Abstract
Intermediate filaments (IFs) are principal components of the cytoskeleton, a dynamic integrated system of structural proteins that provides the functional architecture of metazoan cells. They are major contributors to the elasticity of cells and tissues due to their high mechanical stability and intrinsic flexibility. The basic building block for the assembly of IFs is a rod-like, 60-nm-long tetrameric complex made from two antiparallel, half-staggered coiled coils. In low ionic strength, tetramers form stable complexes that rapidly assemble into filaments upon raising the ionic strength. The first assembly products, “frozen” by instantaneous chemical fixation and viewed by electron microscopy, are 60-nm-long “unit-length” filaments (ULFs) that apparently form by lateral in-register association of tetramers. ULFs are the active elements of IF growth, undergoing longitudinal end-to-end annealing with one another and with growing filaments. Originally, we have employed quantitative time-lapse atomic force and electron microscopy to analyze the kinetics of vimentin-filament assembly starting from a few seconds to several hours. To obtain detailed quantitative insight into the productive reactions that drive ULF formation, we now introduce a “stopped-flow” approach in combination with static light-scattering measurements. Thereby, we determine the basic rate constants for lateral assembly of tetramers to ULFs. Processing of the recorded data by a global fitting procedure enables us to describe the hierarchical steps of IF formation. Specifically, we propose that tetramers are consumed within milliseconds to yield octamers that are obligatory intermediates toward ULF formation. Although the interaction of tetramers is diffusion controlled, it is strongly driven by their geometry to mediate effective subunit targeting. Importantly, our model conclusively reflects the previously described occurrence of polymorphic ULF and mature filaments in terms of their number of tetramers per cross section.
Introduction
Intermediate filaments (IFs) are obligatory structural constituents of metazoan cells. Originally, they were considered to provide passive structural support and to serve as mechanical stress absorbers, thus constituting a central element of cell elasticity (1). This view has been extended in recent years, as it became clear that IFs are intimately connected with the two other major filament systems of the cell, i.e., microtubules and actin filaments, to form a dynamically interacting cytoskeleton (2, 3). More recently, new functions were described for IFs, highlighting their role in events as diverse as cell division, organelle positioning, and physiological stress buffering (4). New studies have also linked the IF cytoskeleton to fibroblast motility and the regulation of lamellipodia formation. In fact, these latter functions were found to depend on the phosphorylation-controlled assembly and disassembly of the IF network (5).
In contrast to microtubules and actin filaments, IFs are resistant to treatment with high salt buffers and detergents, and therefore, they have to be subjected to high concentrations of denaturants for solubilization, purification, and in vitro reconstitution. Microtubules and actin filaments grow by the addition of globular subunits, i.e., tubulin dimers and actin monomers, to “seeds” and filaments. In contrast, the assembly of cytoplasmic IFs begins by lateral association of highly charged rod-like tetrameric complexes to full-width “minifilaments” termed “unit-length filaments” (ULFs). This assembly has originally been observed for vimentin but has later been found with all cytoplasmic IF proteins, including keratins and neurofilament proteins (6, 7, 8, 9). During in vitro reconstitution, the antiparallel, half-staggered tetrameric complexes are formed during refolding of the urea-denatured monomers by dialysis into low ionic strength buffers. In fact, they already form coiled-coil dimers in the presence of 6–7 M urea. In 5–6 M urea, dimers in a structurally precise manner further associate into apolar tetramers with the amino-terminal segments overlapping in the center and the carboxy-terminal ends sticking out (10, 11, 12). Tetramer formation is actually the first step in IF assembly, leading to an intermediate that is stable at low ionic strength. During the first phase of salt-induced assembly, tetramers spontaneously associate laterally into ULFs (10, 11, 13). In a second phase, ULFs longitudinally anneal end-to-end, thereby yielding short filaments containing different numbers of ULFs. As a consequence, free ULFs are eventually consumed (14, 15, 16). Notably, the ends of growing filaments are also able to anneal longitudinally with the ends of other filaments, thereby giving rise to longer filaments in a nonpolar fashion. In a third phase, i.e., during progressive elongation, vimentin IFs radially compact into mature filaments with a diameter of 10–12 nm (10, 17, 18). Mass measurements of filament segments by scanning transmission electron microscopy (STEM) revealed for human vimentin that the main numbers of molecules per filament cross section were 8, 10, and to some extent also 12 tetramers for both mature IFs and ULFs (10, 13). These values indicated some degree of polymorphism for recombinant IF proteins but did perfectly recapitulate a previously documented polymorphism occurring with in vitro assembled authentic keratins (19, 20).
Using “time-lapse” electron microscopy (EM) with conventional pipetting procedures, the lateral assembly process of IF protein tetramers can be followed from about 1 s onward, as the addition of chemical fixing agents such as glutaraldehyde inhibits further assembly instantaneously (13). By this approach, ULFs are prominently visualized; however, subunits smaller than ULFs are not resolved precisely in a quantitative manner, and thus, classification of individual particles during the early assembly stages is not possible. Similarly, in a recent time-resolved light-scattering study, the initial phase of assembly was not accessible to the instrumental setup, as assembly data were only obtained from 18 to 30 s onward (21). At these time points, however, practically all vimentin tetramers are consumed for the formation of ULFs, and elongation to filaments is underway (14). Therefore, to follow the assembly process on a timescale below 1 s and to quantitate the increase in molecular weight of particles over the course of subunit association until they reach a mature ULF state, we used a stopped-flow device in combination with static light scattering. The transport time of the solution from the mixer of the stopped-flow module to the observation cuvette is in the millisecond range and thus small enough to follow the reaction in the subsecond range. Moreover, as the intensity of scattered light is proportional to the molecular mass, it is possible to model the measured light scattering curves numerically. Notably, because tetramers and ULFs are both 60 nm in length and as we have not observed elongation of subcomplexes to occur in high-resolution EM, we are confident that intermediates such as octameric and 16-mer complexes are 60 nm long as well, thereby allowing for efficient modeling of the light-scattering data.
With a stopped-flow setup, we use an established biophysical method to follow the immediate early phase of IF formation. Employing previously established modeling algorithms, we show that the lateral association of soluble IF-protein complexes proceeds very rapidly such that tetrameric units are largely consumed within 1 s. Octameric subunits constitute the next major association species from which ULFs are rapidly formed in various distinct steps.
Materials and Methods
Protein chemical methods
Human vimentin was expressed and purified as described previously (22). Purified proteins were stored at −80°C in 8 M urea, 5 mM Tris-HCl (pH 7.5), 1 mM dithiothreitol, 1 mM EDTA, 0.1 mM EGTA, and 10 mM methyl ammonium chloride. The day before use, vimentin was renatured at room temperature by successive dialysis against dialysis buffer (5 mM Tris-HCl (pH 8.4), 1 mM EDTA, 0.1 mM EGTA, and 1 mM dithiothreitol) containing 6, 4, 2, and 1 M urea, respectively. Dialysis against a large volume of dialysis buffer was continued overnight at 4°C. The next day, dialysis was continued into tetramer buffer (5 mM Tris-HCl (pH 8.4)) for ∼1 h at room temperature. After dialysis, the tetramer samples were filtered through 0.22 μm filters (Millex-GV, polyvinylidene difluoride membrane), and the concentration of the samples was determined by measuring the absorption at 280 nm with ε = 24,900 cm−1M−1 (11). All buffers were filtered (0.22 μm, Millex-GS, mixed cellulose ester membrane) and degassed extensively immediately before use. Analytical ultracentrifugation experiments were carried out in a Beckman analytical ultracentrifuge (model Optima XLA) equipped with an ultraviolet absorption optical system in tetramer buffer at 20°C and a protein concentration of 0.4 g/L. Absorbance scans were recorded at 280 nm. Sedimentation velocity runs were essentially performed and analyzed as described (11). Data were analyzed with the program DCDT+ (version 2.4.0) (23).
