Abstract
Several studies have shown that low expression of hZIP1 is closely associated with many human cancers, including clear cell renal cell carcinoma (ccRCC). In this study, we aimed to explore the potential mechanism responsible for hZIP1 silencing and revealed a novel regulatory pathway in the pathogenesis of ccRCC. Here, miR-223 was predicted and experimentally validated to be a regulator of hZIP1, and its expression was negatively correlated with the mRNA levels of hZIP1 in primary tumors. Upregulation of hZIP1 inhibited cell proliferation, cell cycle progression, and invasion and induced apoptosis, while inhibition of miR-223 showed the opposite effect on cellular processes. Moreover, GAS5 interacted with miR-223 and was markedly downregulated in tumors. Knockdown of GAS5 partially reversed the effect of the miR-223 inhibitor on cell proliferation, cell cycle distribution, apoptosis and invasion. In addition, GAS5 acted as a molecular sponge to positively regulate the mRNA and protein levels of hZIP1 via regulating miR-223. The tumorigenicity of ccRCC cells was enhanced by silencing GAS5 but diminished by overexpression of hZIP1 in vivo. Clinically, the low expression of hZIP1 was significantly correlated with advanced clinical stage and Fuhrman stage. Downregulation of GAS5 indicated tumor progression and recurrence and was independently associated with disease-free survival of patients. Taken together, our results suggest that GAS5 may act as a competing endogenous RNA (ceRNA) to regulate hZIP1 by sponging miR-223 in the progression of ccRCC and that targeting the GAS5/miR-223/hZIP1 axis may serve as a therapeutic strategy for patients.
Keywords: hZIP1, miR-223, GAS5, clear cell renal cell carcinoma, proliferation, apoptosis, invasion
Introduction
Renal cell carcinoma (RCC) is the third most common urological tumor, representing 3% of all cancers in adults [1]. Among all RCC cases, clear cell renal cell carcinoma (ccRCC) is the major histological subtype (70%~80%). In 2013, an estimated 67,000 newly diagnosed cases in China occurred, with the reported incidence rate increasing by 2.5% annually [2]. Therefore, acquiring a better understanding of the pathogenesis underlying ccRCC is important and may provide new therapeutic strategies and effective diagnostic and prognostic biomarkers.
Several studies have demonstrated that hZIP1, a zinc uptake transporter, is frequently silenced in prostate cancer [3]. Overexpression of hZIP1 inhibits the malignant potential of prostate cancer cells [4]. A recent study reported that hZIP1 had persistently low expression in mucinous carcinomas, including ovarian, colon, stomach, and pulmonary carcinoma [5]. Our previous data also found that the protein levels of hZIP1 were significantly downregulated in ccRCC and correlated with tumor stage, Fuhrman stage, and recurrence [6]. However, the underlying mechanism by which hZIP1 is dysregulated in the progression of ccRCC has not yet been studied.
MicroRNAs (miRNAs) are widely accepted to play critical roles in the progression and metastasis of tumors, including ccRCC [7-9]. MiRNAs bind to the 3’-untranslated region (3’-UTR) of target mRNAs via complementarily base pairing and thus act as oncogenes or tumor suppressors in human malignancy [10]. For example, miR-144-3p contributes to cell growth, migration, invasion, and chemoresistance in ccRCC by targeting ARID1A [11]. miR-122 can function as a tumor suppressor in gastric cancer by targeting CREB1 [12]. miR-93-3p promotes the progression of ccRCC by regulating PEDF and may serve as a prognostic factor for patients [13]. Furthermore, increasing studies have shown that long noncoding RNAs (lncRNAs) influence the suppressive effect of miRNAs on protein-coding genes by acting as molecular sponges or competing endogenous RNAs (ceRNAs). For example, downregulation of lncRNA MALAT1 can suppress cell growth by increasing miR-124 and reducing STAT3 expression in lung cancer [14]. The lncRNA XIST exerts a suppressive role on cellular proliferation and metastasis by downregulating miR-23a and subsequently enhancing the expression of RKIP in prostate cancer [15]. In the present study, we aimed to reveal the potential lncRNAs and miRNAs that may be responsible for the dysregulation of hZIP1 in ccRCC.
In this study, we found that miR-223 could affect the mRNA and protein levels of hZIP1 in ccRCC cells. The downregulation of hZIP1 was inversely correlated with miR-223 in tumors. Functional analyses confirmed that overexpression of hZIP1 inhibited proliferation, cell cycle progression, and invasion and induced apoptosis in vitro, which could be reversed by miR-223. Upon further study, we predicted and validated GAS5 as a ceRNA for miR-223. Inhibition of GAS5 attenuated the effect of the miR-223 inhibitor on cell growth, apoptosis, and invasion. In addition, knockdown of GAS5 facilitated tumor growth in vivo, which was abolished by overexpression of hZIP1. Taken together, our data provide a novel mechanism responsible for the downregulation of the hZIP1 and GAS5/miR-223/hZIP1 axis as a new therapeutic strategy for ccRCC.
Materials and methods
Cell culture and patients
Human ccRCC cell lines A498 and 786-O were purchased from the Cell Biology of the Chinese Academy of Sciences (Shanghai, China). All ccRCC cell lines were maintained in DMEM (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS) (Sigma, St. Louis, MO, USA), 100 U/mL penicillin and 100 mg/mL streptomycin at 37°C in a humidified chamber supplemented with 5% CO2.
A total of 55 paired ccRCC tissues and adjacent nontumor kidney tissues were obtained from patients who had undergone surgery in the Department of Urology at the First Hospital of China Medical University from 2011-2012. None of the patients received radio- or chemotherapy before surgery. All protocols in this study were approved by the Institutional Ethics Committee of China Medical University, and informed consent was signed by patients before surgery. Clinical information was obtained from medical records. All tissue samples were frozen immediately in liquid nitrogen and then stored at -80°C until use. Histopathological diagnoses of all specimens were validated by a pathologist. A lack of tumor recurrence during the follow-up period was regarded as disease-free survival (DFS).
