with significant FDG changes in rats with MCAO, generated by either the global mean value or the unbiased mean value, were overlaid on an MRI T2WI image of a rat brain in Paxinos & Watson space (Fig. 4). The hypo-metabolic regions generated by the unbiased mean value were exclusively related to the left MCA, and were similar to those generated by the global mean value. However, the hyper-metabolic regions generated by the unbiased mean value were far fewer than those generated by the global mean value.
Fig. 4.
Superimposition of brain regions with significant FDG changes in rats with MCAO on a corresponding structural rat brain image in Paxinos & Watson space, generated by either the global mean value (light blue and yellow) or the unbiased mean value (dark blue and red) (P < 0.05 with FWE correction and clusters > 50 voxels). Eight coronal and an axial planes are presented. The results of the statistical analysis are presented in pseudo-color, while the structural brain image is presented in gray-scale as background. A warm pseudo-color indicates hyper-metabolic regions of MCAO compared with healthy controls, while a cold pseudo-color indicates hypo-metabolic regions.
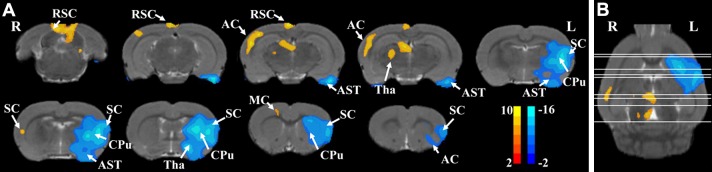
The voxel-wise analysis results from MCAO and control rats generated based on the unbiased mean value are shown in Figure 5, in which nine coronal (Fig. 5A) and an axial (Fig. 5B) plane are presented. The hypo-metabolic voxels lying in regions of the motor cortex, somatosensory cortex, caudate-putamen, thalamus, amygdaloid body, and auditory cortex of the MCAO rats might be associated with the infarction damage of the lesioned hemisphere. The hyper-metabolic voxels lay in regions of the contralateral (unlesioned) motor cortex, somatosensory cortex, thalamus, retrosplenial cortex, and auditory cortex of the MCAO rats compared with the healthy control rats.
Fig. 5.
Voxel-wise analysis results in rats with and without MCAO generated based on the unbiased scale factor (P < 0.05 with FWE correction and clusters > 50 voxels). Nine coronal (A) and an axial plane (B) are presented. The results of the statistical analysis are color-scaled, while the structural brain image is presented in gray-scale as background. A warm pseudo-color indicates hyper-metabolic regions for MCAO compared with healthy controls, while a cold pseudo-color indicates hypo-metabolic regions. The color bar indicates the t-value of each significant voxel in Paxinos and Watson space. AC, auditory cortex; AST, amygdaloid body; CPu, caudate-putamen; L, left; MC, motor cortex; R, right; RSC, retrosplenial cortex; SC, somatosensory cortex; Tha, thalamus.
Discussion
In this study, a data-driven method of generating an unbiased scale factor for intensity normalization in FDG-PET imaging analysis was proposed for stroke studies. With this method, almost all of the dysfunctional voxels, comprising both hyper-metabolic and hypo-metabolic voxels, were automatically excluded from the unbiased scale factor calculation, with no need for prior knowledge. Although using the global mean value as the initial scale factor excluded several normal functional voxels, it was similar to the concept of selecting a specified reference region for scale factor calculation, in that only certain normal functional voxels were used.
For evaluation, simulated data sets were constructed by decreasing voxel values [15]. To simulate cellular dysfunction and recovery after stroke, we used 7 decreasing levels. In order to address this problem as straightforwardly as possible, only ischemia was simulated, so that the unbiased scale factor could be evaluated by both hypo-metabolic detection and false-positive reduction. As illustrated in Figure 3 and Table 1, in the detection of hypo-metabolic regions, the sensitivity and accuracy of our proposed unbiased scale factor were better than the global mean value. Especially, when the voxel value was decreased by > 30%, the accuracy of hypo-metabolic region detection was improved by > 5% by our proposed unbiased scale factor. At all these 7 decreasing levels, no false-positive regions were generated by our unbiased scale factor, while there were many more false-positive regions with the global mean value. Therefore, both sensitivity and accuracy were improved by our unbiased scale factor.
A set of real MCAO model data at the acute stage was also used for qualitative evaluation. As shown in Figure 4, similar to the evaluation results with the simulated data sets, our unbiased scale factor was comparable with the global mean value in the detection of hypo-metabolic regions, but more focused in hyper-metabolic regions. At the acute stage of stroke, these severely hypo-metabolic regions are mainly caused by ischemia and edema, which were specific and macroscopic in FDG-PET images, so that both factors performed well. On the other hand, functional compensation also occurs at the acute stage of stroke, but its mechanisms and structural locations are still unclear; this is also one of the desired objectives of MCAO model studies. The hyper-metabolic regions generated by the unbiased scale factor were more focused than with the global mean value. This focus is important for stroke studies.