Atomic force microscopy
Atomic force microscopy (AFM) was performed in the “scanning in air” mode essentially as described previously (24). Vimentin (0.4 g/L) was assembled at 37°C for 75 min. Reactions were stopped by 10-fold dilution with assembly buffer (25 mM Tris-HCl (pH 7.5) and 50 mM NaCl) followed by fixation with an equal volume of 0.2% glutaraldehyde in assembly buffer for 2 min. Of this solution, 40 μL was deposited to freshly cleaved mica. After 1 min, the specimens were washed with double-distilled water and dried with a steady stream of nitrogen.
Size-exclusion chromatography–multiangle light-scattering analysis of soluble vimentin complexes
Size-exclusion chromatography–multiangle light-scattering (SEC-MALS) analysis was done as described previously (25). Vimentin in tetramer buffer (1 g/L) was centrifuged at 50,000 rpm for 60 min in an Optima MAX ultracentrifuge (Beckman Coulter, Brea, CA). Then, 100 μL of the sample was subjected to size-exclusion chromatography (SEC) using a Superose 6 increase 10/300 column (GE Healthcare, Little Chalfont, UK) connected to a cold ÄKTA basic system (GE Healthcare) at a flow rate of 0.5 mL/min, 4°C. Static multiangle light scattering (MALS) was measured in a DAWN Heleos 8+ instrument (Wyatt Technology, Santa Barbara, CA) by detectors 2–7. The diffraction index was measured in an SEC-3010 refractometer (WGE Dr. Bures). The molecular mass Mn was calculated by the Astra 6.1 software (Wyatt Technology) using the Zimm light-scattering model with dn/dc = 0.185 mL/g. Mn gives the molecular weight average in the selected elution time interval from 16.32 to 17.62 min.
Stopped-flow experiments and data management
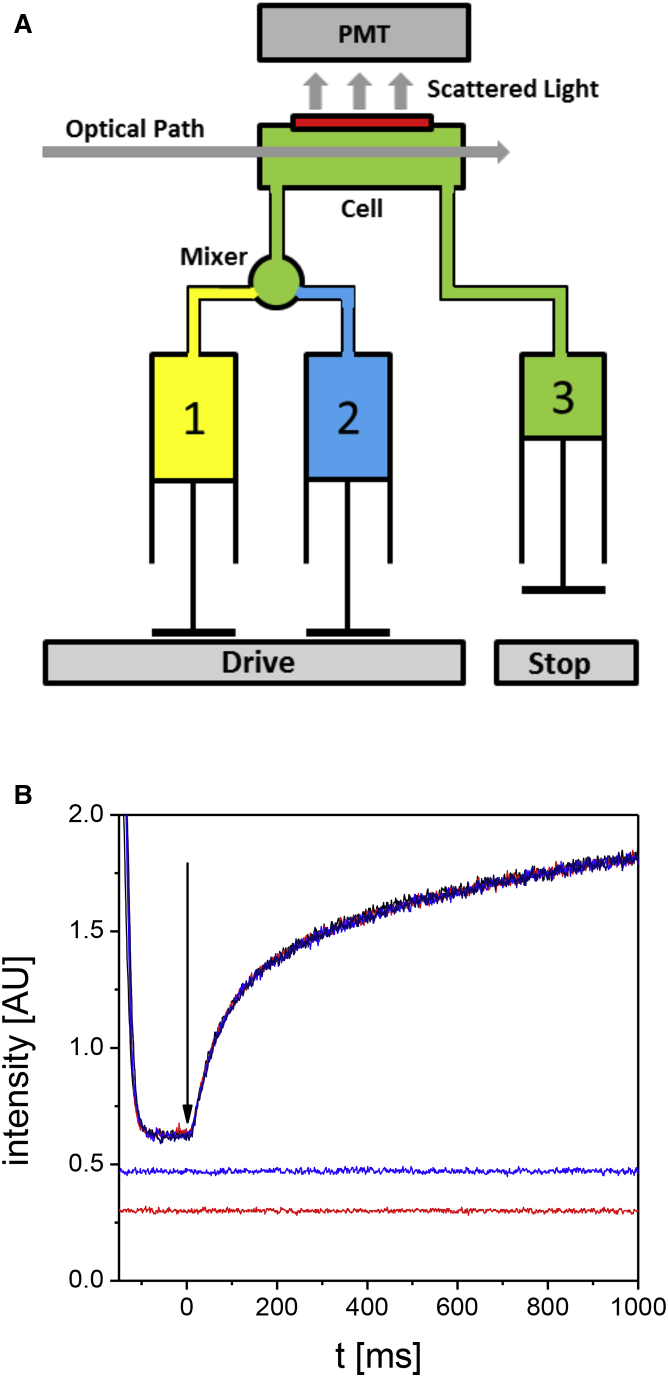
Stopped-flow measurements were performed in a stopped-flow spectrophotometer SF-61 (Hi-Tech Scientific, Salisbury, UK) (26, 27), as depicted schematically in Fig. 1. A solution containing the soluble pool of protein (0.1–1.2 g/L) was filled in a first syringe (1), and assembly start buffer (45 mM Tris-HCl (pH 7.0) and 100 mM NaCl) was filled into a second syringe (2) and equilibrated at 37°C for at least 5 min. Syringes (1) and (2) were driven simultaneously by a drive plate powered by a burst pressure of 400 kPa; the temperature was kept at 37°C. Equal volumes of both solutions were rapidly mixed 1:1 to initiate assembly (“jump-start”). As the fresh assembly mixture was driven into the detection cell, the solution of the previous run was transferred into a third syringe (3) (Fig. 1 B, upper curves). The flow was stopped after 200 μL, as the plunger of the third syringe encountered a stopping block; this event concurrently triggered the data acquisition system (Fig. 1 B, arrow). Before each assembly experiment, both syringes (1) and (2) were rinsed with several milliliters of ddH2O followed by tetramer buffer until no change in the detected scattered light was observed (Fig. 1 B, lower line). This buffer signal served as the baseline in the measurements. In a second step, the intensity of the tetramer signal was measured by mixing a solution of proteins in tetramer buffer with tetramer buffer to the desired protein concentration (Fig. 1 B, middle line). Routinely, 200 μL of protein assembly solution was injected for ∼130 ms into the detection cell to assure that it was completely loaded. These conditions correspond to a flow rate of 1.5 mL/s. Under routine conditions (37°C, 400 kPa), the dead time, which is the time needed for the sample to flow from the sample mixer to the detection cell, was measured to be 3 ms. The optical system consisted of a 150 W Xenon-Hg lamp and a power supply ALX-210 followed by a BH-10 monochromator (Biologic, Claix, France) for measurements at wavelengths of 365 and 436 nm, respectively. Measurements at 594 nm were performed with a diode-pumped solid-state laser (Mambo 25 mW; Cobolt, Vretenvägen, Sweden). The light was transmitted via a multimode fiber and focused into the observation chamber. The scattered-light intensity was detected by a photomultiplier tube (Hi-Tech Scientific) at 90° combined with the electronic unit from Hi-Tech, which consists of the photomultiplier power supply, PSP-60, and the signal conditioning unit, SC-60. The signal of the photomultiplier was electronically filtered by a unity gain amplifier with an internal low-pass filter (time-constant: 1 ms).
Figure 1.
(A) Schematic representation of the stopped-flow apparatus. The three syringes marked 1, 2, and 3 provide substrate in tetramer buffer (1) and assembly start buffer (2) and serve as a reservoir for emptying the cell (3). (B) Recorded intensities for tetramer buffer (lower curve), vimentin tetramers in tetramer buffer (middle curve), and five consecutive assembly reactions (upper curves) are shown. The time point of injection of assembly start buffer is marked by an arrow. PMT, photomultiplier tube; the rectangle on top of the cell represents an aperture; the intensity of the signal is given in arbitrary units (AU). To see this figure in color, go online.
