Abstract
Phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2 or PIP2), is a key component of the inner leaflet of the plasma membrane in eukaryotic cells. In model membranes, PIP2 has been reported to form clusters, but whether these locally different conditions could give rise to distinct pools of unclustered and clustered PIP2 is unclear. By use of both fluorescence self-quenching and Förster resonance energy transfer assays, we have discovered that PIP2 self-associates at remarkably low concentrations starting below 0.05 mol% of total lipids. Formation of these clusters was dependent on physiological divalent metal ions, such as Ca2+, Mg2+, Zn2+, or trivalent ions Fe3+ and Al3+. Formation of PIP2 clusters was also headgroup-specific, being largely independent of the type of acyl chain. The similarly labeled phospholipids phosphatidylcholine, phosphatidylethanolamine, phosphatidylserine, and phosphatidylinositol exhibited no such clustering. However, six phosphoinositide species coclustered with PIP2. The degree of PIP2 cation clustering was significantly influenced by the composition of the surrounding lipids, with cholesterol and phosphatidylinositol enhancing this behavior. We propose that PIP2 cation-bridged cluster formation, which might be similar to micelle formation, can be used as a physical model for what could be distinct pools of PIP2 in biological membranes. To our knowledge, this study provides the first evidence of PIP2 forming clusters at such low concentrations. The property of PIP2 to form such clusters at such extremely low concentrations in model membranes reveals, to our knowledge, a new behavior of PIP2 proposed to occur in cells, in which local multivalent metal ions, lipid compositions, and various binding proteins could greatly influence PIP2 properties. In turn, these different pools of PIP2 could further regulate cellular events.
Introduction
Phosphatidylinositol 4,5-bisphosphate, or PI(4,5)P2 (PIP2), is the major phosphoinositide phosphate species in mammalian cells. Located mainly at the plasma membrane (PM) inner leaflet at ∼1–2 mol% (1, 2), PIP2 is a key player in numerous cellular signaling pathways (3). For example, PIP2 hydrolysis is catalyzed by phospholipase C to generate two second messengers: diacylglycerol and inositol 1,4,5-triphosphate (4, 5). PIP2 is also the precursor of another important lipid second messenger, PI(3,4,5)P3 (6). PIP2 is known to interact with hundreds of different proteins, playing important roles in a broad spectrum of cellular functions, including exocytosis/endocytosis (7), endosomal trafficking (8, 9), cytoskeleton assembly (10), cell polarization, cell migration (3), and ion channel control (11). PIP2 also facilitates assembly of viruses such as human immunodeficiency virus 1 (12, 13, 14). What seem to be local membrane enrichments of PIP2 have led to the hypothesis that distinct pools of PIP2 exist in cells (8, 9, 15).
Although PIP2 appears to form clusters in vitro (16), an understanding of this process has remained elusive, in part because the physical basis of this molecular aggregation is unclear. Many previous studies were based on nonphysiological conditions, for example, PIP2 concentrations much greater than the 1–2 mol% in the PM or else bulk lipid compositions, e.g., only phosphatidylcholine (PC) not reflecting the PM inner leaflet where most PIP2 is found in cells (17, 18, 19). One type of PIP2 cluster involves hydrogen-bonding networks revealed by nuclear magnetic resonance studies (20, 21, 22). A different type of PIP2 cluster is based on divalent metal ions Ca2+ or Mg2+ bridging headgroup phosphates (18, 19, 23, 24, 25, 26, 27). Complementing these experimental approaches, molecular dynamics simulations also showed that Ca2+ altered PIP2 properties and induced PIP2 cluster formation (28, 29).
Some evidence suggests that PIP2 cluster formation also occurs in cells or in unfixed (30, 31) or rapidly frozen (32) membrane sheets prepared from cells. PIP2 microdomains have been visualized indirectly primarily by PIP2-specific binding proteins with fluorescent tags, e.g., the pleckstrin homology domain of phospholipase C δ1 or by PIP2 antibodies (31, 33). Most PIP2 studies in cells have focused on PIP2 association driven by cellular PIP2-binding proteins (16, 34, 35), such as myristoylated alanine-rich C-kinase substrate, which appears to modulate the PIP2 distribution at the inner leaflet during various cellular events (2, 16, 36, 37). But whether PIP2 forms clusters in cells without the influence of binding proteins or antibodies is unexplored.
No direct experimental data have been reported that show maximal solubility of free PIP2 in model membranes, above which clustering occurs. Such high-order aggregation is well known for micelle formation (38). As we show, characterizing PIP2 behavior starting at concentrations of >0.1 mol% misses the origin of PIP2-PIP2 association. Here, we used a traditional approach of surface and colloid chemistry to study micelle formation or phase separation (39). We started with a very low PIP2 concentration and examined its self-aggregation behavior over a wide concentration range from 0.01 up to 2 mol%, exploiting the sensitive assays of self-quenching and Fӧrster resonance energy transfer (FRET). Self-association showed a distinct starting concentration that we term the “critical PIP2 concentration” (CPC). The CPC was remarkably low, i.e., 0.02–0.05 mol% of total lipids, in well-defined, chemically simplified model lipid bilayer mixtures that mimic the inner leaflet lipid composition. Multivalent metal ions were absolutely required for this type of PIP2-PIP2 association. Without special precautions, such metal ions typically contaminate buffers and are leached from glass, complicating interpretations of previous reports on PIP2 behavior.
Materials and Methods
Phospholipids and fluorescent probes
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), L-α-phosphatidylinositol (bovine liver PI), L-α-phosphatidylinositol-4,5-bisphosphate (bovine brain-PI(4,5)P2), brain-PI4P, 18:1/18:1 PI3P, PI4P, PI5P, PI(3,4)P2, PI(3,5)P2, PI(4,5)P2, PI(3,4,5)P3, 18:0/20:4 PI(4,5)P2, and fluorescently labeled TopFluor (TF)-PI(4,5)P2 (1-oleoyl-2-{6-[4-(dipyrrometheneboron difluoride)butanoyl]amino}hexanoyl-sn-glycero-3-phosphoinositol-4,5-bisphosphate), TMR-PI(4,5)P2 (1-oleoyl-2-(6-((4,4-difluoro-1,3-dimethyl-5-(4-methoxyphenyl)-4-bora-3a,4a-diaza-s-indacene-2-propionyl)amino)hexanoyl)-sn-glycero-3-phosphoinositol-4.5-bisphosphate; see Fig. S1 for both chemical structures), and similarly chain-labeled TF- and TMR-labeled phosphatidylethanolamine (PE), phosphatidylserine (PS), PC, and phosphatidylinositol (PI) were purchased from Avanti Polar Lipids (Alabaster, AL). 16:0/16:0 PI(4,5)P2 was purchased from Echelon Biosciences (Salt Lake City, UT). Cholesterol was purchased from Nu-Chek Prep (Elysian, MN). Cholesterol stock solution was prepared by standard gravimetric procedures to 0.2% error. Concentrations of all phospholipid stocks were determined to 1% error by inorganic phosphate assay (40). The working stocks of TF- and TMR-labeled PE, PS, and PC were prepared in chloroform; working stocks of TF- and TMR-labeled PI and PIP2 were prepared in chloroform:methanol:H2O = 20:9:1. Fluorescent probe extinction coefficients were obtained from lot certificates of analysis: 97,000 M−1 cm−1 at 496 nm for TF and 56,000 M−1 cm−1 at 544 nm for TMR. Probe concentrations were determined in methanol by absorption spectroscopy using an HP 8452A spectrophotometer (Hewlett-Packard, Palo Alto, CA). Lipid purity of ≥99.5% was confirmed by thin-layer chromatography (TLC). TLC was performed on washed, activated silica gel plates (Alltech, Deerfield, IL) developed with chloroform:methanol:water = 65:25:4 for most phospholipids and for cholesterol with petroleum ether:diethyl ether:chloroform = 7:3:3. Especially useful, TLC plates for PIPs were prerun with 10% K2C2O4 + 2 mM EDTA and then activated at 100°C for 30 min before use. TLC plates for PIPs were developed with chloroform:methanol:4 N NH4OH = 45:35:10 (41, 42).
