Abstract
The brain has very high energy requirements and consumes 20% of the oxygen and 25% of the glucose in the human body. Therefore, the molecular mechanism underlying how the brain metabolizes substances to support neural activity is a fundamental issue for neuroscience studies. A well-known model in the brain, the astrocyte-neuron lactate shuttle, postulates that glucose uptake and glycolytic activity are enhanced in astrocytes upon neuronal activation and that astrocytes transport lactate into neurons to fulfill their energy requirements. Current evidence for this hypothesis has yet to reach a clear consensus, and new concepts beyond the shuttle hypothesis are emerging. The discrepancy is largely attributed to the lack of a critical method for real-time monitoring of metabolic dynamics at cellular resolution. Recent advances in fluorescent protein-based sensors allow the generation of a sensitive, specific, real-time readout of subcellular metabolites and fill the current technological gap. Here, we summarize the development of genetically encoded metabolite sensors and their applications in assessing cell metabolism in living cells and in vivo, and we believe that these tools will help to address the issue of elucidating neural energy metabolism.
Keywords: Energy metabolism, Astrocyte, Neuron, Genetically encoded fluorescent sensor, Real time monitoring
Introduction
The brain is a highly energy-demanding organ, and although it composes only 2% of the body’s mass, the brain consumes 20% of the oxygen and 25% of the glucose in the human body [1]. Thus, the bioenergetics of the brain has attracted considerable attention in neuroscience research. Neurons take a dominant share of the brain energy consumption, approximately up to 80%, and the rest is used by glial cells [2, 3]. Neurons spend energy on maintenance of the resting potential, action potentials, and synaptic potentials, as well as the generation of neurotransmitters. Among these, the synaptic potential appears to be the main energy-consuming process [4], whereas the action potential has evolved to be energy efficient [5].
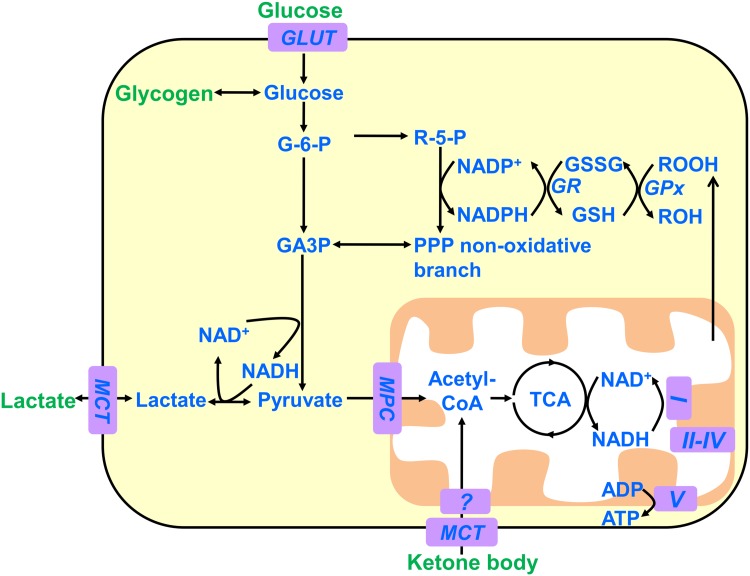
Glucose is the obligatory energy substrate of the adult brain [1, 6] and is catabolized into pyruvate, adenosine triphosphate (ATP), and reduced nicotinamide adenine dinucleotide (NADH) in the cytoplasm (Fig. 1). Pyruvate can be transported into mitochondria for oxidation through the tricarboxylic acid (TCA) cycle and produces much more NADH together with flavin adenine dinucleotide. Oxidation of these reduced coenzymes via the respiratory chain establishes the proton gradient across the mitochondrial membrane and then releases a large number of ATPs. Under anaerobic conditions, pyruvate is converted into lactate by lactate dehydrogenase, with the concurrent conversion of NADH into NAD+. The ratio of pyruvate to lactate is generally balanced by the ratio of [NAD+] to [NADH] in metabolic steady state. Lactate is secreted into extracellular compartments through monocarboxylate transporters. Alternatively, glucose runs through the pentose phosphate pathway with the generation of reduced nicotinamide adenine dinucleotide phosphate (NADPH), which can be utilized for biosynthetic reactions and antioxidant defense responses.
Fig. 1.
The main pathways of neural energy metabolism. Depicted are the main substrates and pathways of cellular energy metabolism in the adult brain. In general, the brain utilizes glucose as an obligatory fuel and makes use of ketone bodies under certain conditions such as fasting or strenuous physical activity. Neurons also metabolize glucose through the pentose phosphate pathway to generate NADPH, which protects neurons from oxidative stress. Astrocytes can synthesize glycogen from glucose and mobilize it to support the energy demands of both astrocytes and neurons. GLUT, glucose transporter; MCT, monocarboxylate transporter; GR, glutathione reductase; Gpx, glutathione peroxidase; MPC, mitochondrial pyruvate carrier; TCA, tricarboxylic acid cycle; G-6-P, glucose 6-phosphate; GA3P, glyceraldehyde-3-phosphate; R-5-P, ribulose 5-phosphate; PPP, pentose phosphate pathway; GSSG, oxidized glutathione; GSH, reduced glutathione; ROOH, hydroperoxides.
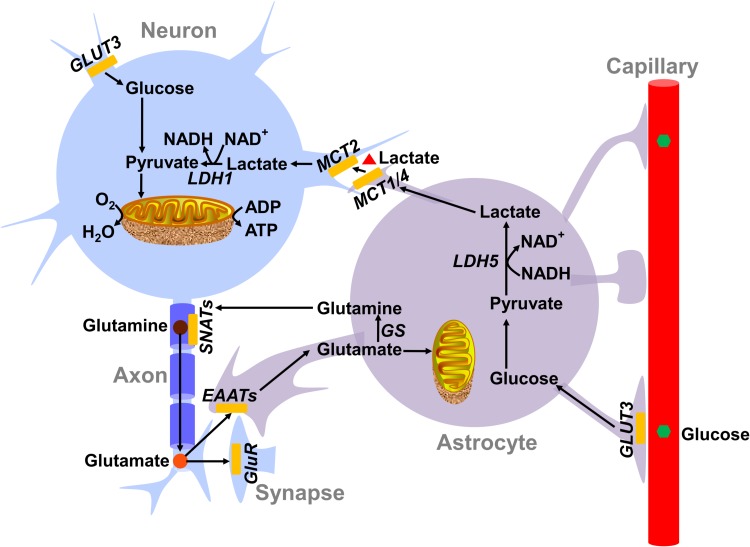
In addition to glucose, various metabolites in the brain can be oxidized for energy production, e.g., lactate, pyruvate, glutamine, glutamate, or acetate [7]. Among them, lactate as an energy substrate has been intensively investigated. A hypothetical model, named the astrocyte-neuron lactate shuttle (ANLS), proposes that glutamate is released into the synaptic cleft upon neuronal activation and stimulates the glucose uptake and aerobic glycolysis of astrocytes, subsequently leading to the enhanced secretion of lactate [1, 8]. Neurons absorb the lactate to sustain their energy charge [1, 8–10] (Fig. 2). The ANLS hypothesis has been experimentally interrogated over the last two decades. A clear consensus has yet to be reached; however, a substantial part of the experiments have declared inconsistent conclusions that are sharply contradictory [11, 12]. The novel idea that lactate acts as a signal transmitter in the metabolic state rather than an energy substrate makes the controversy more complicated [13]. Glycogen serves as the glucose storage form and can be used by astrocytes but not by neurons in the brain [14].
Fig. 2.
