Abstract
Background
Biofilms, or colonies of uropathogen growing on the surface of indwelling medical devices, can inflict obstinate or recurring infection, thought-provoking antimicrobial therapy.
Methods
This prospective analysis included 105 urine samples from catheterized patients receiving intensive care. Ensuing phenotypic identification, antibiotic sensitivity test was performed by modified Kirby–Bauer disc diffusion method following CLSI guidelines; MDR isolates were identified according to the combined guidelines of the European Centre for Disease Prevention and Control (ECDC) and the Centers for Disease Control and Prevention (CDC). Biofilm-forming uropathogens were detected by the tissue culture plate (TCA) method.
Results
The predominant uropathogen in catheter-associated UTIs (CAUTIs) was Escherichia coli 57%, followed by Klebsiella pneumonia 15%, Pseudomonas aeruginosa 12%, Staphylococcus aureus 8%, Enterobacter spp. 3%, Enterococcus faecalis, Acinetobacter spp., and Proteus mirabilis 1.5%, of which 46% isolates were biofilm producers. Prime biofilm producers were Escherichia coli 33%, followed by Klebsiella pneumoniae 30%, Pseudomonas aeruginosa 20%, Staphylococcus aureus 10%, Acinetobacter, and Enterobacter 3.33%. Multidrug resistance associated with biofilm producers were greater than biofilm nonproducers. The Gram-negative biofilm producers found 96.15%, 80.76%, 73.07%, 53.84%, 53.84%, 46.15%, 19.23%, and 11.5% resistant to amoxyclave, ceftazidime, tetracycline, gentamicin, meropenem, nitrofurantoin, amikacin, imipenem, and fosfomycin, respectively. Gram-positive biofilm producers, however, were found 100% resistant to tetracycline, cloxacillin, and amoxyclave: 66.67% resistant to ampicillin while 33.33% resistant to gentamicin, ciprofloxacin, and nitrofurantoin.
Conclusion
High antimicrobial resistance was observed in biofilm producers than non-biofilm producers. Of recommended antimicrobial therapies for CAUTIs, ampicillin and amoxicillin-clavulanate were the least active antibiotics, whereas piperacillin/tazobactam and imipenem were found as the most effectual for gram-negative biofilm producer. Likewise, amoxicillin-clavulanate and tetracycline were the least active antibiotics, whereas vancomycin, fosfomycin, piperacillin-tazobactam, and meropenem were found as the most effective antibiotic for Gram-positive biofilm producer. In the limelight, the activity fosfomycin was commendable against both Gram-positive and Gram-negative biofilm producers.
1. Background
Of nearly 40 percentile of all health care-associated infections, urinary tract infections (UTIs) are the foremost cause of infections; out of these, a bulky proportion, 80%, involve catheter-associated urinary tract infections (CAUTIs) [1]. The urinary catheters are routinely used in urology practice; albeit, advances in design and materials used, UTIs persist as the major snags, owing to the contamination of such indwelling devices [2]. Approximately, prior admission, 12 to 16% of adult hospital inpatients have an indwelling urinary catheter, however, known to be associated with high morbidity, high mortality, increased the length of hospital stay, and increased the cost of treatment [1–3]. Furthermore, the catheter-associated biofilm producers, preceding drug resistivity, and their thought-provoking infection control procedures have been reported in aforementioned studies, which raises our concern on CAUTIs and biofilm producers in our settings [4, 5].
Biofilms are the sessile polymicrobial communities attached to the substratum of biotic and abiotic surfaces and are sheathed within a self-produced extracellular polymeric matrix, that is, polysaccharides intercellular adhesin [2, 5, 6]. The extracellular matrix facilitates communications among the cells through biochemical signals—acyl-homoserine lactone in Gram-negative bacteria and oligopeptides in Gram-positive bacteria—in a phenomenon called as quorum sensing [7]. Not only the matrix precludes the pathogen against host defense but also attributes antimicrobial resistance, by subordinating antibiotic penetration, horizontal transmission of plasmid-associated drug-resistant gene, and altered microenvironment [6, 7]. In this standpoint, early detection of biofilm producers is crucial, to reduce the irrational antimicrobial burden proceeding antimicrobial resistance in the patient; hence, it would be an auxiliary in controlling device-associated infections in medical centers.”
The rationale of our study was to explicate bacterial etiologies, illuminate biofilm-associated resistivity patterns, and to endorse suitable antimicrobial therapy against biofilm producers in CAUTIs.