Quantitative real-time PCR (qRT-PCR)
Total RNA from cells and tissues was isolated using TRIzol reagent (Invitrogen) or the mirVana miRNA Isolation Kit (Applied Biosystems, Foster City, CA, USA). M-MLA reverse transcriptase (Invitrogen) was used for lncRNA reverse transcription. The PrimeScript RT reagent Kit (Takara, Japan) and the SYBR PrimeScript miRNA RT-PCR Kit were used to generate complementary DNA of mRNA and miRNA, respectively. The expression of lncRNA and mRNA was measured using SYBR Green Master Mix (Applied Biosystem) and performed on an ABI 7500 system, and GAPDH was used as the internal control. The expression of miRNA was measured using TaqMan MicroRNA Assays (Thermo, Waltham, MA, USA), and U6 was used as an internal control. The PCR primers were as follows: GAS5 forward, 5’-CCCAAGGAAGGATGAG-3’ and reverse, 5’-ACCAGGAGCAGAACCA-3’; hZIP1 forward, 5’-GCTGTTGCAGAGCCACCTTA-3’ and reverse, 5’-CATGCCCTCTAGCACAGACTG-3’. miR-223 forward, 5’-GTGCAGGGTCCGAGGT-3’ and reverse, 5’-CGGGCTGTCAGTTTGTCA-3’. Quantitative analysis was calculated using the 2-ΔΔCt method.
Cell transfection
Cell transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. A hZIP1 overexpression plasmid was generated by subcloning hZIP1 cDNA (NCBI RefSeq record: NM_014437) into the expression plasmid pcDNA™5/FRT/TO (Invitrogen, Xhol-EcoRV restriction sites) and named pc-hZIP1. The empty vectors were used as controls and named pc-NC. The miRNA-223 mimic, mimic negative control (mim-NC), miRNA-223 inhibitor, and inhibitor negative control (inh-NC) were purchased from GeneChem (Shanghai, China). The specific interference sequence (5’-CTTGCCTGGACCAGCTTAATT-3’) for GAS5 (si-GAS5) was used to knockdown the expression of GAS5, and the control siRNA was named si-NC. The shRNA against GAS5 (sh-GAS5, 5’-CUUGCCUGGACCAGCUUAAUU-3’) was ligated into the pGPU6/GFP/Neo vector. Empty vectors were used as a negative control and named sh-NC. Stable cell lines were created through selection by means of geneticin (G418; Sigma, St Louis, MO, USA) and used in xenograft tumor analysis.
Cell proliferation assay
The MTT assay was used to assess cell proliferation ability. Transfected cells (3000 cells/well) were seeded into 96-well plates, and 20 μL of the MTT solution (5 mg/mL, Sigma) was added to each well at the indicated time point for 4 h. Then, 150 μL DMSO was added to stop the reaction, and the absorbance was read at 490 nm on a microplate reader (Bio Tek, Winooski, VT, USA).
Cell cycle and apoptosis analysis
For cell cycle analysis, cells were harvested and fixed in 70% ethanol overnight. Then, cells were added with RNase (50 μg/mL) and PI (50 μg/mL) (BD, San Jose, CA, USA). After incubation for 30 min, samples were subjected to flow cytometry (FACScan, BD) for analysis. Data were analyzed using CELL Quest 3.0 software (BD).
Apoptosis was detected using an Annexin V-FITC Apoptosis Detection Kit (Beyotime, China). Transfected cells were collected, centrifuged, and resuspended in PBS. After removal of the supernatant, Annexin V binding buffer was used to resuspend the cells. Following double staining with Annexin-V/FITC (5 μL) and PI (10 μL) and incubating at room temperature in the dark for 15 min, the cells were analyzed using a FACSCalibur cytometer (Becton Dickinson, Franklin, NJ, USA).
Cell invasion assay
After 48 h of transfection, ccRCC cells were incubated without FBS in the upper chamber of an insert, which was precoated with Matrigel (BD). The lower chamber contained medium with 10% FBS. After incubation for 24 h, the cells on the upper surface were removed, and the cells on the lower surface were fixed and stained with 0.1% crystal violet in a dark room. Strained cells from at least five random fields were counted.
Luciferase reporter assay
The 3’-UTR of hZIP1 that contained the wild-type or mutant putative binding site of miR-223 was inserted into the psiCHECK2 vector (Promega, Madison, WI, USA). A total of 2.0×104 786-O cells were seeded into 96-well plates and cotransfected with wild-type or mutant hZIP1 reporter plasmid and the miR-223 mimic or mim-NC using Lipofectamine 2000 (Invitrogen). To test the relationship between GAS5 and miR-223, cells were cotransfected with either the miR-223 mimic or mim-NC, psiCHECK Luciferase Expression Reporter (Promega) containing the wild-type (or mutant) 3’-UTR of GAS5. Luciferase activities were measured 48 h after transfection by Dual-Luciferase Assay (Promega) according to the manufacturer’s manual.
Animal experiment
The animal study protocol was approved by the Animal Experimentation Ethics Committee of China Medical University. Briefly, twenty male athymic BALB/c nude mice (4-5 weeks old) were provided by Beijing Laboratory Animal Research Center (Beijing, China), housed in standard conditions, and randomly assigned to the following groups: blank control group, sh-GAS5 group, pc-hZIP1 group, and sh-GAS5+pc-hZIP1 group. Each group included five mice. A total of 5×106 transfected 786-O cells were subcutaneously inoculated into the flanks of mice. The tumor volume was monitored weekly for 4 weeks and calculated using the formula Volume (mm3) = 0.5 × Length × Width2. Finally, the mice were sacrificed, and the tumors were extracted to determine tumor weight.
Western blot analysis
Cells were lysed in SDS lysis buffer (Beyotime, Shanghai, China), and proteins were separated using 12% SDS-PAGE. Then, proteins were transferred onto PVDF membranes (Bio-Rad, Hercules, CA, USA) and blocked for 30 min at room temperature. Membranes were incubated with primary antibodies against hZIP1 (1:2000; ab105416; Abcam, Cambridge, MA, USA) and GAPDH (1:1,000; ab9485; Abcam) at 4°C overnight followed by incubation with HRP-conjugated secondary antibodies. Blots were detected using an ECL detection system.
