Abstract
Exosomes are nanoparticles that transfer cargos from donor cells to recipient cells where they elicit changes in gene expression and metabolism. Evidence suggests that exosomes and their cargos are also absorbed from dietary sources such as bovine milk, and bovine exosomes promote the growth of myofibers in murine C2C12 myotube cell cultures. The aim of the current study was to determine whether the dietary intake of bovine milk exosomes alters strength, gene expression and amino acid profiles in murine skeletal muscles. Male and female C57BL/6 mice, age three weeks, were fed an AIN93G-based, exosome and RNA-depleted (ERD) diet for six weeks; controls were fed an exosome and RNA-sufficient (ERS) diet. Variables of feeding behavior, metabolism, grip strength, liver and kidney function, amino acid profiles, and gene expression patterns were analyzed by using metabolic cages, grip strength analyzers, clinical chemistry analyzers, targeted LC/MS-MS, and RNA sequencing analysis. The diets had no effect on food and water intake, respiratory exchange rate, physical activity, grip strength, markers of liver and kidney dysfunction, and amino acid profiles in muscle. Only twelve and nine mRNAs were differentially expressed in skeletal muscle from female and male mice, respectively, fed ERD and ERS diets. The modest effect of the ERD diet on gene expression and levels of free amino acids in skeletal muscle is consistent with observations that bovine milk exosomes and their cargos accumulate in tissues other than skeletal muscle.
Keywords: amino acids, exosomes, gene expression, milk, skeletal muscle
1. Introduction
Extracellular vesicles (EVs) are membranous, cargo-containing vesicles secreted by virtually all cell types. Originally considered mediators of cellular waste disposal, EVs are now widely recognized for their role in cell-to-cell communication [1]. A distinct subset of EVs, known as exosomes, are up to 150 nm in size, and originate from endosomes and further processing in multivesicular bodies [1–3]. Upon release into the extracellular environment by donor cells, exosomes can be taken up by recipient cells and deliver their cargos [1–4].
Exosome cargos include proteins, lipids, DNA, and various RNA species, such as mRNAs, long non-coding RNAs, and microRNAs. MicroRNAs are of particular interest because of their role in post-transcriptional regulation of gene expression via translation inhibition or target mRNA degradation [5, 6]. The loading of microRNA cargos into exosomes is a non-random process and involves sorting mechanisms [7]. The encapsulation of RNA cargos in exosomes is critical, both for protecting its cargos under degradative conditions such as low pH and RNase-rich environments [8, 9] and for conferring a pathway for cellular uptake by endocytosis [10–12].
The following line of discoveries in our laboratory challenged the paradigm that exosomes and their cargos are derived exclusively from endogenous synthesis, and suggested that exosomes and their cargos can also be obtained from dietary sources such as bovine milk across species boundaries (reviewed in [13]). For example, human intestinal and vascular endothelial cells and rat intestinal cells transport bovine milk exosomes by endocytosis and secrete microRNA cargos across the basolateral membrane [14, 15]. Humans absorb microRNAs from bovine milk [16, 17]; the microRNAs are delivered to circulating immune cells where they alter gene expression [16]. While the bioavailability of dietary microRNAs is still somewhat controversial (see Discussion), independent laboratories have confirmed that bovine milk exosomes and their cargos are bioavailable and deliver functional RNA cargos [18–20]. Our preliminary studies suggest that dietary depletion of exosomes and their RNA cargos elicits phenotypes such as aberrant purine metabolism and impaired spatial learning and memory [21, 22]. MicroRNAs are the prime, but not sole, candidates for causing these effects, because bovine milk contains more than 400 microRNAs, more than 60% of human genes are regulated by human endogenous microRNAs, and the vast majority of bovine microRNAs have nucleotide sequences similar to those of their human orthologs [23–25].
A recent paper by Mobley et al. suggests that supplementation of C2C12 myotube cell cultures with bovine milk exosomes promotes myotube growth, indicating a possible link between milk intake and muscle protein accretion [26]. Also, exosomes appear to facilitate a systemic response to endurance exercise [27]. The main objective of this study was to determine whether bovine milk exosomes also promote muscle protein accretion and muscle strength in a whole animal. As a secondary objective, we assessed potential confounders such as differences in food intake, physical activity, and liver and kidney toxicity between mice fed exosome and RNA-depleted (ERD) and exosome and RNA-sufficient (ERS) diets. The latter is of potential relevance in the context of efforts to use milk exosomes as vehicles for drug delivery [19, 20].