Data acquisition was achieved with a multifunction data acquisition card, PCI-6023E (National Instruments, München, Germany), and a software tool based on the driver software package NI-DAQmx. Output voltages from the signal conditioning unit as well as the stop signal were recorded with a 1 ms sample time. The data acquisition was triggered by the stop signal plus a forerun of several milliseconds to record the full scattering signal. The stopped-flow data were analyzed with the software Origin (V7.0; OriginLab, Northampton, MA). Measurements that exhibited extreme peak values, as caused by dust or air bubbles, were discarded. To reduce noise, mean values of several successive shots were calculated (five shots for protein concentrations below 0.4 g/L and two shots for higher protein concentrations) and were then used for data fitting. The scattering intensity of tetrameric vimentin was measured in tetramer buffer from the intensity of a solution of tetramers at the respective concentration for some seconds. The buffer signal and the tetramer signal were smoothed to remove noise before the mean value and the SD were calculated. Assembly signals were collected from termination of the sample injection onward. The data from the first 7 ms were discarded in all runs because in this very early time interval, the recorded data curves exhibited some irregularities probably because of flow turbulences. The starting point of assembly was further corrected for the experimentally determined dead time of 3 ms. Consequently, all experiments were followed from exactly 10 ms onward after the assembly was started.
Static light scattering
The assembly kinetics of tetramers to ULFs was recorded by static light scattering in a time window, in which filament elongation does not occur substantially (less than 3%). The particles encountered here are distinctly smaller than the wavelength of the light, i.e., the tetramer rods are 60 nm long with a radius a of ∼1 nm, and their lateral assembly product, the ULF, has the same length and an average radius a of ∼7 nm, which is much smaller than the wavelength of light used in this study (see above). Thus, the data can be treated according to Rayleigh scattering, i.e., the scattered light intensity is proportional to the molecular mass and to a first approximation independent of particle shape (28):
| (1) |
with I being the scattered light of the protein, I0 being the intensity of the incident beam, n0 being the refractive index of the solution, dn/dC being the refractive index increment, λ being the wavelength of the light, r being the distance from the reaction cuvette to the detector, NA being the Avogadro number, C being the concentration (g/L), and M being the molecular weight. For a tetramer, we can write this equation as follows:
| (2) |
and for multiple tetrameric complexes:
| (3) |
Consequently, a protein solution of the same concentration C in the form of octamers will scatter twice as much light as tetramers. ULFs consisting of eight tetramers will scatter eight times more light than eight single tetramers.
Data processing
To calculate the reaction constants of the assembly intermediates, we use the program Berkeley Madonna (V 8.3.18, https://www.berkeleymadonna.com). It optimizes a set of parameters (here, the rate constants) in a least-square sense by integrating the reaction kinetics as an initial value problem and comparing the resulting curves to measured data. The basic reaction schemes have originally been established and applied for monitoring the longitudinal assembly reaction of IFs and were adapted here for the description of the ULF assembly process (14). In particular, the primary reactions reflect the association of two tetramers to one octamer (k1), that of two octamers to one 16-mer (k2), and further, that of two 16-mers to one ULF (k3). To consider potential elongation reactions of ULFs with ULFs and IFs, we introduce the reaction constant k4. Accordingly, the following set of differential equations is given for the respective species: T, tetramer; O, octamer; H, 16-mer; U, ULF; U2, ULF-dimer; U3, ULF-trimer; U4, ULF-tetramer:
To allow for the reactions leading to polymorphic forms of the ULFs, we have to consider the addition of octamers to the various oligomers as well as the addition of 16-mers to 24-mers and the reaction of two 24-mers with each other. The corresponding reactions are as follows for the respective species, i.e., V, 24-mer; U(32), 32-mer; U(40), 40-mer; U(48), 48-mer, where U is the sum of U(32), U(40), and U(48):
In particular, the amounts of the individual ULF species, U(32), U(40), and U(48), are calculated by the following terms:
The scattering intensities of each species are defined as multiples of the intensity of the tetramer (Eq. 3). The intensities of all assembly intermediates at each time point are summed up and compared with the time-dependent increase of the measured intensities in the measurements. To obtain robust results, we perform simultaneous curve fittings (global fit) over various conditions, e.g., different wavelengths and protein concentrations. This procedure enables us to select only measurements in which dust and aggregates do not disturb significantly according to the following exclusion criteria: 1) the tetramer intensities are not fixed in the global fit, and 2) the fitted tetramer intensities are calibrated with the measured tetramer intensities, and runs are discarded when the fitted tetramer intensities differ by more than two times the SD with regard to the previously measured tetramer intensities. The integration time step dt in the program Berkeley Madonna is set to 50 μs. Going to lower time steps does not change the results.
Results
Construction of a model for ULF assembly
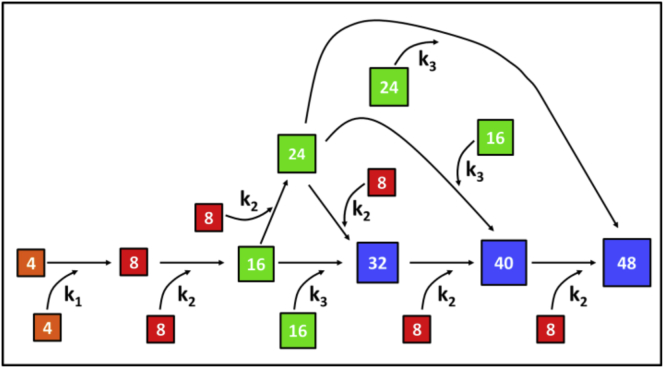
In fact, assembly of IFs starts already during renaturation of monomers by dialysis from 8 M urea into low ionic strength buffer, whereby stable tetrameric complexes are formed as determined by sedimentation equilibrium ultracentrifugation (10, 11). The immediate increase of the ionic strength (“jump-start”) initiates the ordered lateral association of tetramers with each other (k1) and yields octamers as first reaction products (Fig. 2). Octamers further laterally associate into 16-mers (k2), which then can associate to 32-mers (k3). This is the basic scenario leading to the formation of ULFs, consisting of 32 monomers per filament cross section (Fig. 2). However, because IFs have been measured by STEM to exhibit a distinct mass-per-length polymorphism, additional reactions have to be considered (10, 19, 20). In particular, octamers may associate with 16-mers to 24-mers (k2), and two 24-mers may associate to 48-mers (k3). Yet, as a further reaction, 24-mers may associate with 16-mers to 40-mers (k3), which can also be formed by the addition of an octamer to a 32-mer (k2). Also, 40-mers can accept octamers to yield 48-mers (k2). Additionally, the reaction of a 24-mer with an octamer will yield a ULF as well (Fig. 2). All associations in which octamers are engaged can be described by one reaction constant, k2; likewise, all further reactions of 16-mers and 24-mers can be fitted with k3. We think that this description represents a certain simplification but can indeed serve as a valid first approximation assumption. In summary, all these potential interactions will lead to ULFs harboring 32, 40, and 48 monomers, respectively. The original STEM mass determinations revealed these three types of ULFs in roughly equal amounts in all IFs (10, 13).
Figure 2.
Schematic model depicting the assembly of ULFs. By lateral association of tetramers (squares designated 4), octamers (squares designated 8), 16- and 24-mers (squares designated 16 and 24), ULFs harboring 32, 40, and 48 monomers (squares designated 32, 40, and 48) are formed. The reaction constants k1, k2, and k3 are indicated for the respective reactions. To see this figure in color, go online.