Statistical analysis
Data were expressed as the mean ± SEM from at least 3 independent experiments. The expression levels of hZIP1, miR-223, and GAS5 in tumors and nontumor tissues were compared using the paired-samples t test. A Cox proportional hazards model was used to assess the prognostic value of the clinicopathological variables for DFS. P<0.05 was considered statistically significant.
Results
The expression of hZIP1 was regulated by miR-223 in ccRCC cells
Bioinformatics databases (TargetScan and miRDB) were used to predict putative miRNAs that may regulate the expression of hZIP1. As shown in Figure 1A, miR-223 was predicted as a potential regulatory factor for hZIP1. The luciferase activity assay showed that the miR-223 mimic significantly reduced wild-type hZIP1 3’-UTR reporter activity compared with the mimic negative control but had no suppressive effect on mutant hZIP1 3’-UTR reporter activity. Cells overexpressing miR-223 exhibited significant downregulation of hZIP1 expression at the mRNA level and the protein level compared with cells transfected with mimic negative control (Figure 1C and 1D). These results implied that miR-223 suppressed hZIP1 expression by binding to the 3’-UTR of hZIP1 mRNA.
Figure 1.
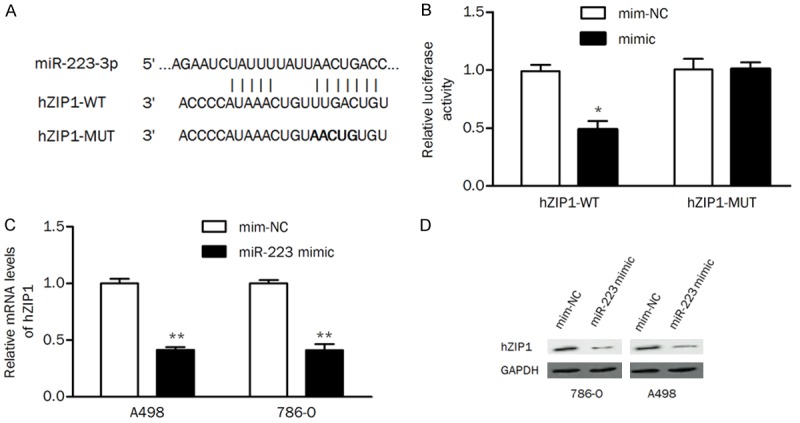
hZIP1 is a direct target of miR-223. A. The putative miR-223 binding site in the 3’-UTR region of hZIP1. B. 786-O cells were cotransfected with miR-223 (or mim-NC) and hZIP1-WT (or hZIP1-MUT) 3’-UTR luciferase reporter constructs, and the luciferase activity was tested. C. hZIP1 mRNA levels were examined by qRT-PCR. D. Protein levels of hZIP1 were detected by western blot in ccRCC cells transfected with the miR-223 mimic. mim-NC, mimic negative control; *P<0.05, **P<0.01 compared with the control group.
miR-223 was inversely correlated with hZIP1 in ccRCC tissues
A QRT-PCR assay was performed to validate the expression of miR-223 and hZIP1 in ccRCC clinical samples. The mRNA expression of hZIP1 was significantly lower in primary tumor tissues than in adjacent normal tissues (P<0.001, Figure 2A). The expression of miR-223 was significantly upregulated in tumors compared with that in nontumor tissues (P=0.003, Figure 2B). The mRNA levels of hZIP1 were significantly negatively correlated with miR-223 expression in tumors (Pearson r=-0.46, P=0.0004; Figure 2C), suggesting that miR-223 may regulate hZIP1 expression in the development of ccRCC.
Figure 2.
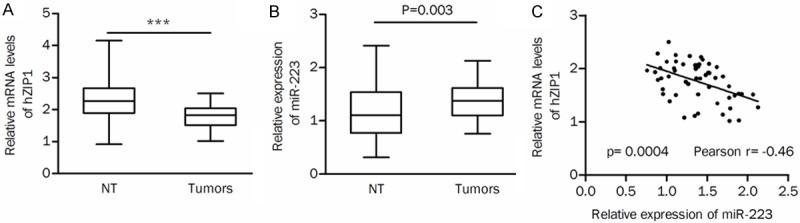
The expression of hZIP1 and miR-223 in clinical tissue samples. A. The mRNA levels of hZIP1 were significantly decreased in tumors. B. The expression of miR-223 was markedly upregulated in tumors compared with that in adjacent nontumor tissues. C. hZIP1 mRNA levels were negatively correlated with miR-223 levels in tumors. NT, nontumor tissues; ***P<0.001.
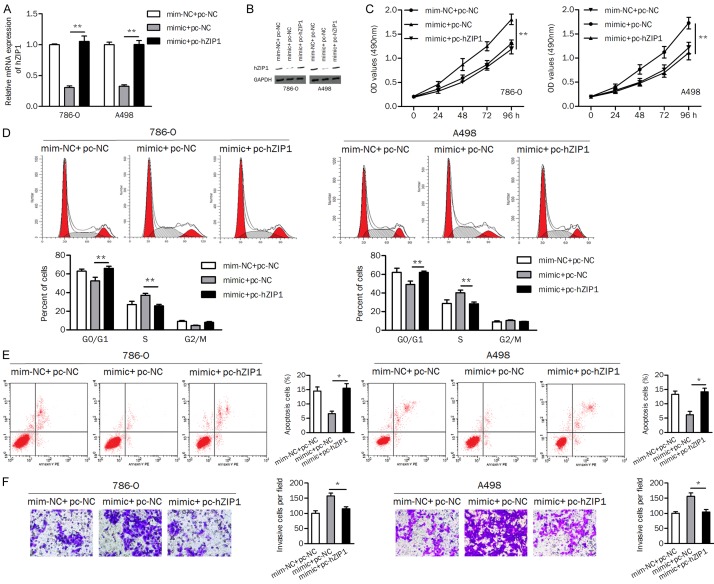
Overexpression of hZIP1 reverses the promotive effect of miR-223 on cellular processes
In this study, we investigated whether miR-223 regulates biological processes by targeting hZIP1. The results of qRT-PCR and western blotting showed that ccRCC cells transfected with the miR-223 mimic had markedly decreased hZIP1 expression, while hZIP1 expression was restored in cells cotransfected with the miR-223 mimic plus the hZIP1 plasmid (Figure 3A and 3B).