2. Materials and methods
2.1. Mouse husbandry and diets
Male and female C57BL/6 mice (Jackson Laboratories, stock number 000664), age 3 weeks, were fed an exosome and RNA-depleted (ERD) diet described and referred to previously as ExoMinus diet [16] for 6 weeks; controls were fed an exosome and RNA-sufficient (ERS) diet which was previously denoted ExoPlus diet (n=5 per group and sex). Briefly, the diet has the same nutrient composition as the AIN-93G diet (Box 1, Supplemental Table 1), but is modified to contain RNA-depleted exosomes from sonicated bovine milk (ERD diet) or RNA-normal exosomes from non-sonicated milk (ERS diet) [16]. The amount of milk consumed by mice is the equivalent of 0.5 liter milk consumed by an adult human. The content of proteins, lipids, carbohydrates, micronutrients, and the number and size of exosomes is the same in ERD and ERS diets; the diets differ only in their content of RNA encapsulated in exosomes and a greater diversity of the size of exosomes in ERD milk ([16], S. Sukreet et al., unpublished observation). Mice were housed in open box cages (for about 4 1/2 weeks) and metabolic cages (for 9 days) at 22°C and a 12-hour light/dark cycle. Mice had free access to food and water. One ERD-fed male mouse stopped eating and drinking at age eight weeks for unknown reasons and was euthanized.
Box 1. ERS and ERD diets.
Exosome and RNA-depleted (ERD) and exosome and RNA-sufficient (ERS) diets are based on the AIN-93G formulation [16, 40]. In the diets, lyophilized milk powder (and soy protein) substitutes for milk casein in the AIN-93G to eliminate dairy exosomes present in the AIN-93G formulation. The milk added to the diets provides the equivalent of 0.5 L milk consumed by a human adult per day, adjusted by body weight in mice. The milk used to prepare the powder for the ERD diet is ultrasonicated for 1.5 h and incubated for 1 h at 37°C prior to lyophilization; the milk used to prepare the powder for the ERS is not ultrasonicated. Ultrasonication leads to a transient disruption of exosome membranes and a >98% depletion of RNA cargos in exosomes, 20% decrease in exosome count (9.1×1012±7.1×1011 exosomes/mL in ERS milk vs. 7.3×1012±3.5×1011 exosomes/mL in ERD milk) and >60% decrease in intestinal exosome transport rates (unpublished). Diet ingredients other than milk are not ultrasonicated, i.e., nutrients other than exosomes and their RNA cargos are the same in ERD and ERS diets.
2.2. Grip strength, metabolic cages and sample collection
After four weeks of feeding (age seven weeks), front forelimb grip strength was measured using an automated animal grip strength system (San Diego Instruments, Inc.). Each mouse received a training session followed by an experimental session, each consisting of three consecutive runs. The maximum force (grams) was recorded for each run. Subsequently, mice were housed individually in metabolic cages (TSE Systems, Inc.) for nine days (three days of acclimation followed by up to six days of data recording). The following variables were recorded: respiratory exchange ratio (VCO2/VO2), activity levels, and food and water intake. Physical activity was measured in units of mice breaking beams of light in x and y planes (counts) per hour. Body weight was measured before and after housing the mice in the metabolic cages. Mice were euthanized using CO2 and blood was collected by cardiac puncture. Skeletal muscle (right and left quadriceps femoris) was harvested, flash frozen in liquid nitrogen and stored at −80°C until analysis.
2.3. Blood chemistry
Blood samples (n=4 per group and sex) were allowed to clot for 30 minutes at room temperature in red-top vacutainer collection tubes (BD Biosciences, Inc.). Serum was separated by centrifugation at 1,500 g at 4°C for 10 minutes and stored at −20°C until analysis. The following markers of liver and kidney health were determined using a Vitros-250 Analyzer (BlockScientific, Inc.): albumin, alanine aminotransferase, blood urea nitrogen, calcium, creatinine, and total protein. The enzyme unit (U) is defined as amount of enzyme that converts 1 μmole of pyruvate per minute at 37°C (1.0 U/l = 0.017 μkat/l).