2. Material and Methods
2.1. Study Design and Setup
The cross-sectional study was conducted at the Department of Microbiology, Janamaitri Foundation Institute of Health Science (JFIHS), Nepal, over a period of six months. The study hospital is a referral centre with medical, surgical, gynecological, pediatric, geriatric, and other specialties.
2.2. Inclusion and Exclusion Criteria
The urine sample of all catheterized patients irrespective of gender and age between 12 and 70 years who met the criteria of CAUTI were included in the study. Nevertheless, noncatheterized patients, either nursed in ward or formerly under antimicrobial therapy at least 48 h prior catheter insertion and no more than two types of organism grown from the clinical sample, were considered as contaminated and consequently, excluded from the study.
2.3. Laboratory Methods
CAUTI was defined using a combination of clinical signs and symptoms and laboratory criteria as described by Stamm [2]. A total of 105 urine sample from the catheterized patient, admitted in intensive care units, were processed semiquantitatively by inoculating 0.001 ml of the specimen (by using a calibrated wire loop) onto the Cystine Lactose Electrolyte Deficient (CLED) agar for the isolation and identification of significant uropathogens [8]. Following the inoculation, the plates were incubated for 24 hours at 37°C in an aerobic atmosphere. The growth of a single organism with a count of ≥102 colony-forming units (CFU)/ml was considered to represent as CAUTIs and was identified using appropriate routine identification methods including colony morphology, Gram stain, and an in-house set of biochemical tests [8].
2.4. Antimicrobial Susceptibility Testing
The susceptibility of bacterial isolates against recommended antibiotics was tested by the modified Kirby–Bauer disk diffusion method on Mueller Hinton agar (HiMedia, India) following guidelines of Clinical and Laboratory Standards Institute (CLSI), Wayne, USA [9]. Antibiotics that were tested in our study include amoxycillin clavulanate (amc 20/10 μg), ampicillin (amp 10 μg), amikacin (ak 30 μg), ceftazidime (caz 30 μg), ceftazolin (30 μg), cefuroxime (cfm 30 μg), ciprofloxacin (cip 5 μg), cloxacillin (cox 30 μg), cotrimoxazole (cot 25 μg), fosfomycin (fo 200 μg), gentamicin (gen 10 μg), imipenem (imp 10 μg), meropenem (mrp 10 μg), nitrofurantion (300 μg), piperacillin-tazobactam (pit 100/10 μg), teteracycline (te 30 μg), and vancomycin (VAN 30 μg) (HiMedia Laboratories, India). Further, elucidations of antibiotic susceptibility results were made conferring to the zone size interpretative standards of CLSI [9]. MDR isolates, resistant to at least one antimicrobial from three different groups of first-line drugs, tested were identified according to the combined guidelines of the European Centre for Disease Prevention and Control (ECDC) and the Centers for Disease Control and Prevention (CDC) [10]. Escherichia coli ATCC 25922, Staphylococcus aureus (ATCC 25923), and Pseusdomonas aeruginosa (ATCC 27853) were used as a control organism for antibiotic susceptibility testing.
2.5. Detection of Biofilm Production
The detection of biofilm was done by tissue culture method/microtiter plate method (TCA), the gold standard method, as described as Christensen et al. [11]. In brief, the bacterial isolates from fresh agar plates were inoculated in 2 ml of BHI broth and incubated for 24 h at 37°C. The cultures were then diluted 1 : 40 with fresh medium (BHI broth supplemented with 1% glucose); 200 µl of the sample was dispensed in the individual microtitration plate (AD Touch, apDianv) and incubated further 24 h at 37°C. With a gentle tapping, the content was removed further with a subsequent washing with phosphate buffer saline (pH 7.2) three times to remove free floating sessile bacteria. The adherent bacteria, biofilm producer, were fixed with sodium acetate (2%) and stained with crystal violet (0.1% w/v) for 10–15 min. The unbound crystal violet solution was removed with a triplicate washing with PBS, and the plate, then, was kept for drying. Finally, all wells were filled with 200 µl ethanol (95%) to release dye from the well and Optical Density (OD) was taken at the wavelength of 630 nm. For a precision, the experiment was performed in triplicate two times. Average OD values of each test strain and negative control were calculated, and OD cut-off values (ODc) were assessed as described by Stepanovie et al. [12].