Figure 3.
miR-223 exerts an oncogenic role by targeting hZIP1 in ccRCC cells. (A and B) The mRNA and protein levels of hZIP1 were verified by qRT-PCR (A) and western blotting (B) assays, respectively. (C) Overexpression of miR-223 promoted cell proliferation, and overexpression of hZIP1 reversed this effect. (D) Upregulation of hZIP1 abolished the stimulatory effect of miR-223 on cell cycle progression. (E) The apoptosis of ccRCC cells was decreased by miR-223 but increased by hZIP1. (F) hZIP1 attenuated the miR-223-induced increase in cell invasion. pc-hZIP1, hZIP1 overexpression vector; pc-NC, empty vector control; mim-NC, mimic negative control; *P<0.05, **P<0.01.
As shown in Figure 3C, overexpression of miR-223 increased the cell proliferation rate, which was reversed by cotransfection with miR-223 and hZIP1 in ccRCC cells. miR-223 mimic induced the cell cycle transition from G1 phase to S phase, while cotransfection with the miR-223 mimic and hZIP1 induced cell cycle arrest at the G1 phase (Figure 3D). Moreover, overexpression of hZIP1 in 786-O and A498 cells attenuated the inhibitory effect of miR-223 on apoptosis (Figure 3E). In addition, miR-223 increased the number of invasive cells, while hZIP1 displayed the opposite effect (Figure 3F). These data suggested that miR-223 exerted an oncogenic role in ccRCC by downregulating hZIP1.
GAS5 is a direct target of miR-223
Increasing studies have shown that lncRNAs act as molecular “sponges” or ceRNAs to regulate miRNAs and downstream protein-coding genes. Thus, we searched for putative interacting lncRNAs based on the online database StarBase v2.0. GAS5 was predicted to be one of the potential interacting lncRNAs of miR-223 (Figure 4A). As shown in Figure 4B, miR-223 repressed the luciferase activity of wild-type GAS5 but not that of the mutant, indicating a direct binding between GAS5 and miR-223. Furthermore, we found that knockdown of GAS5 significantly increased the expression of miR-223 in ccRCC cells (Figure 4C). In contrast, the expression of GAS5 was inhibited by the miR-223 mimic but enhanced by the miR-223 inhibitor (Figure 4D). The expression of GAS5 was also detected in tumors and normal tissues. The results of qRT-PCR revealed that the expression of GAS5 was significantly decreased in tumor tissues compared to that in normal control tissues (Figure 4E). The expression of GAS5 was inversely correlated with the expression levels of miR-223 in tumors (Pearson r=-0.303, P=0.024; Figure 4F). Collectively, these results indicated that GAS5 was a direct target of miR-223.
Figure 4.
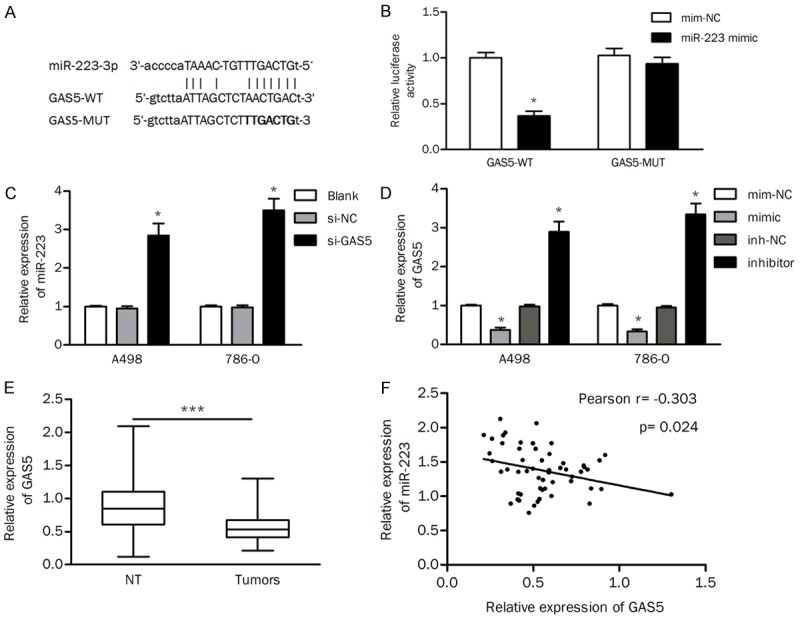
GAS5 regulates the expression of miR-223. A. The binding site between GAS5 and miR-223 was predicted. B. Luciferase activity of reporters containing the GAS5-WT or GAS5-MUT sequence in 786-O cells transfected with the miR-223 mimic or mimic negative control (mim-NC). C. Effects of GAS5 knockdown on the expression of miR-223. D. The expression of GAS5 in ccRCC cells transfected with the miR-223 mimic or inhibitor. E. The expression of GAS5 was reduced in tumors compared with that in nontumor tissues. F. Pearson correlation analysis was performed to detect the relationship between GAS5 and miR-223 in cancer tissues. inh-NC, inhibitor negative control; NT, nontumor tissues; *P<0.05; ***P<0.001.