Liposome preparation
A total of 250 nmol lipid mixtures, including an amount of PIP2 or other PIP for each experiment, were dispensed into each borosilicate culture tube using glass syringes (Hamilton, Reno, NC). Liposomes were prepared using rapid solvent exchange to directly transfer lipid mixtures from organic solvent to aqueous buffer without drying during preparation (43). Samples were sealed under argon at a final lipid concentration of 0.5 mM. Liposomes were then incubated at 55°C for 30 min and then slowly cooled to 23°C overnight with a temperature process controller (Love Controls, series 16A; Dwyer Instruments, Michigan City, IN). Samples were measured the next day. All liposome samples used, except in Fig. 7, were composed of the inner leaflet model POPE/POPS/Chol = 34/30/36.
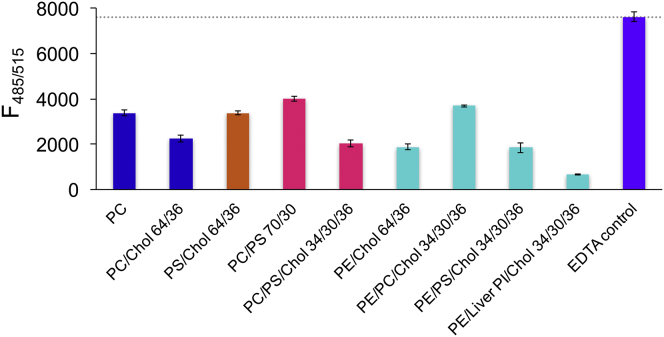
Figure 7.
Lipid composition strongly affects PIP2-cation cluster formation. Bars show fluorescence from a fixed 0.3 mol% TF-PIP2 in model membranes with different lipid compositions. Phospholipid chains were 16:0/18:1 (PO), except for liver PI. Samples were prepared in pure buffer + 0.5 mM Mg2+ and 3 μM each of Al3+, Ca2+, Fe3+, and Zn2+, except for the 1 mM EDTA control. Means and SDs from at least three independent measurements are shown. An EDTA control was measured for every membrane composition; the right-side purple bar shows the average fluorescence of all EDTA controls. To see this figure in color, go online.
Buffer preparation and metal ion measurement
All buffers used were based on 100 mM KCl, 20 mM HEPES (pH = 7.2). “Standard buffers” were prepared with KCl (99%; Sigma-Aldrich, St. Louis, MO), HEPES (99%; Fisher Scientific, Hampton, NH), stored in borosilicate glass bottles, and used in the experiments shown in Figs.1, 2, and S2–S4. What we refer to as “pure buffers” were prepared with KCl (99.999%; Sigma-Aldrich or ACROS (Geel, Belgium)), HEPES (99.5%; Sigma-Aldrich), and stored in Teflon fluorinated ethylene propylene bottles (Nalgene). Water was purified to 18.2 MΩ by passage through a Barnstead MicroPure system (Thermo Fisher, Waltham, MA). Buffers used in the experiments shown in Figs. 3, 4, 5, 6, 7, 8, S5, and S6 were “pure buffers.” Micromolar levels of Al3+, Ca2+, Fe3+, Zn2+, and Mg2+ were prepared from 100 mM stock solutions stored at pH = 2–3. These stocks were made with aluminum chloride (99.999%), calcium chloride (99%), iron (III) chloride (99.9%), zinc chloride (99.995%) and magnesium chloride hexahydrate (99%), all from Sigma-Aldrich. Disodium EDTA (99%; Sigma-Aldrich) was prepared and stored as a 500 mM stock solution at pH = 7.2. Ion concentrations and purities of all stocks and freshly prepared buffers were confirmed by inductively coupled plasma optical emission spectroscopy (ICP-OES) at the Cornell Nutrient Analysis Laboratory using Spectro Arcos ICP-OES (44). The sample size was small enough that no digestion was necessary.
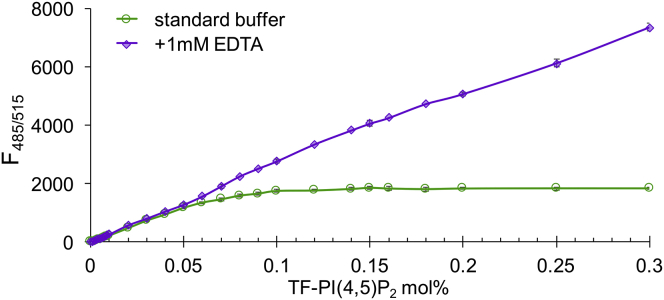
Figure 1.
PIP2 clusters form at very low concentrations, but only in the presence of multivalent cations. Self-quenching occurs when a fluorescent TF-PIP2 concentration reaches CPC. Inner leaflet model membranes were POPE/POPS/Chol (34/30/36). Fluorescence was measured in standard buffer (green) or the same buffer plus 1 mM EDTA (purple) at 23°C. Assays were performed in triplicate with error bars of SDs from the means. Ex/Em slits were 5/5 nm in all experiments except as noted.
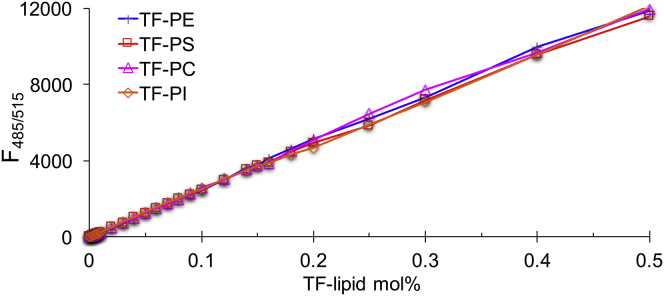
Figure 2.
Other TF-labeled phospholipids do not form multivalent cation-dependent clusters. Fluorophore-labeled lipids were incorporated into inner leaflet model membranes with conditions as in Fig. 1. TF-PE (blue), TF-PS (red), TF-PC (magenta), TF-PI (orange) are shown.
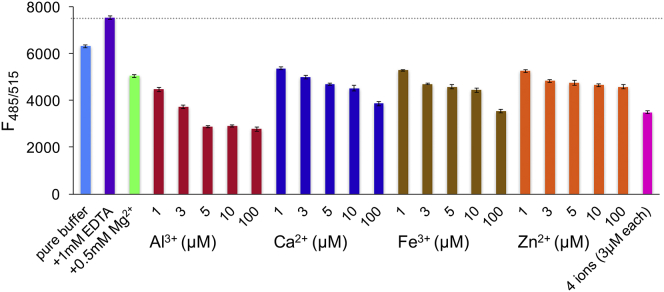
Figure 3.
Various multivalent metal ions drive TF-PIP2 to form clusters. Each bar shows fluorescence from a fixed 0.3 mol% of TF-PIP2 in inner leaflet model membranes as in Fig. 1 except for differences in ionic conditions. See Materials and Methods for a description of highly purified “pure buffer.” Pure buffer, or pure buffer plus the following was used: 1 mM EDTA; 0.5 mM Mg2+; 1, 3, 5, 10, or 100 μM each of Al3+, Ca2+, Fe3+, and Zn2+; or a combination of 3 μM each of Al3+, Ca2+, Fe3+, and Zn2+. Error bars show SDs from the mean from at least three independent measurements.
Figure 4.
