Dear Editor,
It is known that genetic factors play important roles in the pathophysiology of major depressive disorder (MDD). However, its genetic mechanism is still unknown. Recently, the CONVERGE consortium performed whole-genome sequencing in a homogenous Chinese sample (5303 MDD patients and 5337 controls) and one locus near the silent mating type information regulation 2 homolog 1 gene (SIRT1) was identified at a genome-wide significant level [1]. At the mRNA level, we found that SIRT1 expression is significantly down-regulated in the peripheral blood of patients with MDD compared with healthy participants (decreased by 37%) [2]. As such, this suggests that SIRT1 is a novel MDD risk gene in Han Chinese.
The rs3758391 polymorphism is located in the 5’ flanking region of SIRT1, and this single nucleotide polymorphism (SNP) has been reported to affect SIRT1 mRNA expression in healthy Han Chinese [3]. In this study, we assessed the association of the SIRT1 rs3758391 polymorphism with MDD in a Han Chinese population.
A total of 702 patients were recruited from Wenzhou Kangning Hospital and Jinhua Second Hospital in Zhejiang Province. All patients were diagnosed with MDD according to the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV) criteria. The inclusion and exclusion criteria were as reported in our previous publications [4–7]. The standard diagnostic assessments were supplemented with clinical information obtained by a review of medical records and interviews with family informants. Seven hundred and eleven healthy controls were recruited from a group of blood donors in the same regions and were not psychiatrically screened. The control participants were self-reported to be free of psychiatric disorders, alcohol dependence, drug abuse, or a family history of psychiatric disorders. All procedures were reviewed and approved by the Institutional Review Boards of both participating institutions. This study was performed in accordance with the guidelines laid out in the Declaration of Helsinki as revised in 1989. All participants were of Han Chinese origin and provided written informed consent before any study-related procedures were performed.
Genomic DNA was isolated from whole blood using a Tiangen DNA isolation kit (Tiangen Biotech, Beijing, China). The SNP rs3758391 was genotyped using TaqMan assays, with details as described previously [8–11]. Subsequently, we conducted a meta-analysis to evaluate the association of rs3758391 with MDD. The literature was searched in PubMed (http://www.ncbi.nlm.nih.gov/pubmed/) and SCOPUS (http://www.scopus.com) with the keywords “SIRT1 or silent mating type information regulation 2 homolog 1”, “polymorphism or variant”, “rs3758391”, and “depression or MDD” in various combinations. Bibliographies or citations from the retrieved articles were also checked. All papers were published before March, 2018. Eligible studies for our meta-analysis met all of the following criteria: (1) were association studies written in English; (2) described the genotyping method and used commonly-accepted diagnostic criteria, such as the DSM; (3) contained independent data; and (4) presented sufficient data to calculate the odds ratio (OR) with confidence interval (CI) and P value. Major exclusion criteria included deviation from Hardy-Weinberg equilibrium, overlap with previous studies, or insufficient information for extraction. Authors of the studies were contacted when needed.
As MDD is reasonably considered to originate from aberrant brain functions [12], we used the brain eQTL (expression quantitative trait loci) database (http://caprica.genetics.kcl.ac.uk/BRAINEAC/), a large exon-specific data set covering ten human brain regions [13], for eQTL analysis.
Hardy-Weinberg equilibrium and allele and genotype frequencies were analyzed using SHEsis (http://analysis.bio-x.cn) [14]. Power analysis was performed using Quanto 1.2.3 (http://hydra.usc.edu/GxE). All P values were two-tailed, and P < 0.05 was considered statistically significant after Bonferroni correction.