2.6. Data Analysis
The information regarding patients' profile and the results were entered into a computer program. Data analysis was carried out using the Statistical Package for Social Sciences (SPSS™) version 20.0 (IBM, Armonk, NY, USA) and presented in percentage base distribution.
2.7. Ethical Consideration
Written approval was taken from the Institutional Review Committee (IRC) of Janamaitri Foundation Institute of Health Science (JFIHS) after submitting and presenting the research proposal. Written informed consent was taken from every patient or their guardians before enrollment in the study.
3. Results
3.1. Patient Demographics
During the study period, a total of 105 urine specimens from the patients suspected with catheter-associated UTIs were processed. Among total clinical specimens, 61.9% (65/105) were found with a growth of at least one significant pathogen confirming the urinary tract infection (UTI). Female (43, 66.2%) was the most affected group, with predominant etiologies as Escherichia coli 56.9% (37/65) followed by Klebsiella pneumoniae 10 (15%), Pseudomonas aeroginosa 8 (12%), Staphylococcus aureus 5 (8%), Enterobacter spp 2 (3%), and Enterococcus faecalis, Acinetobacter spp., and Proteus mirabilis 1 (1.5% each) (Table 1).
Table 1.
Etiological agent of catheter-associated UTIs.
| Organism | Frequency | Percent |
|---|---|---|
| Escherichia coli | 37 | 56.9 |
| Klebsiella pneumoniae | 10 | 15.4 |
| Pseudomonas aeroginosa | 8 | 12.3 |
| Staphyloccous aureus | 5 | 7.7 |
| Enterobacter spp. | 2 | 3.1 |
| Enterococcus faecalis | 1 | 1.5 |
| Proteus mirabilis | 1 | 1.5 |
| Acinetobacter spp. | 1 | 1.5 |
| Total | 65 | 100.0 |
3.2. Detection of Biofilm Producers
In the current study, 30 (46%) strains were in vitro positive for the biofilm production and 35 (54%) were negative for the biofilm production. Out of which (n=7, 9, and 14) were strong, moderate, and weak biofilm producer. Escherichia coli (33.33%) was found to be more biofilm producer followed by Klebsiella pneumoniae (30%), Pseudomonas aeroginosa (20%), Staphylococcus aureus (10%), and Acinetobacter spp. and Enterobacter spp. (3.33% each) (Table 2).
Table 2.
Degree of biofilm production by the bacterial isolates.
| Organisms | Nonproducer (%) | Biofilm producer | Total biofilm producer (%) | ||
|---|---|---|---|---|---|
| Strong producer | Moderate producer | Weak producer | |||
| Escherichia coli | 27 (77.14) | 1 | 4 | 5 | 10 (33.33) |
| Klebsiella pneumonia | 1 (2.86) | 2 | 3 | 4 | 9 (30.00) |
| Enterococcus faecalis | 1 (2.86) | 0 | 0 | 0 | 0 (0.00) |
| Pseudomonas aeroginosa | 2 (5.71) | 3 | 1 | 2 | 6 (20.00) |
| Proteus mirabilis | 1 (2.86) | 0 | 0 | 0 | 0 (0.00) |
| Staphylococcus aureus | 2 (5.71) | 0 | 0 | 3 | 3 (10.00) |
| Acinetobacter spp. | 0 (0.00) | 1 | 0 | 0 | 1 (3.33) |
| Enterobacter spp. | 1 (2.86) | 0 | 1 | 0 | 1 (3.33) |
| Total | 35 (100) | 7 | 9 | 14 | 30 (100) |
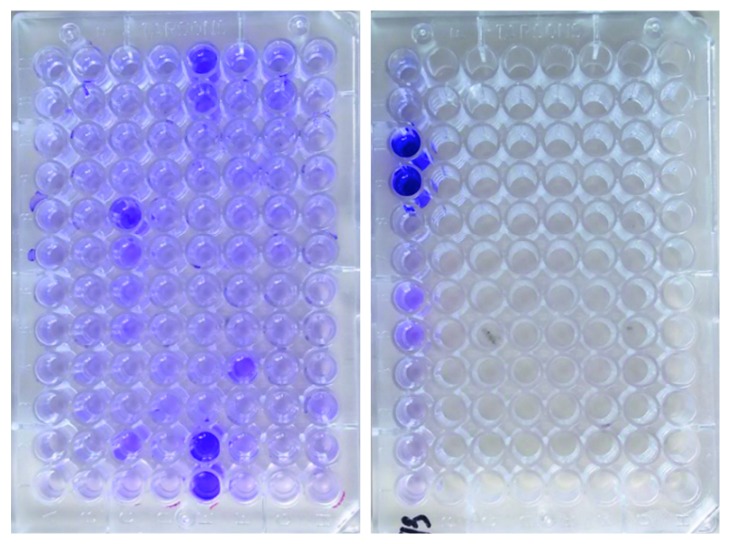
Microtitration plate shows the biofilm production by TCA method as shown in Figure 1.