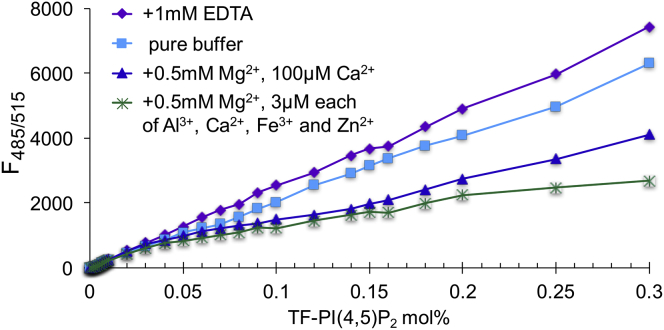
Physiological metal ion conditions can also cause strong PIP2 clustering. Analysis as in Fig. 1 measured in three buffer conditions is shown: pure buffer +1 mM EDTA (purple, diamond), pure buffer (light blue, square), pure buffer + 0.5 mM Mg2+ and 100 μM Ca2+ (dark blue, triangle), and pure buffer + 0.5 mM Mg2+ and 3 μM each of Al3+, Ca2+, Fe3+, and Zn2+ (green, asterisk).
Figure 5.
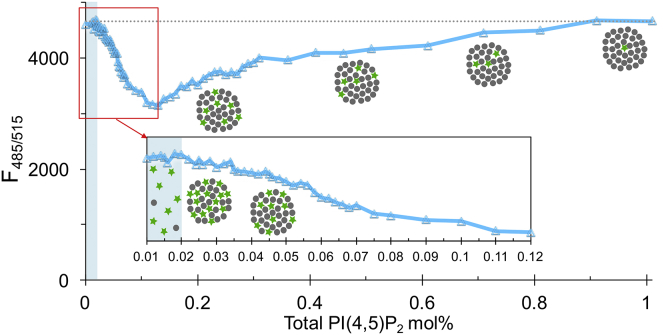
PIP2 cluster formation starts at a low, well-defined PIP2 concentration. Each point is the fluorescence from a fixed 0.01 mol% TF-PIP2 in POPE/POPS/Chol (34/30/36) with additional unlabeled brain-PIP2 from 0.002 to 1 mol%. The X axis indicates total PIP2 from 0.01 up to 1.01 mol%. All samples are in pure buffer with 0.5 mM Mg2+ and 3 μM of each Al3+, Ca2+, Fe3+, and Zn2+. The inset shows expansion of the 0.01–0.12 mol% data. Schematic illustrations show that PIP2 stays dispersed at very low concentrations and then forms clusters just above 0.02 mol%. At that point, most of the fluorescence is from the free TF-PIP2, and fluorescence from clusters is significantly quenched. As more brain-PIP2 (black circles) is added, TF-PIP2 (green stars) is diluted, leading to dequenching in each cluster. The assay was repeated three times independently with similar results.
Figure 6.
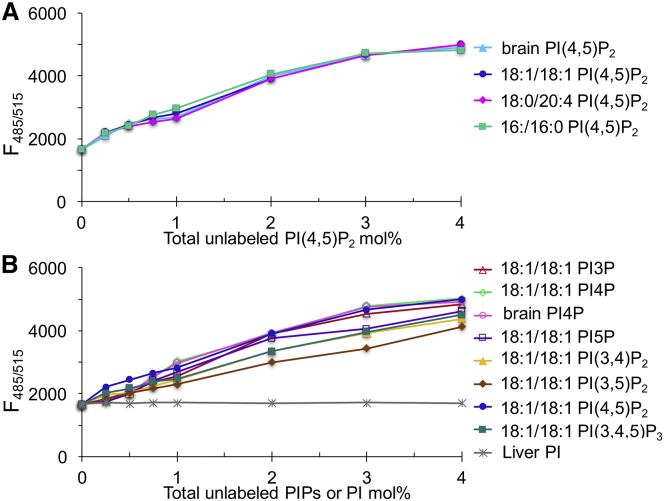
Effects of acyl chain type and PIP headgroup on coclustering with PIP2. (A) Acyl chains do not influence PIP2 incorporation into clusters. Each line corresponds to a fixed 0.2 mol% TF-PIP2 in POPE/POPS/Chol (34/30/36), with increasing amounts of unlabeled PIP2 in pure buffer plus 0.5 mM Mg2+ and 3 μM each of Al3+, Ca2+, Fe3+, and Zn2+, brain-PI(4,5)P2 (cyan), 18:1-PI(4,5)P2 (blue), 18:0/20:4-PI(4,5)P2 (magenta), and 16:0-PI(4,5)P2 (green). (B) Other PIP species enter PIP2 clusters, but PI does not. Assay as in (A) but with additional unlabeled PI or PIPs is shown; liver PI (gray), brain-PI4P (pink), or the 18:1/18:1 species PI3P (red), PI4P (lime), PI5P (purple), PI(3,4)P2 (yellow), PI(3,5)P2 (brown), PI(4,5)P2 (blue), and PI(3,4,5)P3 (green) are shown. Small differences in PIPs coclustering were reproducible in independent measurements.
Figure 8.
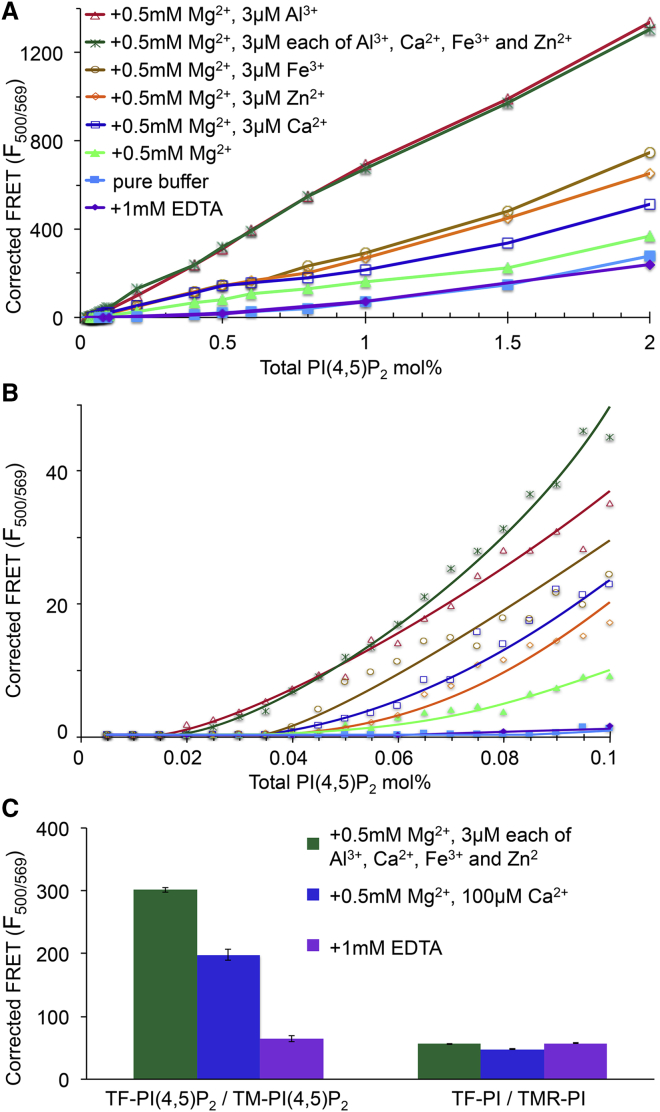
FRET detects cation dependence of PIP2 cluster formation. (A) Corrected FRET is shown (see Materials and Methods) as a function of PIP2 concentration in POPE/POPS/Chol (34/30/36). PIP2 mixtures were 98% brain-PIP2 + 1% of the donor TF-PIP2 and 1% of the acceptor TMR-PIP2. Ex/Em slits were 5/10 nm with a temperature of 23°C. (B) Expansion is shown of the low concentration region of (A) with PIP2 from 0.005 to 0.1 mol%. (C) Physiological levels of Ca2+ and Mg2+ lead to strong clustering of 2 mol% PIP2 but not PI. Corrected FRET signals for 2 mol% total PIP2 measured as in (A), in parallel with 2% total PIP2 and PI, of which 98% was brain-PIP2 and 1% each was TF-PI and TMR-PI. Buffer conditions were 0.5 mM Mg2+ and 3 μM each of Al3+, Ca2+, Fe3+, and Zn2+ (green), 0.5 mM Mg2+ and 100 μM Ca2+ (blue), and 1 mM EDTA (purple). Ex/Em slits were 5/5 nm. Bars represent averages from at least three independent measurements; error bars represent SDs from the mean.