The astrocyte-neuron lactate shuttle model. The astrocyte-neuron lactate shuttle model highlights the important role of lactate secreted by astrocytes as the fuel for neurons. As proposed in this model, the neurotransmitter glutamate released upon neuronal excitation is taken up by astrocytes. On one side, astrocytic glutamate is used to synthesize glutamine and recycled in neurons for facilitating glutamate regeneration; on the other side, glutamate stimulates the glycolytic activity and lactate secretion of astrocytes. Thereafter, the lactate is transported into neurons to supply the intense energy consumption of neuronal activities. The metabolite transporters and enzymes predominantly expressed in neurons and astrocytes are listed. GluR: glutamate receptor; GS: glutamine synthetase; EAAT: excitatory amino acid transporter; GLUT1 and GLUT3: glucose transporter 1 and 3; LDH1 and LDH5: lactate dehydrogenase 1 and 5; MCT1, MCT2, and MCT4: monocarboxylate transporter 1, 2, and 4; SNATs: sodium-coupled amino acid transporters.
Fatty acids generally make no contribution to brain fuel due to the existence of the blood-brain barrier and the restriction of the blood oxygen supply [15]; however, their metabolic intermediates, ketone bodies, can be quickly mobilized to supply brain energy under certain physiological conditions, such as a fasting diet or strenuous physical exercise [16].
Traditional Assays for Neural Energy Metabolism
There are two main functional brain imaging techniques to assess the metabolic state in vivo: positron emission tomography (PET) and functional magnetic resonance imaging (fMRI) [11, 17–19]. PET utilizes positron-emitting metabolites such as glucose and oxygen as tracers and allows the determination of the cerebral metabolic rate of glucose or oxygen consumption, while fMRI mostly measures brain oxygenation [1].
There are a few accepted methods for assaying metabolites in vitro, including enzymatic cycling assays, chromatography, and mass spectrometry. However, these methods require cell lysis and are incompatible with studying the dynamics in intact cells. While metabolic flux analysis, which requires a stable-isotope-labeled metabolite precursor, is invaluable to the detailed understanding of the underlying metabolic pathways, it focuses solely on an often abridged, predefined metabolic network and fails to quantify reactions in specific compartments [20]. Similar to metabolomics, metabolic flux analysis is also invasive and not suitable for the study of metabolite dynamics in living cells. To monitor the intracellular metabolic state, researchers have utilized single-photon or dual-photon microscopy and imaged the weak endogenous fluorescence of NAD(P)H [10, 21, 22]. Redox reactions catalyzed by various NAD(H)-dependent dehydrogenases are vital in energy metabolism, such as glycolysis, the TCA cycle, and oxidative phosphorylation; therefore, the intracellular NAD+/NADH redox state is often considered a very useful cellular metabolic readout [23–25]. However, these methods have drawbacks of low sensitivity, cell injury caused by ultraviolet irradiation, and insufficient resolution between NADH and NADPH. In addition, cytosolic signals are very difficult to be separated and measured from intense mitochondrial signals because intrinsic NAD(P)H fluorescence signals mostly originate from the mitochondria.
New Tools for Imaging Neural Energy Metabolism
Recently, genetically encoded fluorescent sensors have been extensively applied to the real-time monitoring of various metabolic processes [26–29]. These sensors are usually chimera proteins containing a ligand-sensing domain and a fluorescent protein (FP) or FP pair. Compared to conventional biochemical methods, these sensors offer a few advantages for cell metabolism imaging. First, the dynamics of ligand binding or dissociation are often reversible for FP-based sensors, thus allowing the real-time recording of metabolic activities in vitro and in vivo. Second, these sensors are quite easily introduced into diverse cells by DNA transfection or targeted to different organelles by tagging with organelle-specific signal peptides. Third, the fluorescence of these sensors can be readily recorded by routine equipment such as a microplate reader, fluorescence microscope, or flow cytometer.
At present, a few genetically encoded metabolite sensors have been developed for the detection of cellular energy metabolism in the brain (Table 1). The sensors in the first class report the charges of nicotinamide adenine nucleotides in brain energy metabolism and include the NADH sensors of the Frex family [30]; the NAD+ sensor LigA-cpVenus [31]; the NAD+/NADH ratio sensors Peredox [32], RexYFP [33], and SoNar [34, 35]; the NADPH sensors of the iNap family [36]; and the NADP+ sensor Apollo-NADP+ [37] (Table 1). The sensors in the second class report the adenylate energy charges in brain energy metabolism and include the ATP sensors in the ATeam family [38, 39] and the ATP/ADP ratio sensors in the Perceval family [40, 41] (Table 1). The sensors in the last class report vital energy substrates in the brain and include the glucose sensors in the FLII12Pglu family [42, 43], the lactate sensor Laconic [44], the pyruvate sensor Pyronic [45], and the glutamine sensors in the FLIPQ-TV3.0 family [46] (Table 1).
Table 1.
Fluorescence properties of genetically encoded metabolite sensors.
| Sensor | Species sensed | Kd for analyte | Dynamic range (%) | Detection mode (fluorescent protein) | Rate of binding/ dissociation | pH sensitivity | References |
|---|---|---|---|---|---|---|---|
| Frex | NADH | ~ 3.7 μmol/L, 11 μmol/La | 800 | Ratiometric (cpYFP) | Fast/Fast | Sensitive | [30] |
| LigA-cpVenus | NAD+ | ~ 65 μmol/L | 100 | Ratiometric (cpVenus) | Fast/Fast | Sensitive | [31] |
| Peredox | NAD+/NADH ratio | ~ 330 | 150 | Intensiometric (cpT-Sapphire) | Fast/Slow | Resistant | [32] |
| RexYFP | NAD+/NADH ratio | N.D. | 50 | Intensiometric (cpYFP) | N.D. | Sensitive | [33] |
| SoNar | NAD+/NADH ratio | ~ 40 | 1500 | Ratiometric (cpYFP) | Fast/Fast | Resistantb | [34] |
| iNap1; iNap2; iNap3; iNap4 |
NADPH | ~ 2 μmol/L; ~ 6.4 μmol/L; ~ 25 μmol/L; ~ 120 μmol/L |
900; 1000; 900; 500 |
Ratiometric (cpYFP) | Fast/Fast | Resistantb | [36] |
| Apollo-NADP+ | NADP+ | 0.1–20 µmol/L | 15–20 | Ratiometric (FP) | Fast/Fast | Resistant | [37] |
| AT3.10; AT3.10MGK; AT1.03; AT1.03YEMK; AT1.03NL |
ATP | ~ 7.4 μmol/L; | 100; | FRET (mseCFP and mVenus); | Fast/Fast | Resistant | [38, 39] |
| ~ 14 μmol/L; | N.D. | FRET (mseCFP and mVenus); | |||||
| ~ 3.3 mmol/L; | 130; | FRET (mseCFP and cpmVenus); | |||||
| ~ 1.2 mmol/L; | N.D. | FRET (mseCFP and cpmVenus); | |||||
| ~ 1.77 mmol/L | 90 | FRET (mseCFP and cpmVenus) | |||||
| Perceval; PercevalHR |
ATP/ADP ratio | 0.2; 3.5 |
100; 700 |
Ratiometric (cpmVenus) | Fast/Fast | Sensitive | [40, 41] |
| FLII12Pglu-600µ; FLII12Pglu-10aa; FLII12Pglu-15aa; FLII12Pglu-δ4; FLII12Pglu-700µδ6 |
Glucose | ~ 583 μmol/L; ~ 1480 μmol/L; ~ 2650 μmol/L; ~ 600 μmol/L; ~ 660 μmol/L; |
65; 30; 45; 55; 50 |
FRET (eCFP and citrine-eYFP) | N.D. | Resistant | [42, 43] |
| FGBP1 mM | Glucose | ~ 1000 μmol/L; | 690 | Ratiometric (cpYFP) | Fast/N.D. | Sensitive | [57] |
| Laconic | Lactate | ~ 8 μmol/L; ~ 830 μmol/L |
8; 11 |
FRET (mTFP and Venus) | N.D. | Sensitive | [44] |
| Pyronic | Pyruvate | ~ 107 μmol/L | 20 | FRET (mTFP and Venus) | N.D. | Resistant | [45] |
| FLIPQ-TV3.0_R75K; FLIPQ-TV3.0_R75M; FLIPQ-TV3.0_D157N; |
Glutamine | ~ 1.5 μmol/L; ~ 50 μmol/L; ~ 130 μmol/L; |
37; 10; 35; 14; 13 |
FRET (mTFP1 and Venus) | N.D. | Resistant | [46] |
| FLIPQ-TV3.0_R75MY86A; | ~ 1.6 mmol/L; | ||||||
| FLIPQ-TV3.0_R75MW220A | ~ 7.6 mmol/L |
Genetically encoded metabolite sensors available are listed and recommended for the spatiotemporal imaging of cellular energy metabolism. N.D., not determined. FP, fluorescent protein. aThe apparent Kd of Frex is ~ 3.7 μmol/L at pH 7.4 and ~ 11 μmol/L at pH 8.0. bFluorescence of SoNar (iNap) when excited at 420 nm, dynamic range, and are pH-resistant.
