Abstract
The P-cluster is a unique iron-sulfur center that likely functions as a dynamic electron (e−) relay site between the Fe-protein and the catalytic FeMo-cofactor in nitrogenase. The P-cluster has been shown to undergo large conformational changes upon 2-e− oxidation which entail the coordination of two of the Fe centers to a Ser side chain and a backbone amide nitrogen, respectively. Yet, how and if this 2-e− oxidized state (POX) is involved in catalysis by nitrogenase is not well established. Here, we present the crystal structures of reduced and oxidized MoFe-protein (MoFeP) from Gluconacetobacter diazotrophicus (Gd), which natively possesses an Ala residue in the position of the Ser ligand to the P-cluster. While reduced Gd-MoFeP is structurally identical to previously characterized counterparts around the FeMo-cofactor, oxidized Gd-MoFeP features an unusual Tyr coordination to its P-cluster along with ligation by a backbone amide nitrogen. EPR analysis of the oxidized Gd-MoFeP P-cluster confirmed that it is a 2-e− oxidized, integer-spin species. Importantly, we have found that the sequence positions corresponding to the Ser and Tyr ligands are almost completely covariant among Group I nitrogenases. These findings strongly support the possibility that the POX state is functionally relevant in nitrogenase catalysis and that a hard, O-based anionic ligand serves to stabilize this state in a switchable fashion.
Graphical abstract

Nitrogenase is the only enzyme known to catalyze nitrogen fixation.1–4 It is a two-component complex composed of the iron-protein (FeP) and the molybdenumiron protein (MoFeP) in the most well-studied form of nitrogenase. FeP is an homodimeric (γ2) ATPase containing a [4Fe:4S] cluster and couples ATP hydrolysis to the transfer of e− to the catalytic MoFeP, an α2β2 heterotetramer. The e− transfer (ET) chain in nitrogenase extends over a distance of over 30 Å from the [4Fe:4S] of FeP to iron-molybdenum cofactor (FeMoco) of MoFeP,5 the [7Fe:1Mo:9S:1C:homocitrate] 6–8 cluster responsible for N2 reduction. Positioned in the middle of this chain is the P-cluster, an [8Fe:7S] cluster bridging the αβ-dimer interface of MoFeP that likely mediates ET between FeP and FeMoco. Despite its modest reduction potential, FeP is the only known biological agent that can support catalysis by MoFeP.1–4 This exclusivity of FeP has been attributed to its ability to form specific, nucleotide-state-dependent interactions with MoFeP, which modulate the ET distance between the [4Fe:4S] and the P-cluster,9 and decrease the reduction potential of the FeP [4Fe:4S] cluster.10,11 Importantly, several lines of evidence suggest that ATP-bound FeP also must be able to gate ET between the P-cluster and FeMoco, necessarily through long-distance effects induced by specific FeP-MoFeP docking interactions.2,4,12-15 However, the structural details of these conformational gating events have not been elucidated.