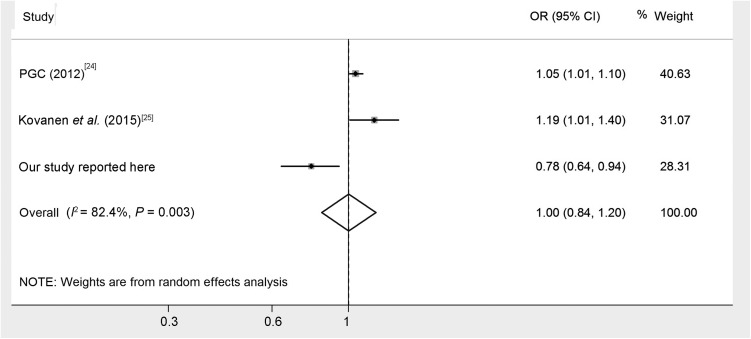
The genotype and allele frequencies for rs3758391 polymorphism among the 702 MDD patients and 711 healthy controls are shown in Table 1. The distribution of rs3758391 polymorphism was consistent with Hardy-Weinberg equilibrium in both the MDD and control groups, and rs3758391 showed significant genotypic and allelic differences between the case and control groups (P = 0.025 and P = 0.01, respectively). The frequency of the C allele in rs3758391 was higher in patients than in controls (OR = 1.29; 95% CI: 1.06–1.57). With the false-positive rate controlled as 0.05, the statistical power to detect the OR value as 1.29 for the risk allele was expected to be 0.79 in our samples on considering the 2% prevalence of MDD among Han Chinese [15] and under an additive genetic model. We analyzed the linkage disequilibrium between the rs12415800 and rs3758391 polymorphisms using http://archive.broadinstitute.org/mpg/snap/ and estimated that there was no disequilibrium (Table S1; D’ = 0.818, r2 = 0.098). To obtain a more comprehensive view of any potential association of rs3758391 with MDD, we conducted a meta-analysis by pooling our data with those of two previously published articles that met the inclusion criteria (Table S2). In total, three case-control association studies were included for meta-analysis. We found a significant heterogeneity in the homogeneity analysis (I2 = 82.4%, P = 0.003), then calculated the meta-analysis using the random-effects method, and found no significant difference in allelic distribution of the rs3758391 polymorphism between MDD and controls (Fig. 1, Z = 0.04, P = 0.97).
Table 1.
Distribution of rs3758391 genotype and alleles in MDD cases and healthy controls.
| SNP | n | Genotype, n (%) | P | Allele, n (%) | P | OR (95% CI) | |||
|---|---|---|---|---|---|---|---|---|---|
| rs3758391 | C/C | C/T | T/T | C | T | ||||
| Case | 702 | 31 (4.4) | 207 (29.5) | 464 (66.1) | 0.025 | 269 (19.2) | 1135 (80.8) | 0.01 | 1.29 (1.06-1.57) |
| Control | 711 | 16 (2.3) | 189 (26.6) | 506 (71.2) | 221 (15.5) | 1201 (84.5) | |||
Fig. 1.
Forest plot for overall association of rs3758391 polymorphism with MDD.
T allele versus C allele, square with size by weight and extending line for each study present the OR and 95% CI, respectively. Weight reflects the contribution of each study to the pooled estimate; the latter is shown by a diamond.
We then performed an eQTL analysis to investigate whether the SNP rs3758391 influences SIRT1 expression in the brain. As shown in Fig. S1, we found a significant association between rs3758391 and SIRT1 expression in the occipital cortex (P = 0.003, P = 0.03 after Bonferroni correction). Carriers with the CC genotype had significantly lower levels of SIRT1 expression in the occipital cortex than those with the TT genotype.
SIRT1 has been identified as a risk gene contributing to MDD at a genome-wide significant level [1] and is reported to mediate a chronic stress-elicited depression-like phenotype at a preclinical level [16]. Our previous work also indicated that SIRT1 plays an important role in the etiology of MDD [2]. In this study, we reported an association between the SIRT1 rs3758391 polymorphism and MDD in Han Chinese. However, our meta-analysis did not replicate this result. There was a wide divergence of the T allele frequency of rs3758391 among different ethnic populations (27% in Caucasians versus 85% in Han Chinese), implying that rs3758391 is likely to be an ethnicity-dependent polymorphism. Consequently, genetic heterogeneity may have affected the results of our meta-analysis.
SIRT1, a member of the sirtuin protein family, has been shown to mediate diverse cellular events including inflammation, apoptosis, autophagy, and cell growth [17, 18]. There is evidence that SIRT1 plays an important role in regulating inflammatory responses by modulating interleukin-6 (IL-6) expression [19], and the literature has documented that inhibition of SIRT1 promotes the secretion of IL-6 [20]. Intriguingly, we recently found a significantly increased level of IL-6 mRNA in patients with MDD [4]. Thus, our findings suggest that the involvement of SIRT1 in MDD may be associated with its role in regulating the inflammatory response.