2.4. Muscle amino acids
Skeletal muscle tissue (17.5 mg; n=5 ERS and n=4 ERD males and females) was extracted using methanol, dried, and stored at −80°C until analyzed as previously described with minor modifications, namely the use of a 5-μm Luna NH2 column (150 × 2 mm) at a flow rate of 0.4 ml/min [28]. Prior to mass spectrometry, samples were re-suspended in 40 μl of 85% acetonitrile and 20 μl were injected. Targeted metabolite quantification was performed using a QTRAP 6500 mass spectrometer (AB Sciex LLC) coupled to a Shimadzu Nexera X2 HPLC system (Shimadzu Co., Japan). Peak analysis was performed using Analyst Software (AB Sciex LLC). Multiple reaction monitoring transition data were collected for each amino acid metabolite (Supplemental Table 2).
2.5. Gene expression
Total RNA was extracted from 30 mg skeletal muscle (n=3 per group and sex) using RNeasy Fibrous Tissue Mini Kit (Qiagen, Inc.) according to manufacturer’s instructions. RNA integrity was confirmed in the DNA Sequencing Core at the University of Nebraska Medical Center by using a Fragment Analyzer Automated CE System (Advanced Analytical Technologies, Inc.). Libraries were prepared for each sample using the TrueSeq RNA Library Prep Kit v2 (Illumina, Inc.) and sequenced using an Illumina HiSeq 2500 platform and a 150 base-pair paired-end protocol (RNA-seq).
Data quality was assessed using FastQC [29]. Adaptor sequences and reads containing ambiguous bases or having average quality score less than 30 were removed. The remaining high quality reads were aligned to the reference sequences in murine RNA [GRCm38, mm 10] by using RSEM [30]. EBSeq [31] was applied to identify the differentially expressed transcripts between two feeding groups. Only transcripts with at least a 2-fold difference in expressions between the two groups were included in the downstream analysis. Based on the differentially expressed mRNAs, enriched gene ontology (GO) and functional pathways (P 0.05) were also obtained utilizing the Database for Annotation, Visualization and Integrated Discovery (DAVID v6.8) [32]. Sequence data were deposited in the BioProject database under accession ID: PRJNA417163.
RNA-seq data were confirmed using quantitative real-time PCR (RT-qPCR) as described previously with minor modifications and analyzed using the comparative CT method and the PCR primers shown in Supplemental Table 3 [33, 34]; expression was normalized using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a housekeeping control. Melting curve analysis was completed to ensure production of a single product (not shown). RNA-seq analysis suggested that the expression of interleukin 6 receptor alpha (I16rα) did not depend on the diet and therefore I16rα was used as negative control in RT-qPCR analysis.
2.6. Western blot analysis
Total protein was extracted from ~20 mg skeletal muscle tissue using ice-cold lysis buffer [20 mM Tris (pH 7.8), 137 mM NaCl, 2.7 mM KC1, 1 mM MgCl2, 1% Triton X-100, 10% (w/v) glycerol, 1 mM EDTA, 1 mM dithiothreitol] with a protease inhibitor cocktail (Sigma-Aldrich, cat. #P8340). Protein concentrations were determined using the bicinchoninic assay (Thermo Fisher Scientific) and samples were stored at −80°C until further analysis. The expression of suppressor of cytokine signaling 2 (Socs2) and β-actin (control) was assessed using rabbit anti-Socs2 polyclonal antibody (Abcam) and rabbit anti-β-actin (Cell Signaling Technology), respectively, and an Odyssey Clx Imaging System (LI-COR Biosciences) as described previously with minor modifications [33].
2.7. Statistical analyses
Levene’s test was used to test for homogeneity of variance. Unpaired, two-tailed Student’s t-test (if variances were homogenous) or Welch’s t-test (if variances were heterogenous) was used when two groups were compared. Multivariate analysis of metabolomics data was completed on Metaboanalyst 3.0 [35]. R version 1.0.136 (R Foundation for Statistical Computing) was used for statistical analyses. Data are expressed as means ± standard errors. Differences were considered statistically significant if P<0.05. Power calculations suggest that our sample size was powered to detect grip strength differences of ~25% in males and ~33% in females with 80% power at an alpha level of 0.05.
3. Results
3.1. Overall metabolism and liver and kidney health
Food and water intake, physical activity and RER were not significantly different between mice fed ERD and ERS diets (0.09<P<0.93; Table 1). Likewise, markers of liver and kidney health were not significantly different between the diet groups (Table 2) and within the normal range in C57BL/6 mice (Supplemental Table 4), except for calcium, total protein, and alanine aminotransferase activities, which were higher than the normal range (see Discussion).
