Abstract
HNSCC is the sixth most common cancer worldwide and is characterized as an aggressive, malignant tumor. MiR-34a-5p (miR-34a) expression has been strongly linked to HNSCC development. However, the exact target gene of miRNA-34a-as well as its biological and mechanistic pathways-are unclear. It is critical that the clinical value of HNSCC receive further study. We conducted a continuous variable, meta-analysis from data found in the cancer literature as well as that provided by the Cancer Genome Atlas (TCGA) to estimate miR-34a expression in HNSCC. Next, TCGA database, microarray, and MiRWalk were all used to predict the target gene of miR-34a in HNSCC. Then, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were used to explore the underlying molecular mechanism of miR-34a in HNSCC. Finally, we used Spearman’s analysis and survival curves to identify the roles of related target genes involved in significant pathways during the development of HNSCC. Expression levels of miR-34a in HNSCC were significant lower than those in normal tissues (P<0.05) and summary receiver operating characteristic (sROC) was 0.72. The collective results obtained from KEGG and GO indicated that miR-34a may be involved in the development of HNSCC via known cancer pathways, including the p53 and/or PI3K-Akt signalling pathways. Our results suggested miR-34a has potential use as a novel, non-invasive and highly sensitive biomarker for diagnostic in HNSCC. Finally, it likely plays an essential role in the deterioration and ultimate tumorigenesis of HNSCC through determined cancer pathways.
Keywords: miR-34a-5p, HNSCC, meta-analysis, bioinformatics, TCGA
Introduction
MicroRNAs (miRNAS) include such family members as miR-34, which itself has several isoforms (miR-34a, miR-34b, and miR-34c). Of these, miR-34b and miR-34c share a common original transcript, while miR-34a is encoded by its own transcript [1]. Recently, work has focused on the unique miR-34a-5p (miR-34a), which has become an oncological research hotspot [2]. Moreover, miR-34a is encoded in the chromosomal location 1p36.22, and is associated with various tumors [3,4]. As a consistent tumor suppressor, it has been shown to regulate tumor cell apoptosis [5,6]. Previous studies have shown that miR-34a expression is related to multiple cancer types, including colorectal, osteosarcoma, lung, breast, and pancreatic [7-11].
Head and neck squamous cell carcinoma (HNSCC) is a malignant cancer that develops in the squamous epithelium of the mouth, pharynx (nasopharynx, oropharynx, hypopharynx), and larynx. HNSCC is the sixth most common cancer worldwide and is characterized as an aggressive, malignant tumor. Alarmingly, it has now become the fifth leading cause of cancer-related deaths worldwide [12]. Approximately 50,000 new cases of HNSCC are reported each year, resulting in the deaths of 13000 people annually in the United States alone [13]. Advances in surgical treatment as well as radiotherapeutic and chemotherapeutic approaches have significantly increased the survival rate for the disease. However, the overall survival rate for HNSCC has remained stagnant over the past 20 years.
It has been reported that there are approximately 50 miRNAs that are known to have altered expression in HNSCC [14]. For instance, Bhavna and colleagues [15] demonstrated that dysregulation of miR-34a expression was related to the development of HNSCC. However, the exact target gene of this miRNA-as well as its biological and mechanistic pathways-are unclear. Given this, it is critical that the clinical value of HNSCC receive further study.
Here, we performed a continuous variable, meta-analysis from data obtained from the cancer literature as well as the Cancer Genome Atlas (TCGA). These data were used to evaluate miR-34a expression in HNSCC. In addition, the potential diagnostic value of miR-34a was estimated using a diagnostic meta-analysis. In addition, the possible molecular mechanisms for miR-34a in HNSCC were analyzed comprehensively using both the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) enrichment analyses.