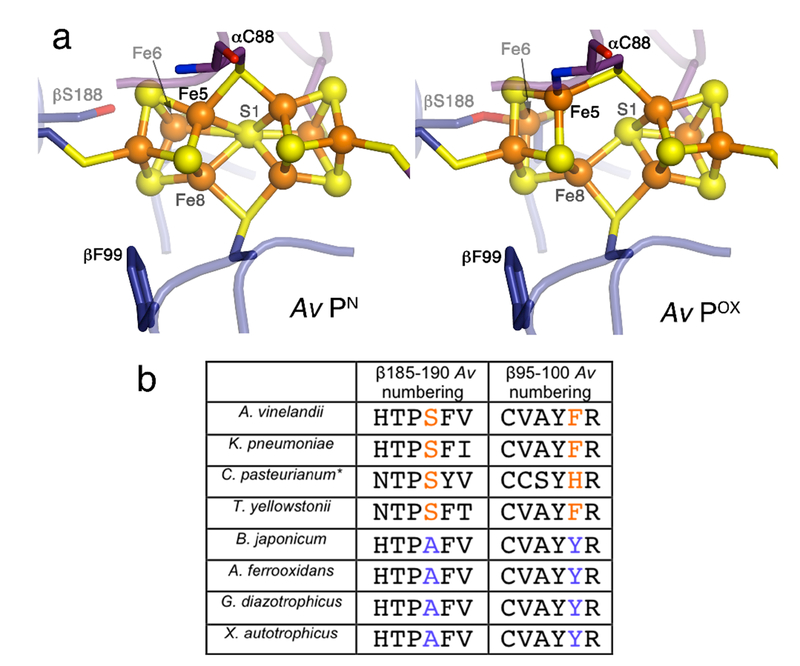
While MoFeP has been crystallographically characterized in many different forms, including various FeP-complexed states5,9,16,17 or from different organisms,7,18,19 no apparent structural changes have been detected between the P-cluster and FeMoco suggestive of a conformational gate. The only notable conformational variation is between the all-ferrous (PN) and the 2-e− oxidized (POX) states of the P-cluster (Figure 1a):20 in the PN state, the P-cluster has a structure that can be considered as a fusion between two closed [4Fe:4S] cubane units which share a single, hexacoordinate sulfur, S1. In the POX state, one of the cubane units opens up, whereby Fe5 forms a new bond to the backbone amide N of α-C88 (Azotobacter vinelandii or Av numbering), Fe6 becomes coordinated to β-S188, and both Fe5 and Fe6 dissociate from S1.20 Since both the α-C88 backbone N and β-S188 hydroxyl group likely have to be in their deprotonated, anionic forms to coordinate Fe ions, they could stabilize the P-cluster in an oxidized state, thereby lowering its reduction potential and rendering it capable of delivering electrons to FeMoco.
Figure 1.
a) Structures of the Av-MoFeP P-cluster in PN and POX states (PDB IDs: 3MIN and 2MIN).20 b) Alignment of selected amino acid sequence segments (β185-190 and β95-100 in Av-MoFeP numbering) surrounding the P-cluster in Group I nitrogenases. *C. pasteurianum MoFeP, a Group II species, is also included. For a more extensive alignment, see Table S1.
Accordingly, any structural changes remotely induced by FeP that favor ligation by β-S188 and the α-C88 backbone nitrogen could provide a basis for conformational redox gating in nitrogenase and explain the exclusivity of FeP as the biological e− donor for MoFeP. However, there is no consensus that the 2-e− oxidized POX state is a mechanistically relevant intermediate, since ATP-dependent ET processes in the nitrogenase complex are believed to proceed through 1-e− steps,4,21-23 with the all-ferrous PN and the 1-e− oxidized P1+ states as operative species. Importantly, β-S188 is not strictly conserved; in particular, it is frequently replaced by an Ala residue in Group I nitrogenases (Fig. 1b and Table S1).24 In order to investigate the importance of Ser ligation in nitrogenase catalysis and its potential involvement in conformationally gated ET, we set out to structurally characterize MoFeP from G. diazotrophicus, which features an Ala in place of β-S188.
G. diazotrophicus is an obligate aerobic bacterium that lives in close association with the roots of many agriculturally important plants (e.g., sugarcane and coffee).25 Fisher and Newton previously isolated Gd-MoFeP and FeP,26 which respectively carry ~60% and 72% sequence identity to their Av-MoFeP counterparts. Initial biochemical characterization of these proteins confirmed their α2β2 and γ2 compositions and that the Gd-MoFeP displayed a specific catalytic activity that is approximately half of that observed for Av-MoFeP26 Through modification of the original procedure, we were able to culture G. diazotrophicus (Fig. S1) and obtain an optimized yield of 2 mg/L of purified Gd-MoFeP (Fig. S2). We determined the maximum specific activity of Gd-MoFeP for the reduction the alternative substrate C2H2 to be ca. 1400 nmol C2H4.min−1.mg protein−1, which accords with the previously reported values.26
The crystal structure of the dithionite (DT)-reduced Gd-MoFeP was determined at 1.83-Å resolution with the final refinement parameters of Rcryst = 0.129 and Rfree = 0.151 (Table S2). An overlay of Gd-MoFeP with Av-MoFeP structures reveals a very high similarity with an overall RMSD of 0.7 Å based on all α-C atoms (Fig. S3 and S4). The coordination and the secondary-sphere environments of FeMoco are identical to one another in both species (Fig. S4), including the orientation of functionally important and conserved residues such as α-His211, α-V86, and α-R112 (α-H195, α-V70, and α-R96 in Av-MoFeP) that are proposed to be important for substrate interactions.27 The P-cluster of DT-reduced Gd-MoFeP is also perfectly superimposable on that of Av-MoFeP in the PN state, as are the intervening residues between the P-cluster and FeMoco (Fig. S4).