Self-quenching and FRET
A 1.8 mL volume of buffer was added to 0.2 mL of a 0.5 mM liposome sample to yield 50 μM total lipid in the cuvette. Fluorescence was collected on a Hitachi F-7000 FL spectrofluorimeter (Hitachi High Technologies America, Schaumburg, IL) at 23°C. Wavelengths used for self-quenching studies were (ex/em) as follows: TF (485/515 nm), TMR (544/569 nm), and light scattering (440/420 nm). For FRET studies, corrections were made for the non-FRET contributions of direct excitation of TMR acceptors and donor TF fluorescence bleedthrough as well as light scattering by the vesicle suspension. Data were collected with slits for ex/em = 5/5 nm and a 10 s integration time, except for 10/10 nm slits for experiments shown in Fig. 5 and 5/10 nm for experiments shown in Fig. 8, A and B.
Results
PIP2 cluster formation is driven by multivalent metal ions
We exploited fluorescence self-quenching to detect PIP2 cluster formation with inner leaflet lipid model composition using POPE/POPS/Chol (34/30/36). We used the synthetic fluorescent PIP2 analog TF-PIP2 (see Fig. S1 for chemical structure) to examine PIP2 distribution in membranes. As shown by (19), self-quenching of TF-PIP2 can reveal PIP2-PIP2 association. In our standard buffer, increasing the concentration of TF-PIP2 led to a linear increase of fluorescence only up to ∼0.02 mol% of total lipids (Fig. 1). From 0.03 to 0.04 mol%, fluorescence increased weakly, implying fluorescence quenching via fluorophore-fluorophore interactions, and above 0.1% fluorescence increased by a little, apparently because of strong self-quenching. We interpret these results to reflect formation of tightly packed PIP2 in clusters. In stark contrast, the presence of 1 mM EDTA caused TF-PIP2 self-quenching to disappear completely, even up to 0.3 mol%, with only a linear increase in fluorescence being observed as the TF-PIP2 fraction increased. This EDTA effect implies that multivalent metal ions are present in the standard buffer and are required to induce such PIP2 cluster formation.
To confirm that cluster formation is specific to PIP2, we examined the self-quenching profiles of TF-labeled PE, PS, PC, and PI in the same standard buffer (Fig. 2). Increasing the concentrations of these labeled lipids from 0 to 0.5 mol% resulted in a linear increase of fluorescence, i.e., a lack of self-quenching and thus no cluster formation in this concentration range. EDTA only eliminated the self-quenching of TF-PIP2, but has a negligible effect on the fluorescence profile of other lipids such as TF-PC at these concentrations (Fig. S2). This behavior of TF-labeled lipids other than PIP2 means that the appearance of TF-PIP2 self-quenching in Fig. 1 at low concentrations is specific to PIP2 and not caused by the acyl chain fluorophore. TF-PC did undergo self-quenching above 2–3 mol% (Fig. S3). To examine whether unlabeled PIP2 exhibits similar cation-bridged clustering, we diluted TF-PIP2 twofold with brain-PIP2 or with 18:/18:1- or 18:0/20:4-PIP2 (Fig. S4). TF-PIP2 still showed a CPC at ∼0.02–0.03 mol%, which was consistent with unlabeled PIP2 forming clusters just like TF-labeled PIP2. As further confirmation, TMR-PIP2 (see Fig. S1 for chemical structure), which carries a slightly different acyl chain fluorescent label, also exhibited strong self-quenching at the same concentration as TF-PIP2, whereas again other TMR-labeled lipids did not self-quench (Fig. S5). These results clearly demonstrate that cluster formation is an intrinsic property of PIP2 rather than an artifact due to the TF or TMR moiety.
The striking effect of EDTA to eliminate PIP2 self-quenching shows that multivalent metal ions are crucial to PIP2 cluster formation. To examine which metal ions were present in the standard buffer prepared with reagent-grade chemicals, we used ICP-OES (44) to identify and quantitate these metal ions. The primary multivalent metal ions in the standard buffer were Ca2+ (57 μM), Al3+ (10 μM), Zn2+ (6 μM), and Fe3+ (0.1 μM), shown in Table 1. We then examined the effects of these four candidate metal ions on PIP2 cluster formation, individually and in combination. These experiments required buffers free of multivalent metal ions down to the submicromolar level, so we prepared a “pure buffer” from ultrapure chemicals, storing this in Teflon bottles to avoid leaching of metal ions from glass (45). ICP-OES analysis showed that contaminating metal ions in pure buffer stored in Teflon bottles was <0.1 μM (Table 1). This pure buffer was used in experiments shown in Figs. 3, 4, 5, 6, 7, and 8, with or without the addition of multivalent metal ions or EDTA.
Table 1.
Multivalent Metal Ion Analysis of Buffers by ICP-OES
| Metal ion [μM] | Al3+ | Ca2+ | Fe3+ | Mg2+ | Zn2+ |
|---|---|---|---|---|---|
| MQ H2O | – | – | – | – | – |
| Standard buffer (in glass) | 10 | 57 | 0.1 | – | 6 |
| Pure buffer (in glass) | 0.2 | 7 | – | 16 | 0.2 |
| Pure buffer (in teflon) | – | 0.1 | – | – | 0.1 |
| Pure buffer+ 0.5 mM Mg2+/3 μM each of Al3+, Ca2+, Fe3+, and Zn2+ (in teflon) | 3 | 3 | 3 | 490 | 3 |
| 0.5 mM Mg2+ | – | – | – | 500 | – |
| 1 mM Al3+ | 1050 | 0.1 | – | – | 0.1 |
| 1 mM Ca2+ | – | 980 | – | – | 0.1 |
| 1 mM Fe3+ | 0.5 | 0.5 | 1020 | – | 0.1 |
| 1 mM Zn2+ | – | 0.1 | 1 | – | 1010 |
Standard buffer was prepared with 99% KCI, 99% HEPES; pure buffer was prepared with 99.999% KCI, 99.5% HEPES. Multivalent metal ions are diluted from 100 mM stocks. –, undetectable (<0.1 μM).
We tested 1, 3, 5, 10, and 100 μM of Al3+, Ca2+, Fe3+, and Zn2+ added to pure buffer. Because the cytosol contains ∼0.5 mM free Mg2+ in many cell types (46), we also tested this Mg2+ concentration. Based on the self-quenching experiment shown in Fig. 1 for TF-PIP2, we chose a defined PIP2 concentration of 0.3 mol% at which to compare each metal ion for PIP2 cluster formation (Fig. 3). We interpret a reduced fluorescence relative to the maximal fluorescence of the EDTA control to reflect PIP2 cluster formation. In this assay, 0.3 mol% TF-PIP2 in pure buffer exhibited 84% of the fluorescence observed in the EDTA control, which was consistent with the submicromolar concentrations of multivalent metal ions still present in this buffer. Fig. 3 shows that as little as an additional 1 μM of either Al3+, Ca2+, Fe3+, or Zn2+ caused significant further self-quenching. Among the four candidate multivalent metal ions tested, Al3+ at 5 μM promoted PIP2 cluster formation most strongly, giving the lowest fluorescence. Combining 3 μM each of Al3+, Ca2+, Fe3+, and Zn2+ yielded similar PIP2 cluster formation compared with that of 3 μM Al3+ alone. With 0.5 mM Mg2+ as the only multivalent cation, TF-PIP2 exhibited 67% of the maximal fluorescence, implying that physiological levels of Mg2+ cause PIP2 to form loose clusters. We also tested the effect of Mg2+ from 0.05 to 0.5 mM in the presence of 0.5 mM EGTA to eliminate all other multivalent metal ions. TF-PIP2 cluster promotion by Mg2+ was found to be concentration dependent, i.e., the higher the [Mg2+], the greater the TF-PIP2 self-quenching observed (Fig. S6).