Materials and methods
Literature search and screening
A search was conducted on both English and Chinese medical literature using PubMed, Science Direct, Web of Science, Wiley Online Library, EMBASE, the China National Knowledge Infrastructure Database (CNKI), the Chinese Biomedical Literature Database (CBM), the China Science and Technology Journal Database (VIP), the Wanfang Database. All publications related to miR-34a expression and HNSCC up to February 20, 2018 were included. The retrieval strategy for this study was as follows: (SCC OR hnscc OR “squamous cell carcinoma” OR “squamous cell cancer”) AND (oropharyngeal OR oropharynx OR “head and neck” OR “oral cavity” OR nasopharynx OR hypopharynx OR laryngopharynx OR oral OR larynx OR laryngopharyngeal OR laryngeal OR pharyngeal OR laryngeal OR tongue OR tonsil OR tonsillar OR nose OR “nasal sinus” OR lip OR cheek OR “nasal cavity” OR “paranasal sinuses” OR palatal OR buccal) AND (miR-34a OR miRNA-34a OR microRNA-34a OR miR34a OR miRNA34a OR microRNA34a OR “miR 34a” OR “miRNA 34a” OR “microRNA 34a” OR miR-34a-5p OR miRNA-34a-5p OR microRNA-34a -5p).
Our meta-analysis results were evaluated by two independent researchers (JX Li and DN Yao) who used the same multistep process. First, the title and abstract were read to exclude any irrelevant literature. The remaining studies were then separately examined by the researchers. Finally, any controversial issues were addressed by a third reviewer. The study was determined to be eligible if it met the following inclusion criteria: (1) Clinical samples were taken from HNSCC and normal tissue or cell lines; (2) Reverse transcription quantitative PCR (RT-qPCR) was used to determine miR-34a expression levels; and (3) Study provided sufficient data to determine relevant descriptive statistics (mean and standard deviation, SD) as well as 95% confidence intervals (95% CI). A study was excluded if it met one of the following conditions: (1) It was a duplicate studies, (2) It had insufficiently detailed data to calculate the needed descriptive and inferential statistics, and/or (3) It was a review, letter, case report, or conference article. If a study lacked sufficient data for our analysis, we made all attempts to contact the author for the original dataset. Finally, we extracted pivotal information from all eligible studies, including author name, published time, country, sample size, analysis method, and type of samples used.
TCGA data mining
The expression levels and clinical parameter data for miR-34a were extracted using the OncoLnc website (http://www.oncolnc.org/) [16]. Correlations between all tested clinical parameters and miR-34a expression levels was determined using a commercially available statistical package (SPSS 22.0) (Nanning, Guangxi Zhuang Autonomous Region, China).
Statistical analysis
STATA 12.0 software was used to analyze the resulting data culled from our meta-analysis as well as our TCGA data (Nanning, Guangxi Zhuang Autonomous Region, China). We used standard mean difference (SMD) along with 95% CI to assess miR-34a expression values. GraphPad Prism 6 (Nanning, Guangxi Zhuang Autonomous Region, China) was used to generate scatter diagrams for miR-34a expression levels in both normal and HNSCC samples. The potential heterogeneity of the selected study data were evaluated using I2 index. When I2>50% or P<0.05, the fixed-effect model was used; otherwise, the random-effect model was applied. SPSS 22.0 was utilized for all statistical analysis. All measurement data were shown using mean ± SD (X ± s) and t-tests were used for comparisons between groups. Rates for count data were given as percentages (%) and compared using χ2 tests. When determining the diagnostic value of miR-34a, according to true positive (TP), false positive (FP), false negative (FN) and true negative (TN), and then we use meta-Disc software to calculate the Specificity; Sensitivity; Positive likelihood ratio (PLR); Negative likelihood ratio (NLR); Diagnostic Odds ratio (DOR) and summary receiver operating characteristic (sROC) curve. Moreover, all expression data of miR-34a, including TCGA and studies, have been normalized to log2. P<0.05 was taken as a statistically significant difference for all analyses.