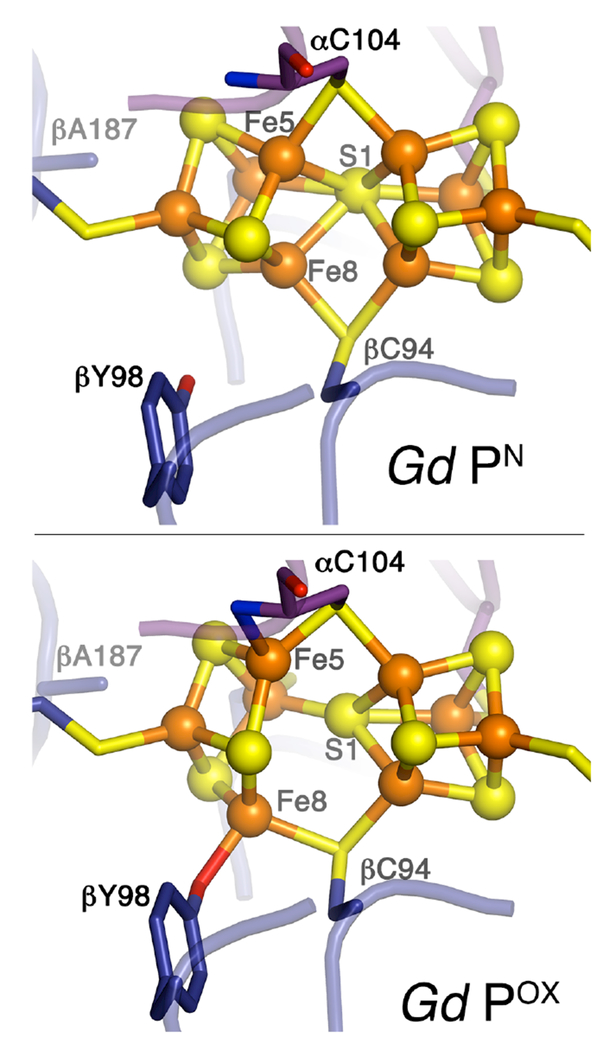
We next determined the crystal structure of indigo disulfonate (IDS)-oxidized Gd-MoFeP at 2.6-Å resolution (Rcryst = 0.169 and Rfree = 0.207) (Table S2) to compare it to those from organisms containing a coordinating Ser ligand to POX.7,18,19 While the FeMoco environment remains unaltered upon oxidation of Gd-MoFeP (Fig. S5), as expected, there is a striking difference in the coordination of the oxidized P-cluster (Fig. 2 and S6). In oxidized Gd-MoFeP, Fe5 is ligated to the backbone amide N of α-C104 (α-C88 in Av-MoFeP numbering) as in the other species (see Table 1 for a comparison of coordination metrics) and dissociated from the central S1. Yet, in place of the Ser ligand, which coordinates Fe6 in Av-, Kp-, and Cp-MoFeP but is replaced by Ala in Gd-MoFeP, we observed Fe8 to be ligated to the O atom of the β-Y98 side chain (positional equivalent to β-F99 in Av-MoFeP), with a bond distance of 2.1 Å (Fig. 2, S6, and Table 1). Tyr-ligated Fe8 also appears to be fully dissociated from S1 (dFe8-S1 = 3.4 Å) like the Ser-coordinated Fe6 in other species (dFe6-S1>3.6 Å). Another notable difference is that Fe8 is coordinated to a bridging Cys (β-C94) in Gd-MoFeP, whereas Fe6 is coordinated to a terminal one in other species (β-C153 in Av-MoFeP). Thus, while chemical oxidation gives rise to an opened 4Fe-3S unit with two O- and N-based ligands in all nitrogenase species, the exact mechanism by which this is achieved and the relative orientations of the participating ligands with respect to the αβ-dimer interface are quite distinct.