Table 1.
Effect of ERS and ERD diets on food and water intake, RER, and activity levels in male and female C57BL/6 mice. Values are means ± standard errors (n=5 mice per group and sex, except for ERD males (n=4)).
| Females | Males | ||||
|---|---|---|---|---|---|
| Variable | Cycle | ERD | ERS | ERD | ERS |
| Food Intake (g/day) | -- | 3.9 ± 0.2 | 3.7 ± 0.2 | 3.9 ± 0.4 | 4.5 ± 0.1 |
| Water Intake (ml/day) | -- | 4.1 ± 0.6 | 3.1 ± 0.1 | 2.9 ± 0.1 | 3.2 ± 0.3 |
| Respiratory exchange | Light | 0.91 ± 0.02 | 0.89 ± 0.01 | 0.90 ± 0.03 | 0.92 ± 0.02 |
| ratio (VCO2/VO2) | Dark | 0.97 ± 0.03 | 0.96 ± 0.03 | 0.95 ± 0.03 | 0.96 ± 0.02 |
| Physical activity | Light | 431± 109 | 509± 103 | 166 ± 38 | 286 ± 45 |
| (counts/hour) | Dark | 2094 ± 419 | 2280 ± 477 | 1649 ± 264 | 1572±292 |
Table 2.
Effect of ERS and ERD diets on liver and kidney function in male and female C57BL/6 mice. Values are means ± standard errors (n=4 mice per group and sex).1
| Females | Males | |||
|---|---|---|---|---|
| Variable | ERD | ERS | ERD | ERS |
| Calcium (mg/dl) | 11.4 ± 0.4 | 11.8 ± 0.2 | 11.3 ± 0.7 | 11.9 ± 0.3 |
| Total protein (g/dl) | 7.1 ± 0.4 | 7.8 ± 0.8 | 7.7 ± 0.9 | 6.2 ± 0.2 |
| Albumin (g/dl) | 3.8 ± 0.3 | 4.5 ± 0.7 | 4.4 ± 0.7 | 3.3 ± 0.1 |
| Alanine aminotransferase (U/l) | 164.0 ± 73.3 | 102.3 ± 49.4 | 199.0 ± 87.0 | 138.8 ± 74.4 |
| Blood urea nitrogen (mg/dl) | 18.3 ± 2.1 | 17.8 ± 1.4 | 23.0 ± 3.2 | 22.8 ± 2.8 |
| Creatinine (mg/dl) | 0.63 ± 0.19 | 0.43 ± 0.13 | 0.50 ± 0.18 | 0.58 ± 0.16 |
1Abbreviation: U, enzyme units
3.2. Muscle physiology
Diets had no statistically significant effect on forearm grip strength in males and females (Figure 1) and amino acid levels in skeletal muscle, supported by principal component analysis and hierarchal clustering analysis showing no separation between the ERD group and the ERS controls (Figure 2).
Figure 1.
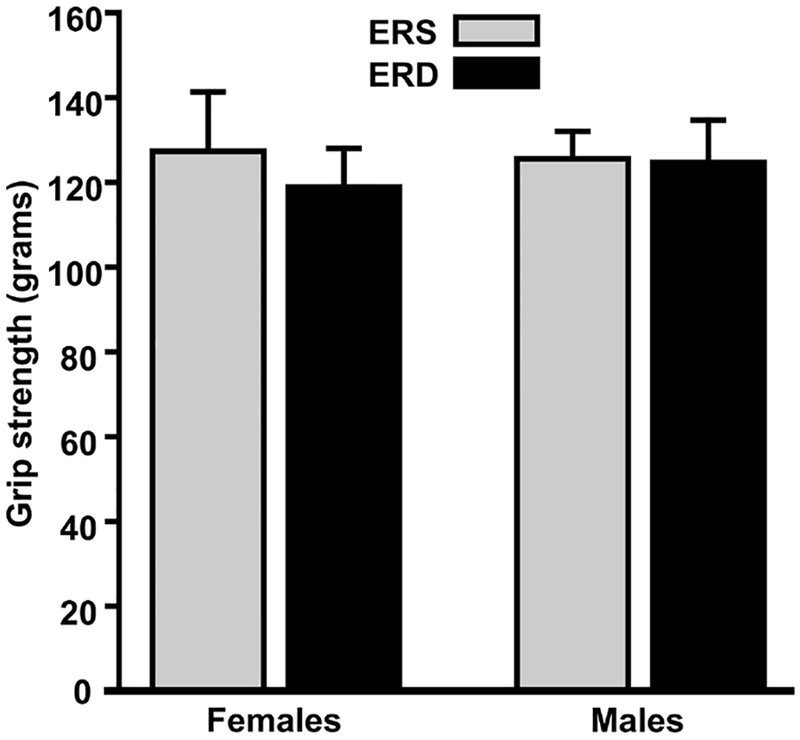
Effect of exosome and RNA-defined diets on front forearm grip strength in C57BL/6 mice fed ERD and ERS diets. Values are means ± standard errors (n=5).