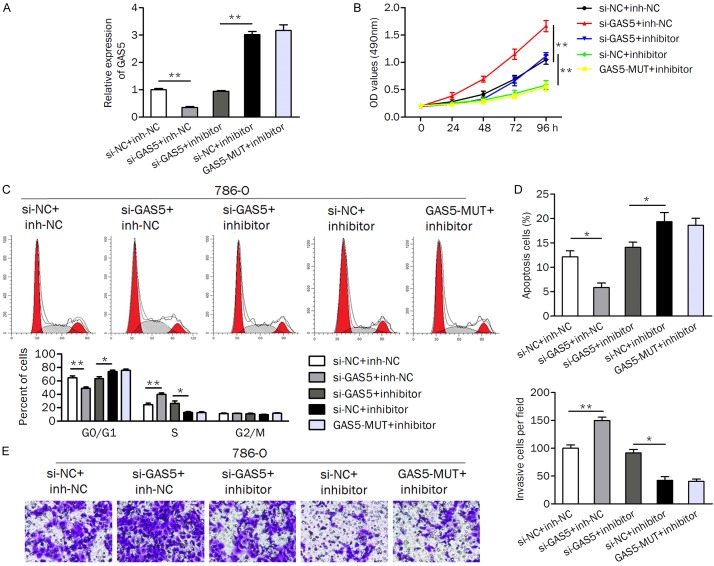
The GAS5/miR-223 axis regulated cell growth, apoptosis, and invasion
786-O cells were cotransfected with si-GAS5 (or si-NC) and the miR-223 inhibitor (or inhibitor control) to evaluate the regulatory effect of GAS5/miR-223 on biological processes. The cells transfected with mutant GAS5 (GAS5-MUT) and the miR-223 inhibitor were used as negative controls. QRT-PCR was performed to determine the expression of GAS5 in cells (Figure 5A). Knockdown of GAS5 significantly upregulated the miR-223 inhibitor-induced decrease in the cell proliferation rate (Figure 5B). Likewise, the introduction of si-GAS5 contributed to cell cycle progression from the G0/G1 phase to S phase and reversed cell cycle arrest at the G0/G1 phase induced by the miR-223 inhibitor (Figure 5C). For apoptosis analysis, knockdown of GAS5 significantly suppressed cell apoptosis regardless of the miR-223 inhibitor (Figure 5D). In addition, si-GAS5 promoted the cell invasion ability (Figure 5E).
Figure 5.
Knockdown of GAS5 attenuates the effect of the miR-223 inhibitor on cellular processes. (A) The expression of GAS5 was measured in cells by qRT-PCR. (B) Inhibition of GAS5 impaired the miR-223 inhibitor-induced reduction in the cell proliferation rate. (C and D) Cell cycle distribution (C) and apoptosis (D) analysis of cells after cotransfection with the miR-223 inhibitor (or inh-NC) and si-GAS5 (si-NC) was detected. (E) Knockdown of GAS5 promoted cell invasion, while inhibition of miR-223 suppressed it. si-GAS5, specific siRNA for knocking down GAS5; inh-NC, inhibitor negative control; si-NC, negative control siRNA. GAS5-MUT, mutant GAS5. *P<0.05; **P<0.01.
GAS5 was positively correlated with hZIP1
We intended to investigate whether GAS5 modulates the expression of hZIP1 through “sponging” miR-223. Inhibition of miR-223 led to an upregulation of the mRNA and protein levels of hZIP1, which could be reversed by si-GAS5 in ccRCC cells (Figure 6A and 6B). The mRNA and protein expression of hZIP1 was also reduced in the si-GAS1+inh-NC group compared to the expression in the si-NC+inh-NC group, implying a positive correlation between GAS5 and hZIP1. Additionally, Pearson’s correlation analysis showed that GAS5 expression was positively correlated with hZIP1 expression in ccRCC tissues, but this correlation failed to attain statistical significance (Pearson r=0.258, P=0.056; Figure 6C). Our findings suggested that GAS5 might positively modulate the expression of hZIP1 by sponging miR-223.
Figure 6.
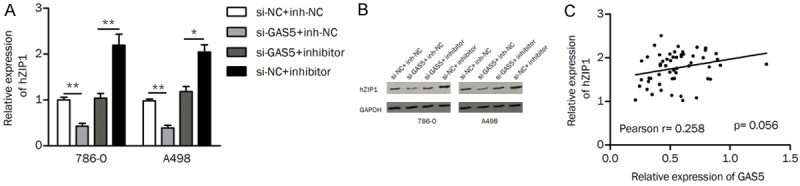
The effect of GAS5 on the expression of hZIP1. A. The mRNA level of hZIP1 was detected by qRT-PCR. B. The protein level of hZIP1 was decreased in the si-GAS5+inhibitor group compared to that in the si-NC+inhibitor group. C. GAS5 was positively correlated with hZIP1 expression in tumor tissues; *P<0.05; **P<0.01.
hZIP1 is a downstream effector of GAS5 in the formation of xenograft tumors
Considering that GAS5 positively regulates hZIP1, we aimed to verify whether hZIP1 is a downstream effector of GAS5 in vivo. After transfection with sh-GAS5, the expression level of GAS5 was significantly reduced in 786-O cells (Figure 7A). Experiments using nude mice indicated that overexpression of hZIP1 inhibited tumor growth in vivo. Knockdown of GAS5 contributed to mouse tumor growth, while cotransfection with hZIP1-expressing plasmids abolished this effect (Figure 7B). The tumor volume and weight of mice in the pc-hZIP1+sh-GAS5 group were significantly smaller than those in the sh-GAS5 group (Figure 7C and 7D).
Figure 7.
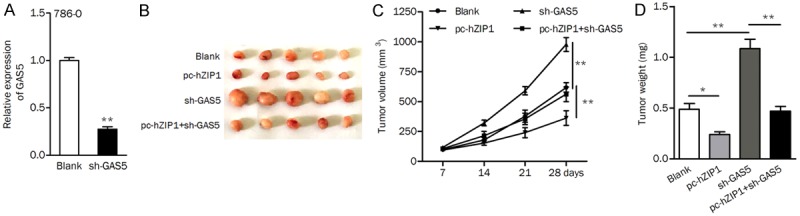
hZIP1 is a downstream effector of GAS5 in tumor growth. (A) The expression of GAS5 was significantly downregulated in cells transfected with sh-GAS5 compared to that in control cells. (B) Photographs showing representative tumors formed. (C and D) Overexpression of hZIP1 not only inhibited tumor growth alone but also abolished the promotive effect of sh-GAS5 on tumor volume (C) and tumor weight (D). *P<0.05, **P<0.01.
The clinical significance of the GAS5/miR-223/hZIP1 axis in the progression of ccRCC
The above results suggested that dysregulated GAS5, miR-223, and hZIP1 might play a role in the development of ccRCC. Thus, we also analyzed the correlations between these dysregulated genes and clinicopathological features. The mean expression levels of each gene in tumors were used as cut-off values to divide cases into high or low groups. Low expression of hZIP1 was significantly associated with advanced tumor stage (P<0.001) and Fuhrman stage (P=0.029, Table 1). Both high expression of miR-223 and low expression of GAS5 were significantly correlated with tumor stage (P=0.04 and 0.01, respectively) but not with Fuhrman stage (Table 1). The low expression of GAS5 was associated with tumor recurrence (P=0.019, Table 1).