Figure 2.
Structures of the Gd-MoFeP P-cluster in PN and Pox states.
Table 1.
Changes in coordination distances of the P-cluster upon oxidation (PN→POX) in different organisms.
| d (S1-Fe6/Fe8) (Å) | d (OSer/Tyr-Fe6/Fe8) (Å) | d (NCys-Fe5) (Å) | |
|---|---|---|---|
| Cp | 2.4 → 3.6 | 3.2 → 2.0 | 3.3 → 2.3 |
| Kp | 2.4 → 3.8 | 3.6 → 2.2 | 3.4 → 2.2 |
| Av | 2.5 → 4.0 | 3.4 → 1.9 | 3.4 → 2.1 |
| Gd | 2.5 → 3.4 | 3.6 → 2.1 | 3.4 → 2.6 |
These observations prompted us to conduct a more detailed sequence analysis, which revealed that the amino acid positions corresponding to β-187 and β-98 in Gd-MoFeP (or β-188 and β-99 in Av-MoFeP) are almost completely covariant among Group I nitrogenases (Fig. 1b and Table S1).28 That is, if MoFeP has a Ser in the first position (like Av- and Kp-MoFeP), it features a Phe in the second. If an organism has an Ala in the first position (like Gd-MoFeP), it has a Tyr in the second.
These findings strongly suggest that the motif of two N- and O-based ligands, which coordinate the oxidized P-cluster, is highly conserved and functionally important.
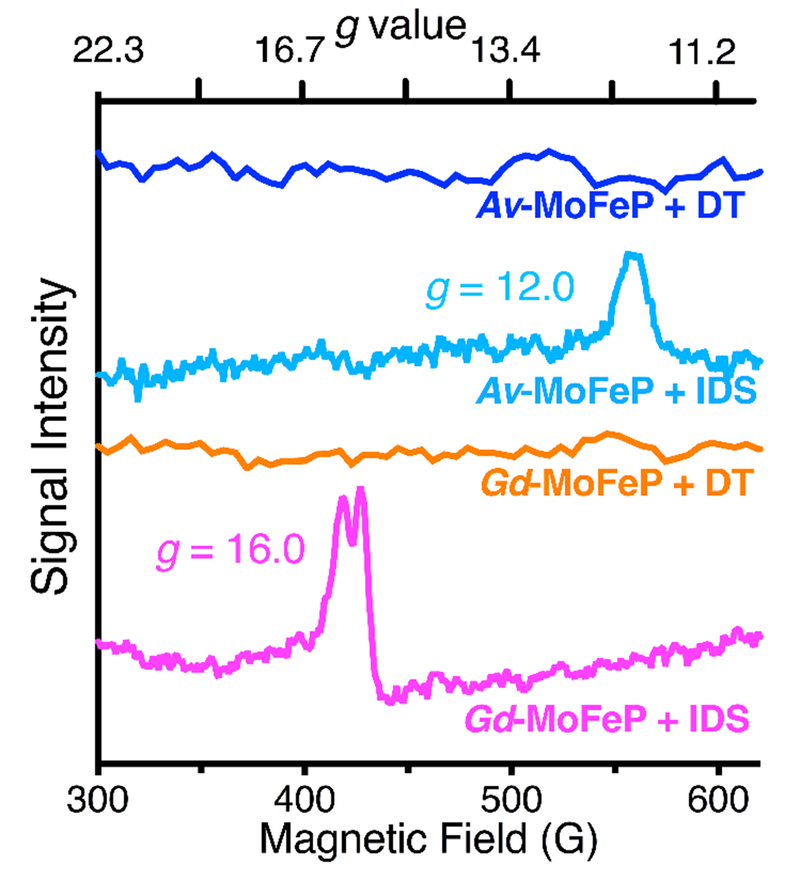
In order to ascertain that the DT-reduced and IDS-oxidized Gd-MoFeP correspond to the PN and POX states observed for Av-MoFeP, we carried out EPR spectroscopy. The X-band EPR spectrum of DT-reduced Gd-MoFeP features a strong, S=3/2 signal (g=4.3., 3.6, and 2.0), which matches resting-state FeMoco (MN) signatures from other species including A. vinelandii (Fig. S7a).29–31 DT-reduced Gd-MoFeP samples do not display any signal associated with the P-cluster (Figure S7a), as expected from the diamagnetic nature of the PN state. In POX, the P-cluster in Av-MoFeP displays a characteristic EPR signal at g=12.0 in parallel mode (Fig. 3), which has been assigned to a non-Kramers, integer-spin system with S≥3.32 We observed that the samples of IDS-oxidized Gd-MoFeP similarly display a low-field EPR signal at g=16, which is only detectable in the parallel collection mode and disappears upon re-reduction with DT (Fig. 3, S7b, and S7c). These observations are consistent with an integer-spin, 2-e− oxidized P-cluster. Interestingly, a similar EPR feature at g=15.6 was reported for the thionine-treated Xanthobacter autotrophicus (Xa) and attributed to the 2-e− oxidized form of the P-cluster with S=3 or 4.32 Strikingly, Xa-MoFeP, like Gd-MoFeP, features an Ala at position β-188 (Av-MoFeP numbering) and a Tyr at position β-99 (Av-MoFeP numbering). With all of the EPR, crystallography, and genetic sequence data in hand, we can thus conclude that the integer-spin, POX state can be achieved with alternative O-based ligands associated with either Fe6 (Ser) or Fe8 (Tyr), which give rise to subtle but significant differences in the P-cluster electronic structure.