Figure 2.
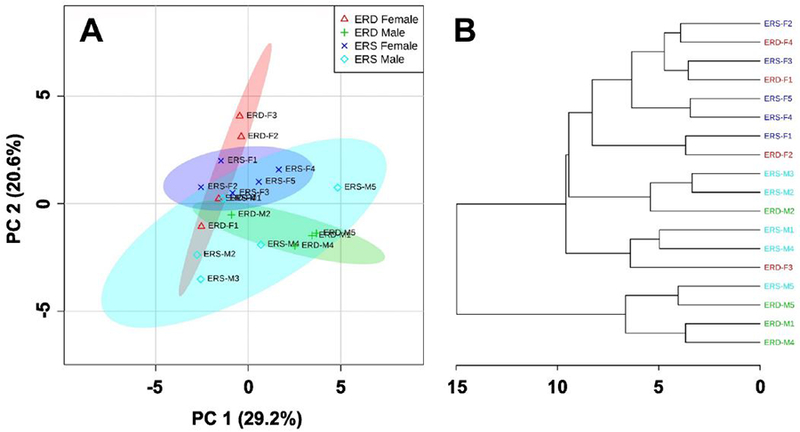
Principal component analysis (A) and hierarchical clustering (B) of amino acid levels in mice fed ERD and ERS diets (n=5 ERS and n=4 ERD males and females).
3.3. Gene expression
Twelve and nine mRNAs were differentially expressed in skeletal muscle from female and male mice, respectively, fed ERD and ERS diets if a 2-fold change was used as cut-off (Figure 3, Supplemental Table 5). For example, the expression of Tmem100 mRNA was 2.47 ± 0.64 fold higher in female mice fed ERD diets than in those fed ERS diets and the expression of Rhobtb1 mRNA and Socs2 mRNA was 2.22 ± 0.16 fold and 2.86 ± 1.11 fold lower, respectively, in ERD females than in ERS females (Supplemental Table 5). The expression of these mRNAs was confirmed with RT-qPCR analysis, using I16rα as negative control (Figure 4). The increased expression of Socs2 mRNA in ERD females compared to ERS females was paralleled by a differential expression of Socs2 protein (Figure 4). If a 1.5-fold change was used as cut-off in RNA-seq analysis, 31 and 13 mRNAs were differentially expressed in skeletal muscle from female and male mice, respectively, fed ERD and ERS diets (not shown). One KEGG pathway and three GO terms were upregulated in females fed the ERD diet compared with females fed the ERS diet (Table 3). All these functional processes are involved in ribosome biology and mRNA translation, consistent with the increased expression of mRNA coding for Rpl36a, Rpl28, and Rps27rt in ERD females compared with ERS females. No KEGG pathway or GO term enrichment was observed in ERD males compared with ERS males.
Figure 3.
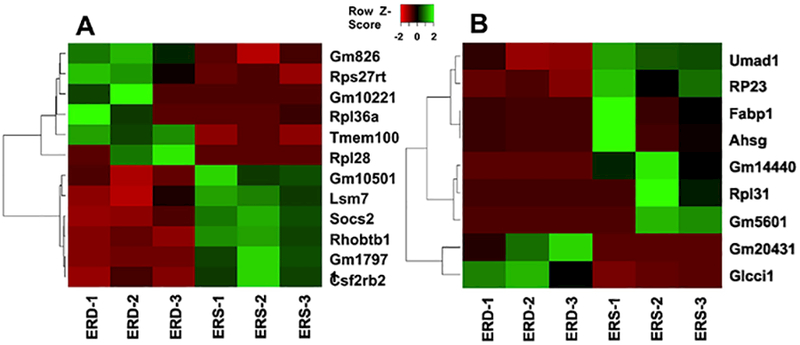
Heat map of differentially expressed genes (≥2-fold change) in ERD vs. ERS females (A) and males (B) (n=3).