NADH Sensors, Frex Family
The Frex sensor was developed by inserting a circularly permuted yellow fluorescent protein (cpYFP) between the tandem structure of the NADH-binding transcription factor Rex from Bacillus subtilis (B-Rex) [30]. Frex has a 518 nm emission peak when excited at either 420 nm or 500 nm. Upon NADH binding, Frex exhibits a 9-fold increase in fluorescence with 500 nm excitation and an almost constant fluorescence with 420 nm excitation, making it an intrinsically ratiometric sensor (ratio of 500/420 nm) [30]. This sensor exclusively binds to NADH with a Kd of ~ 3.7 μmol/L at pH 7.4 and 11 μmol/L at pH 8.0 (Table 1), reflecting the reliable measurement range of the NADH concentration from 0.15 μmol/L to 90 μmol/L [29]. The Frex sensor selectively binds to NADH with no response to a physiological concentration of free NAD+, enabling it to accurately measure cellular NADH regardless of the NAD+/NADH ratio [35]. Two Frex variants have also been developed to better measure low cytosolic NADH levels and high mitochondrial NADH levels; one is the higher-affinity FrexH sensor with an apparent Kd of 40 nmol/L at pH 7.4 and the other is the lower-affinity C3L194K sensor with an apparent Kd of 50 μmol/L at pH 8.0 [30]. Frex sensors respond to physiological pH fluctuations (pH 6.0–8.0) since they are based on the pH-sensitive cpYFP. In addition, the fluorescence of Frex sensors is also weak, often 10-fold lower than that of cpYFP [30, 34]. The Frex family sensors have been used to investigate the NADH dynamics affected by NADH transport, glycolysis, mitochondrial activity, and redox perturbation [30], as well as the malate-aspartate NADH shuttle activity in cancer cells [47].
NAD+ Sensor, LigA-cpVenus
The NAD+ sensor LigA-cpVenus is composed of the NAD+-binding DNA ligase from bacteria (LigA) and circularly-permuted Venus fluorescent protein (cpVenus) [31]. LigA-cpVenus shows a major excitation peak at 500 nm and a minor peak at 405 nm, with a single emission peak at 520 nm. Upon NAD+ binding, the fluorescence of LigA-cpVenus excited at 488 nm decreases by 50%, while its fluorescence excited at 405 nm remains constant, rendering a change in the fluorescence ratio (488 nm/405 nm) up to ~ 100%. The sensor exhibits high specificity and a reversible response to NAD+ with an apparent Kd of 65 μmol/L (Table 1). Similar to cpVenus, the sensor is also sensitive to pH effects. Using this sensor, Xiaolu A. Cambronne et al. reported that cytosolic NAD+ can be transported to mitochondria [31].
NAD+/NADH Ratio Sensor, Peredox
Peredox is constructed by inserting a circularly-permutated T-sapphire (cpT-sapphire) fluorescent protein between two truncated units of the thermophilic Rex from Thermus aquaticus (T-Rex) [32]. Peredox has a high affinity toward NADH with no response to NAD+; however, NAD+ competes with NADH for binding and lowers the fluorescence response of Peredox to NADH. Thus, Peredox reports the NAD+/NADH ratio rather than the NADH level. However, Peredox’s fluorescence response is not strictly dependent on the NAD+/NADH ratio because a 3-fold change in the NAD+ pool size in the physiological range produces a 2-fold change in the sensor midpoint of the NAD+/NADH ratio [32]. The affinity of Peredox with a of 330 makes it easily saturated in some cellular contexts. Peredox has a 150% dynamic range, which is much smaller than that of Frex and SoNar sensors (Table 1). Peredox is an intensiometric sensor and has an excitation maximum at 400 nm and an emission maximum at 510 nm [32]. To achieve a ratiometric measurement, Peredox is fused with the red fluorescent protein mCherry. The fusion sensor, however, is prone to aggregation in mammalian cells [32]. Among the NAD+/NADH ratio sensors, Peredox is the only pH-resistant sensor, enabling its application under some conditions in which pH fluctuates [32]. Currently, this sensor has been exploited to monitor the cytosolic NAD+/NADH response to PI3K signaling pathway inhibition [32] and mitochondrial Ca2+ loading [48].
NAD+/NADH Ratio Sensor, RexYFP
RexYFP is engineered by embedding cpYFP in a T-Rex monomer with the short oligopeptide linkers SAG and GT [33]. RexYFP’s spectra have a single excitation peak at 490 nm and a single emission peak at 518 nm, with a 50% dynamic range (Table 1). This sensor can bind to either NADH or NADPH with dissociation constants of 0.18 μmol/L and 6.2 μmol/L, respectively [33]. This property indicates that interference with NADPH at a physiological concentration should be taken into consideration. Similar to Frex and SoNar, RexYFP’s fluorescence is sensitive to pH. This sensor has been used to study the influence of mitochondrial activities on the cellular NAD+/NADH redox state [33, 49].
NAD+/NADH Ratio Sensor, SoNar
SoNar is a recently-developed NAD+/NADH sensor [34, 35]. It is also the smallest sensor among the NAD+/NADH sensors described above and consists of a cpYFP insertion behind residue 189 of the truncated T-Rex via the short oligopeptide linkers SAG and G. SoNar is an intrinsically ratiometric sensor with two excitation maxima, at 418 nm and 500 nm, and one emission maximum at 518 nm. SoNar exhibits an opposite directional change in the presence of NADH or NAD+, with an apparent Kd of 0.2 μmol/L for NADH and 5 μmol/L for NAD+ at pH 7.4, making it a separate sensor for NADH or NAD+ in vitro when only one of the forms exists. Unlike Peredox, SoNar’s response strictly depends on the NAD+/NADH ratio rather than the absolute concentration of NAD+ or NADH [34] in physiological conditions. The sensor has an apparent of 40 (Table 1), making it able to report a wide range of NAD+/NADH ratios with little risk of saturation. In comparison with the time resolution of Peredox of few seconds [32], SoNar’s response to a ligand is immediate [34]. SoNar has a 1500% dynamic range, almost 10-fold greater than that of Peredox and 2-fold greater than that of Frex, making it one of the most responsive genetically encoded sensors currently available. SoNar exhibits intense fluorescence, 2- to 100-fold greater than that of Frex, Peredox, and cpYFP [34]. When excited at 485 nm, SoNar’s fluorescence is also affected by pH fluctuations, as other cpYFP-based sensors, whereas the dynamic range and of its fluorescence excited at 420 nm are more pH-resistant [34]. The above ideal properties of SoNar enable it to track subtle metabolic changes over a broad range of NAD+/NADH ratios and perform well in in vivo studies involving mammalian models and high-throughput screening [34, 35, 50, 51].