Moreover, the voxel-wise analysis results generated by our proposed unbiased scale factor are detailed in Figure 5. Compared with healthy controls, the significant hypo-metabolic voxels of the MCAO rats at the acute stage mainly lay in the hemisphere with infarction damage, comprising motor cortex, somatosensory cortex, caudate-putamen, thalamus, amygdaloid body, and auditory cortex, consistent with previous research on animals [23–25] and clinical experiments [26]. At the acute stage of stroke, the hypo-metabolic regions in FDG-PET images not only contain the infarction area, but also the edematous and functionally impaired areas, such as those involved in motor impairment. Among these significantly hypo-metabolic regions, motor cortex, somatosensory cortex, and caudate-putamen are known to be involved in gaze, orientation, and motor skills, which are also known to be relevant to stroke [27–29].
As shown in Figure 5, the significant hyper-metabolic voxels of the MCAO rats at the acute stage were mainly located in the contralateral (unlesioned) motor cortex, somatosensory cortex, thalamus, retrosplenial cortex, and auditory cortex. Previous studies have demonstrated that almost all functional activities are accomplished by bilateral functional areas in healthy controls [30, 31]. This indicates that the contralateral (unlesioned) functional areas may supplement the corresponding lesioned areas, but this may play a negative role in the functional recovery after ischemic stroke [11, 32, 33]. The role of the unlesioned hemisphere might be influenced by the lesion volume, and this is involved in the functional alterations after stroke. A number of studies have proposed that inhibition of the unlesioned hemisphere improves the motor performance after stroke [33, 34]. However, the mechanism of functional compensation after stroke awaits further research, in which our proposed method could provide more accurate analysis of FDG-PET imaging results.
Moreover, our proposed method is not only limited to stroke studies. It can be generalized to studies of other diseases, such as Alzheimer’ disease [35], and individual subject analysis. Similarly, the hyper/hypo-metabolic voxels of patients and individuals can also be excluded iteratively by choosing the global mean value or another reference value as the initial factor. Besides the voxel-wise analysis, semi-quantitative analysis based on regions of interest (ROIs) is another widely-used method in PET image analysis. The standardized uptake value ratio is one of the most widely used semi-quantitative parameters, which is computed using PET counts from a selected ROI relative to reference regions [36, 37]. Our proposed method can be used to objectively generate unbiased reference regions.
In addition, there are several limitations of our study. First, only the acute stage of the MCAO model was simulated, so that the 7 levels of decreasing voxel values started from 10%. However, the functional decline in several other diseases, such as depression, may be < 10%. Second, both the location and degree of hyper-metabolic voxels at the acute stage were unclear, so only hypo-metabolic voxels were simulated. Nevertheless, in the real acute stage of MCAO, there were several hyper-metabolic voxels. Third, as shown in Figure 3, when the voxel value decreased by < 30%, our proposed scale factor performed similar to the global mean value. Therefore, our proposed scale factor could be further optimized in the future.
In conclusion, we propose a data-driven means of generating an unbiased scale factor for intensity normalization in FDG-PET imaging analysis after stroke.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81471741, 81471728, and 81671770).
Compliance with Ethical Standards
Conflict of interest
All authors claim that there are no conflicts of interest.