Table 1.
Clinical characteristics of patients with dysregulated hZIP1/miR-223/GAS5
| Variables | Case | hZIP1 | P | miR-223 | P | GAS5 | P | |||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||
| n=55 | L, n=22 | H, n=33 | L, n=27 | H, n=28 | L, n=33 | H, n=22 | ||||
| Gender | 0.646 | 0.513 | 0.909 | |||||||
| Female | 18 | 8 | 10 | 10 | 8 | 11 | 7 | |||
| Male | 37 | 14 | 23 | 17 | 20 | 22 | 15 | |||
| Age, y | 0.662 | 0.489 | 0.512 | |||||||
| <55 | 23 | 10 | 13 | 10 | 13 | 15 | 8 | |||
| ≥55 | 32 | 12 | 20 | 17 | 15 | 18 | 14 | |||
| Clinical stage | <0.001 | 0.040 | 0.010 | |||||||
| I | 31 | 5 | 26 | 19 | 12 | 14 | 17 | |||
| II, III, IV | 24 | 17 | 7 | 8 | 16 | 19 | 5 | |||
| Fuhrman stage | 0.029 | 0.093 | 0.151 | |||||||
| 1, 2 | 39 | 12 | 27 | 22 | 17 | 21 | 18 | |||
| 3, 4 | 16 | 10 | 6 | 5 | 11 | 12 | 4 | |||
| Recurrence | 0.279 | 0.792 | 0.019 | |||||||
| No | 44 | 16 | 28 | 22 | 22 | 23 | 21 | |||
| Yes | 11 | 6 | 5 | 5 | 6 | 10 | 1 | |||
L, low expression; H, high expression; y, years; P<0.05 presented in bold.
The impact of GAS5, miR-223, and hZIP1 on DFS of ccRCC patients was analyzed using a Cox regression model. Univariate analysis suggested that Fuhrman stage and GAS5 level were correlated with DFS. Multivariate analyses confirmed that higher Fuhrman stage and low expression of GAS5 were independently correlated with DFS with a relative risk of recurrence of 3.523 (P=0.039) and 7.902 (P=0.050), respectively (Table 2).
Table 2.
Disease-free survival analyses
| Variables | Univariate | Multivariate | ||
|---|---|---|---|---|
|
|
|
|||
| P | HR (95%) | P | HR (95%) | |
| Gender (F/M) | 0.962 | 0.969 (0.256-3.670) | ||
| Age (<55/≥55) | 0.873 | 1.106 (0.323-3.787) | ||
| Clinical stage (I/II, III, IV) | 0.196 | 2.209 (0.665-7.337) | ||
| Fuhrman stage (1, 2/3, 4) | 0.018 | 4.227 (1.284-13.921) | 0.039 | 3.523 (1.063-11.671) |
| hZIP1 (H/L) | 0.087 | 2.900 (0.858-9.801) | ||
| miR-223 (L/H) | 0.760 | 1.204 (0.367-3.947) | ||
| GAS5 (H/L) | 0.035 | 9.160 (1.064-72.077) | 0.050 | 7.902 (0.999-62.527) |
F, female; M, male; L, low expression; H, high expression. P<0.05 presented in bold.
Discussion
Our previous study found that the protein levels of hZIP1 were downregulated in ccRCC tissues and indicated a higher tumor stage, Fuhrman stage, recurrence and DFS of patients [6]. Similarly, decreased mRNA expression of hZIP1 was observed in ccRCC tissues and was associated with advanced clinical stage and Fuhrman stage. These data support the critical role of hZIP1 in the development of ccRCC.
A previous study showed that RREB-1 overexpression could inhibit the expression of hZIP1 in prostate cancer [16]. In this study, we explored the mechanism responsible for the dysregulation of hZIP1 in ccRCC progression. The expression of protein-coding genes can be repressed by miRNAs. MiR-223 was predicted and experimentally validated to be an upstream regulator of hZIP1. In addition, miR-182 was found to inhibit the expression of hZIP1 by binding to its 3-UTR [17]. Therefore, hZIP1 may be a critical tumor-related gene and may be regulated by multiple miRNAs in various tumors. Upregulation of miR-223 was observed in tumor tissues and negatively correlated with hZIP1, suggesting that miR-223/hZIP1 might be involved in renal carcinogenesis. miR-223 expression has been demonstrated to be highly expressed in various cancers, including pancreatic, gastric, prostate, and bladder cancers [18-21]. Our data expand our knowledge of miR-223 in the development ccRCC. Upregulation of miR-223 was frequently observed in advanced-stage tumors, which was consistent with other studies [22,23].
Accumulating evidence has validated that lncRNAs may exert their roles in tumorigenesis by competitively binding to cancer-related miRNAs [24]. We investigated the potential lncRNAs that may play a role in regulating miR-223/hZIP1 and tumorigenesis. The luciferase activity of wild-type GAS5 was repressed by miR-223. In addition, miR-223 and GAS5 could affect each other, suggesting a direct interaction between miR-223 and GAS5. We found that the expression of GAS5 was downregulated in ccRCC tissues and higher stage tumors and independently correlated with poor DFS of patients. A previous study also reported that the expression of GAS5 was decreased in ccRCC tissues [25]. Similarly, GAS5 was decreased in gastric cancer tissues and indicated poor overall survival and DFS [26]. Low expression of GAS5 was found to be associated with tumor size, lymph node metastasis and poor prognosis of liver cancer [27]. These data suggested that GAS5 might be utilized in prognosis prediction for cancer patients. Furthermore, the low expression of GAS5 in tumors was accompanied by high expression of miR-223, supporting the negative correlation between GAS5 and miR-223. Knockdown of GAS5 led to downregulation of hZIP1 in ccRCC cells. The expression of GAS5 was positively associated with hZIP1, and the statistical significance nearly approached 0.05. These findings suggested that GAS5 positively regulated the expression of hZIP1 by sponging miR-223.