NADPH Sensor, iNaps
Very recently, we developed a series of highly responsive NADPH sensors by engineering SoNar’s binding site to switch its ligand selectivity from NADH to NADPH [36]. The four members of iNaps, iNap1–4, have similar fluorescent properties but cover a wide range of NADPH detection, with Kd varying from 2 μmol/L to 120 μmol/L (Table 1). Similar to SoNar, iNap1 has two excitation maxima, at ~ 420 nm and 500 nm, and one emission maximum at 515 nm. When binding to NADPH, the fluorescence of iNap1 with 420 nm excitation rises by 3.5-fold and that with 485 nm excitation drops by 2.5-fold, leading to a 9-fold ratiometric fluorescence change. The dynamic range of iNap1 remains constant under temperature variations from 20 °C to 42 °C. iNap sensors have inherited most of their favorable characteristics from SoNar, including a large dynamic range, rapid responsiveness, intense fluorescence, targeting of subcellular organelles, ratiometric imaging, and dynamic measurements in living cells and in vivo [36]. Similar to SoNar, the fluorescence excited at 420 nm but not at 485 nm is pH resistant; therefore, a ratiometric and pH-resistant measurement can be achieved by fusion to the red fluorescent protein mCherry or by normalization to a control FP iNapc [36]. iNap sensors are valuable tools for monitoring NADPH dynamics under physiological and stress conditions in both live cells and zebrafish [36].
NADP+ Sensor, Apollo-NADP+
The NADP+ sensor Apollo-NADP+ was created by fusing a human glucose-6-phosphate dehydrogenase (G6PD) mutant to a fluorescent protein [37]. The G6PD mutant protein is enzymatically inactive and binds exclusively to NADP+ with an apparent Kd varying from 0.1 μmol/L to 20 μmol/L [37] (Table 1). Binding to NADP+ induces the homodimerization of the G6PD mutant, thereby eliciting a FRET (fluorescence resonance energy transfer) signal between homologous fluorescent proteins [37]. The measurements of Apollo-NADP+ sensors are based on steady-state fluorescence anisotropy, which is the ratio of the parallel to perpendicular emission intensity upon polarized light excitation. The anisotropy readout of the sensors is pH-resistant within a pH range of 7.25–8.0. A significant advantage of Apollo-NADP+ sensors is that their fluorescent proteins are tunable, enabling multiplex imaging. The dynamic responses of the sensors, however, are as small as 15%–20%. In practice, the response may be 5% or less in live cells, limiting the efficient application of these sensors under physiological conditions. A possible improvement that may enhance the homologous FRET efficiency is the use of fluorescent proteins smaller than those in the GFP family, for example, the eel fluorescent protein UnaG. Cameron et al. used a Cerulean-tagged version of Apollo-NADP+ to study β-TC3 cells responding to oxidative stress and demonstrated by multiplex imaging that NADPH is significantly depleted before H2O2 accumulation [37].
ATP Sensor, ATeam
ATeam sensors are FRET-based sensors for ATP that are composed of the epsilon subunit of Bacillus subtilis F0F1-ATP synthase and a FRET fluorescent protein pair (the cyan fluorescent protein mseCFP and the yellow fluorescent protein mVenus) [38]. ATeam has four variants (AT3.10, AT3.10MGK, AT1.03YEMK, and AT1.03), whose Kd for ATP ranges from 7.4 μmol/L to 3.3 mmol/L [38] (Table 1). The dissociation constant depends considerably on the temperature, with a 5-fold increase from an elevation of 10 °C. Tsuyama et al. reported the development of a modified version, AT1.03NL, to improve the performance of AT1.03 at relatively low temperatures (20 °C–30 °C) [39]. AT1.03NL detects ATP changes more sensitively than the original AT1.03 in Drosophila melanogaster and Caenorhabditis elegans, which are typically cultured between 20 °C and 25 °C. Upon ATP binding, the fluorescence ratio (mVenus/mseCFP) of AT1.03 increases by ~ 2.3-fold. Compared to the signals of cpYFP-based sensors, ATeam’s FRET signal is resistant to physiological pH disturbances. ATeam sensors respond rapidly and have been used for studying the bioenergetics of cancer cells [52] and the chemical screening of immunogenic cell death [53].
ATP/ADP Sensor, PercevalHR
PercevalHR [41] is an improved version of the original ATP/ADP sensor Perceval [40]. The monomer structure of the sensor was invented by inserting circularly-permuted mVenus into an ATP-binding protein from Methanocaldococcus jannaschii, GlnK1, and the common form comprises a monomer chimera structure with two tandem GlnK1 proteins [40]. The PercevalHR sensor has two distinct excitation peaks, at 420 nm and 500 nm, and one emission peak at 515 nm. ATP binding increases the fluorescence from 500 nm excitation and ADP binding increases the fluorescence from 420 nm excitation, leading to a signal change of ~ 8-fold [41]. The affinity of PercevalHR for ATP or ADP is quite high, with apparent dissociation constants (Kd) of 1–3 μmol/L, thus it is always occupied by either ATP or ADP at their physiological concentrations. This characteristic enables PercevalHR to report the ATP/ADP ratio rather than the ATP or ADP level. PercevalHR shows a half-maximal change (KATP/ADP) of ~ 3.5 and covers a reliable detection range of 0.4 to 40 [41] (Table 1). Like many cpFP-based sensors, the fluorescence signal of PercevalHR also depends on the pH. PercevalHR has been used to visualize the association between the cellular energy state and the opening of K+ channels [41] and to study the influence of axonal mitochondrial transport on the cellular energy state in neurons [54].
Glucose Sensor, FLII12Pglu
The FLII12Pglu glucose sensors, which are modified versions of the original glucose sensor FLIPglu-600µ [55], are composed of the Escherichia coli glucose-binding protein MglB and a FRET pair (eCFP/citrine-eYFP) [42]. The addition of glucose to this sensor induces a decrease in eCFP emission and an increase in Citrine-eYFP emission. FLII12Pglu has five variants (FLII12Pglu-600µ, FLII12Pglu-10aa, FLII12Pglu-15aa, FLII12Pglu-δ4, and FLII12Pglu-700µδ6), whose Kd for glucose ranges from ~ 0.6 mmol/L to ~ 2.65 mmol/L in vitro. Upon glucose binding, the ratios of FLII12Pglu sensors change by 30%–65% (Table 1). Compared to the original FLIPglu-600µ, FLII12Pglu-700µδ6 has a higher ratio change (50%) and a wider detection range (0.05 mmol/L–9.6 mmol/L) for glucose [42]. These glucose sensors have been applied to the real-time monitoring of glucose dynamics in yeast and mammalian cells [43, 56]. Very recently, we developed a highly-responsive, cpYFP-based glucose sensor, FGBP1mM, which displays an ~ 700% fluorescence change in vitro, almost 10-fold greater than that of FRET-based glucose sensors [57] (Table 1).
Lactate Sensor, Laconic
Laconic is a FRET-based lactate sensor and composed of the E. coli transcription factor LldR and a FRET pair (mTFP/Venus) [44]. Laconic has two apparent Kd values of ~ 8 μmol/L and ~ 830 μmol/L and can quantify lactate from 1 μmol/L to 10 mmol/L (Table 1). Notably, the sensor can bind to citrate at high concentrations. In addition, pyruvate blocks the response of Laconic to low levels of lactate [44]. Laconic has a very small dynamic range (~ 10%), making the tracking of subtle changes under physiological conditions difficult. This sensor has been used for monitoring the lactate flux under hypoxic stress in cancer cells [58] and in astrocytes [59].
Pyruvate Sensor, Pyronic
Pyronic is a FRET-based pyruvate sensor and similar in design to the lactate sensor Laconic [45]. It is composed of the E. coli transcriptional regulator PdhR and a FRET pair (mTFP/Venus). Pyronic has an apparent Kd of ~ 107 μmol/L for pyruvate and an ~ 20% dynamic range [45] (Table 1). This sensor has high specificity and pH resistance. Pyronic has been used to investigate the effect of ammonium ions on lactate production in astrocytes [59].