Figure 1.
Microtitration plate showing the biofilm production by the TCA method.
3.3. The Antimicrobial Resistant Pattern in Biofilm Producers and Nonproducers
Gram-negative biofilm producers, more than 90%, were resistant to ampicillin and amoxicillin-clavulanate, and nearly 94% of biofilm producer and nonproducer were found to be sensitive to fosfomycin. Besides, the greater percentile of antimicrobial resistivity was found to be associated with biofilm producers than biofilm nonproducers. The Gram-positive biofilm producers, 3 of 3 bacterial isolates, were resistance to amoxicillin-clavulanate, tetracyclin, and cloxacillin; nonetheless, the antibiotics—vancomycin, fosfomycin, meropenem, and piperacillin/tazobactam—were found to be effective even for the biofilm producers (Table 3).
Table 3.
Antibiotic susceptibility of biofilm producer and non-biofilm producers.
| Antibiotics | Biofilm producer isolates (n=26) | % | Biofilm nonproducer isolates (n=33) | % | Resistance of all isolates (n=59) | % |
|---|---|---|---|---|---|---|
| Gram-negatives isolates | ||||||
| Ampicillin | 24 | 92.30 | 28 | 84.84 | 52 | 88.13 |
| Ceftazolin | 20 | 76.92 | 25 | 75.75 | 45 | 76.27 |
| Gentamicin | 14 | 53.84 | 11 | 30.30 | 25 | 42.37 |
| Ciprofloxacin | 19 | 73.07 | 23 | 69.69 | 42 | 71.18 |
| Nitrofurantion | 12 | 46.15 | 5 | 15.15 | 17 | 28.81 |
| Amikacin | 12 | 46.15 | 11 | 33.33 | 23 | 38.98 |
| Amoxicillin-clavulanate | 25 | 96.15 | 24 | 72.72 | 49 | 83.05 |
| Piperacillin-tazobactam | 7 | 26.92 | 7 | 21.22 | 14 | 23.72 |
| Cefuroxime | 20 | 76.92 | 24 | 72.72 | 44 | 74.57 |
| Imipenem | 5 | 19.23 | 1 | 3.03 | 6 | 10.16 |
| Fosfomycin | 3 | 11.5 | 1 | 3.03 | 4 | 6.77 |
| Meropenem | 14 | 53.84 | 12 | 36.36 | 26 | 44.06 |
| Tetracyclin | 19 | 73.07 | 17 | 51.51 | 36 | 61.01 |
| Cotrimoxazol | 17 | 65.38 | 21 | 63.63 | 38 | 64.40 |
| Ceftazidime | 21 | 80.76 | 21 | 63.63 | 42 | 71.18 |
|
| ||||||
| Antibiotics | Biofilm producer isolates (n=3) | % | Biofilm nonproducer isolates (n=3) | % | Resistance of all isolates (n=6) | % |
|
| ||||||
| Gram-positive isolates | ||||||
| Ampicillin | 2 | 66.67 | 1 | 33.33 | 3 | 50 |
| Gentamicin | 1 | 33.33 | 0 | 0.00 | 1 | 16.67 |
| Ciprofloxacin | 1 | 33.33 | 0 | 0.00 | 1 | 16.67 |
| Nitrofurantion | 1 | 33.33 | 0 | 0.00 | 1 | 16.67 |
| Amoxicillin-clavulanate | 3 | 100 | 1 | 33.33 | 4 | 66.67 |
| Piperacillin-tazobactam | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 |
| Meropenem | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 |
| Tetracyclin | 3 | 100 | 2 | 66.67 | 5 | 83.33 |
| Cloxacillin | 3 | 100 | 2 | 66.67 | 5 | 83.33 |
| Vancomycin | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 |
| Fosfomycin | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 |
The multidrug combination, in Gram-positive isolates and Gram-negative isolates, was found to be resistant to ciprofloxacin/cotrimoxazole/ampicillin (75%) and ampicillin/cloxacillin/teicoplanin (66.67%) (Table 4).