References
- 1.Zimmer L, Luxen A. PET radiotracers for molecular imaging in the brain: past, present and future. Neuroimage. 2012;61:363–370. doi: 10.1016/j.neuroimage.2011.12.037. [DOI] [PubMed] [Google Scholar]
- 2.Tousseyn S, Krishnan B, Wang ZI, Wongwiangjunt S, Nayak CS, Mosher JC, et al. Connectivity in ictal single photon emission computed tomography perfusion: a cortico-cortical evoked potential study. Brain. 2017;140:1872–1884. doi: 10.1093/brain/awx123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wollenweber FA, Darr S, Muller C, Duering M, Buerger K, Zietemann V, et al. Prevalence of amyloid positron emission tomographic positivity in poststroke mild cognitive impairment. Stroke. 2016;47:2645–2648. doi: 10.1161/STROKEAHA.116.013778. [DOI] [PubMed] [Google Scholar]
- 4.Heiss WD. PET imaging in ischemic cerebrovascular disease: current status and future directions. Neurosci Bull. 2014;30:713–732. doi: 10.1007/s12264-014-1463-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong Y, Song FH, Ma JJ, He XX, Amer S, Gu WZ, et al. Small-animal PET demonstrates brain metabolic change after using bevacizumab in a rat model of cerebral ischemic injury. Neurosci Bull. 2014;30:834–844. doi: 10.1007/s12264-014-1470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimmer ER, Parent MJ, Souza DG, Leuzy A, Lecrux C, Kim HI, et al. [18F]FDG PET signal is driven by astroglial glutamate transport. Nat Neurosci. 2017;20:393–395. doi: 10.1038/nn.4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitamura A, Kaneta T, Miyata G, Takanami K, Hiraide T, Fukuda H, et al. Positive correlations between tumor uptake on FDG PET and energy expenditure of patients with esophageal cancer. Ann Nucl Med. 2011;25:241–246. doi: 10.1007/s12149-010-0456-9. [DOI] [PubMed] [Google Scholar]
- 8.Busk M, Horsman MR, Jakobsen S, Bussink J, van der Kogel A, Overgaard J. Cellular uptake of PET tracers of glucose metabolism and hypoxia and their linkage. Eur J Nucl Med Mol Imaging. 2008;35:2294–2303. doi: 10.1007/s00259-008-0888-9. [DOI] [PubMed] [Google Scholar]
- 9.Jiang XF, Zhang T, Sy C, Nie BB, Hu XY, Ding Y. Dynamic metabolic changes after permanent cerebral ischemia in rats with/without post-stroke exercise: a positron emission tomography (PET) study. Neurol Res. 2014;36:475–482. doi: 10.1179/1743132814Y.0000000350. [DOI] [PubMed] [Google Scholar]
- 10.Viaro R, Bonazzi L, Maggiolini E, Franchi G. Cerebellar modulation of cortically evoked complex movements in rats. Cereb Cortex. 2017;27:3525–3541. doi: 10.1093/cercor/bhw167. [DOI] [PubMed] [Google Scholar]
- 11.Alia C, Spalletti C, Lai S, Panarese A, Lamola G, Bertolucci F, et al. Neuroplastic changes following brain ischemia and their contribution to stroke recovery: novel approaches in neurorehabilitation. Front Cell Neurosci. 2017;11:76. doi: 10.3389/fncel.2017.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Longa EZ, Weinstein PR, Carlson S, Cummins R. Reversible middle cerebral artery occlusion without craniectomy in rats. Stroke. 1989;20:84. doi: 10.1161/01.STR.20.1.84. [DOI] [PubMed] [Google Scholar]
- 13.Fueger BJ, Czernin J, Hildebrandt I, Tran C, Halpern BS, Stout D, et al. Impact of animal handling on the results of F-18-FDG PET studies in mice. J Nucl Med. 2006;47:999–1006. [PubMed] [Google Scholar]
- 14.Matsumura A, Mizokawa S, Tanaka M, Wada Y, Nozaki S, Nakamura F, et al. Assessment of microPET performance in analyzing the rat brain under different types of anesthesia: comparison between quantitative data obtained with microPET and ex vivo autoradiography. Neuroimage. 2003;20:2040–2050. doi: 10.1016/j.neuroimage.2003.08.020. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Nie B, Zhu H, Liu H, Wang J, Duan S, et al. Factors affecting the voxel-based analysis of diffusion tensor imaging. Chine Sci Bull. 2014;59:4077–4085. doi: 10.1007/s11434-014-0551-8. [DOI] [Google Scholar]
- 16.Friston KJ, Ashburner J, Frith CD, Poline JB, Heather JD, Frackowiak RSJ. Spatial registration and normalization of images. Human Brain Mapping. 1995;3:165–189. doi: 10.1002/hbm.460030303. [DOI] [Google Scholar]
- 17.Paxinos GWC. The Rat Brain in Stereotaxic Coordinates. 5. New York: Academic Press; 2005. [Google Scholar]
- 18.Nie B, Chen K, Zhao S, Liu J, Gu X, Yao Q, et al. A rat brain MRI template with digital stereotaxic atlas of fine anatomical delineations in paxinos space and its automated application in voxel-wise analysis. Hum Brain Mapp. 2013;34:1306–1318. doi: 10.1002/hbm.21511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nie B, Liu H, Chen K, Jiang X, Shan B. A statistical parametric mapping toolbox used for voxel-wise analysis of FDG-PET images of rat brain. PLoS One. 2014;9:e108295. doi: 10.1371/journal.pone.0108295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nie B, Hui J, Wang L, Chai P, Gao J, Liu S, et al. Automatic method for tracing regions of interest in rat brain magnetic resonance imaging studies. J Magn Reson Imaging. 2010;32:830–835. doi: 10.1002/jmri.22283. [DOI] [PubMed] [Google Scholar]