The ULF polymorphism is a direct consequence of the “jump-start” mode of assembly, which is essential for a kinetic measurement setup. When IFs are assembled by dialysis from low against high ionic strength buffer, the mass distribution along mature extended filaments is much more uniform, favoring ULF segments with 32 subunits per cross section (10). However, the fact that ULFs with different numbers of subunits integrate into one and the same IF demonstrates that they are functional for longitudinal assembly.
Characterization of soluble vimentin complexes
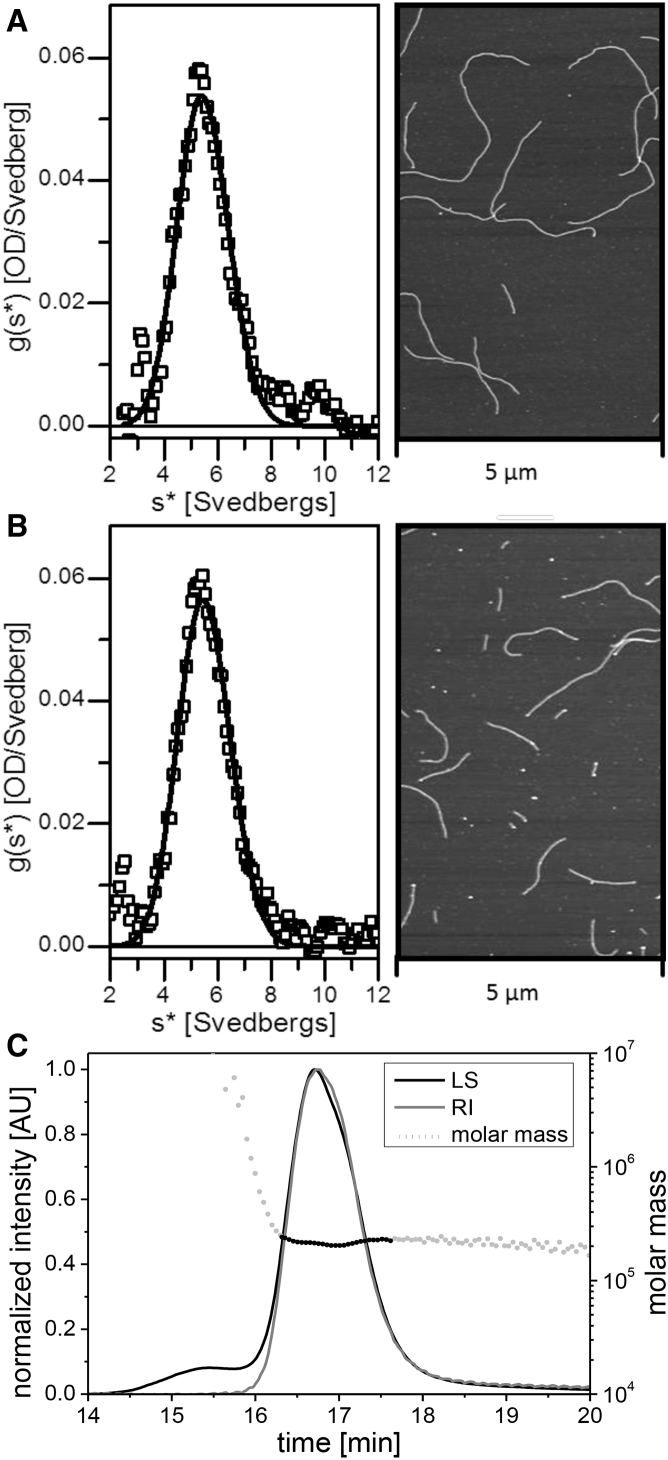
The homogeneity of the protein complexes at the beginning of an assembly experiment is of absolute importance for a uniform association regime to occur. Indeed, depending on the type of preparation, higher-order complexes may be present and result in significant ambiguities during the early assembly phase (21). Therefore, we started our experiments with the characterization of soluble complexes obtained after reconstitution into tetramer buffer by analytical ultracentrifugation. By sedimentation velocity, we observed a sharp, symmetrical peak at 5.5 S (Fig. 3 A, left panel), which compares well with results obtained previously (11). There, we had shown by sedimentation equilibrium that such sedimentation velocity data represented a homogenous tetrameric species in the concentration range we used here. To test for the assembly competence of this preparation, we performed standard “jump-start” assembly and documented filament formation by AFM (Fig. 3 A, right panel). Moreover, to inspect if the subunit tetramers were assembled in a nonaggregated, functional state, we dissolved the assembled filaments in low-ionic strength, i.e., tetramer buffer. The following analysis by analytical ultracentrifugation analysis revealed the existence of uniform 5.5 S complexes (Fig. 3 B, left panel). By shifting the salt concentration again to physiological values, extensive filament formation occurred, indicating that the “jump-start” assembly procedure leaves the tetrameric complexes fully functional (Fig. 3 B, right panel). This experiment provides a way to assemble/disassemble (“cycle”) IFs multiple times.
Figure 3.
Characterization of soluble vimentin complexes in tetramer buffer. (A) Sedimentation velocity ultracentrifugation shows vimentin renatured in tetramer buffer (left panel). After initiation of assembly, filaments were visualized by atomic force microscopy (right panel). (B) Sedimentation velocity ultracentrifugation shows the products obtained after dialysis of the filaments shown in (A) back into tetramer buffer (left panel). These complexes were subjected to another round of assembly, and the resulting filaments were again visualized by atomic force microscopy (right panel). (C) Vimentin renatured into tetramer buffer was subjected to size-exclusion chromatography, and the eluted protein was analyzed by multiangle light scattering (SEC-MALS). Recorded were the light-scattering signal (LS) and the protein-absorption signal (RI), with the left axis in arbitrary units (AU). Dotted line: molar mass. The prepeak between 15 and 16 min is caused by nonspecific particles.
As a second independent method for accurate molecular mass determination of the products obtained by renaturation of urea-solubilized vimentin in tetramer buffer, we subjected the proteins to SEC-MALS experiments. The molecular mass was determined to be 2.14 × 105 and 2.17 × 105 (error <1%) for two runs, which is in excellent agreement with the calculated molecular weight (2.14 × 105) for a vimentin tetramer (Fig. 3 C). The ultracentrifugation step before the SEC-MALS experiments was effective in removing dust particles as well as larger protein aggregates and was therefore routinely employed. The measured radius of gyration, 14.3 and 14.7 nm (10% error), comes close to the expected value of 17 nm for a thin uniform rod with a length of 60 nm. Because the data obtained with the SEC-MALS experiments are in perfect agreement with those obtained by analytical ultracentrifugation, we conclude that the static light-scattering signal obtained with tetramers is well suited to follow the distinct association reactions of tetramers.