Table 4.
Multidrug resistance isolates in biofilm producer and non-biofilm producer and multidrug resistance in Gram-negative isolates.
| Multidrug combination | Biofilm producer isolates (n=12) | % | Biofilm nonproducer isolates (n=22) | % |
|---|---|---|---|---|
| Multidrug resistance in Gram-positive isolates | ||||
| AK, CIP, COT, AMP | 6 | 50 | 7 | 31.81 |
| AK, CIP, COT | 7 | 58 | 8 | 36.36 |
| CIP, COT, AMP | 9 | 75 | 13 | 59.00 |
| COT, AMP, AK | 7 | 58 | 8 | 36.36 |
| AMP, AK, CIP | 7 | 58 | 7 | 31.81 |
|
| ||||
| Multiple drug combination | Biofilm producer isolates (n=3) | % | Biofilm nonproducer isolates (n=3) | % |
|
| ||||
| Multidrug resistance in Gram-positive isolates | ||||
| AMP, COX, TE, NIT | 1 | 33.33 | 0 | 0.00 |
| AMP, COX, TE | 2 | 66.67 | 1 | 33.33 |
| COX, TE, NIT | 1 | 33.33 | 0 | 0.00 |
| TE, NIT, AMP | 1 | 33.33 | 0 | 0.00 |
| NIT, AMP, COX | 1 | 33.33 | 0 | 0.00 |
AK: amikacin; CIP: ciprofloxacin; COT: cotrimoxazole; AMP = ampicillin; COX = cloxacillin; TE = tetracyclin; NIT = nitrofurantoin.
4. Discussion
Most aspects of the diagnosis, treatment, and prevention of CAUTI are influenced by the tenacity of biofilm-associated uropathogens [13]. Meanwhile, in patients with underlying diseases or under intensive care, the relevance of detection of biofilm producers is crucial since CAUTIs is a common nosocomial infection [1]. The prevalence of catheter-associated urinary tract infection was found to be 61.9%, with predominance in female sufferers (43, 66.2%). Similar prevalence was reported in a review of Nicolle from 15 developing countries like ours [14]. This might be due to the anatomical differences of urogenital organs—anal proximity and shorter urethra in female [15].
Our study revealed that the most common bacterial etiology contributing CAUTIs was Escherichia coli 57%, followed by Klebsiella pneumoinae 15%, Pseudomonas aeruginosa 12%, S. aureus 8%, Enterobacter spp. 3%, and Enterococcus faecalis, and Acinetobacter spp. and Proteus mirabilis (1.5% each) which was nearly similar to the findings Seif Elden Salwa et al. who found that Escherichia coli (50%) was predominantly acquired infection followed by Klebsiella spp. (30%), Pseudomonas spp. (8%), Staphylococcus aureus (8.6%), and Proteus spp. (4.6%) [16]. In a study, Niveditha et al. and Pramodhini reported 70% of Escherichia coli were isolated [17, 18]; in contrast, Ahmed Abdallah et al. reported 31% Escherichia coli in their study [19]. The recent prevalence study, however, depicted the predominance of the Escherichia coli attaining 79.1% leading UTIs [20]. Similarly, 46% of the bacterial isolates were detected as biofilm producers in the CAUTIs, ensuing with the TCA method. Similar findings were reported by Ghanwate et al. and Ahmed Abdallah et al. where the biofilm producers were 47.5% and 43.5%, respectively [19, 21]. Niveditha et al. and Reid et al., however, found the associated biofilm producers 60% and 73%, respectively, which matched with our study [17, 22].
In our study, 46% isolates were biofilm producers using the tissue culture plate method. Similar findings were observed by Safia Maqbool et al. and Ahmed Abdallah et al. where 47.5% and 43.5% uropathogenic isolates were biofilm producers, respectively; Niveditha et al. and Reid et al., however, found 60% and 73% biofilm producers as CAUTIs [17, 19, 22, 23]. The uropathogen, Escherichia coli 33%, emerges as a predominant biofilm producer followed by Klebsiella 30%, Pseudomonas spp. 20%, S. aureus 10%, and Acinetobacter and Enterobacter (3.33% each) in our settings. The findings were comparable with Sharma et al. 67.5% and Nivenditha et al. 60% for the predominant isolate—biofilm producing Escherichia coli [17, 24]. However, Ponnusamy et al. in their study revealed that of 100 Escherichia coli strains, 72 were biofilm-positive phenotype [25]. Conferring a degree of biofilm producers, in our study, 11%, 14%, and 21% were strong, moderate, and weak biofilm producers, whereas Mishra et al. disclosed, 1.5%, 10.5%, and 34.3% as strong, moderate, and weak biofilm producers in their study [4].