It has been documented that rs3758391 functionally affects the mRNA expression of SIRT1 [3]. Our eQTL results showed that rs3758391 is associated with SIRT1 expression in the occipital cortex and individuals with the CC genotype have significantly lower levels of SIRT1 expression in the occipital cortex than those with the TT genotype. Emerging evidence from molecular studies and neuroimaging suggests involvement of the occipital cortex in the etiology of MDD [21, 22]. Our previous work also indicated that complement factor H disturbs the occipital cortex and increases the risk of MDD [5]. It is known that this factor is a component of the innate immune system that mediates inflammation by regulating complement [23]. Therefore, we hypothesize that inflammatory activity may underlie the involvement of the occipital cortex in MDD. This needs further investigation.
There are several limitations that should be noted. First, cross-sectional association studies always have the potential for population stratification, even though all participants were demographically and ethnically matched in this study. Therefore, we could not fully exclude the possibility of a population structure effect in our sample. Second, this was an exploratory study performed in a subset of the general Chinese Han population. The sample size is modest and precludes us from making any definitive statements on the association between SIRT1 and MDD. Third, we did not psychiatrically screen the control participants.
In conclusion, we used a comprehensive analysis to explore the potential relationship between a functional SNP rs3758391 within SIRT1 and the risk of MDD in Han Chinese. Our preliminary findings suggest that SIRT1 rs3758391 confers susceptibility to MDD in Han Chinese, whereas this polymorphism seems to be ethnicity-dependent. Therefore, further investigations are required to validate our results.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We are deeply grateful to all participants. This work was supported by the National Natural Science Foundation of China (81471358 and 81671326) and the Key Developing Disciplines of Shanghai Municipal Commission of Health and Family Planning, China (2015ZB0405).
Compliance with Ethical Standards
Conflict of interest
The authors declare that there are no conflicts of interest.
References
- 1.CONVERGE consortium. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature 2015, 523: 588–591. [DOI] [PMC free article] [PubMed]
- 2.Luo XJ, Zhang C. Down-regulation of SIRT1 gene expression in major depressive disorder. Am J Psychiatry. 1046;2016:173. doi: 10.1176/appi.ajp.2016.16040394. [DOI] [PubMed] [Google Scholar]
- 3.Hu YY, Wang LY, Chen SF, Liu XH, Li HF, Lu XF, et al. Association between the SIRT1 mRNA expression and acute coronary syndrome. J Atheroscler Thromb. 2015;22:165–182. doi: 10.5551/jat.24844. [DOI] [PubMed] [Google Scholar]
- 4.Zhang C, Wu ZG, Zhao GQ, Wang F, Fang YR. Identification of IL6 as a susceptibility gene for major depressive disorder. Sci Rep 2016, 6. [DOI] [PMC free article] [PubMed]
- 5.Zhang C, Zhang DF, Wu ZG, Peng DH, Chen J, Ni J, et al. Complement factor H and susceptibility to major depressive disorder in Han Chinese. Br J Psychiatry. 2016;208:446–452. doi: 10.1192/bjp.bp.115.163790. [DOI] [PubMed] [Google Scholar]
- 6.Zhang C, Wu Z, Hong W, Wang Z, Peng D, Chen J, et al. Influence of BCL2 gene in major depression susceptibility and antidepressant treatment outcome. J Affect Disord. 2014;155:288–294. doi: 10.1016/j.jad.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 7.Zhang C, Li ZZ, Wu ZG, Chen J, Wang ZW, Peng DH, et al. A study of N-methyl-D-aspartate receptor gene (GRIN2B) variants as predictors of treatment-resistant major depression. Psychopharmacology. 2014;231:685–693. doi: 10.1007/s00213-013-3297-0. [DOI] [PubMed] [Google Scholar]