Figure 3.
Parallel mode EPR spectra (10 K) of Av-MoFeP and Gd-MoFeP in DT-reduced and IDS-oxidized states.
Like Ser coordination, ligation of a Tyr to an FeS cluster is extremely rare and previously detected in only one other system.33 The role of an O-based (Tyr or Ser) ligand as a redox switch for an FeS-cluster is readily rationalized. The negatively charged tyrosinate or serinate functionalities can both serve to decrease the reduction potential of the P-cluster to induce ET to FeMoco like a cysteinate ligand. Yet, unlike the latter, they are hard ligands (according to the Pearson classification),34 whose coordination to an FeS-cluster would be dependent on whether the Fe center that they coordinate to is in the +3 (hard, favored) or +2 (borderline soft, disfavored) oxidation state, making them suitable as reversible redox switches. The same argument holds for a backbone N ligand, particularly if fully deprotonated. In contrast, the cysteinate group, which is anionic but can be classified as soft, has no strong preference for either Fe oxidation state, rendering it a poor choice as a redox switch. In fact, substitution of β-S188 with Cys in Av-MoFeP predominantly restricts the P-cluster to oxidized states and greatly diminishes activity.35
The finding that simultaneous coordination by an amide N- and a Ser/Tyr O-ligand is highly conserved also argues for the functional importance of the POX state.36 In analogy, recent studies on O2-tolerant NiFe-hydrogenases have established the presence of an unusual [4Fe:3S] cluster, which can assume three oxidation states like the P-cluster.37 Notably, this [4Fe:3S] cluster undergoes conformational changes that are remarkably similar to those observed for the P-cluster, including ligation by a backbone amide N to an Fe-center, stabilizing the 2-e− oxidized state of the cluster.38–40 It has been proposed that the ability of the [4Fe:3S] cluster to rapidly transfer two e− to the catalytic site prevents the accumulation of the inactive “Ni-A” state and enables catalysis in the presence of O2.38–40 Similarly, rapid transfer of multiple e− from the P-cluster to FeMoco could minimize the lifetime of intermediate reduction states that are prone to the loss of e− through H2 evolution. Further experiments are needed to directly probe the involvement of the POX state in nitrogenase catalysis and test this hypothesis.