Antibiotic resistance patterns of biofilm producer and nonproducer were observed; sequentially, high resistance against tested antibiotics was attributed in biofilm producers. In Gram-negative bacterial isolates, the resistance against four groups of antibiotics, ampicillin, nitrofurantoin, tetracycline, and meropenem, was equated in biofilm producer versus non-biofilm producer isolates; the consecutive antibiotics were 92.3% versus 84.84%, 46.15% versus 15.15%, 73.07% versus 51.51%, and 53.84% versus 36.31%, respectively. Similarly, the antibiotics ampicillin, ciprofloxacin, nitrofurantion, and tetracycline were 66.67% versus 33.33%, 33.33% versus 0%, 33.33% versus 0%, and 100% versus 66.67%, respectively. Pramodhini et al., nonetheless, disclosed ampicillin (83.3% versus 60%), cephotaxime (73.3% versus 35%), norfloxacin (80% versus 60%), and nalidxic acid (93% versus 70%) from his study [18]. Furthermore, fosfomycin has shown promising in vitro activity against both Gram-positive and Gram-negative biofilm producers, as earlier experienced by Neuner et al., Mihailescu et al. and Marquès et al. in treating urinary tract infections [26–28].
Hence, the antimicrobial resistivity in the isolate possibly attributed with the biofilm productions. In Gram-positive isolates, the resistance against 3 groups of antibiotics amoxicillin-clavulanate, tetracyclin, and cloxacillin was equated in biofilm producer versus non-biofilm producer isolates; the consecutive antibiotics were 100% resistant in biofilm producers while 33.3%, 66.7%, and 66.77% were found to be resistant in non-biofilm producers. Similarly, MRSA, resistant pattern linked with the tenacity of biofilm producers, was observed by Ando et al. [29]. In biofilms, poor antibiotic penetration, nutrient limitation and slow growth, adaptive stress responses, and formation of persister cells are hypothesized to constitute a multilayered defense [30].
The backdrop of antimicrobial resistant in biofilm producers urges seeking of naive therapeutic alternatives despite conventional antibiotic therapies. In the meantime, to prevent or to combat these obstinate biofilm producers, small molecules (N-acetylcysteine, Ca2+ and Mg2+ chelators), and matrix-targeting enzymes (dispersin B, DNase I, proteinase K and trypsin), bactericidal and antiadhesion coatings (trimethylsilane, carboxybetaine methacrylate, organoselenium, heparin, and hyaluronic acid; polymer brush coatings; and furanones) were commenced successfully in developed nations [31]. Ironically, the indorsed therapeutic solutions are farther than reach in the developing countries like ours.
5. Conclusion
Biofilm producing bacteria are responsible for many recalcitrant infections and are notoriously hard to eradicate, owing to the possible acquisition of multidrug status. High antimicrobial resistance was observed in biofilm producing pathogens than nonproducers. Of recommended antimicrobial therapies for CAUTIs, ampicillin and amoxicillin-clavulanate were the least active antibiotics, whereas piperacillin/tazobactam, fosfomycin, and imipenem were found as the most effective antibiotics among Gram-negative biofilm producers. Likewise, among Gram-positive biofilm producer isolates; amoxicillin-clavulanate and tetracycline were the least active, whereas vancomycin, fosfomycin, piperacillin/tazobactam, and meropenem were found as the most effective antibiotic. In the limelight, the activity of fosfomycin was laudable against both Gram-positive and Gram-negative biofilm producers. Hence, an early identification of biofilm producers with subsequent detection of antibiotic resistivity pattern is mandatory, to improve the clinical management of CAUTIs when the patient requires an intensive care.
Acknowledgments
The authors would like to thank Asst. Prof. Shyam Kumar Mishra (Institute of Medicine, TUTH) for his tremendous technical support.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Ethical Approval
Written informed consent was obtained from every patient for granting participation in an interview and to extract pertinent sociodemographic and clinical data from their respective clinical files, respecting confidentiality.
Consent
Written informed consent was obtained from the patients for relevant investigations, and publication of the findings was taken from every patient and coauthors.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Govinda Maharjan and Priyatam Khadka conceived the study, designed the manuscript, and reviewed the literature. Ganesh Chapagain, Gomik Siddhi Shilpakar, and Guna Raj Dhungana reviewed the manuscript and gave the concept of the research paper and critically reviewed the manuscript. All authors read the final version of the manuscript and approved for publication.