Figure 4.
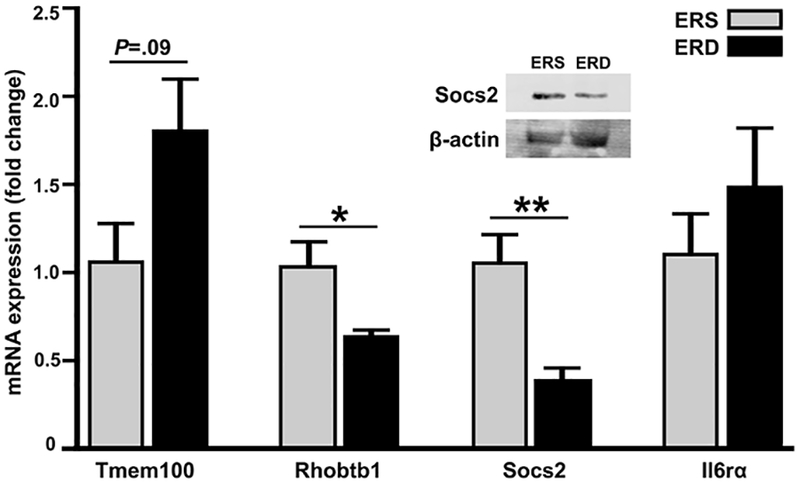
Relative mRNA expression of Tmem100, Rhobtbb1, Socs2, and I16rα (negative control) and protein expression of Socs2 and β-actin (control) in skeletal muscle in female C57BL/6 mice fed ERS and ERD diets. Values are means ± standard errors of RT-qPCR analysis. *P<0.05, **P<0.01 vs. ERS-fed mice (n=4).
Table 3.
Functional pathway and GO terms enriched in ERD female samples (n=3).
| Category | Enriched Terms | P-value |
|---|---|---|
| GO Molecular Function | GO:0003735~structural constituent of ribosome | 0.005 |
| KEGG Pathway | mmu03010: Ribosome | 0.005 |
| GO Biological Process | GO: 0006412~translation | 0.010 |
| GO Cellular Component | GO:0022625~cytosolic large ribosomal subunit | 0.032 |
4. Discussion
This paper advances the field of milk exosomes in three areas of great significance for studies of dietary exosomes and their cargos. First, this is the paper investigating the effects of bovine milk exosomes on gene expression and amino acid levels in skeletal muscle and grip strength in a whole animal model. Our data suggest that dietary milk exosomes have only a modest effect on gene expression and metabolism in skeletal muscle in young adult mice. This observation is consistent with recent and ongoing studies suggesting that the majority of milk exosomes and their RNA cargos accumulate in tissues other than skeletal muscle [19, 36]. These studies are not mutually exclusive with milk exosome-dependent growth of C2C12 myotubes, which was a rationale for conducting this study, but underline the importance of testing biological effects of treatments in whole organisms [26].
Second, this paper provides evidence that food and water consumption, RER and physical activity were the same in mice fed ERD and ERS diets. This is an important consideration in studies of phenotypes other than those related to skeletal muscle. For example, our preliminary studies suggest that dietary depletion of exosomes and their RNA cargos elicits phenotypes such as aberrant purine metabolism, loss of fecundity, loss of spatial learning and memory and changes in the gut microbiome [21, 22, 37, 38]. Thus, the findings reported in this paper suggest that the phenotypes in these studies were not confounded by food and water consumption and physical activity.
Third, efforts are underway to use milk exosomes as scalable vehicles for the delivery of drugs that are unstable or poorly bioavailable [19, 20]. In this context, it will be important to demonstrate that the intake of bovine milk exosomes is safe. Bovine milk exosomes did not cause liver and kidney dysfunction, if administered in a dietary matrix. Some samples exhibited moderate hemolysis, which cause artificially high readings for serum calcium, total protein and alanine aminotransferase activity [39]. Thus, the moderately higher values for these variables compared with reference values were probably caused by sample hemolysis.