In sum, the results presented here provide new evidence for the functional importance of a hard, anionic O-based ligand in the conformational redox gating events in nitrogenase. The mechanism by which FeP induces conformational changes in the P-cluster still remains to be determined, however, it is possible the mechanical strain generated by nucleotide-dependent structural changes in FeP can be transmitted to the P-cluster to modulate its coordination-dependent redox equilibria. In addition, our study highlights the utility of investigating covariance patterns among nitrogenase sequences as well as studying nitrogenases from a broader range of organisms.
Supplementary Material
Acknowledgments.
We thank J. Rittle, D. Suess, T. Spatzal, J. Kaiser and J. Howard for helpful discussions and A. Borovik for the use of EPR instrumentation. This work was funded by an NIH grant (GM099813) and a Frasch Foundation Award (735-HF12) to F.A.T. C.P.O. is the recipient of a postdoctoral fellowship from the USDA (2015-67012-22895).
Footnotes
Supporting Information. Experimental details, additional figures and tables related to biochemical, crystallographic and EPR analysis (PDF). The Supporting Information is available free of charge on the ACS Publications website.
REFERENCES
- (1).Burgess BK; Lowe DJ Chem. Rev 1996, 96, 2983. [DOI] [PubMed] [Google Scholar]
- (2).Howard JB; Rees DC Chem. Rev 1996, 96, 2965. [DOI] [PubMed] [Google Scholar]
- (3).Rees DC; Tezcan FA; Haynes CA; Walton MY; Andrade S; Einsle O; Howard JB Philos. Trans. Royal Soc. A 2005, 363, 971. [DOI] [PubMed] [Google Scholar]
- (4).Hoffman BM; Lukoyanov D; Yang Z-Y; Dean DR; Seefeldt LC Chem. Rev. 2014, 114, 4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Schindelin H; Kisker C; Schlessman JL; Howard JB; Rees DC Nature 1997, 387, 370. [DOI] [PubMed] [Google Scholar]
- (6).Einsle O; Tezcan FA; Andrade SL; Schmid B; Yoshida M; Howard JB; Rees DC Science 2002, 297, 1696. [DOI] [PubMed] [Google Scholar]
- (7).Spatzal T; Aksoyoglu M; Zhang L; Andrade SLA; Schleicher E; Weber S; Rees DC; Einsle O Science 2011, 334, 940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Lancaster KM; Roemelt M; Ettenhuber P; Hu Y; Ribbe MW; Neese F; Bergmann U; DeBeer S Science 2011, 334, 974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Tezcan FA; Kaiser JT; Mustafi D; Walton MY; Howard JB; Rees DC Science 2005, 309, 1377. [DOI] [PubMed] [Google Scholar]
- (10).Watt GD; Wang ZC; Knotts RR Biochemistry 1986, 25, 8156. [Google Scholar]
- (11).Lanzilotta WN; Seefeldt LC Biochemistry 1997, 36, 12976. [DOI] [PubMed] [Google Scholar]
- (12).Roth LE; Nguyen JC; Tezcan FA J. Am. Chem. Soc 2010, 132, 13672. [DOI] [PubMed] [Google Scholar]
- (13).Roth LE; Tezcan FA J. Am. Chem. Soc. 2012, 134, 8416. [DOI] [PubMed] [Google Scholar]
- (14).Danyal K; Mayweather D; Dean DR; Seefeldt LC; Hoffman BM J. Am. Chem. Soc. 2010, 132, 6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Danyal K; Dean DR; Hoffman BM; Seefeldt LC Biochemistry 2011, 50, 9255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Tezcan FA; Kaiser JT; Howard JB; Rees DC J. Am. Chem. Soc. 2015, 137, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Chiu H-J; Peters JW; Lanzilotta WN; Ryle MJ; Seefeldt LC; Howard JB; Rees DC Biochemistry 2001, 40, 641. [DOI] [PubMed] [Google Scholar]