Functionally, hZIP1 overexpression exerted tumor suppression by inhibiting cell viability and motility in vitro and tumor growth in vivo. In our previous study, knockdown of hZIP1 promoted cell proliferation and invasion. The tumor suppressor role of hZIP1 in ccRCC tissues was in accordance with its low expression status in tumor tissues. Overexpression of hZIP1 results in the inhibition of NF-κB activity as well as a decrease in downstream pro-metastatic and antiapoptotic gene expression in prostate cancer [4]. Although the inhibitory effect of hZIP1 on tumor-related biological processes has not been widely reported, the expression of hZIP1 is decreased in mucinous carcinomas from a variety of organs [5], implying that hZIP1 might have an important role in human malignancy.
In the present study, inhibition of miR-223 suppressed cell growth, cell cycle progression, and invasion and enhanced apoptosis, and these effects were reversed by knockdown of GAS5. Overexpression of miR-223 abolished the suppressive effect of hZIP1. Thus, GAS5 might exert a tumor suppressor role by sponging miR-223, ultimately upregulating the expression of hZIP1 in ccRCC. GAS5 may act as a ceRNA to regulate miRNAs. For example, GAS5 suppresses tumor malignancy by downregulating miR-222 in glioma [28]. GAS5 suppresses cell growth and motility in osteosarcoma by regulating the miR-221/ARHI pathway [29]. GAS5 acts as a ceRNA for miR-222-3p and modulates the expression of PTEN and the downstream AKT pathway in papillary thyroid carcinoma [30]. In addition, GAS5 has frequently been found to be a negative regulator of miR-21 in different types of tumors [31-33]. GAS5 also functions as a sponge for miR-18a, inhibiting its capability to repress CTGF protein levels [34]. These observations imply that GAS5 may interact with different miRNAs and regulate various genes in different tumors. In addition, a xenograft model experiment demonstrated that hZIP1 is a downstream effector of GAS5 and could attenuate the tumor growth induced by GAS5 silencing.
In conclusion, we demonstrate that decreased hZIP1 expression is a common event underlying ccRCC, indicating that hZIP1 may play a key role in the progression of ccRCC. Further analysis of the potential mechanism provided the first evidence that GAS5 exerted a tumor suppressor function in the progression of ccRCC by modulating the miR-223/hZIP1 pathway.
Acknowledgements
This study was supported by the the Doctoral Scientific Research Foundation of Liaoning Province (No. 201501015).
Disclosure of conflict of interest
None.
References
- 1.Slaby O, Jancovicova J, Lakomy R, Svoboda M, Poprach A, Fabian P, Kren L, Michalek J, Vyzula R. Expression of miRNA-106b in conventional renal cell carcinoma is a potential marker for prediction of early metastasis after nephrectomy. J Exp Clin Cancer Res. 2010;29:90. doi: 10.1186/1756-9966-29-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 3.Franklin RB, Feng P, Milon B, Desouki MM, Singh KK, Kajdacsy-Balla A, Bagasra O, Costello LC. hZIP1 zinc uptake transporter down regulation and zinc depletion in prostate cancer. Mol Cancer. 2005;4:32. doi: 10.1186/1476-4598-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Golovine K, Makhov P, Uzzo RG, Shaw T, Kunkle D, Kolenko VM. Overexpression of the zinc uptake transporter hZIP1 inhibits nuclear factor-kappaB and reduces the malignant potential of prostate cancer cells in vitro and in vivo. Clin Cancer Res. 2008;14:5376–5384. doi: 10.1158/1078-0432.CCR-08-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desouki MM, Franklin RB, Costello LC, Fadare O. Persistent low expression of hZip1 in mucinous carcinomas of the ovary, colon, stomach and lung. J Ovarian Res. 2015;8:40. doi: 10.1186/s13048-015-0169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong X, Kong C, Zhang Z, Liu X, Zhan B, Chen Z, Shi D. hZIP1 that is down-regulated in clear cell renal cell carcinoma is negatively associated with the malignant potential of the tumor. Urol Oncol. 2014;32:885–892. doi: 10.1016/j.urolonc.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 7.Yan J, She Q, Shen X, Zhang Y, Liu B, Zhang G. Potential role of microRNA-375 as biomarker in human cancers detection: a meta-analysis. Biomed Res Int. 2017;2017:1875843. doi: 10.1155/2017/1875843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang C, Tabatabaei SN, Ruan X, Hardy P. The dual regulatory role of MiR-181a in breast cancer. Cell Physiol Biochem. 2017;44:843–856. doi: 10.1159/000485351. [DOI] [PubMed] [Google Scholar]
- 9.Fujii N, Hirata H, Ueno K, Mori J, Oka S, Shimizu K, Kawai Y, Inoue R, Yamamoto Y, Matsumoto H, Shimabukuro T, Udoh K, Hoshii Y, Dahiya R, Matsuyama H. Extracellular miR-224 as a prognostic marker for clear cell renal cell carcinoma. Oncotarget. 2017;8:109877–109888. doi: 10.18632/oncotarget.22436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santarpia L, Nicoloso M, Calin GA. MicroRNAs: a complex regulatory network drives the acquisition of malignant cell phenotype. Endocr Relat Cancer. 2010;17:F51–75. doi: 10.1677/ERC-09-0222. [DOI] [PubMed] [Google Scholar]
- 11.Xiao W, Lou N, Ruan H, Bao L, Xiong Z, Yuan C, Tong J, Xu G, Zhou Y, Qu Y, Hu W, Gao Y, Ru Z, Liu L, Xiao H, Chen K, Yang H, Zhang X. Mir-144-3p promotes cell proliferation, metastasis, sunitinib resistance in clear cell renal cell carcinoma by downregulating ARID1A. Cell Physiol Biochem. 2017;43:2420–2433. doi: 10.1159/000484395. [DOI] [PubMed] [Google Scholar]