While this paper has a great impact on the above areas of study, some uncertainties remain. For example, it is unknown whether the minor changes in pathways and biological processes have their origins in skeletal muscle or whether they are due to cross-talk between other tissues or the gut microbiome with the skeletal muscle. Note that bovine milk exosomes do not accumulate in skeletal muscle [19, 36]. It is also unknown if dietary exosomes had a stronger effect on skeletal muscle if mice suffered from sarcopenia. These uncertainties are currently addressed in our laboratory by exploring the crosstalk between dietary milk exosomes and the gut microbiome, and by studying the effects of dietary exosomes on muscle protein accretion and function in mice suffering from sarcopenia. Notwithstanding these ongoing studies we conclude that studies in tissues other than skeletal muscle might be more fruitful for future research of phenotypes caused by the dietary intake of bovine milk exosomes.
Supplementary Material
Acknowledgements
The authors acknowledge the use of the Biomedical and Obesity Research Core and the assistance of Dr. Steven Kachman in the Nebraska Center for the Prevention of Obesity Disease through Dietary Molecules (NIH 1P20GM104320) at the University of Nebraska-Lincoln, and the services provided by the DNA Sequencing Core at the University of Nebraska Medical Center (NIH P20GM103427, 1P30GM110768 and P30CA036727). The authors acknowledge the Holland Computing Center at the University of Nebraska-Lincoln for providing computational support.
Funding: This work was supported by the National Institute of Food and Agriculture (NIFA), U.S. Department of Agriculture, under award number 2015-67017-23181, National Institutes of Health (NIH) grant 1P20GM104320, and NIFA2016-67001-25301/NIH R01 DK107264, the Gerber Foundation, the Egg Nutrition Center, the University of Nebraska Agricultural Research Division (Hatch Act), USDA multistate group W3002 (all to JZ), and NIH award P30GM103335 (JA). JZ serves as a consultant for PureTech Health, Inc.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
The authors declare no conflict of interest.
References
- [1].Maas SLN, Breakefield XO, Weaver AM. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017;27:172–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Abels ER, Breakefield XO. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell Mol Neurobiol. 2016;36:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Edgar JR. Q&A: What are exosomes, exactly? BMC Biol. 2016;14:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mattick JS, Makunin IV. Non-coding RNA. Human Mol Genet. 2006;15:R17–R29. [DOI] [PubMed] [Google Scholar]
- [7].Stevanato L, Thanabalasundaram L, Vysokov N, Sinden JD. Investigation of Content, Stoichiometry and Transfer of miRNA from Human Neural Stem Cell Line Derived Exosomes. PLoS One. 2016;11:e0146353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Benmoussa A, Lee CH, Laffont B, Savard P, Laugier J, Boilard E, et al. Commercial dairy cow milk microRNAs resist digestion under simulated gastrointestinal tract conditions. J Nutr. 2016;146:2206–15. [DOI] [PubMed] [Google Scholar]
- [9].Izumi H, Kosaka N, Shimizu T, Sekine K, Ochiya T, Takase M. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J Dairy Sci. 2012;95:4831–41. [DOI] [PubMed] [Google Scholar]
- [10].Tian T, Wang Y, Wang H, Zhu Z, Xiao Z. Visualizing of the cellular uptake and intracellular trafficking of exosomes by live-cell microscopy. J Cell Biochem. 2010;111:488–96. [DOI] [PubMed] [Google Scholar]
- [11].Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tian T, Zhu YL, Zhou YY, Liang GF, Wang YY, Hu FH, et al. Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J Biol Chem. 2014;289:22258–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zempleni J, Aguilar-Lozano A, Sadri M, Sukreet S, Manca S, Wu D, et al. Biological activities of extracellular vesicles and their cargos from bovine and human milk in humans and implications for infants. JNutr. 2017;147:3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wolf T, Baier SR, Zempleni J. The intestinal transport of bovine milk exosomes is mediated by endocytosis in human colon carcinoma caco-2 cells and rat small intestinal IEC-6 cells. JNutr. 2015;145:2201–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kusuma Jati R, Manca S, Friemel T, Sukreet S, Nguyen C, Zempleni J. Human vascular endothelial cells transport foreign exosomes from cow’s milk by endocytosis. Am J Physiol Cell Physiol. 