Exploring miR-34a as an important gene in HNSCC
The differential expression of genes in HNSCC was based on TCGA data and was extracted using GEPIA (http://http://gepia.cancerpku.cn). Any upregulated genes with log2-fold change (FC) >2 were subsequently selected. In addition, MiRWalk 2.0 [17] was applied to determine possible miR-34a target genes. Any upregulated genes with SUM ≥3 were selected. Moreover, we also used existing data of GSE59617 chip from Gene Expression Omnibus (GEO), which was transfected with miR-34a into HNSCC cell lines. Genes with log2FC >1 were then selected. Finally, we utilized VENNY figure (https://omictools.com/venny-tool) to identify overlap in the three network platforms that were used to select the upregulated genes that were potential target genes of miR-34a in HNSCC.
Bioinformatics analysis based on predicted miR-34a target genes in HNSCC
Gene ontology (GO) [18] consisted of three parts: Biological process (BP), molecule function (MF), and cellular component (CC). Kyoto Encyclopedia of Genes and Genomes (KEGG) [19] was used in conjunction with DAVID 6.8 (https://david.ncifcrf.gov/summary.jsp) and R (version 3.3.3) and was based on promising miR-34a target genes. This approach was used in order to investigate the underlying mechanism of miR-34a in HNSCC. BINGO and the Enrichment Map plug-in Cytoscape were used to visualize the results of the GO and KEGG pathway analyses. STRING (https://string-db.org/) was also applied to build the protein-protein interaction (PPI) network, allowing us to visualize the connections between the target genes involved in important pathways.
Gene validation involved in significant signalling pathway
UALCOULD [20] (http://ualcan.path.uab.edu/index.html) was used to determine the expression levels of potential target genes in both HNSCC and normal tissue as well as their relationships with prognosis. Spearman’s correlation analysis was applied in LinkedOmics [21] (http://www.linkedomics.org/) to verify the correlations between miR-34a and identified target genes involved in significant pathways.
Results
Study retrieval and data extraction of studies and TCGA
On the basis of our retrieval strategy, we selected 37 papers identified using PubMed, EMBASE, Wiley Online Library, Science Direct, Web of Science, the Wanfang Database, CBM, VIP, and CNKI. After reading the summary and full-text review, Bhavna K (PMID: 22629428), Manikanda [22] (PMID: 26625772), and Li W [23] (PMID: 24482044) were included based on our criteria for eligibility (Figure 1). We then extracted relevant data for miR-34a expression levels in HNSCC from the TCGA database. Collectively, four studies comprising 635 cases were collected. Additional, detailed information is shown in Table 1.
Figure 1.
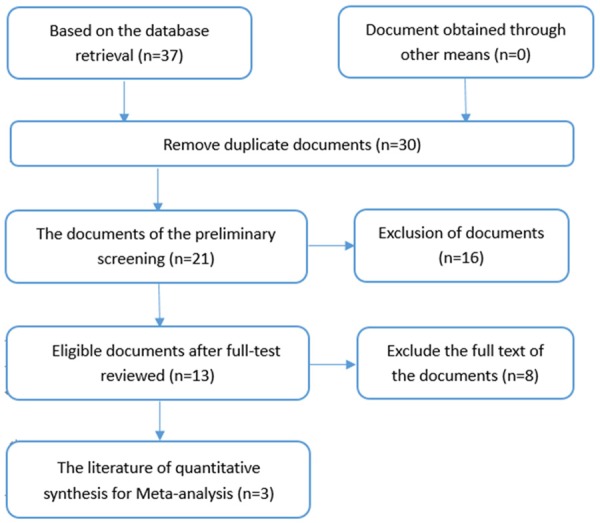
The flow chart of the search and selection of documents.
Table 1.