- 12.Rao M, Zhu Y, Zhou Y, Cong X, Feng L. MicroRNA-122 inhibits proliferation and invasion in gastric cancer by targeting CREB1. Am J Cancer Res. 2017;7:323–333. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Wang L, Yang G, Zhu X, Wang Z, Wang H, Bai Y, Sun P, Peng L, Wei W, Chen G, Li G, Zamyatnin AA Jr, Glybochko PV, Xu W. miR-93-3p inhibition suppresses clear cell renal cell carcinoma proliferation, metastasis and invasion. Oncotarget. 2017;8:82824–82834. doi: 10.18632/oncotarget.20458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S, Mei Z, Hu HB, Zhang X. The lncRNA MALAT1 contributes to non-small cell lung cancer development via modulating miR-124/STAT3 axis. J Cell Physiol. 2018;233:6679–6688. doi: 10.1002/jcp.26325. [DOI] [PubMed] [Google Scholar]
- 15.Du Y, Weng XD, Wang L, Liu XH, Zhu HC, Guo J, Ning JZ, Xiao CC. LncRNA XIST acts as a tumor suppressor in prostate cancer through sponging miR-23a to modulate RKIP expression. Oncotarget. 2017;8:94358–94370. doi: 10.18632/oncotarget.21719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou J, Milon BC, Desouki MM, Costello LC, Franklin RB. hZIP1 zinc transporter down-regulation in prostate cancer involves the overexpression of ras responsive element binding protein-1 (RREB-1) Prostate. 2011;71:1518–1524. doi: 10.1002/pros.21368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mihelich BL, Khramtsova EA, Arva N, Vaishnav A, Johnson DN, Giangreco AA, Martens-Uzunova E, Bagasra O, Kajdacsy-Balla A, Nonn L. miR-183-96-182 cluster is overexpressed in prostate tissue and regulates zinc homeostasis in prostate cells. J Biol Chem. 2011;286:44503–44511. doi: 10.1074/jbc.M111.262915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rachagani S, Macha MA, Menning MS, Dey P, Pai P, Smith LM, Mo YY, Batra SK. Changes in microRNA (miRNA) expression during pancreatic cancer development and progression i n a genetically engineered KrasG12D;Pdx1-Cre mouse (KC) model. Oncotarget. 2015;6:40295–40309. doi: 10.18632/oncotarget.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou X, Jin W, Jia H, Yan J, Zhang G. MiR-223 promotes the cisplatin resistance of human gastric cancer cells via regulating cell cycle by targeting FBXW7. J Exp Clin Cancer Res. 2015;34:28. doi: 10.1186/s13046-015-0145-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei Y, Yang J, Yi L, Wang Y, Dong Z, Liu Z, Ou-yang S, Wu H, Zhong Z, Yin Z, Zhou K, Gao Y, Yan B, Wang Z. MiR-223-3p targeting SEPT6 promotes the biological behavior of prostate cancer. Sci Rep. 2014;4:7546. doi: 10.1038/srep07546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, Sevignani C, Byrne D, Negrini M, Pagano F, Gomella LG, Croce CM, Baffa R. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007;25:387–392. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 22.Li ZW, Yang YM, Du LT, Dong Z, Wang LL, Zhang X, Zhou XJ, Zheng GX, Qu AL, Wang CX. Overexpression of miR-223 correlates with tumor metastasis and poor prognosis in patients with colorectal cancer. Med Oncol. 2014;31:256. doi: 10.1007/s12032-014-0256-5. [DOI] [PubMed] [Google Scholar]
- 23.Jacob H, Stanisavljevic L, Storli KE, Hestetun KE, Dahl O, Myklebust MP. Identification of a sixteen-microRNA signature as prognostic biomarker for stage II and III colon cancer. Oncotarget. 2017;8:87837–87847. doi: 10.18632/oncotarget.21237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma Y, Lu Y, Lu B. MicroRNA and Long non-coding RNA in ovarian carcinoma: translational insights and potential clinical applications. Cancer Invest. 2016;34:465–476. doi: 10.1080/07357907.2016.1227446. [DOI] [PubMed] [Google Scholar]
- 25.Qiao HP, Gao WS, Huo JX, Yang ZS. Long non-coding RNA GAS5 functions as a tumor suppressor in renal cell carcinoma. Asian Pac J Cancer Prev. 2013;14:1077–1082. doi: 10.7314/apjcp.2013.14.2.1077. [DOI] [PubMed] [Google Scholar]
- 26.Sun M, Jin FY, Xia R, Kong R, Li JH, Xu TP, Liu YW, Zhang EB, Liu XH, De W. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer. 2014;14:319. doi: 10.1186/1471-2407-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tu ZQ, Li RJ, Mei JZ, Li XH. Down-regulation of long non-coding RNA GAS5 is associated with the prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol. 2014;7:4303–4309. [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao X, Wang P, Liu J, Zheng J, Liu Y, Chen J, Xue Y. Gas5 exerts tumor-suppressive functions in human glioma cells by targeting miR-222. Mol Ther. 2015;23:1899–1911. doi: 10.1038/mt.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye K, Wang S, Zhang H, Han H, Ma B, Nan W. Long noncoding RNA GAS5 suppresses cell growth and epithelial-mesenchymal transition in osteosarcoma by regulating the miR-221/ARHI pathway. J Cell Biochem. 2017;118:4772–4781. doi: 10.1002/jcb.26145. [DOI] [PubMed] [Google Scholar]
- 30.Zhang XF, Ye Y, Zhao SJ. LncRNA Gas5 acts as a ceRNA to regulate PTEN expression by sponging miR-222-3p in papillary thyroid carcinoma. Oncotarget. 2017;9:3519–3530. doi: 10.18632/oncotarget.23336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C, Xu M, Wu F, Mo YY. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013;20:1558–1568. doi: 10.1038/cdd.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W, Zhai L, Wang H, Liu C, Zhang J, Chen W, Wei Q. Downregulation of LncRNA GAS5 causes trastuzumab resistance in breast cancer. Oncotarget. 2016;7:27778–27786. doi: 10.18632/oncotarget.8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu L, Pang X, Shang W, Xie H, Feng Y, Feng G. Long non-coding RNA GAS5 sensitizes renal cell carcinoma to sorafenib via miR-21/SOX5 pathway. Cell Cycle. 2018 doi: 10.1080/15384101.2018.1475826. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Li M, Xie Z, Wang P, Li J, Liu W, Tang S, Liu Z, Wu X, Wu Y, Shen H. The long noncoding RNA GAS5 negatively regulates the adipogenic differentiation of MSCs by modulating the miR-18a/CTGF axis as a ceRNA. Cell Death Dis. 2018;9:554. doi: 10.1038/s41419-018-0627-5. [DOI] [PMC free article] [PubMed] [Google Scholar]