2016;310:C800–C7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Baier SR, Nguyen C, Xie F, Wood JR, Zempleni J. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow’s milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures, and mouse livers. J Nutr. 2014;144:1495–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wang L, Sadri M, Giraud D, Zempleni J. RNase H2-Dependent Polymerase Chain Reaction and Elimination of Confounders in Sample Collection, Storage, and Analysis Strengthen Evidence That microRNAs in Bovine Milk Are Bioavailable in Humans. J Nutr. 2018;148:153–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Izumi H, Tsuda M, Sato Y, Kosaka N, Ochiya T, Iwamoto H, et al. Bovine milk exosomes contain microRNA and mRNA and are taken up by human macrophages. J Dairy Sci. 2015;98:2920–33. [DOI] [PubMed] [Google Scholar]
- [19].Munagala R, Aqil F, Jeyabalan J, Gupta RC. Bovine milk-derived exosomes for drug delivery. Cancer Lett. 2016;371:48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Agrawal AK, Aqil F, Jeyabalan J, Spencer WA, Beck J, Gachuki BW, et al. Milk-derived exosomes for oral delivery of paclitaxel. Nanomedicine. 2017;13:1627–36. [DOI] [PubMed] [Google Scholar]
- [21].Aguilar-Lozano A, Baier SR, Adamec J, Sadri M, Giraud D, Zempleni J. Depletion of dietary microRNAs from cow’s milk causes an increase of purine metabolites in human body fluids and mouse livers. FASEB J. 2016;30 (supplement 1): 127–1. [peer-reviewed meeting abstract]. [Google Scholar]
- [22].Mutai E, Zhou F, Zempleni J. Depletion of dietary bovine milk exosomes impairs sensorimotor gating and spatial learning in C57BL/6 mice. FASEB J. 2017;31:150–4. [peer-reviewed meeting abstract]. [Google Scholar]
- [23].Sun J, Aswath K, Schroeder SG, Lippolis JD, Reinhardt TA, Sonstegard TS. MicroRNA expression profiles of bovine milk exosomes in response to Staphylococcus aureus infection. BMC Genomics. 2015;16:806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mobley CB, Mumford PW, McCarthy JJ, Miller ME, Young KC, Martin JS, et al. Whey protein-derived exosomes increase protein synthesis and hypertrophy in C2-C12 myotubes. J Dairy Sci. 2017;100:48–64. [DOI] [PubMed] [Google Scholar]
- [27].Safdar A, Saleem A, Tarnopolsky MA. The potential of endurance exercise-derived exosomes to treat metabolic diseases. Nat Rev Endocrinol. 2016;12:504–17. [DOI] [PubMed] [Google Scholar]
- [28].Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc. 2012;7:872–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Babraham Bioinformatics. FastQC. 2017; http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [accessed 8/24/2017].
- [30].Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011; 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BM, et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29:1035–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Dennis G Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- [33].Pestinger V, Wijeratne SSK, Rodriguez-Melendez R, Zempleni J. Novel histone biotinylation marks are enriched in repeat regions and participate in repression of transcriptionally competent genes. J Nutr Biochem. 2011;22:328–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. [DOI] [PubMed] [Google Scholar]
- [35].Xia J, Wishart DS. Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. Curr Protoc Bioinformatics. 2016;55:14 0 1–0 91. [DOI] [PubMed] [Google Scholar]
- [36].Manca S, Giraud D, Zempleni J. The bioavailability and distribution of bovine milk exosomes is distinct from that of their cargos in mice. FASEB J. 2017;31:148–2. [peer-reviewed meeting abstract].27677546 [Google Scholar]
- [37].Sadri M, Xie F, Wood J, Zempleni J. Dietary depletion of cow’s milk microRNAs impairs fecundity in mice. FASEB J. 2016;30:673–5. [peer-reviewed meeting abstract]. [Google Scholar]
- [38].Zhou F, Paz AH, Sadri M, Fernando CS, Zempleni J. A diet defined by its content of bovine milk exosomes alters the composition of the intestinal microbiome in C57BL/6 mice. FASEB J. 2017;31:965–24. [peer-reviewed meeting abstract].27920150 [Google Scholar]
- [39].Calgary Laboratory Services. Effects of hemolysis on clinical specimens. 2017 ed Calgary, Alberta: 2017; http://www.calgarylabservices.com/lab-services-guide/specimen-collection/hemolysis.aspx [accessed 10/20/2017]. [Google Scholar]
- [40].Reeves PG, Nielsen FH, Fahey GC Jr. AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J Nutr. 1993;123:1939–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