References
- 1.Syed M. A., Manzoor U., Shah I., Bukhari H. A. Antibacterial effects of tungsten nanoparticles on the Escherichia coli strains isolated from catheterized urinary tract infection (UTI) cases and Staphylococcus aureus. New Microbiologica. 2010;33(4):329–335. [PubMed] [Google Scholar]
- 2.Stamm W. E. Catheter-associated urinary tract infections: epidemiology, pathogenesis, and prevention. American Journal of Medicine. 1991;91(3):S65–S71. doi: 10.1016/0002-9343(91)90345-x. [DOI] [PubMed] [Google Scholar]
- 3.Weinstein J. W., Mazon D., Pantelick E., Cirincione R., Dembry L. M., Hierholzer W. J. A decade of prevalence surveys in a tertiary-care center: trends in nosocomial infection rates, device utilization, and patient acuity. Infection Control and Hospital Epidemiology. 2014;20(8):543–548. doi: 10.1086/501675. [DOI] [PubMed] [Google Scholar]
- 4.Mishra S. K., Basukala P., Basukala O., Parajuli K., Pokhrel B. M., Rijal B. P. Detection of biofilm production and antibiotic resistance pattern in clinical isolates from indwelling medical devices. Current Microbiology. 2014;70(1):128–134. doi: 10.1007/s00284-014-0694-5. [DOI] [PubMed] [Google Scholar]
- 5.Sabir N., Ikram A., Zaman G., et al. Bacterial biofilm-based catheter-associated urinary tract infections: causative pathogens and antibiotic resistance. American Journal of Infection Control. 2017;45(10):1101–1105. doi: 10.1016/j.ajic.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 6.Watnick P., Kolter R. Biofilm, city of microbes. Journal of Bacteriology. 2000;182(10):2675–2679. doi: 10.1128/jb.182.10.2675-2679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J., Park H., Chung S. Microfluidic approaches to bacterial biofilm formation. Molecules. 2012;17(8):9818–9834. doi: 10.3390/molecules17089818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Isenberg H. D. Clinical Microbiology Procedures Handbook. 2nd. Washington, DC, USA: ASM Press; 2004. [Google Scholar]
- 9.CLSI. Performance Standards for Antimicrobial Disk Susceptibility Tests; Approved Standard. 12th. Vol. 35. Wayne, PA, USA: Clinical & Laboratory Standards Institute; 2015. M02-A12. [Google Scholar]
- 10.Magiorakos A., Srinivasan A., Carey R. B., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clinical Microbiology and Infection. 2011;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 11.Christensen G. D., Simpson W. A., Bisno A. L., Beachley E. H. Adherence of slim-producing strains of staphylococcus epidermidis to smooth surfaces. Infection and Immunity. 1982;37(1):318–326. doi: 10.1128/iai.37.1.318-326.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stepanovic S., Vukovic D., Hola V., Di Bonaventura G., Djukic S., Cirkovic I. R. F. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by Staphylococci. APMIS. 2007;115(8):891–899. doi: 10.1111/j.1600-0463.2007.apm_630.x. [DOI] [PubMed] [Google Scholar]
- 13.Trautner B., Darouiche R. Role of biofilm in catheter-associated urinary tract infection. American Journal of Infection Control. 2004;32(3):177–183. doi: 10.1016/s0196-6553(03)00799-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicolle L. E. Catheter associated urinary tract infections. Antimicrobial Resistance and Infection Control. 2014;3(1):1–8. doi: 10.1186/2047-2994-3-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forbes B., Sham D. W. A. Bailey and Scott’s Diagnostic Microbiology. 12th. Maryland Heights, MO, USA: Mosby Elsevier; 2007. [Google Scholar]
- 16.Seif Elden Salwa S., El-Temawy Abd El khalek A. A. E. H., Ahmed E. H. Biofilm formation by E. Coli causing catheter associated urinary tract infection (CAUTI) in Assiut University Hospital. Egyptian Journal of Medical Microbiology. 2013;22(4):101–110. doi: 10.12816/0004967. [DOI] [Google Scholar]