Switching to pure buffer containing known amounts of multivalent metal ions, we found TF-PIP2 self-quenching profile was similar to that shown in Fig. 1, which was carried out in the standard buffer. We tested the combination of 0.5 mM Mg2+ with 3 μM each of Al3+, Ca2+, Fe3+, and Zn2+. With this mixture, TF-PIP2 exhibited strong self-quenching starting at 0.02 mol%. Although Al3+ is not a cytosolic ion of interest, we examined it here because of its strong promotion of PIP2 clustering. We also tested a more physiological buffer containing 0.5 mM Mg2+ together with 100 μM Ca2+, which mimics the transient calcium influx environment near the inner leaflet of the PM (Fig. 4). Physiological levels of Ca2+ and Mg2+ are sufficient to cause strong TF-PIP2 quenching, confirming that the PIP2 clustering we are observing here is biologically relevant. Considering that the buffer containing 0.5 mM Mg2+ and 3 μM each of Al3+, Ca2+, Fe3+, and Zn2+ still causes the strongest PIP2 clustering, we used this mixed ion buffer for experiments shown in Figs. 5, 6, 7, and 8 to maximize the self-quenching and FRET signals.
High-resolution examination of PIP2 cluster formation
A simple model to describe cluster formation posits that at very low concentrations, PIP2 exists essentially as individual molecules freely diffusing in the membrane. At the CPC, these PIP2 self-associate, analogous to micelle or phase formation. To test this model, we developed a dilution experiment based on reversible fluorescence self-quenching. We reasoned that if unlabeled PIP2 behaves just like TF-PIP2, then gradually increasing the concentration of unlabeled PIP2 in the presence of a constant TF-PIP2 concentration would have these effects. 1) First, the unlabeled PIP2 would dilute the unclustered TF-PIP2 without changing the fluorescence when total PIP2 is still below the CPC, i.e., a perfectly horizontal line of fluorescence versus total PIP2 concentration. 2) Then, upon reaching the CPC, the unlabeled PIP2 would drive self-association, abruptly reducing fluorescence. 3) Finally, at higher concentrations, the unlabeled PIP2 would dilute the fluorescent PIP2 molecules that are in the clusters, relieving self-quenching and therefore leading to an increase in fluorescence. In the experiment shown in Fig. 5, each of the 68 points represents a liposome sample with a fixed 0.01 mol% of TF-PIP2, together with increasing concentrations of brain-PIP2, from 0 to 1.0 mol%. The initial 0.01 mol% TF-PIP2 was not in clusters, still being dispersed in the membranes (Fig. 4). As unlabeled brain PIP2 was added, up to a total PIP2 concentration of 0.02 mol% (Fig. 5, shaded region in inset), TF-PIP2 fluorescence was constant, still at its maximal unquenched level, as shown pictorially by the dispersed green dots in the cartoon. When total PIP2 exceeded that concentration, TF-PIP2 self-quenching began, as evidenced by decreasing fluorescence, implying cluster formation (Fig. 5, circle of green and black dots in cartoon). Between 0.02 and 0.13 mol%, TF fluorescence continuously decreased, indicating significant self-quenching even with the ratio of labeled to unlabeled PIP2 within the cluster being in the range of 0.1. When the concentration of total PIP2 reached ∼0.13 mol%, additional brain PIP2 diluted the TF-PIP2 in each cluster enough that fluorescence again increased. Eventually the fluorescence fully recovered to exactly its initial unquenched value. This unique self-quenching profile supports the conclusion that PIP2 self-associates, forming cation-bridged clusters.
Self-association of PIP2 is headgroup-specific
To study whether acyl chain types affect PIP2 cluster formation, we compared four PIP2 species differing in acyl chain length and saturation: 18:1/18:1, 18:0/20:4, 16:0/16:0, and the natural brain-PIP2, which contains a mixture of acyl chains (Fig. 6 A). We made use of the finding shown in Fig. 1 and Fig. 4 that at 0.2 mol%, TF-PIP2 is highly quenched in the presence of multivalent metal ions. Each point in Fig. 6 A represents an individual liposome sample that contained 0.2 mol% of TF-PIP2 together with additional unlabeled PIP2 species, ranging in concentration from 0.25 to 4 mol% of total lipids. If the unlabeled PIP2 were to cocluster with TF-PIP2, then TF-PIP2 would be diluted in each cluster by the unlabeled PIP2. All four unlabeled PIP2 species diluted the highly quenched TF-PIP2 in exactly the same way. These results imply that the clustering behavior is not influenced by the hydrophobic portion of the PIP2 molecule.
We also tested whether other PIP species can cocluster with PIP2 (Fig. 6 B). PI phosphate (PI3P, PI4P, PI5P), PI bisphosphate (PI(3,4)P2, PI(3,5)P2, PI(4,5)P2), and PI trisphosphate (PI(3,4,5)P3) were studied using the same dilution assay as in Fig. 6 A, with a fixed 0.2 mol% of TF-PIP2 and varying concentrations of PI or the seven PIP species. PI did not dilute the TF-PIP2 signal and thus did not cocluster with PIP2. In contrast, all PIP species coclustered with PIP2 to similar degrees. We found these small differences to be reproducible in independent measurements. That PI phosphate can cocluster but PI cannot shows that one additional phosphate group on the inositol ring is sufficient for cation-bridged clustering to occur. In summary, coclustering with PIP2 requires only PIP headgroups and is insensitive to acyl chains.
Membrane composition affects PIP2 cation cluster formation
To explore the PIP2 mixing behavior in different lipid environments, we examined PIP2 cluster formation in mixtures besides POPE/POPS/Chol. We compared the self-quenching of 0.3 mol% PIP2 in different model membranes in pure buffer containing 0.5 mM Mg2+ together with 3 μM each of Ca2+, Al3+, Fe3+, and Zn2+ (Fig. 7). Similar to the experiments shown in Fig. 3, the lower the fluorescence at this fixed mol% of TF-PIP2 compared to the maximal fluorescence in the EDTA control, the stronger the PIP2 cluster formation. The results showed that PIP2 cation cluster formation was indeed dependent on bulk lipid composition. We found notable promotion of PIP2 cation clustering by cholesterol and by PI: 1) the presence of 36 mol% cholesterol in mixtures with POPC and POPC/POPS favored cluster formation. That cholesterol strongly enhances cluster formation implies either of two quite different mechanisms: 1) cholesterol does not enter the PIP2 cation clusters but instead promotes immiscibility of PIP2 by increasing its chemical potential, decreasing the fraction of free, unquenched TF-PIP2; or 2) cholesterol does enter and condense PIP2 cation clusters, promoting fluorescence quenching. The latter is less likely, given the polyunsaturation of brain-PIP2 and the monounsaturation of all other lipids except for liver-PI in this experiment and the known preference of cholesterol to be surrounded by saturated acyl chains. We note that studies in the absence of multivalent cations favor the interpretation that cholesterol binds to and stabilizes multivalent cation-free PIP2 clusters (20); in POPE/X/Chol (34/30/36), the strongest self-quenching was observed when X = liver-PI with a value of 9% maximal fluorescence. We interpret this result to imply an especially low PIP2 solubility in the presence of PI, which is a known major component of the PM cytosolic leaflet. Compared with the simple PC composition used by many researchers, the sixfold greater quenching of TF-PIP2 fluorescence in POPE/liver-PI/Chol shows that bulk membrane composition is a major factor in the formation of PIP2 cation clusters.