- 21.Casteels C, Vermaelen P, Nuyts J, Van Der Linden A, Baekelandt V, Mortelmans L, et al. Construction and evaluation of multitracer small-animal PET probabilistic atlases for voxel-based functional mapping of the rat brain. J Nucl Med. 2006;47:1858–1866. [PubMed] [Google Scholar]
- 22.Gutierrez DF, Zaidi H. Automated analysis of small animal PET studies through deformable registration to an atlas. Eur J Nucl Med Mol Imaging. 2012;39:1807–1820. doi: 10.1007/s00259-012-2188-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cha J, Kim ST, Jung WB, Han YH, Im GH, Lee JH. Altered white matter integrity and functional connectivity of hyperacute-stage cerebral ischemia in a rat model. Magn Reson Imaging. 2016;34:1189–1198. doi: 10.1016/j.mri.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Brown CE, Aminoltejari K, Erb H, Winship IR, Murphy TH. In vivo voltage-sensitive dye imaging in adult mice reveals that somatosensory maps lost to stroke are replaced over weeks by new structural and functional circuits with prolonged modes of activation within both the peri-infarct zone and distant sites. J Neurosci. 2009;29:1719–1734. doi: 10.1523/JNEUROSCI.4249-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohajerani MH, Aminoltejari K, Murphy TH. Targeted mini-strokes produce changes in interhemispheric sensory signal processing that are indicative of disinhibition within minutes. Proc Natl Acad Sci U S A. 2011;108:E183–E191. doi: 10.1073/pnas.1101914108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park CH, Chang WH, Ohn SH, Kim ST, Bang OY, Pascual-Leone A, et al. Longitudinal changes of resting-state functional connectivity during motor recovery after stroke. Stroke. 2011;42:1357–1362. doi: 10.1161/STROKEAHA.110.596155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erlich JC, Bialek M, Brody CD. A cortical substrate for memory-guided orienting in the rat. Neuron. 2011;72:330–343. doi: 10.1016/j.neuron.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coull JT, Hwang HJ, Leyton M, Dagher A. Dopamine precursor depletion impairs timing in healthy volunteers by attenuating activity in putamen and supplementary motor area. J Neurosci. 2012;32:16704–16715. doi: 10.1523/JNEUROSCI.1258-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bedard P, Sanes JN. Brain representations for acquiring and recalling visual-motor adaptations. Neuroimage. 2014;101:225–235. doi: 10.1016/j.neuroimage.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hetu S, Gregoire M, Saimpont A, Coll MP, Eugene F, Michon PE, et al. The neural network of motor imagery: an ALE meta-analysis. Neurosci Biobehav Rev. 2013;37:930–949. doi: 10.1016/j.neubiorev.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 31.Liang X, Zou Q, He Y, Yang Y. Topologically reorganized connectivity architecture of default-mode, executive-control, and salience networks across working memory task loads. Cereb Cortex. 2016;26:1501–1511. doi: 10.1093/cercor/bhu316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward NS, Brown MM, Thompson AJ, Frackowiak RSJ. Neural correlates of motor recovery after stroke: a longitudinal fMRI study. Brain. 2003;126:2476–2496. doi: 10.1093/brain/awg245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mansoori BK, Jean-Charles L, Touvykine B, Liu A, Quessy S, Dancause N. Acute inactivation of the contralesional hemisphere for longer durations improves recovery after cortical injury. Exp Neurol. 2014;254:18–28. doi: 10.1016/j.expneurol.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 34.Khedr EM, Abdel-Fadeil MR, Farghali A, Qaid M. Role of 1 and 3 Hz repetitive transcranial magnetic stimulation on motor function recovery after acute ischaemic stroke. Eur J Neurol. 2009;16:1323–1330. doi: 10.1111/j.1468-1331.2009.02746.x. [DOI] [PubMed] [Google Scholar]
- 35.Pan X, Chen Z, Gei G, Pan S, Bao W, Ren S, et al. Long-term cognitive improvement after benfotiamine administration in patients with Alzheimer’s disease. Neurosci Bull. 2016;32:591–596. doi: 10.1007/s12264-016-0067-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pourdehnad M, Basu S, Duarte P, Okpaku AS, Saboury B, Hustinx R, et al. Reduced grey matter metabolism due to White matter edema allows optimal assessment of brain tumors on F-18-FDG-PET. Hell J Nucl Med. 2011;14:219–223. [PubMed] [Google Scholar]
- 37.Rabb CH. Nylon monofilament for intraluminal middle cerebral artery occlusion in rats. Stroke. 1996;27:151-151. [PubMed] [Google Scholar]