ULF formation monitored in a stopped-flow device
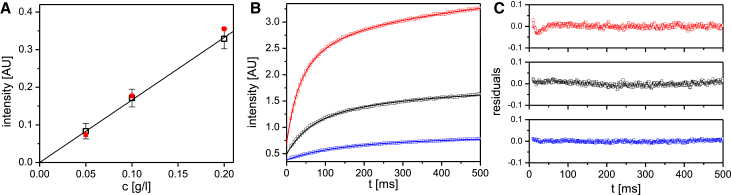
First, we determined the light-scattering intensity signals for vimentin complexes at different protein concentrations in tetramer buffer. These values are needed to evaluate the time-dependent changes in scattered light intensity after initiation of assembly in the stopped-flow device. After filling the cell, a stable signal was obtained at each protein concentration investigated, i.e., 0.05, 0.10, and 0.20 g/L, with a linear increase in signal intensity over the three different concentrations used (Fig. 4 A). These results document the fact that the individual particles do not interact under these conditions, which is in agreement with data from analytical ultracentrifugation (11) and SEC-MALS (see above). With these data at hand, we proceeded to determine the assembly kinetics. Assembly was initiated by adding assembly start buffer to the tetramers. After the observation cell was filled with the assembly mixture, the flow was stopped, and the scattering signal was recorded (Fig. 4 B). The rapid rise of the intensity in the first 100 ms reflected the strong molecular interaction of tetramers to higher-order oligomers; the increase continues to 500 ms although the slope is slightly decreasing over time. By EM, ULFs were observed in abundance when the assembly was stopped by fixation with glutaraldehyde after less than 1 s of assembly, the first time point for a measurement with this kind of “time-lapse” method (10, 13). In addition, the EM analysis revealed ∼60-nm-long particles of lower diameter than standard 32-mer ULFs, as expected for precursors harboring only 16- or 24-mers. At all three protein concentrations, the residuals to the fit exhibited minimal deviations from the values derived from the experimental data, indicating that no systematic deviations occurred and that the assembly took place in a highly coordinated manner (Fig. 4 C).
Figure 4.
Intensity plots from stopped-flow experiments at three protein concentrations. (A) Initial signal intensities of tetramers in tetramer buffer are shown as a function of the protein concentration in arbitrary units (AU). Open squares: mean values of the tetramer intensity signals. The error bars designate the plus-minus SDs from the means. Filled squares: values obtained from the global fit to all data points for the given protein concentration. (B) Time-dependent changes of the scattering intensities are shown in arbitrary units (AU) after addition of assembly start buffer to the three protein concentrations depicted in (A). Mean values were calculated from the measurements at five successive time points and plotted as follows: lower curve: 0.05 g/L; middle curve: 0.10 g/L; upper curve: 0.20 g/L. The lines represent values obtained by a global fit to the whole set of data obtained for the three protein concentrations. (C) Residuals of the scattering curves shown in (B). The upper, middle, and lower panels relate to the upper, middle, and lower curves shown in (B). To see this figure in color, go online.
Dissection of ULF formation
Over the course of our study, we systematically varied two parameters: 1) the wavelength (365, 436, and 594 nm) and 2) the protein concentration (from 0.05 to 0.6 g/L). Under all conditions, we observed very similar results (Table 1). Therefore, Rayleigh scattering can be reliably used at these short timescales and in the protein concentration range employed.
Table 1.
Determination of Rate Constants
| c [g/L] | ʎ [nm] | k1a | k2a | k3a | k4a | χ2b |
|---|---|---|---|---|---|---|
| 0.05/0.1/0.2 | 436 | 158 | 128 | 13 | 1.1 | 3/6/3 |
| 0.05/0.1/0.2c | 436 | 179 | 118 | 15 | 0.5 | 4/6/6 |
| 0.4 | 365/436/594 | 253 | 109 | 16 | 0.5 | 5/8/13 |
| 0.4d | 594 | 191 | 116 | 16 | 0.5 | 8/7/5 |
| 0.6e | 594 | 214 | 97 | 13 | 0.4 | 5/5 |
| Mean ± SD | 199 ± 33 | 114 ± 10 | 14.6 ± 1.4 | 0.6 ± 0.3 |
Global fitting-derived rate constants in μM−1s−1.
χ2, chi-squared test.
Results do not change significantly if k4 is set to 0.5 μM−1s−1, as determined at the higher protein concentrations listed below.
3 × independent measurements.
2 × independent measurements.
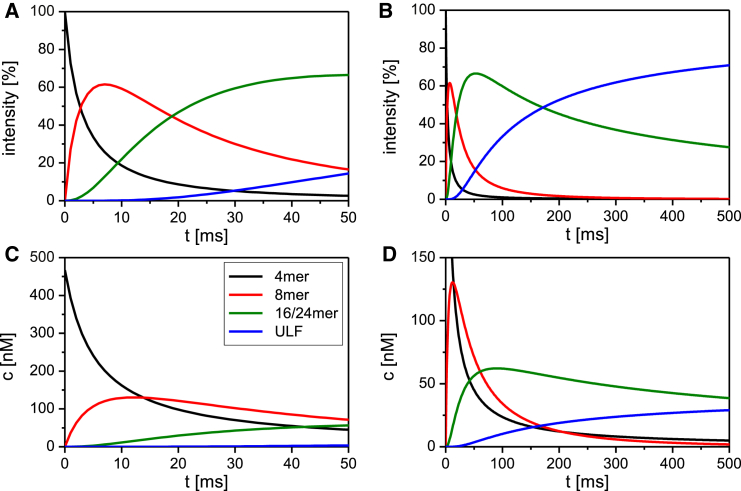
By taking the mean values of the derived rate constants (see Fig. 2), we were able to calculate, employing the equations presented in Materials and Methods, the contributions of the individual complexes to the observed signal at any given time point after initiation of assembly, all the way from tetramers to ULFs, for instance, as shown here at 0.1 g/L (Fig. 5, A and B). The intensity signal reveals a vigorous reduction of tetramers within the first milliseconds. We estimate that after ∼3 ms, the tetramer signal is down to 50%, and the octamer signal is up to 50%. Correspondingly, hardly any signal is coming from 16- and 24-mers or from ULFs yet (Fig. 5 A). By 50 ms, the tetramer signal has decreased steadily down to ∼3%. The octamer signal peaks at 7 ms with ∼60% and drops to ∼17% by 50 ms. The signal contributed by 16-mers and 24-mers is climbing up to 50% by 66 ms (Fig. 5 B). In parallel with the formation of 16- and 24-mers, ULFs contribute increasingly to the signal intensity from 50 ms onward, at which time they have 14% and later 50% by 180 ms (Fig. 5 B). Tetramers and octamers are practically used up by 100 and 200 ms, respectively (Fig. 5 B). The situation is in principle very similar when the concentrations of the assembly intermediates are plotted versus time: starting with 0.5 μM tetramers, corresponding to 2 μM monomers (0.1 mg/mL, black curve in Fig. 4 B), we observe a fast decay of the tetramer concentration to 50 nM within the first 40 ms (Fig. 5 C, black line). At the same time, 16- and 24-mers increase to ∼50 nM (Fig. 5 C), whereas ULFs are present at only 2 nM. By 500 ms, the tetramer concentration has dropped to ∼5 nM, corresponding to roughly 1% of the initial concentration (Fig. 5 D). At 700 ms, the concentration of ULF and the sum of 16- and 24-mers is nearly the same at around 30 nM (data not shown). However, the concentration of ULF is steadily increasing over the first 500 ms, whereas that of 16- and 24-mers is slowly decreasing from ∼80 ms on (Fig. 5 D, blue versus green line).
Figure 5.
(A and B) Signal intensity profiles (%) of the various assembly intermediates over (A) 50 ms and (B) 500 ms. (C and D) Molar distributions of the assembly intermediates over (C) 50 ms and (D) 500 ms are shown. The designation of the colored lines to assembly intermediates is depicted in (C). 16- and 24-mers are treated as one species. The calculations for the intensity profiles (A and B) and the molar distributions of assembly intermediates (C and D) over time were done for 0.1 g/L with the equations specified in Materials and Methods.