PIP2 self-association is detected with high sensitivity by FRET
We used an independent experimental method, FRET, to study PIP2 cation-bridged cluster formation (47). This technique is sufficiently sensitive to detect PIP2 at the low concentrations below the CPC. The FRET assay has several advantages. First, most of the PIP2 is unlabeled brain-PIP2, with the donor fluorophore TF-PIP2 and the acceptor fluorophore TMR-PIP2 each comprising only 1% of the total PIP2. Second, the form of the FRET signal enables a more precise identification of the CPC where PIP2 self-association abruptly starts. Liposome samples were prepared with varying fractions of total PIP2, each with a constant ratio of 98/1/1 = brain-PIP2/TF-PIP2/TMR-PIP2. At these low fluorophore fractions, the donor TF-PIP2 and the acceptor TMR-PIP2 showed no self-quenching (data not shown). Eight different ionic conditions were studied (Fig. 8 A). We observed modest but clear PIP2 cluster formation by 0.5 mM Mg2+ with a CPC of 0.04 mol%. This effect of Mg2+ is consistent with that observed by self-quenching (Fig. 3). Adding an additional 3 μM of any of the multivalent metal ions to the 0.5 mM Mg2+ resulted in a stronger FRET signal and lower CPC, which was again consistent with the self-quenching data (Fig. 8 B). With 3 μM Al3+ and 0.5 mM Mg2+, the strongest FRET signals were observed, and the lowest CPC = 0.02 mol% of total lipids. Combining 3 μM each of Al3+, Ca2+, Fe3+, and Zn2+ with 0.5 mM Mg2+ did not further increase the FRET or lower the CPC. These FRET results reveal two different kinds of evidence of PIP2 cation interactions. First, the magnitude of the FRET signal at 2 mol% of total PIP2 is different for the different multivalent metal ions (Fig. 8 A), with the largest FRET signals found in the presence of Al3+ and 100 μM Ca2+ and a far smaller FRET signal from Mg2+. A greater FRET signal might indicate closer packing of PIP2 fluorophores within a tight cation-bridged cluster compared to a looser cluster. Second, the remarkably low PIP2 CPC of 0.02–0.04 mol% indicates high-affinity interaction of the PIP2 with the cation. Consistently, Al3+ showed the highest affinity for PIP2. At the physiological level of 2 mol% total PIP2, we also measured FRET between TF-PIP2 and TMR-PIP2 in the physiological buffer that contains 0.5 mM Mg2+ + 10 μM Ca2+ (in Fig. 8 C, note that narrower slits were used than in Fig. 8, A and B). This FRET signal is ∼3-fold greater than that in the 1 mM EDTA control buffer, slightly lower than the maximal FRET signal in 0.5 mM Mg2+ + 3 μM each of the four other ions. Under these near-physiological conditions, FRET is in agreement with the self-quenching results shown in Fig. 4 for PIP2-forming clusters. As a control FRET experiment in which clustering is not expected, TF-PI and TMR-PI replaced the two similarly labeled PIP2 species in each buffer condition. The PI FRET pair showed the same background FRET signals in all three buffers, i.e., no effect of EDTA or metal ion, confirming that PI does not form clusters, which was consistent with the self-quenching results of Fig. 2. We emphasize that the background FRET signals with EDTA we observed here (above 0.5 mol% total PIP2) might arise from loose clusters formed by hydrogen-bonding networks, as suggested by previous studies (20, 21, 22). In summary, the quantitative agreement of the self-quenching and FRET assays indicates that they are reporting on the same phenomenon of cation-bridged cluster formation.
Discussion
We have discovered a fundamental feature of PIP2 behavior that occurs in a wide range of model lipid bilayers and that is likely to be important for understanding PIP2 function in cells: in the presence of micromolar concentrations of multivalent cations or of a physiological 0.5 mM Mg2+ concentration, PIP2 molecules freely diffuse in the bilayer only up to 0.02–0.05 mol% of total lipid. Starting at this low CPC, additional PIP2 molecules form clusters bridged by multivalent metal ions, in equilibrium with free PIP2 in the bilayer. An abrupt onset of PIP2-PIP2 contacts at the same PIP2 concentration appears in both the self-quenching experiment of Fig. 5 and the FRET experiment of Fig. 8. This sudden start of self-association is a characteristic of high-order aggregation, as in micelle formation or phase separation (39). However, with the data obtained so far, we cannot distinguish whether PIP2 self-association represents a phase within the bilayer or else a high-order aggregation, as with micelles. PIP2 cluster formation does not depend on acyl chain species and excludes the phospholipids PE, PC, PS, and PI, which themselves show little or no clustering. In contrast, the six other known phosphoinositide species cocluster with the cation-bridged PIP2 clusters.
An abrupt onset of FRET signals implies that some type of cluster has formed distinct from the bulk lipid and into which the previously very dilute dyes have become concentrated. Although the clusters form abruptly at low PIP2 concentrations of 0.02–0.05 mol%, the incremental FRET increase with added PIP2 gradually reaches a linear increase at > ∼0.1 mol% up to the physiological range of 1–2 mol%. A similar result is found for self-quenching. For both FRET and self-quenching, the linear increase with increasing PIP2 mol% above the CPC implies that all additional PIP2 has the same properties, with larger or more numerous clusters but of the same material.
Thus, although the PIP2 concentration in the PM cytosolic leaflet is reported to be 1–2 mol%, if our model membranes reflect the behavior of cell membranes in the presence of multivalent cations, a majority of this PIP2 could be tied up in PIP2 cation clusters. The chemical potential of PIP2 in this cluster would be low. That is, the PIP2 tendency to bind or react should be equal to that of the free PIP2, which is at a concentration of only 2–5 molecules per 10,000 lipids, almost two orders of magnitude lower than might be expected based on its fraction within the PM cytosolic leaflet. Or, from another point of view, a change in the local concentration of multivalent cations such as Ca2+, from submicromolar to perhaps tens of micromolar, can be viewed in two ways. 1) In that region of the PM where PIP2 cation clusters form, the thermodynamic activity of PIP2 drops significantly, e.g., 25- to 50-fold from 1 to 0.04 or 0.02 mol%. (2) In that same region of the PM, cation-bridged clusters form, on which some proteins assemble, for example, viral Gag proteins. In other words, some kinds of PIP2-controlled behavior would decrease significantly, but other kinds could increase when PIP2 forms clusters.
For several reasons, previous studies with model membranes were not able to uncover the remarkable PIP2 behavior described here. First, those studies focused on membranes containing PIP2 at, or in many cases above, the 1–2 mol% present in the PM cytosolic leaflet. According to our findings, PIP2 at greater than ∼0.1 mol% of total lipids in the presence of multivalent metal ions would exist in these PIP2 cation clusters. Only very sensitive techniques, such as those based on fluorescence, are able to report on PIP2 behavior in the range of concentrations where we have shown PIP2 self-association to begin. Second, many other studies did not account for the effects of multivalent metal ions, which typically are present as contaminants in the micromolar range and which we now know to be critical in promoting clustering. Some published experiments, especially those in the presence of EDTA, studied what one might refer to as “free PIP2.” Such PIP2 could still be hydrogen bonded in a network but would be expected to be less tightly associated than the PIP2 cation clusters that we describe here.