Glutamine Sensors, FLIPQ-TV3.0 Family
The sensors in the FLIPQ-TV3.0 family are improved FRET-based glutamine sensors and consist of the E. coli glutamine-binding protein glnH and a FRET fluorescent protein pair (monomeric teal fluorescent protein mTFP1 and Venus) [46]. FLIPQ-TV3.0_R75K, R75M, D157N, R75MY86A, and R75MW220A variants have an apparent Kd for glutamine of 1.5 μmol/L, 50 μmol/L, 130 μmol/L, 1.6 mmol/L, and 7.6 mmol/L, respectively (Table 1). Mammalian glutamine concentrations are expected to be 1–22 mmol/L [46]; thus, the R75MY86A and R75MW220A mutants are more suitable for detection of physiological levels of glutamine. Similar to most FRET-based sensors, these two sensors have a small dynamic range (~ 13%), which considerably decreases with increases in pH, especially in the physiological pH range (pH 7.0–8.0), making them barely applicable to mitochondria (pH 8.0). Using these sensors, Gruenwald et al. demonstrated that glutamine transporter activity can easily be analyzed in living cells [46].
A Practical Guide to Metabolite Sensors
In the applications of the sensors described above, careful consideration should be given to the following issues, namely, the sensor’s affinity, fluorescence readout, and pH sensitivity. Metabolism is highly compartmentalized in cells, and the cytoplasm and mitochondria are the main locations of energy metabolism. Thus, an important issue for sensor usage is that the affinity of the sensor must match the physiological concentration of metabolite in the organelles or cells of interest.
The free NADH level in cytosol is ~ 0.12 μmol/L and in mitochondria ~ 30 μmol/L [30]. Thus, we recommend SoNar (0.2 μmol/L), RexYFP (0.18 μmol/L), FrexH (40 nmol/L), and Peredox (the first version, < 5 nmol/L), with high affinity for NADH, for detection of the cytosolic NAD+/NADH redox state, and the Frex (11 μmol/L, pH 8.0) and C3L194K (50 μmol/L, pH 8.0) sensors, with low affinity for NADH, for mitochondrial NADH detection (Table 1). The free NAD+ level in cytosol is ~ 106 μmol/L and in mitochondria ~ 230 μmol/L [31]. The LigA-cpVenus sensor has an apparent Kd of 65 μmol/L for NAD+, which is compatible with the cytosolic NAD+ pool in mammalian cells. However, LigA-cpVenus may be mostly saturated by NAD+ in mitochondria, so it works less well under NAD+-boosting conditions.
The free NADPH concentration in the cytosol is ~ 3 μmol/L and in mitochondria ~ 37 μmol/L [36]. The iNap family, with different affinities for NADPH, covers the physiological range of NADPH. We recommend iNap1 (2 μmol/L), with high affinity for NADPH, for cytosolic NADPH detection and iNap3 (25 μmol/L), with low affinity for NADPH, for mitochondrial NADPH detection (Table 1). Apollo-NADP+ sensors have an apparent Kd of 0.1 to 20 μmol/L and are compatible with cytosolic applications. We should note that Apollo-NADP+ cannot easily target other organelles owing to its specific properties.
The ATP concentration in the cytosol of mammalian cells reaches millimolar concentrations [4], and the ADP concentration is 3- to 10-fold lower [60]. Interestingly, the ATP levels in mitochondria are significantly lower than those in the cytosol [38]. Thus, AT1.03 (3.3 mmol/L), with a low affinity for ATP, is recommended for cytosolic ATP measurements, and AT1.03YEMK (2.1 mmol/L), with a high affinity for ATP, for mitochondrial ATP measurements (Table 1). PercevalHR can report an increase or decrease in the ATP/ADP ratio in the cytosol. However, as we have seen, the PercevalHR sensor has not been tested in mitochondria. In view of the sensor affinity, FLII12Pglu-700μδ6, Laconic, Pyronic, and FLIPQ-TV3.0 are more suitable for cytosolic glucose, lactate, pyruvate, and glutamine detection, respectively.
Once an appropriate sensor is chosen, the next consideration is the readout of the sensor’s fluorescence. Typically, ratiometric measurements are preferred because their readouts are not dependent on the sensor expression. The Frex family, LigA-cpVenus, SoNar, the iNap family, and PercevalHR are single fluorescent protein-based sensors, and their ratiometric measurements are based on the ratio of the outputs from the two excitation channels. ATeam, FLII12Pglu-700μδ6, Laconic, Pyronic, and FLIPQ-TV3.0 are FRET-based sensors, and their ratiometric measurements are based on the ratio of the outputs from the two emission channels. RexYFP and Peredox are intensiometric sensors. Their ratiometric measurements are dependent on the fusion of the red fluorescent protein mCherry. For Apollo-NADP+ sensors, the imaging of steady-state fluorescence anisotropy is heavily dependent on sophisticated instrumentation that is not readily available in most laboratories.
The sensor’s pH sensitivity should be carefully considered, especially in contexts where the pH fluctuates, such as the mitochondrial matrix. Among the sensors discussed above, Peredox, Apollo-NADP+, ATeam, FLII12Pglu-700μδ6, Pyronic, and FLIPQ-TV3.0 are relatively pH-resistant, whereas the Frex family, SoNar, RexYFP, LigA-cpVenus, iNap family, PercevalHR, and Laconic are highly pH-sensitive. Typically, the pH effects on metabolite sensors may be corrected by measuring the fluorescence of the metabolite sensor and the control sensor in parallel because of their very similar pH responses. Taking the iNap1 sensor as an example of correction of pH effects with iNapc, we suggest that users always measure the fluorescence ratio of iNapc and iNap1 in parallel experiments and then calculate the pH-corrected ratio (R′iNap1) of cells as follows: R′iNap1 = RiNap1/RiNapc (RiNap1 and RiNapc represent the excitation ratio 420/485 nm for iNap1 and iNapc, respectively).
Application to the Study of Neural Energy Metabolism
A few important questions on neural energy metabolism remain unsolved [61–63]. The most controversial issue is whether the energy charge of neurons depends on their oxidization of glucose or on the lactate from astrocytes during neuronal activity. Another question is how neuronal activity rewires the energy metabolism to support neuronal functions. Finally, since energy is consumed mostly at synaptic sites that are remote from cell bodies, how is the local energy supply realized in synapses? We summarize the direct evidence related to these questions, focusing on the application of the sensors noted above.
Previous studies of NAD(P)H autofluorescence [10] and a fluorescent glucose tracer [64] have suggested that brain activation enhances more glycolytic metabolism in astrocytes than in neurons, but the visualization of lactate metabolism in the brain was not realized until recently by the application of the lactate sensor Laconic and the pyruvate sensor Pyronic [65]. These sensors were specifically expressed in neurons or astrocytes in the primary somatosensory cortex of the mouse brain. Under anesthesia, an intravenous infusion of lactate raises the Laconic signal more in neurons than in astrocytes, and pyruvate infusion lowers the Laconic signal in neurons much less than in astrocytes. Combined with the biochemical assays of lactate in blood, these results define a lactate concentration gradient from astrocytes to neurons under resting conditions. The lactate sensor was further used to elucidate the lactate shuttle during physiological neural activity. In adult mice, electrical stimulation induces an increase in the cytosolic lactate level [66]. Exogenous K+ [66] or NH4+ [59] leads to an apparent elevation of the lactate level in 293T cells when astrocytes are co-cultured with 293T cells, implicating the ion-induced lactate shuttle from astrocytes. Collectively, these studies substantially support the astrocyte-neuron lactate shuttle model (Fig. 3, pathway ①).