- 17.Niveditha S., Pramodhini S., Umadevi S., Kumar S., Stephen S. The isolation and the biofilm formation of uropathogens in the patients with catheter associated urinary tract infections (UTIs) Journal of Clinical and Diagnostic Research. 2012;6(9):1478–1482. doi: 10.7860/jcdr/2012/4367.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pramodhini S. Antiobiotic resistance pattern of biofilm forming uropathogens isolated from catheterised patients in Pondicherry, India. Australasian Medical Journal. 2012;5(7):344–348. doi: 10.4066/amj.2012.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahmed Abdallah N. M., Elsayed S. B., Yassin Mostafa M. M., El-Gohary G. M. Biofilm forming bacteria isolated from urinary tract infection,relation to catheterization and susceptibility to antibiotics. International Journal for Biotechnology and Molecular. 2011;2(10):172–178. [Google Scholar]
- 20.Pradhan B. P. S. Prevalence of urinary tract infection and antibiotic susceptibility pattern to urinary pathogens in Kathmandu Medical College and Teaching Hospital. Birat Journal of Health Sciences. 2017;2(1):134–137. doi: 10.3126/bjhs.v2i1.17290. [DOI] [Google Scholar]
- 21.Niraj Ghanwate A., Thakare P. V., Bhis P. R., Dhanke A., Apotikar S. Prevention of biofilm formation in urinary catheter by coating enzymes/gentamycin/EDTA. International Journal of Biotechnology and Bioengineering. 2012;6(10):487–489. [Google Scholar]
- 22.Reid G., Charbonneau-Smith R. N., Lam D., Kang Y. S., Lacerte M., Hayes K. C. Bacterial biofilm formation in the urinary bladder of spinal cord injured patients. Paraplegia. 1992;30:711–717. doi: 10.1038/sc.1992.138. [DOI] [PubMed] [Google Scholar]
- 23.Nandini M. S., Madhusudan K. Bacteriological profile of catheter associated urinary tract infection and its antimicrobial susceptibility pattern in a Tertiary Care Hospital. Journal of Pharmaceutical Sciences and Research. 2016;8(4):204–207. [Google Scholar]
- 24.Sharma M., Aparna, Yadav S., Chaudhary U. Biofilm production in uropathogenic Escherichia coli. Indian Journal of Pathology & Microbiology. 2009;52(2):p. 294. doi: 10.4103/0377-4929.48960. [DOI] [PubMed] [Google Scholar]
- 25.Ponnusamy P., Natarajan V., Sevanan M. The in vitro biofilm formation by the uropathogenic Escherichia coli and their antimicrobial susceptibility patterns. Asian Pacific Journal of Tropical Medicine. 2012;5(3):210–213. doi: 10.1016/s1995-7645(12)60026-1. [DOI] [PubMed] [Google Scholar]
- 26.Neuner E. A., Sekeres J., Hall G. S., Van Duin D. Experience with fosfomycin for treatment of urinary tract infections due to multidrug-resistant organisms. Antimicrobial Agents and Chemotherapy. 2012;56(11):5744–5748. doi: 10.1128/aac.00402-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marquès C., Tasse J., Pracros A., et al. Effects of antibiotics on biofilm and unattached cells of a clinical Staphylococcus aureus isolate from bone and joint infection. Journal of Medical Microbiology. 2015;64(9):1021–1026. doi: 10.1099/jmm.0.000125. [DOI] [PubMed] [Google Scholar]
- 28.Mihailescu R., Tafin U. F., Corvec S., et al. High activity of fosfomycin and rifampin against methicillin-resistant Staphylococcus aureus biofilm in vitro and in an experimental foreign-body infection model. Antimicrobial Agents and Chemotherapy. 2014;58(5):2547–2553. doi: 10.1128/aac.02420-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ando E., Monden K., Mitsuhata R., Kariyama R., Kumon H. Biofilm formation among methicillin-resistant Staphylococcus aureus isolates from patients with urinary tract infection. Acta Med Okayama. 2004;58(4):207–214. doi: 10.18926/AMO/32090. [DOI] [PubMed] [Google Scholar]
- 30.Stewart P. S. Mechanisms of antibiotic resistance in bacterial biofilms. International Journal of Medical Microbiology. 2002;292(2):107–113. doi: 10.1078/1438-4221-00196. [DOI] [PubMed] [Google Scholar]
- 31.Chen M., Yu Q., Sun H. Novel strategies for the prevention and treatment of biofilm related infections. International Journal of Molecular Sciences. 2013;14(9):18488–18501. doi: 10.3390/ijms140918488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.