PIP2 self-association is robust in the sense of not being dependent on a particular multivalent cation. Al3+ had the strongest influence on cluster formation, and Mg2+ had the weakest, but the variety of cations that promote cluster formation, which include Ca2+, Zn2+, and Fe3+, implies that the two-dimensional array of the many PIP2 phosphate groups creates a strong cation-binding matrix. Physiological levels of 100 μM Ca2+ and 0.5 mM Mg2+ are sufficient to cause strong PIP2 clustering, which implies that PIP2 is most likely to be clustered during transient cellular calcium influx. We do not yet have a molecular model to explain the mechanism by which multivalent metal ions promote clustering, with some ions such as Al3+ being more effective than others such as Mg2+. One possibility is that the quantitatively lesser self-quenching and FRET promoted by Mg2+, compared with some other multivalent ions, might reflect more loosely packed PIP2, with other lipid molecules in the cluster hindering direct contact between PIP2 molecules. Another possibility is that the cluster size differs depending on the ion. Fluorescent PIP2 molecules at the periphery of a cluster presumably would be less quenched, and the fraction of molecules at the periphery would be larger for clusters with dimensions of a few nanometers than for ones with micron size. However, it is important to note that the critical concentration at which clustering begins is similar for all of the ions tested, i.e, in the range of 0.02–0.05 mol% of total lipid. In forming a stable cation-bridged cluster, PIP2 cation behavior is like the well-known inorganic chemical behavior of the strong association of multivalent cations with phosphate itself (48). It is noteworthy that even with the repulsive effect of the approximately four negative charges under physiological conditions (21, 49, 50), PIP2 molecules still strongly attract each other by multivalent metal ion bridging of headgroup phosphates. This attraction would affect PIP2 physical chemical properties and spatial distribution in membranes.
Much previous research was based on especially simple lipid compositions. Our finding that bulk lipid composition affects clustering has consequences for the behavior of PIP2 in cells. That PIP2 cation clustering depends on the type and concentration of multivalent cations and on the composition of the surrounding lipids has an interesting implication. The concentration at which clusters form is also the concentration of free PIP2 in the bilayer in equilibrium with clustered PIP2. If compositionally distinct regions occur in the cytosolic leaflet of the PM and if multivalent metal ion type or concentration is spatially localized, for example as in transient Ca2+ or Zn2+ influx at certain sites (51, 52), different local concentrations of free PIP2 would result. Consistent with this idea, the original suggestion of “membrane rafts” was based on experimental findings of distinct compositional regions, apical and basolateral, in intestinal epithelial cell PMs (53).
Understanding how PIP2 exists as “spatially separated pools” in cellular membranes will be an important underpinning of analysis of function. A large number of cellular proteins interact with PIP2. The proteins that bind to free PIP2 would experience decreased function when PIP2 cation-bridged clusters form, whereas those that bind preferentially to such clusters would experience enhanced function. Carefully designed experiments with model membranes and purified proteins could define such preferences for individual proteins. Elucidating the status of the PIP2 molecules in the PM is critical to understanding the cell biological behavior resulting from protein binding. The methods that we report here can be used to examine even better and more complex models of the PM, including PIP2 within a membrane model with additional components, such as PE/PS/PI/PC/Chol, and including the polyunsaturated acyl chains abundant in natural PE and PS.
Author Contributions
Y.W., V.M.V., and G.W.F. designed the research. Y.W. conducted all experiments and analyzed the data. Y.W., V.M.V., and G.W.F. wrote the manuscript.
Acknowledgments
We thank the Cornell Nutrient Analysis Laboratory and Michael A. Rutzke and Tatyana Dokuchayeva for help with ICP-OES data collection and analysis. We thank Robert A. Dick for providing helpful advice.
This work was supported by National Institutes of Health grants 2R01GM107013 (V.M.V.) and R01GM105684 (G.W.F.).
Editor: Kalina Hristova.
Footnotes
Six figures and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)30573-3.
Supporting Material
References
- 1.Ferrell J.E., Jr., Huestis W.H. Phosphoinositide metabolism and the morphology of human erythrocytes. J. Cell Biol. 1984;98:1992–1998. doi: 10.1083/jcb.98.6.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McLaughlin S., Wang J., Murray D. PIP(2) and proteins: interactions, organization, and information flow. Annu. Rev. Biophys. Biomol. Struct. 2002;31:151–175. doi: 10.1146/annurev.biophys.31.082901.134259. [DOI] [PubMed] [Google Scholar]
- 3.Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhee S.G. Regulation of phosphoinositide-specific phospholipase C. Annu. Rev. Biochem. 2001;70:281–312. doi: 10.1146/annurev.biochem.70.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berridge M.J., Irvine R.F. Inositol trisphosphate, a novel second messenger in cellular signal transduction. Nature. 1984;312:315–321. doi: 10.1038/312315a0. [DOI] [PubMed] [Google Scholar]
- 6.Cantley L.C. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 7.Martin T.F. PI(4,5)P2-binding effector proteins for vesicle exocytosis. Biochim. Biophys. Acta. 2015;1851:785–793. doi: 10.1016/j.bbalip.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simonsen A., Wurmser A.E., Stenmark H. The role of phosphoinositides in membrane transport. Curr. Opin. Cell Biol. 2001;13:485–492. doi: 10.1016/s0955-0674(00)00240-4. [DOI] [PubMed] [Google Scholar]
- 9.Martin T.F. PI(4,5)P(2) regulation of surface membrane traffic. Curr. Opin. Cell Biol. 2001;13:493–499. doi: 10.1016/s0955-0674(00)00241-6. [DOI] [PubMed] [Google Scholar]
- 10.Raucher D., Stauffer T., Meyer T. Phosphatidylinositol 4,5-bisphosphate functions as a second messenger that regulates cytoskeleton-plasma membrane adhesion. Cell. 2000;100:221–228. doi: 10.1016/s0092-8674(00)81560-3. [DOI] [PubMed] [Google Scholar]
- 11.Suh B.C., Hille B. Regulation of ion channels by phosphatidylinositol 4,5-bisphosphate. Curr. Opin. Neurobiol. 2005;15:370–378. doi: 10.1016/j.conb.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Ono A., Ablan S.D., Freed E.O. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc. Natl. Acad. Sci. USA. 2004;101:14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saad J.S., Muriaux D.M. Editorial: role of lipids in virus assembly. Front. Microbiol. 2015;6:410. doi: 10.3389/fmicb.2015.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mücksch F., Laketa V., Kräusslich H.-G. Synchronized HIV assembly by tunable PIP2 changes reveals PIP2 requirement for stable Gag anchoring. eLife. 2017;6:e25287. doi: 10.7554/eLife.25287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinchliffe K.A., Ciruela A., Irvine R.F. PIPkins1, their substrates and their products: new functions for old enzymes. Biochim. Biophys. Acta. 1998;1436:87–104. doi: 10.1016/s0005-2760(98)00140-4. [DOI] [PubMed] [Google Scholar]
- 16.Brown D.A. PIP2Clustering: from model membranes to cells. Chem. Phys. Lipids. 2015;192:33–40. doi: 10.1016/j.chemphyslip.2015.07.021. [DOI] [PubMed] [Google Scholar]
- 17.Redfern D.A., Gericke A. pH-dependent domain formation in phosphatidylinositol polyphosphate/phosphatidylcholine mixed vesicles. J. Lipid Res. 2005;46:504–515. doi: 10.1194/jlr.M400367-JLR200. [DOI] [PubMed] [Google Scholar]