Fig. 3.
Diverse proposed models of neural energy metabolism. At least four models of neural energy metabolism have been proposed based on the current evidence. The first model (①) is the ANLS hypothesis, which proposes that neuronal activity augments astrocytic glycolysis and lactate secretion and feeds neurons with lactate as an energy substrate. The second model (②) suggests that the lactate shuttle noted above occurs only under resting rather than stimulated conditions. The third model (③) postulates the direct uptake of glucose by neurons from the interstitium. The last model (④) states that lactate is transported from astrocytes to oligodendrocytes through gap junctions and that oligodendrocytes nurture and recharge axons with lactate.
Evidence opposing the ANLS hypothesis has been accumulating. In an acute hippocampal slice, the metabolic response of neurons to stimulation was evaluated by the NAD+/NADH sensor Peredox and the glucose sensor Sweeties [67]. Synaptic stimulation immediately triggered a small decrease in the neuronal glucose level and then a large increase in the NADH/NAD+ level [67]. Inhibition of neither lactate transport nor the mitochondrial NADH shuttle pathway affected the transient NADH/NAD+ increase, while an ionotropic glutamate transporter inhibitor or a glycolytic inhibitor mitigated this increase. These findings demonstrate that neuronal activation elicits a transient increase in the glycolytic flux, which is largely independent of lactate uptake. Of note is that monocarboxylate transporter inhibitors give rise to a small but discernable decline in NADH/NAD+ when neurons are in a resting state, suggesting that the lactate shuttle occurs under baseline conditions [67] (Fig. 3, pathway ②). Similarly, the infrared-labeled glucose tracer 2DG-RI supports the viewpoint of the direct uptake of glucose by neurons [68] (Fig. 3, pathway ③). In contrast, luciferase-based imaging of ATP dynamics and functional studies have noted the importance of the energy expenditure by synaptic vesicle cycling [4] and the axonal energy supply by oligodendroglial cells [63, 69, 70] (Fig. 3, pathway ④).
In summary, diverse cellular mechanisms for neural energy metabolism have been proposed. Recently, a novel proposal has emerged that lactate is a preferential energy substrate over glucose in cancer [71] as well as in physiological conditions [72]. Genetically-encoded fluorescent sensors are a promising tool to address questions on neural energy metabolism.
Conclusions and Outlook
Genetically-encoded fluorescent sensors allow the spatiotemporal imaging of metabolite dynamics in living cells and in vivo. These sensors not only can be applied to the quantification of metabolite concentrations at the single-cell level and even the subcellular level; more importantly, they are valuable tools for the systematic analysis of regulatory networks affecting metabolite flux, i.e., uptake, efflux, and metabolism. Therefore, these sensors provide good alternatives to traditional biochemical methods. There is no doubt that a few challenges still lie ahead. At present, the sensors for NAD(P)+/NAD(P)H, ATP/ADP, and glucose show good performance (i.e., large dynamic range, appropriate affinity, and intense fluorescence) in practical application, while there is a pressing need for the optimization of sensors for lactate, pyruvate, and glutamine due to their small dynamic changes, inappropriate affinity, or pH sensitivity [44–46]. For example, the lactate sensor Laconic may become saturated by lactate in highly glycolytic cells, which produce large amounts of lactate; furthermore, lactate-induced acidification also may introduce an error in lactate determination during glycolysis because of the sensor’s pH sensitivity. All these factors make it difficult for Laconic to track subtle changes under physiological conditions. Glycolysis-produced lactate may be taken up into cells, converted back to pyruvate, and processed by the TCA cycle and oxidative phosphorylation in mitochondria. The pH-derived artifact is greater for the poorly responsive pyruvate sensor Pyronic, especially in mitochondria, whose pH is labile depending on the energy metabolism state and reactive oxygen species generation. In addition, the glucose sensors in the FLII12Pglu family, the lactate sensor Laconic, the pyruvate sensor Pyronic, and glutamine sensors are based on FRET and have similar colors; thus, they cannot operate in the same subcellular location. Thus, the development of sensors with various colors, especially red, is very important, and such sensors will be favorable for the multiplex imaging of metabolites at the single-cell level and between different cell types. When they combine the advantages of metabolomics and metabolic flux analysis in delineating global metabolite profiling and metabolic networks, these sensors will provide deep insight into metabolic regulation and expand our knowledge of metabolism in health and disease.
Acknowledgements
This review was supported by the National Key Research and Development Program of China (2017YFA050400 and 2017YFC0906900), the National Natural Science Foundation of China (31722033, 91649123, 31671484, 31225008, and 31470833), the Shanghai Science and Technology Commission (14XD1401400, 16430723100, and 15YF1402600), Young Elite Scientists Sponsorship Program by China Association for Science and Technology (to YZ), Shanghai Young Top-notch Talent (to YZ), the State Key Laboratory of Bioreactor Engineering (to YY), and Fundamental Research Funds for the Central Universities (to YY and YZ).
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no competing interests.
Contributor Information
Yuzheng Zhao, Email: yuzhengzhao@ecust.edu.cn.
Yi Yang, Email: yiyang@ecust.edu.cn.
References
- 1.Belanger M, Allaman I, Magistretti PJ. Brain energy metabolism: focus on astrocyte-neuron metabolic cooperation. Cell Metab. 2011;14:724–738. doi: 10.1016/j.cmet.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 2.Harris JJ, Jolivet R, Attwell D. Synaptic energy use and supply. Neuron. 2012;75:762–777. doi: 10.1016/j.neuron.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 3.Hyder F, Rothman DL, Bennett MR. Cortical energy demands of signaling and nonsignaling components in brain are conserved across mammalian species and activity levels. Proc Natl Acad Sci U S A. 2013;110:3549–3554. doi: 10.1073/pnas.1214912110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rangaraju V, Calloway N, Ryan TA. Activity-driven local ATP synthesis is required for synaptic function. Cell. 2014;156:825–835. doi: 10.1016/j.cell.2013.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alle H, Roth A, Geiger JR. Energy-efficient action potentials in hippocampal mossy fibers. Science. 2009;325:1405–1408. doi: 10.1126/science.1174331. [DOI] [PubMed] [Google Scholar]
- 6.Mergenthaler P, Lindauer U, Dienel GA, Meisel A. Sugar for the brain: the role of glucose in physiological and pathological brain function. Trends Neurosci. 2013;36:587–597. doi: 10.1016/j.tins.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zielke HR, Zielke CL, Baab PJ. Direct measurement of oxidative metabolism in the living brain by microdialysis: a review. J Neurochem. 2009;109(Suppl 1):24–29. doi: 10.1111/j.1471-4159.2009.05941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pellerin L, Magistretti PJ. Glutamate uptake into astrocytes stimulates aerobic glycolysis: a mechanism coupling neuronal activity to glucose utilization. Proc Natl Acad Sci U S A. 1994;91:10625–10629. doi: 10.1073/pnas.91.22.10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pellerin L, Magistretti PJ. Neuroscience. Let there be (NADH) light. Science. 2004;305:50–52. doi: 10.1126/science.1100428. [DOI] [PubMed] [Google Scholar]