- 8.Zhang C, Zhang J, Fan J, Cheng W, Du Y, Yu S, et al. Identification of ANKK1 rs1800497 variant in schizophrenia: new data and meta-analysis. Am J Med Genet B Neuropsychiatr Genet. 2014;165B:564–571. doi: 10.1002/ajmg.b.32259. [DOI] [PubMed] [Google Scholar]
- 9.Zhang C, Cai J, Zhang J, Li Z, Guo Z, Zhang X, et al. Genetic modulation of working memory deficits by ankyrin 3 gene in schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2014;50:110–115. doi: 10.1016/j.pnpbp.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 10.Cai J, Zhang W, Yi Z, Lu W, Wu Z, Chen J, et al. Influence of polymorphisms in genes SLC1A1, GRIN2B, and GRIK2 on clozapine-induced obsessive-compulsive symptoms. Psychopharmacology (Berl) 2013;230:49–55. doi: 10.1007/s00213-013-3137-2. [DOI] [PubMed] [Google Scholar]
- 11.Lu WH, Zhang C, Yi ZH, Li ZZ, Wu ZG, Fang YR. Association between BDNF Val66Met polymorphism and cognitive performance in antipsychotic-naive patients with schizophrenia. J Mol Neurosci. 2012;47:505–510. doi: 10.1007/s12031-012-9750-4. [DOI] [PubMed] [Google Scholar]
- 12.Zhang ZF, Ni JL, Zhang JT, Tang WX, Li X, Wu ZG, et al. A haplotype in the 5’-upstream region of the NDUFV2 gene is associated with major depressive disorder in Han Chinese. J Affect Disord. 2016;190:329–332. doi: 10.1016/j.jad.2015.10.034. [DOI] [PubMed] [Google Scholar]
- 13.Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17:1418–1428. doi: 10.1038/nn.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–98. doi: 10.1038/sj.cr.7290286. [DOI] [PubMed] [Google Scholar]
- 15.Gu L, Xie J, Long J, Chen Q, Pan R, Yan Y, et al. Epidemiology of major depressive disorder in mainland china: a systematic review. PLoS One. 2013;8:e65356. doi: 10.1371/journal.pone.0065356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abe-Higuchi N, Uchida S, Yamagata H, Higuchi F, Hobara T, Hara K, et al. Hippocampal Sirtuin 1 signaling mediates depression-like behavior. Biol Psychiatry. 2016;80:815–826. doi: 10.1016/j.biopsych.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 17.Yoshizaki T, Schenk S, Imamura T, Babendure JL, Sonoda N, Bae EJ, et al. SIRT1 inhibits inflammatory pathways in macrophages and modulates insulin sensitivity. Am J Physiol Endocrinol Metab. 2010;298:E419–E428. doi: 10.1152/ajpendo.00417.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rahman S, Islam R. Mammalian Sirt1: insights on its biological functions. Cell Commun Signal. 2011;9:11. doi: 10.1186/1478-811X-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang L, Chen Q, Meng Z, Sun L, Zhu L, Liu J, et al. Suppression of Sirtuin-1 increases IL-6 expression by activation of the Akt pathway during allergic asthma. Cell Physiol Biochem. 2017;43:1950–1960. doi: 10.1159/000484119. [DOI] [PubMed] [Google Scholar]
- 20.Volonte D, Zou H, Bartholomew JN, Liu Z, Morel PA, Galbiati F. Oxidative stress-induced inhibition of Sirt1 by caveolin-1 promotes p53-dependent premature senescence and stimulates the secretion of interleukin 6 (IL-6) J Biol Chem. 2015;290:4202–4214. doi: 10.1074/jbc.M114.598268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maciag D, Hughes J, O’Dwyer G, Pride Y, Stockmeier CA, Sanacora G, et al. Reduced density of calbindin immunoreactive GABAergic neurons in the occipital cortex in major depression: relevance to neuroimaging studies. Biol Psychiatry. 2010;67:465–470. doi: 10.1016/j.biopsych.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hasler G, Northoff G. Discovering imaging endophenotypes for major depression. Mol Psychiatry. 2011;16:604–619. doi: 10.1038/mp.2011.23. [DOI] [PubMed] [Google Scholar]
- 23.de Cordoba Rodriguez. S, Esparza-Gordillo J, Goicoechea de Jorge E, Lopez-Trascasa M, Sanchez-Corral P. The human complement factor H: functional roles, genetic variations and disease associations. Mol Immunol. 2004;41:355–367. doi: 10.1016/j.molimm.2004.02.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.