Assessment of the model
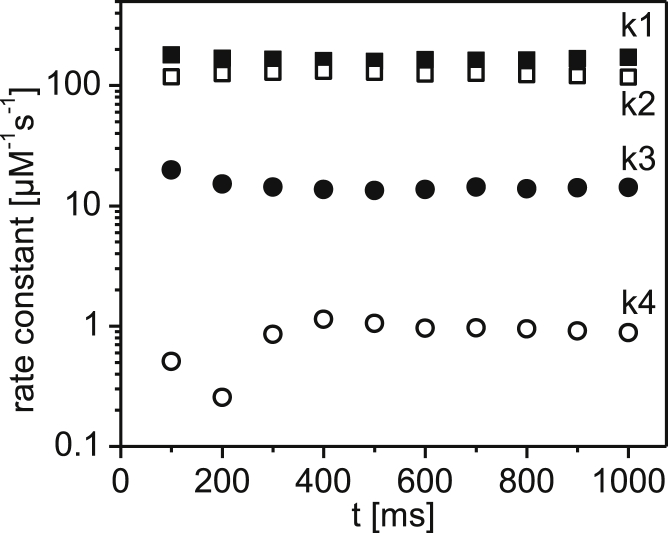
In the next step, we assessed how the rate constants develop when the data are fitted beyond 500 ms such that ULF formation but not filament elongation is the dominant feature. Therefore, we fitted the rate constants from 100 ms up to 1 s (Fig. 6). Here, both k1 (and k2) and k3 stay constant. Moreover, when we also consider the beginning of the longitudinal association of ULFs, introducing k4, we find this to be 100-fold lower than k1. For further analysis of the data in the range of several seconds, the elongation of ULF requires the introduction of a form factor, which makes the whole calculation more complex. However, for now, we want to present only the initial formation of ULFs, i.e., assembly time below 500 ms, and leave the treatment of the elongation reaction for a future study.
Figure 6.
Global fit-derived values for the rate constants k1 (filled squares), k2 (open squares), k3 (filled circles), and k4 (open circles).
Discussion
The kinetics of supermolecular fiber formation for self-assembly systems such as actin filaments and microtubules has been analyzed by shifting the incubation conditions from those favoring the soluble state to those inducing supermolecular assembly of their globular subunits, actin, and tubulin, respectively. Hence, the polymerization of monomeric actin is initiated by a sudden increase of the salt concentration (29). Likewise, for the α-/β-tubulin dimer, the addition of salt and glycerol as well as a change in temperature from 4 to 37°C are key parameters to “switch on” assembly (30, 31). The presence of nucleotides, such as ATP and GTP, enhances the assembly process significantly, as both actin and tubulin exhibit nucleotidase activity during filament assembly. Moreover, “seeds” are needed to mediate a lag-phase-free assembly process (32). In stark contrast, cytoplasmic IF proteins do not exhibit nucleotidase activity, and for filament assembly activity to occur, one has to start from tetrameric complexes, as no monomers and dimers are obtained under nondenaturing, near-physiological conditions. Tetramers assemble immediately when the ionic strength is increased, and hence, they are the principal assembly precursors for the formation of cytoplasmic IFs (8).
With the establishment of a model of the vimentin tetramer at atomic resolution (33, 34), we have a quantitative description of the complex at hand that sets the geometrical limits for the treatment of data derived from light scattering and EM (14, 21, 35). Moreover, for the interpretation of the stopped-flow data, several pieces of information derived from EM have to be implemented. First, when tetramers are assembled in a kinetic mode by the instantaneous increase of the ionic strength (“jump-start”), massive ULF formation has already occurred by the time assembly is stopped through addition of glutaraldehyde immediately after its initiation. This procedure yields results for time points from two seconds onward (13). It clearly documented that ULFs exist abundantly at this time together with complexes of the same length but with smaller diameters (7, 10, 14). Second, STEM mass measurements of IFs revealed three major types of subsegments along individual filaments differing by one octameric unit each, with values close to four, five, and six octamers per filament cross section. With another powerful technique called small-angle x-ray scattering, octamers have been demonstrated to be a major soluble species at high protein concentration and slightly elevated ionic strength, indicating that it needs only a small shift in conditions to cause tetramers to interact with each other (35).
Different from the time-lapse fixation method, stopped-flow in combination with light scattering provides an ultrafast procedure, during which the assembly can be continuously followed from 10 ms onward without the need for fixation or deposition of the sample on an EM grid. Exactly these dynamic intermediate assembly products that disturb STEM mass measurement on a grid can be addressed directly via their scattering signal. For the treatment of the light-scattering data obtained by stopped-flow experiments, we theoretically explored which interactions would yield the three major octamer-derived ULF types with four, five, or six octamers per filament cross section. Furthermore, we could determine which reaction constants were needed to describe these assembly reactions: k1 for the interaction of tetramers to octamers; k2 for any interaction in which an octamer may engage, be it another octamer or a 16-, 24-, 32-, or 40-mer; and k3 for the interaction of 16- and 24-mers with each other. In addition, the beginning longitudinal annealing of ULF can be described with k4, although during the first 500 ms of assembly, k4 is 100-fold smaller than k1 and k2. In summary, the model depicted in Fig. 2 is able to precisely describe the data generated with vimentin for concentrations from 1 to 10 μM. These values are actually within the range of vimentin concentrations found in cells (36).
Filament elongation following ULF formation
Our previous work (16) analyzed longitudinal IF assembly by generating time-dependent length distribution profiles, which were then evaluated by numeric models. Because the length distributions were measured on timescales of seconds to minutes, these models focused on longitudinal IF growth. Only rough estimates of the kinetic parameters for lateral growth were used, and these were selected such that this part of IF assembly was not rate limiting (14). Following IF assembly on the millisecond timescale enabled us to describe the early assembly phase with much higher precision. We now determined the rate constant for the lateral vimentin assembly of two tetramers under standard conditions directly from the light-scattering measurements. Therefore, our results for the lateral assembly, which to our knowledge are new, now complement our earlier studies and enable us to describe the immediate IF assembly process, i.e., the formation of ULFs on a timescale from milliseconds to hours (14, 16, 37, 38). The model (see Fig. 2) also allows us to describe the concentration course of all intermediate species participating in the assembly process, i.e., tetramers, octamers, and higher-order complexes over time (Fig. 5). Accordingly, more than 99% of the tetramers are consumed throughout the first second of assembly. In particular, the concentration of tetramers is down to 50% after 4 ms, corresponding to an increase of the octamer concentration to 50%, as hardly any larger higher complex is formed at that time point. This behavior is in stark contrast to the assembly of actin filaments and microtubules from actin monomers and tubulin dimers. In both cases, a significant lag phase in the assembly is observed unless “nuclei”—the equivalent of ULFs—are employed.
Previously, the lateral assembly of vimentin has been investigated by small-angle x-ray scattering in combination with microfluidics (39). Note the principle difference between our experimental design using stopped-flow and diffusive mixing, as employed in these microfluidics experiments. Stopped-flow offers the great advantage of exhibiting virtually no lag time for mixing in the beginning. Here, the time resolution is given by the minimal exposure time needed per time point. In contrast, diffusive mixing and continuous flow projects each time point on a different position in the flow channel and thus allows for exposure times much longer than the time gap Δt between two measured time points (40). Hence, both methods are highly complementary. In the stopped-flow approach, we show that the first lateral vimentin assembly, i.e., the assembly of two tetramers, is one to two orders of magnitude slower than the diffusion-limited association of two uniformly reactive spheres. Moreover, the calculated reaction rate is significantly slower when only a limited segment of the rod-like tetramers is assumed to be reactive in the specific interaction of two tetramers (41). In particular, reactive segments smaller than 1 nm describe the observed reaction constant k1 in a satisfactory manner. In a recent hydrogen-deuterium exchange (HDex) study of vimentin, it was actually shown that a short segment at the start of coil 2 of vimentin is instrumental in the formation of octamers from tetramers (42).