- 10.Kasischke KA, Vishwasrao HD, Fisher PJ, Zipfel WR, Webb WW. Neural activity triggers neuronal oxidative metabolism followed by astrocytic glycolysis. Science. 2004;305:99–103. doi: 10.1126/science.1096485. [DOI] [PubMed] [Google Scholar]
- 11.Magistretti PJ, Allaman I. A cellular perspective on brain energy metabolism and functional imaging. Neuron. 2015;86:883–901. doi: 10.1016/j.neuron.2015.03.035. [DOI] [PubMed] [Google Scholar]
- 12.Dienel GA. Lack of appropriate stoichiometry: Strong evidence against an energetically important astrocyte-neuron lactate shuttle in brain. J Neurosci Res. 2017;95:2103–2125. doi: 10.1002/jnr.24015. [DOI] [PubMed] [Google Scholar]
- 13.Bergersen LH, Gjedde A. Is lactate a volume transmitter of metabolic states of the brain? Front Neuroenergetics. 2012;4:5. doi: 10.3389/fnene.2012.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magistretti PJ, Pellerin L. Cellular mechanisms of brain energy metabolism and their relevance to functional brain imaging. Philos Trans R Soc Lond B Biol Sci. 1999;354:1155–1163. doi: 10.1098/rstb.1999.0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schonfeld P, Reiser G. Why does brain metabolism not favor burning of fatty acids to provide energy? Reflections on disadvantages of the use of free fatty acids as fuel for brain. J Cereb Blood Flow Metab. 2013;33:1493–1499. doi: 10.1038/jcbfm.2013.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lutas A, Yellen G. The ketogenic diet: metabolic influences on brain excitability and epilepsy. Trends Neurosci. 2013;36:32–40. doi: 10.1016/j.tins.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reddan MC, Wager TD. Modeling pain using fMRI: from regions to biomarkers. Neurosci Bull. 2018;34:208–215. doi: 10.1007/s12264-017-0150-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim E, Park H. Pairwise classifier ensemble with adaptive sub-classifiers for fMRI pattern analysis. Neurosci Bull. 2017;33:41–52. doi: 10.1007/s12264-016-0077-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guan X, Xu X, Zhang M. Region-specific iron measured by MRI as a biomarker for Parkinson’s disease. Neurosci Bull. 2017;33:561–567. doi: 10.1007/s12264-017-0138-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiller K, Metallo CM. Profiling metabolic networks to study cancer metabolism. Curr Opin Biotechnol. 2013;24:60–68. doi: 10.1016/j.copbio.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Patterson GH, Knobel SM, Arkhammar P, Thastrup O, Piston DW. Separation of the glucose-stimulated cytoplasmic and mitochondrial NAD(P)H responses in pancreatic islet beta cells. Proc Natl Acad Sci U S A. 2000;97:5203–5207. doi: 10.1073/pnas.090098797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eto K, Tsubamoto Y, Terauchi Y, Sugiyama T, Kishimoto T, Takahashi N, et al. Role of NADH shuttle system in glucose-induced activation of mitochondrial metabolism and insulin secretion. Science. 1999;283:981–985. doi: 10.1126/science.283.5404.981. [DOI] [PubMed] [Google Scholar]
- 23.Lin SJ, Guarente L. Nicotinamide adenine dinucleotide, a metabolic regulator of transcription, longevity and disease. Curr Opin Cell Biol. 2003;15:241–246. doi: 10.1016/S0955-0674(03)00006-1. [DOI] [PubMed] [Google Scholar]
- 24.Houtkooper RH, Canto C, Wanders RJ, Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev. 2010;31:194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Canto C, Menzies KJ, Auwerx J. NAD metabolism and the control of energy homeostasis: a balancing act between mitochondria and the nucleus. Cell Metab. 2015;22:31–53. doi: 10.1016/j.cmet.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsien RY. Constructing and exploiting the fluorescent protein paintbox (Nobel Lecture) Angew Chem Int Ed Engl. 2009;48:5612–5626. doi: 10.1002/anie.200901916. [DOI] [PubMed] [Google Scholar]
- 27.Mehta S, Zhang J. Reporting from the field: genetically encoded fluorescent reporters uncover signaling dynamics in living biological systems. Annu Rev Biochem. 2011;80:375–401. doi: 10.1146/annurev-biochem-060409-093259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Y, Yang Y. Profiling metabolic states with genetically encoded fluorescent biosensors for NADH. Curr Opin Biotechnol. 2015;31C:86–92. doi: 10.1016/j.copbio.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Y, Yang Y. Real-time and high-throughput analysis of mitochondrial metabolic states in living cells using genetically encoded NAD+/NADH sensors. Free Radic Biol Med. 2016;100:43–52. doi: 10.1016/j.freeradbiomed.2016.05.027. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Y, Jin J, Hu Q, Zhou HM, Yi J, Yu Z, et al. Genetically encoded fluorescent sensors for intracellular NADH detection. Cell Metab. 2011;14:555–566. doi: 10.1016/j.cmet.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cambronne XA, Stewart ML, Kim D, Jones-Brunette AM, Morgan RK, Farrens DL, et al. Biosensor reveals multiple sources for mitochondrial NAD(+) Science. 2016;352:1474–1477. doi: 10.1126/science.aad5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hung YP, Albeck JG, Tantama M, Yellen G. Imaging cytosolic NADH-NAD(+) redox state with a genetically encoded fluorescent biosensor. Cell Metab. 2011;14:545–554. doi: 10.1016/j.cmet.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bilan DS, Matlashov ME, Gorokhovatsky AY, Schultz C, Enikolopov G, Belousov VV. Genetically encoded fluorescent indicator for imaging NAD(+)/NADH ratio changes in different cellular compartments. Biochim Biophys Acta. 2014;1840:951–957. doi: 10.1016/j.bbagen.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Y, Hu Q, Cheng F, Su N, Wang A, Zou Y, et al. SoNar, a highly responsive NAD+/NADH sensor, allows high-throughput metabolic screening of anti-tumor agents. Cell Metab. 2015;21:777–789. doi: 10.1016/j.cmet.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Y, Wang A, Zou Y, Su N, Loscalzo J, Yang Y. In vivo monitoring of cellular energy metabolism using SoNar, a highly responsive sensor for NAD(+)/NADH redox state. Nat Protoc. 2016;11:1345–1359. doi: 10.1038/nprot.2016.074. [DOI] [PubMed] [Google Scholar]
- 36.Tao R, Zhao Y, Chu H, Wang A, Zhu J, Chen X, et al. Genetically encoded fluorescent sensors reveal dynamic regulation of NADPH metabolism. Nat Methods. 2017;14:720–728. doi: 10.1038/nmeth.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cameron WD, Bui CV, Hutchinson A, Loppnau P, Graslund S, Rocheleau JV. Apollo-NADP(+): a spectrally tunable family of genetically encoded sensors for NADP(+) Nat Methods. 2016;13:352–358. doi: 10.1038/nmeth.3764. [DOI] [PubMed] [Google Scholar]
- 38.Imamura H, Nhat KP, Togawa H, Saito K, Iino R, Kato-Yamada Y, et al. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc Natl Acad Sci U S A. 2009;106:15651–15656. doi: 10.1073/pnas.0904764106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsuyama T, Kishikawa J, Han YW, Harada Y, Tsubouchi A, Noji H, et al. In vivo fluorescent adenosine 5′-triphosphate (ATP) imaging of Drosophila melanogaster and Caenorhabditis elegans by using a genetically encoded fluorescent ATP biosensor optimized for low temperatures. Anal Chem. 2013;85:7889–7896. doi: 10.1021/ac4015325. [DOI] [PubMed] [Google Scholar]
- 40.Berg J, Hung YP, Yellen G. A genetically encoded fluorescent reporter of ATP:ADP ratio. Nat Methods. 2009;6:161–166. doi: 10.1038/nmeth.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tantama M, Martinez-Francois JR, Mongeon R, Yellen G. Imaging energy status in live cells with a fluorescent biosensor of the intracellular ATP-to-ADP ratio. Nat Commun. 2013;4:2550. doi: 10.1038/ncomms3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takanaga H, Chaudhuri B, Frommer WB. GLUT1 and GLUT9 as major contributors to glucose influx in HepG2 cells identified by a high sensitivity intramolecular FRET glucose sensor. Biochim Biophys Acta. 2008;1778:1091–1099. doi: 10.1016/j.bbamem.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hou BH, Takanaga H, Grossmann G, Chen LQ, Qu XQ, Jones AM, et al. Optical sensors for monitoring dynamic changes of intracellular metabolite levels in mammalian cells. Nat Protoc. 2011;6:1818–1833. doi: 10.1038/nprot.2011.392. [DOI] [PubMed] [Google Scholar]