- (18).Mayer SM; Lawson DM; Gormal CA; Roe SM; Smith BE J. Mol. Biol. 1999, 292, 871. [DOI] [PubMed] [Google Scholar]
- (19).Zhang L-M; Morrison CN; Kaiser JT; Rees DC Acta Crystallogr. D 2015, 71, 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Peters JW; Stowell MHB; Soltis SM; Finnegan MG; Johnson MK; Rees DC Biochemistry 1997, 36, 1181. [DOI] [PubMed] [Google Scholar]
- (21).Thorneley RN; Lowe D Biochem. J 1984, 224, 903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Wilson P; Nyborg A; Watt G Biophys. Chem. 2001, 91, 281. [DOI] [PubMed] [Google Scholar]
- (23).Hoffman BM; Lukoyanov D; Dean DR; Seefeldt LC Acc. Chem. Res 2013, 46, 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Howard JB; Kechris KJ; Rees DC; Glazer AN PloS One 2013, 8, e72751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Muñoz-Rojas J; Caballero-Mellado J Microb. Ecol 2003, 46, 454. [DOI] [PubMed] [Google Scholar]
- (26).Fisher K; Newton WE Biochim. Biophys. Acta 2005, 1750, 154. [DOI] [PubMed] [Google Scholar]
- (27).Yang Z-Y; Dean DR; Seefeldt LC J. Biol. Chem. 2011, 286, 19417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Among the 45 Group I NifK sequences, there are only two exceptions: Geobacter sulfurreducens and Pelobacter carbinolicus simultaneously have a Ser and Tyr in these positions. Among all 74 nitrogenase sequences analyzed, there is only one species, Methanococcus aeolicus Nankai-3, with an Ala-Ala pair, which does not appear have a coordinating O-based ligand in either of these positions. We note that neither the structures nor the specific catalytic activities of these three nitrogenase variants are known.
- (29).Münck E; Rhodes H; Orme-Johnson W; Davis L; Brill W; Shah V Biochim. Biophys. Acta 1975, 400, 32. [DOI] [PubMed] [Google Scholar]
- (30).Zimmermann R; Münck E; Brill WJ; Shah VK; Henzl MT; Rawlings J; Orme-Johnson WH Biochim. Biophys. Acta 1978, 537, 185. [DOI] [PubMed] [Google Scholar]
- (31).Huynh B; Henzl M; Christner J; Zimmermann R; Orme-Johnson W; Münck E Biochim. Biophys. Acta 1980, 623, 124. [DOI] [PubMed] [Google Scholar]
- (32).Surerus KK; Hendrich MP; Christie PD; Rottgardt D; Orme-Johnson WH; Münck E J. Am. Chem. Soc. 1992, 114, 8579. [Google Scholar]
- (33).Nicolet Y; Rohac R; Martin L; Fontecilla-Camps JC Proc. Natl. Acad. Sci. USA 2013, 110, 7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Pearson RG J. Am. Chem. Soc. 1963, 85, 3533. [Google Scholar]
- (35).Chan JM; Christiansen J; Dean DR; Seefeldt LC Biochemistry 1999, 38, 5779. [DOI] [PubMed] [Google Scholar]
- (36).Rupnik K; Hu YL; Lee CC; Wiig JA; Ribbe MW; Hales BJ J. Am. Chem. Soc. 2012, 134, 13749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Fritsch J; Lenz O; Friedrich B Nat. Rev. Microbiol. 2013, 11, 106. [DOI] [PubMed] [Google Scholar]
- (38).Shomura Y; Yoon K-S; Nishihara H; Higuchi Y Nature 2011, 479, 253. [DOI] [PubMed] [Google Scholar]
- (39).Fritsch J; Scheerer P; Frielingsdorf S; Kroschinsky S; Friedrich B; Lenz O; Spahn CM Nature 2011, 479, 249. [DOI] [PubMed] [Google Scholar]
- (40).Volbeda A; Amara P; Darnault C; Mouesca J-M; Parkin A; Roessler MM; Armstrong FA; Fontecilla-Camps JC Proc. Natl. Acad. Sci. USA 2012, 109, 5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.