The in vivo assembly of vimentin IFs
For both the actin and the α-/β-tubulin complex systems of cotranslational folding, the factors centering around the chaperonin containing TCP-1 have been described (43). Their action is absolutely required for the delivery of fully folded, functional protein into the cytoplasm. With IF proteins, much less is known; however, a strong impact of small heat shock proteins for the integrity of the muscle-specific desmin filament system has been documented, and an impact of mutated αB-crystallin on growing desmin IFs was demonstrated (44). In addition, earlier work reported so-called “dynamic cotranslation,” during which IF-messenger-RNA is translated during transport along microtubules, and the resulting IF proteins are picked up during the process and transported to the respective assembly sites. It was hypothesized that these IF particles (termed squiggles) contain tetramers or already higher-ordered structures up to ULFs (45). Whatever the case is, a pool of soluble IF precursors such as tetramers has been experimentally documented to exist in cells. Indeed, this pool appears to be very small, probably because of a very rapid incorporation of tetramers into filaments (46).
Outlook
In contrast to the in vitro IF assembly system, in which disassembly of filaments is negligible and no “treadmilling-type” or “catastrophic-disassembly” reactions analogous to actin filaments or microtubules occur (47), the situation in cultured cells and in tissues is different for various functional reasons. Here, posttranslational modifications and, in particular, phosphorylation play a major role in facilitating a dynamic turnover of filaments and cytoskeletal compartments in different phases of the cell cycle or under distinct physiological situations (48, 49, 50, 51). By using the stopped-flow system presented here, it will now be feasible to investigate the modulatory potential of posttranslational modifications of specific IF-proteins on the lateral assembly of their tetramers to ULFs. Furthermore, future stopped-flow studies will be directed to the investigation of the assembly properties of tetrameric species of other cytoplasmic IF proteins, such as the muscle-specific desmin, the neurofilament triplet proteins, and the keratins.
Author Contributions
U.A., J.L., and H.H. developed the original concept. N.M., S.W., and R.K. developed the experimental procedures and designed the data processing routines. W.H.G. set up a second stopped-flow device and gave essential experimental instructions. N.M. and J.K. optimized experimental routines. N.M. and L.K. performed the majority of the stopped-flow experiments, and N.M. analyzed the data. M.M. purified the protein and assisted L.K. with her work in the lab. J.B. and E.H. performed and analyzed the SEC-MALS experiments. K.T. calculated the form factors. N.M. and H.H. wrote the manuscript.
Acknowledgments
We thank Tatjana Wedig (B062, German Cancer Research Center) and Alessio Falzone (B040, German Cancer Research Center) for excellent technical assistance.
This work was supported by the German Research Foundation (DFG) through grants LA 500/17-1 (J.L.), KI 1464/2-1 (R.K.), GO 598/9-1 (W.H.G.), as well as HE 1853/4-3 and HE 1853/11-1 (H.H.).
Editor: Markus Buehler.
Footnotes
Jörg Langowski, deceased May 6, 2017.
References
- 1.Fuchs E., Weber K. Intermediate filaments: structure, dynamics, function, and disease. Annu. Rev. Biochem. 1994;63:345–382. doi: 10.1146/annurev.bi.63.070194.002021. [DOI] [PubMed] [Google Scholar]
- 2.Herrmann H., Bär H., Aebi U. Intermediate filaments: from cell architecture to nanomechanics. Nat. Rev. Mol. Cell Biol. 2007;8:562–573. doi: 10.1038/nrm2197. [DOI] [PubMed] [Google Scholar]
- 3.Ma X., Foo Y.H., Wohland T. Fluorescence cross-correlation spectroscopy (FCCS) in living cells. Methods Mol. Biol. 2014;1076:557–573. doi: 10.1007/978-1-62703-649-8_25. [DOI] [PubMed] [Google Scholar]
- 4.Toivola D.M., Strnad P., Omary M.B. Intermediate filaments take the heat as stress proteins. Trends Cell Biol. 2010;20:79–91. doi: 10.1016/j.tcb.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Helfand B.T., Mendez M.G., Goldman R.D. Vimentin organization modulates the formation of lamellipodia. Mol. Biol. Cell. 2011;22:1274–1289. doi: 10.1091/mbc.E10-08-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herrmann H., Hesse M., Magin T.M. Functional complexity of intermediate filament cytoskeletons: from structure to assembly to gene ablation. Int. Rev. Cytol. 2003;223:83–175. doi: 10.1016/s0074-7696(05)23003-6. [DOI] [PubMed] [Google Scholar]
- 7.Herrmann H., Aebi U. Intermediate filament assembly: fibrillogenesis is driven by decisive dimer-dimer interactions. Curr. Opin. Struct. Biol. 1998;8:177–185. doi: 10.1016/s0959-440x(98)80035-3. [DOI] [PubMed] [Google Scholar]
- 8.Herrmann H., Aebi U. Intermediate filaments: molecular structure, assembly mechanism, and integration into functionally distinct intracellular Scaffolds. Annu. Rev. Biochem. 2004;73:749–789. doi: 10.1146/annurev.biochem.73.011303.073823. [DOI] [PubMed] [Google Scholar]
- 9.Ip W., Hartzer M.K., Robson R.M. Assembly of vimentin in vitro and its implications concerning the structure of intermediate filaments. J. Mol. Biol. 1985;183:365–375. doi: 10.1016/0022-2836(85)90007-5. [DOI] [PubMed] [Google Scholar]
- 10.Herrmann H., Häner M., Aebi U. Structure and assembly properties of the intermediate filament protein vimentin: the role of its head, rod and tail domains. J. Mol. Biol. 1996;264:933–953. doi: 10.1006/jmbi.1996.0688. [DOI] [PubMed] [Google Scholar]
- 11.Mücke N., Wedig T., Herrmann H. Molecular and biophysical characterization of assembly-starter units of human vimentin. J. Mol. Biol. 2004;340:97–114. doi: 10.1016/j.jmb.2004.04.039. [DOI] [PubMed] [Google Scholar]
- 12.Wu K.C., Bryan J.T., Steinert P.M. Coiled-coil trigger motifs in the 1B and 2B rod domain segments are required for the stability of keratin intermediate filaments. Mol. Biol. Cell. 2000;11:3539–3558. doi: 10.1091/mbc.11.10.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrmann H., Häner M., Aebi U. Characterization of distinct early assembly units of different intermediate filament proteins. J. Mol. Biol. 1999;286:1403–1420. doi: 10.1006/jmbi.1999.2528. [DOI] [PubMed] [Google Scholar]
- 14.Kirmse R., Portet S., Langowski J. A quantitative kinetic model for the in vitro assembly of intermediate filaments from tetrameric vimentin. J. Biol. Chem. 2007;282:18563–18572. doi: 10.1074/jbc.M701063200. [DOI] [PubMed] [Google Scholar]
- 15.Winheim S., Hieb A.R., Mücke N. Deconstructing the late phase of vimentin assembly by total internal reflection fluorescence microscopy (TIRFM) PLoS One. 2011;6:e19202. doi: 10.1371/journal.pone.0019202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Portet S., Mücke N., Herrmann H. Vimentin intermediate filament formation: in vitro measurement and mathematical modeling of the filament length distribution during assembly. Langmuir. 2009;25:8817–8823. doi: 10.1021/la900509r. [DOI] [PubMed] [Google Scholar]
- 17.Georgakopoulou S., Möller D., Aebi U. Near-UV circular dichroism reveals structural transitions of vimentin subunits during intermediate filament assembly. J. Mol. Biol. 2009;386:544–553. doi: 10.1016/j.jmb.2008.12.053. [DOI] [PubMed] [Google Scholar]
- 18.Wickert U., Mücke N., Herrmann H. Characterization of the in vitro co-assembly process of the intermediate filament proteins vimentin and desmin: mixed polymers at all stages of assembly. Eur. J. Cell Biol. 2005;84:379–391. doi: 10.1016/j.ejcb.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 19.Steven A.C., Trus B.L., Steinert P.M. Conformity and diversity in the structures of intermediate filaments. Ann. N. Y. Acad. Sci. 1985;455:371–380. doi: 10.1111/j.1749-6632.1985.tb50423.x. [DOI] [PubMed] [Google Scholar]