- 44.San Martin A, Ceballo S, Ruminot I, Lerchundi R, Frommer WB, Barros LF. A genetically encoded FRET lactate sensor and its use to detect the Warburg effect in single cancer cells. PLoS One. 2013;8:e57712. doi: 10.1371/journal.pone.0057712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.San Martin A, Ceballo S, Baeza-Lehnert F, Lerchundi R, Valdebenito R, Contreras-Baeza Y, et al. Imaging mitochondrial flux in single cells with a FRET sensor for pyruvate. PLoS One. 2014;9:e85780. doi: 10.1371/journal.pone.0085780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gruenwald K, Holland JT, Stromberg V, Ahmad A, Watcharakichkorn D, Okumoto S. Visualization of glutamine transporter activities in living cells using genetically encoded glutamine sensors. PLoS One. 2012;7:e38591. doi: 10.1371/journal.pone.0038591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang H, Zhou L, Shi Q, Zhao Y, Lin H, Zhang M, et al. SIRT3-dependent GOT2 acetylation status affects the malate-aspartate NADH shuttle activity and pancreatic tumor growth. EMBO J. 2015;34:1110–1125. doi: 10.15252/embj.201591041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marcu R, Wiczer BM, Neeley CK, Hawkins BJ. Mitochondrial matrix Ca(2)(+) accumulation regulates cytosolic NAD(+)/NADH metabolism, protein acetylation, and sirtuin expression. Mol Cell Biol. 2014;34:2890–2902. doi: 10.1128/MCB.00068-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen H, Xu H, Potash S, Starkov A, Belousov VV, Bilan DS, et al. Mild metabolic perturbations alter succinylation of mitochondrial proteins. J Neurosci Res. 2017;95:2244–2252. doi: 10.1002/jnr.24103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oldham WM, Clish CB, Yang Y, Loscalzo J. Hypoxia-mediated increases in L-2-hydroxyglutarate coordinate the metabolic response to reductive stress. Cell Metab. 2015;22:291–303. doi: 10.1016/j.cmet.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Titov DV, Cracan V, Goodman RP, Peng J, Grabarek Z, Mootha VK. Complementation of mitochondrial electron transport chain by manipulation of the NAD+/NADH ratio. Science. 2016;352:231–235. doi: 10.1126/science.aad4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roesch A, Vultur A, Bogeski I, Wang H, Zimmermann KM, Speicher D, et al. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell. 2013;23:811–825. doi: 10.1016/j.ccr.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Menger L, Vacchelli E, Adjemian S, Martins I, Ma Y, Shen S, et al. Cardiac glycosides exert anticancer effects by inducing immunogenic cell death. Sci Transl Med. 2012;4:143ra199. doi: 10.1126/scitranslmed.3003807. [DOI] [PubMed] [Google Scholar]
- 54.Zhou B, Yu P, Lin MY, Sun T, Chen Y, Sheng ZH. Facilitation of axon regeneration by enhancing mitochondrial transport and rescuing energy deficits. J Cell Biol. 2016;214:103–119. doi: 10.1083/jcb.201605101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deuschle K, Okumoto S, Fehr M, Looger LL, Kozhukh L, Frommer WB. Construction and optimization of a family of genetically encoded metabolite sensors by semirational protein engineering. Protein Sci. 2005;14:2304–2314. doi: 10.1110/ps.051508105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bermejo C, Haerizadeh F, Takanaga H, Chermak D, Frommer WB. Optical sensors for measuring dynamic changes of cytosolic metabolite levels in yeast. Nat Protoc. 2011;6:1806–1817. doi: 10.1038/nprot.2011.391. [DOI] [PubMed] [Google Scholar]
- 57.Hu HY, Wei YF, Wang DC, Su N, Chen XJ, Zhao YZ, et al. Glucose monitoring in living cells with single fluorescent protein-based sensors. Rsc Advances. 2018;8:2485–2489. doi: 10.1039/C7RA11347A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jamali S, Klier M, Ames S, Barros LF, McKenna R, Deitmer JW, et al. Hypoxia-induced carbonic anhydrase IX facilitates lactate flux in human breast cancer cells by non-catalytic function. Sci Rep. 2015;5:13605. doi: 10.1038/srep13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lerchundi R, Fernandez-Moncada I, Contreras-Baeza Y, Sotelo-Hitschfeld T, Machler P, Wyss MT, et al. NH4(+) triggers the release of astrocytic lactate via mitochondrial pyruvate shunting. Proc Natl Acad Sci U S A. 2015;112:11090–11095. doi: 10.1073/pnas.1508259112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Erecinska M, Silver IA. Ions and energy in mammalian brain. Prog Neurobiol. 1994;43:37–71. doi: 10.1016/0301-0082(94)90015-9. [DOI] [PubMed] [Google Scholar]
- 61.Dienel GA. Brain lactate metabolism: the discoveries and the controversies. J Cereb Blood Flow Metab. 2012;32:1107–1138. doi: 10.1038/jcbfm.2011.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schurr A. Cerebral glycolysis: a century of persistent misunderstanding and misconception. Front Neurosci. 2014;8:360. doi: 10.3389/fnins.2014.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morrison BM, Lee Y, Rothstein JD. Oligodendroglia: metabolic supporters of axons. Trends Cell Biol. 2013;23:644–651. doi: 10.1016/j.tcb.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chuquet J, Quilichini P, Nimchinsky EA, Buzsaki G. Predominant enhancement of glucose uptake in astrocytes versus neurons during activation of the somatosensory cortex. J Neurosci. 2010;30:15298–15303. doi: 10.1523/JNEUROSCI.0762-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Machler P, Wyss MT, Elsayed M, Stobart J, Gutierrez R, von Faber-Castell A, et al. In vivo evidence for a lactate gradient from astrocytes to neurons. Cell Metab. 2016;23:94–102. doi: 10.1016/j.cmet.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 66.Sotelo-Hitschfeld T, Niemeyer MI, Machler P, Ruminot I, Lerchundi R, Wyss MT, et al. Channel-mediated lactate release by K(+)-stimulated astrocytes. J Neurosci. 2015;35:4168–4178. doi: 10.1523/JNEUROSCI.5036-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Diaz-Garcia CM, Mongeon R, Lahmann C, Koveal D, Zucker H, Yellen G. Neuronal stimulation triggers neuronal glycolysis and not lactate uptake. Cell Metab. 2017;26(361–374):e364. doi: 10.1016/j.cmet.2017.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lundgaard I, Li B, Xie L, Kang H, Sanggaard S, Haswell JD, et al. Direct neuronal glucose uptake heralds activity-dependent increases in cerebral metabolism. Nat Commun. 2015;6:6807. doi: 10.1038/ncomms7807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee Y, Morrison BM, Li Y, Lengacher S, Farah MH, Hoffman PN, et al. Oligodendroglia metabolically support axons and contribute to neurodegeneration. Nature. 2012;487:443–448. doi: 10.1038/nature11314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Funfschilling U, Supplie LM, Mahad D, Boretius S, Saab AS, Edgar J, et al. Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature. 2012;485:517–521. doi: 10.1038/nature11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Faubert B, Li KY, Cai L, Hensley CT, Kim J, Zacharias LG, et al. Lactate metabolism in human lung tumors. Cell. 2017;171(358–371):e359. doi: 10.1016/j.cell.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hui S, Ghergurovich JM, Morscher RJ, Jang C, Teng X, Lu W, et al. Glucose feeds the TCA cycle via circulating lactate. Nature. 2017;551:115–118. doi: 10.1038/nature24057. [DOI] [PMC free article] [PubMed] [Google Scholar]