- 18.Levental I., Christian D.A., Janmey P.A. Calcium-dependent lateral organization in phosphatidylinositol 4,5-bisphosphate (PIP2)- and cholesterol-containing monolayers. Biochemistry. 2009;48:8241–8248. doi: 10.1021/bi9007879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarmento M.J., Coutinho A., Fernandes F. Ca(2+) induces PI(4,5)P2 clusters on lipid bilayers at physiological PI(4,5)P2 and Ca(2+) concentrations. Biochim. Biophys. Acta. 2014;1838:822–830. doi: 10.1016/j.bbamem.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 20.Jiang Z., Redfern R.E., Gericke A. Cholesterol stabilizes fluid phosphoinositide domains. Chem. Phys. Lipids. 2014;182:52–61. doi: 10.1016/j.chemphyslip.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kooijman E.E., King K.E., Gericke A. Ionization properties of phosphatidylinositol polyphosphates in mixed model membranes. Biochemistry. 2009;48:9360–9371. doi: 10.1021/bi9008616. [DOI] [PubMed] [Google Scholar]
- 22.Graber Z.T., Jiang Z., Kooijman E.E. Phosphatidylinositol-4,5-bisphosphate ionization and domain formation in the presence of lipids with hydrogen bond donor capabilities. Chem. Phys. Lipids. 2012;165:696–704. doi: 10.1016/j.chemphyslip.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y.H., Collins A., Janmey P.A. Divalent cation-induced cluster formation by polyphosphoinositides in model membranes. J. Am. Chem. Soc. 2012;134:3387–3395. doi: 10.1021/ja208640t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slochower D.R., Wang Y.H., Janmey P.A. Counterion-mediated pattern formation in membranes containing anionic lipids. Adv. Colloid Interface Sci. 2014;208:177–188. doi: 10.1016/j.cis.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y.H., Slochower D.R., Janmey P.A. Counterion-mediated cluster formation by polyphosphoinositides. Chem. Phys. Lipids. 2014;182:38–51. doi: 10.1016/j.chemphyslip.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levental I., Cebers A., Janmey P.A. Combined electrostatics and hydrogen bonding determine intermolecular interactions between polyphosphoinositides. J. Am. Chem. Soc. 2008;130:9025–9030. doi: 10.1021/ja800948c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carvalho K., Ramos L., Picart C. Giant unilamellar vesicles containing phosphatidylinositol(4,5)bisphosphate: characterization and functionality. Biophys. J. 2008;95:4348–4360. doi: 10.1529/biophysj.107.126912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slochower D.R., Huwe P.J., Janmey P.A. Quantum and all-atom molecular dynamics simulations of protonation and divalent ion binding to phosphatidylinositol 4,5-bisphosphate (PIP2) J. Phys. Chem. B. 2013;117:8322–8329. doi: 10.1021/jp401414y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ellenbroek W.G., Wang Y.H., Liu A.J. Divalent cation-dependent formation of electrostatic PIP2 clusters in lipid monolayers. Biophys. J. 2011;101:2178–2184. doi: 10.1016/j.bpj.2011.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Honigmann A., van den Bogaart G., Jahn R. Phosphatidylinositol 4,5-bisphosphate clusters act as molecular beacons for vesicle recruitment. Nat. Struct. Mol. Biol. 2013;20:679–686. doi: 10.1038/nsmb.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van den Bogaart G., Meyenberg K., Jahn R. Membrane protein sequestering by ionic protein-lipid interactions. Nature. 2011;479:552–555. doi: 10.1038/nature10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujita A., Cheng J., Fujimoto T. A distinct pool of phosphatidylinositol 4,5-bisphosphate in caveolae revealed by a nanoscale labeling technique. Proc. Natl. Acad. Sci. USA. 2009;106:9256–9261. doi: 10.1073/pnas.0900216106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Várnai P., Balla T. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J. Cell Biol. 1998;143:501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laux T., Fukami K., Caroni P. GAP43, MARCKS, and CAP23 modulate PI(4,5)P(2) at plasmalemmal rafts, and regulate cell cortex actin dynamics through a common mechanism. J. Cell Biol. 2000;149:1455–1472. doi: 10.1083/jcb.149.7.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McLaughlin S., Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- 36.Glaser M., Wanaski S., McLaughlin S. Myristoylated alanine-rich C kinase substrate (MARCKS) produces reversible inhibition of phospholipase C by sequestering phosphatidylinositol 4,5-bisphosphate in lateral domains. J. Biol. Chem. 1996;271:26187–26193. doi: 10.1074/jbc.271.42.26187. [DOI] [PubMed] [Google Scholar]
- 37.Gambhir A., Hangyás-Mihályné G., McLaughlin S. Electrostatic sequestration of PIP2 on phospholipid membranes by basic/aromatic regions of proteins. Biophys. J. 2004;86:2188–2207. doi: 10.1016/S0006-3495(04)74278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis B.M., Richens J.L., O’Shea P. Label-free critical micelle concentration determination of bacterial quorum sensing molecules. Biophys. J. 2011;101:245–254. doi: 10.1016/j.bpj.2011.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Israelachvili J.N. Academic press; Cambridge, MA: 2011. Intermolecular and Surface Forces. [Google Scholar]
- 40.Kingsley P.B., Feigenson G.W. The synthesis of a perdeuterated phospholipid: 1,2-dimyristoyl-Sn-glycero-3-phosphocholine-D72. Chem. Phys. Lipids. 1979;24:135–147. [Google Scholar]
- 41.Gonzalez-Sastre F., Folch-Pi J. Thin-layer chromatography of the phosphoinositides. J. Lipid Res. 1968;9:532–533. [PubMed] [Google Scholar]
- 42.Andrews W.V., Conn P.M. Measurement of inositol phospholipid metabolites by one-dimensional thin-layer chromatography. Methods Enzymol. 1987;141:156–168. doi: 10.1016/0076-6879(87)41064-1. [DOI] [PubMed] [Google Scholar]
- 43.Buboltz J.T., Feigenson G.W. A novel strategy for the preparation of liposomes: rapid solvent exchange. Biochim. Biophys. Acta. 1999;1417:232–245. doi: 10.1016/s0005-2736(99)00006-1. [DOI] [PubMed] [Google Scholar]
- 44.Rutzke M.A. Encyclopedia of Geochemistry. Second Edition. Springer International Publishing; 2017. Atomic absorption, inductively coupled plasma optical emission spectroscopy, and infrared spectroscopy; pp. 1–8. [Google Scholar]
- 45.Jang H.K., Chung Y.D., Lee S. Effects of chemical etching with hydrochloric acid on a glass surface. J. Vac. Sci. Technol. A. 2000;18:2563–2566. [Google Scholar]
- 46.Romani A. Regulation of magnesium homeostasis and transport in mammalian cells. Arch. Biochem. Biophys. 2007;458:90–102. doi: 10.1016/j.abb.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 47.Loura L.M., Prieto M. FRET in membrane biophysics: an overview. Front. Physiol. 2011;2:82. doi: 10.3389/fphys.2011.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rashchi F., Finch J.A. Polyphosphates: a review their chemistry and application with particular reference to mineral processing. Miner. Eng. 2000;13:1019–1035. [Google Scholar]
- 49.Toner M., Vaio G., McLaughlin S. Adsorption of cations to phosphatidylinositol 4,5-bisphosphate. Biochemistry. 1988;27:7435–7443. doi: 10.1021/bi00419a039. [DOI] [PubMed] [Google Scholar]
- 50.van Paridon P.A., de Kruijff B., Wirtz K.W. Polyphosphoinositides undergo charge neutralization in the physiological pH range: a 31P-NMR study. Biochim. Biophys. Acta. 1986;877:216–219. doi: 10.1016/0005-2760(86)90137-2. [DOI] [PubMed] [Google Scholar]
- 51.Que E.L., Bleher R., O’Halloran T.V. Quantitative mapping of zinc fluxes in the mammalian egg reveals the origin of fertilization-induced zinc sparks. Nat. Chem. 2015;7:130–139. doi: 10.1038/nchem.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parekh A.B., Penner R. Store depletion and calcium influx. Physiol. Rev. 1997;77:901–930. doi: 10.1152/physrev.1997.77.4.901. [DOI] [PubMed] [Google Scholar]
- 53.Simons K., van Meer G. Lipid sorting in epithelial cells. Biochemistry. 1988;27:6197–6202. doi: 10.1021/bi00417a001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.