- 20.Parry D.A., Steinert P.M. Springer-Verlag; New York: 1995. Intermediate Filament Structure. [Google Scholar]
- 21.Lopez C.G., Saldanha O., Köster S. Lateral association and elongation of vimentin intermediate filament proteins: a time-resolved light-scattering study. Proc. Natl. Acad. Sci. USA. 2016;113:11152–11157. doi: 10.1073/pnas.1606372113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herrmann H., Kreplak L., Aebi U. Isolation, characterization, and in vitro assembly of intermediate filaments. Methods Cell Biol. 2004;78:3–24. doi: 10.1016/s0091-679x(04)78001-2. [DOI] [PubMed] [Google Scholar]
- 23.Philo J.S. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal. Biochem. 2006;354:238–246. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 24.Mücke N., Kreplak L., Langowski J. Assessing the flexibility of intermediate filaments by atomic force microscopy. J. Mol. Biol. 2004;335:1241–1250. doi: 10.1016/j.jmb.2003.11.038. [DOI] [PubMed] [Google Scholar]
- 25.Gaik M., Flemming D., Hurt E. Structural basis for assembly and function of the Nup82 complex in the nuclear pore scaffold. J. Cell Biol. 2015;208:283–297. doi: 10.1083/jcb.201411003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldmann W.H., Guttenberg Z., Ezzell R.M. Analysis of the F-actin binding fragments of vinculin using stopped-flow and dynamic light-scattering measurements. Eur. J. Biochem. 1998;254:413–419. doi: 10.1046/j.1432-1327.1998.2540413.x. [DOI] [PubMed] [Google Scholar]
- 27.Goldmann W.H., Guttenberg Z., Isenberg G. The study of fast reactions by the stopped flow method. In: Isenberg G., editor. Modern Optics, Electronics, and High Precision Techniques in Cell Biology. Springer-Verlag Berlin-Heidelberg; 1998. pp. 161–172. [Google Scholar]
- 28.Van Holde K.E. Prentice Hall; New Jersey: 1998. Principles of Physical Biochemistry. [Google Scholar]
- 29.Wegner A. Head to tail polymerization of actin. J. Mol. Biol. 1976;108:139–150. doi: 10.1016/s0022-2836(76)80100-3. [DOI] [PubMed] [Google Scholar]
- 30.Lee J.C., Timasheff S.N. The reconstitution of microtubules from purified calf brain tubulin. Biochemistry. 1975;14:5183–5187. doi: 10.1021/bi00694a025. [DOI] [PubMed] [Google Scholar]
- 31.Shelanski M.L., Gaskin F., Cantor C.R. Microtubule assembly in the absence of added nucleotides. Proc. Natl. Acad. Sci. USA. 1973;70:765–768. doi: 10.1073/pnas.70.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pollard T.D. Actin and actin-binding proteins. Cold Spring Harb. Perspect. Biol. 2016;8:a018226. doi: 10.1101/cshperspect.a018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aziz A., Hess J.F., Hunt J.F. The structure of vimentin linker 1 and rod 1B domains characterized by site-directed spin-labeling electron paramagnetic resonance (SDSL-EPR) and X-ray crystallography. J. Biol. Chem. 2012;287:28349–28361. doi: 10.1074/jbc.M111.334011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chernyatina A.A., Nicolet S., Strelkov S.V. Atomic structure of the vimentin central α-helical domain and its implications for intermediate filament assembly. Proc. Natl. Acad. Sci. USA. 2012;109:13620–13625. doi: 10.1073/pnas.1206836109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sokolova A.V., Kreplak L., Strelkov S.V. Monitoring intermediate filament assembly by small-angle x-ray scattering reveals the molecular architecture of assembly intermediates. Proc. Natl. Acad. Sci. USA. 2006;103:16206–16211. doi: 10.1073/pnas.0603629103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldman R.D., Grin B., Kuczmarski E.R. Intermediate filaments: versatile building blocks of cell structure. Curr. Opin. Cell Biol. 2008;20:28–34. doi: 10.1016/j.ceb.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mücke N., Winheim S., Herrmann H. In vitro assembly kinetics of cytoplasmic intermediate filaments: a correlative Monte Carlo simulation study. PLoS One. 2016;11:e0157451. doi: 10.1371/journal.pone.0157451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Portet S. Dynamics of in vitro intermediate filament length distributions. J. Theor. Biol. 2013;332:20–29. doi: 10.1016/j.jtbi.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 39.Brennich M.E., Nolting J.F., Köster S. Dynamics of intermediate filament assembly followed in micro-flow by small angle X-ray scattering. Lab Chip. 2011;11:708–716. doi: 10.1039/c0lc00319k. [DOI] [PubMed] [Google Scholar]
- 40.Saldanha O., Brennich M.E., Köster S. The filament forming reactions of vimentin tetramers studied in a serial-inlet microflow device by small angle x-ray scattering. Biomicrofluidics. 2016;10:024108. doi: 10.1063/1.4943916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berg O.G., von Hippel P.H. Diffusion-controlled macromolecular interactions. Annu. Rev. Biophys. Biophys. Chem. 1985;14:131–160. doi: 10.1146/annurev.bb.14.060185.001023. [DOI] [PubMed] [Google Scholar]
- 42.Premchandar A., Mücke N., Dadlez M. Structural dynamics of the vimentin coiled-coil contact regions involved in filament assembly as revealed by hydrogen-deuterium exchange. J. Biol. Chem. 2016;291:24931–24950. doi: 10.1074/jbc.M116.748145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lundin V.F., Leroux M.R., Stirling P.C. Quality control of cytoskeletal proteins and human disease. Trends Biochem. Sci. 2010;35:288–297. doi: 10.1016/j.tibs.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 44.Perng M.D., Wen S.F., Quinlan R.A. Desmin aggregate formation by R120G alphaB-crystallin is caused by altered filament interactions and is dependent upon network status in cells. Mol. Biol. Cell. 2004;15:2335–2346. doi: 10.1091/mbc.E03-12-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang L., Shav-Tal Y., Goldman R.D. Assembling an intermediate filament network by dynamic cotranslation. J. Cell Biol. 2006;172:747–758. doi: 10.1083/jcb.200511033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soellner P., Quinlan R.A., Franke W.W. Identification of a distinct soluble subunit of an intermediate filament protein: tetrameric vimentin from living cells. Proc. Natl. Acad. Sci. USA. 1985;82:7929–7933. doi: 10.1073/pnas.82.23.7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nöding B., Herrmann H., Köster S. Direct observation of subunit exchange along mature vimentin intermediate filaments. Biophys. J. 2014;107:2923–2931. doi: 10.1016/j.bpj.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ivaska J., Pallari H.M., Eriksson J.E. Novel functions of vimentin in cell adhesion, migration, and signaling. Exp. Cell Res. 2007;313:2050–2062. doi: 10.1016/j.yexcr.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 49.Sawant M., Schwarz N., Leube R.E. Threonine 150 phosphorylation of keratin 5 is linked to epidermolysis bullosa simplex and regulates filament assembly and cell viability. J. Invest. Dermatol. 2017;138:627–636. doi: 10.1016/j.jid.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 50.Sawant M.S., Leube R.E. Consequences of keratin phosphorylation for cytoskeletal organization and epithelial functions. Int. Rev. Cell Mol. Biol. 2017;330:171–225. doi: 10.1016/bs.ircmb.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 51.Snider N.T., Omary M.B. Post-translational modifications of intermediate filament proteins: mechanisms and functions. Nat. Rev. Mol. Cell Biol. 2014;15:163–177. doi: 10.1038/nrm3753. [DOI] [PMC free article] [PubMed] [Google Scholar]