The characteristics of literature with inclusion and exclusion criteria
HNSCC miR-34a expression
MiR-34a expression levels in normal and HNSCC tissue are shown in Figure 2. Three of the selected studies indicated that miR-34a expression in HNSCC was lower than in normal tissues (P<0.05). We next performed a meta-analysis to explore mrR-34a expression levels in HNSCC. Due to high heterogeneity (I2=66.9%) (Figure 3A), we used the random-effect model. The combined standard mean difference (SMD) was -0.70 (95% CI: -1.28, -0.11) (Figure 3B). Results indicated that miR-34a was expressed at low levels in HNSCC.
Figure 2.
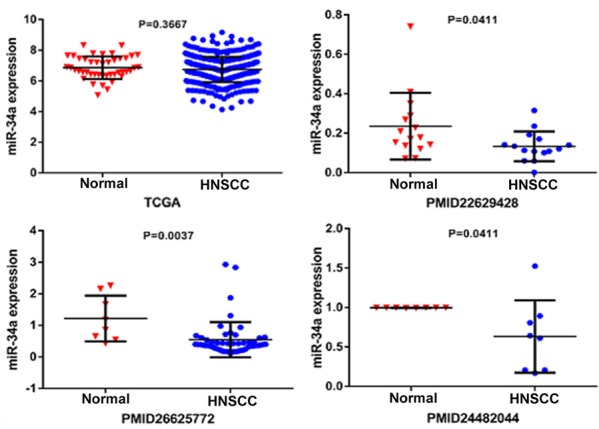
The scatter diagram of miR-34a expression levels in HNSCC incorporate each study.
Figure 3.
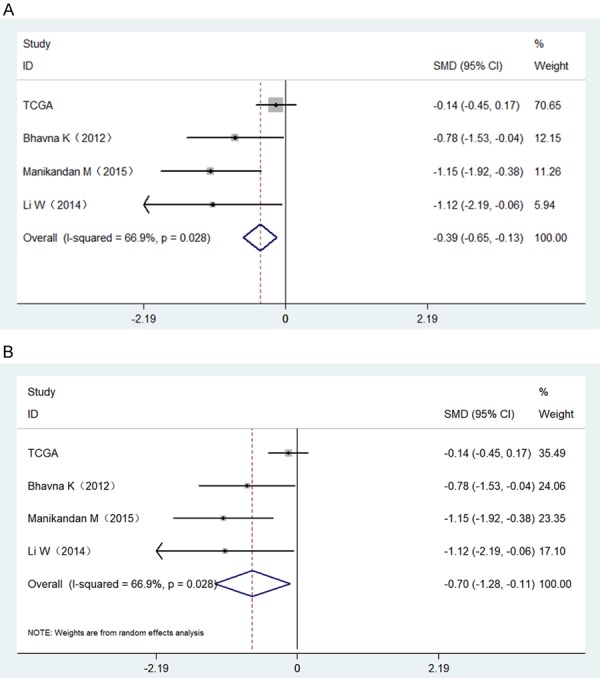
Forest plots evaluating the miR-34a expression between HNSCC and normal tissues based on selected studies and TCGA data. A. A fixed-effect model of Forest plots; B. A random effects model of Forest plots.
Diagnostic value of miR-34a for HNSCC
We next used a random-effects model to analyze the diagnostic value of miR-34a for HNSCC. This was due to the significant heterogeneity in the analysis of DOR, PLR, NLR, sensitivity, and specificity of the included studies. Diagnostic meta-analysis results revealed that the pooled specificity, sensitivity, likelihood ratios (negative and positive), and diagnostic odds ratio were as follows: 0.33 (95% CI: 0.24-0.42), 0.97 (95% CI: 0.95-0.98), 0.64 (95% CI: 0.05-7.76), 1.09 (95% CI: 0.70-1.70), and 1.69 (95% CI: 0.09-32.94), respectively (Figure 4A-E). The ROC curve based on each study is shown in Figure 5. Analysis of the area under curve (AUC) of the sROC curve was 0.72 (95% CI: 0.68, 0.76) (Figure 6).
Figure 4.
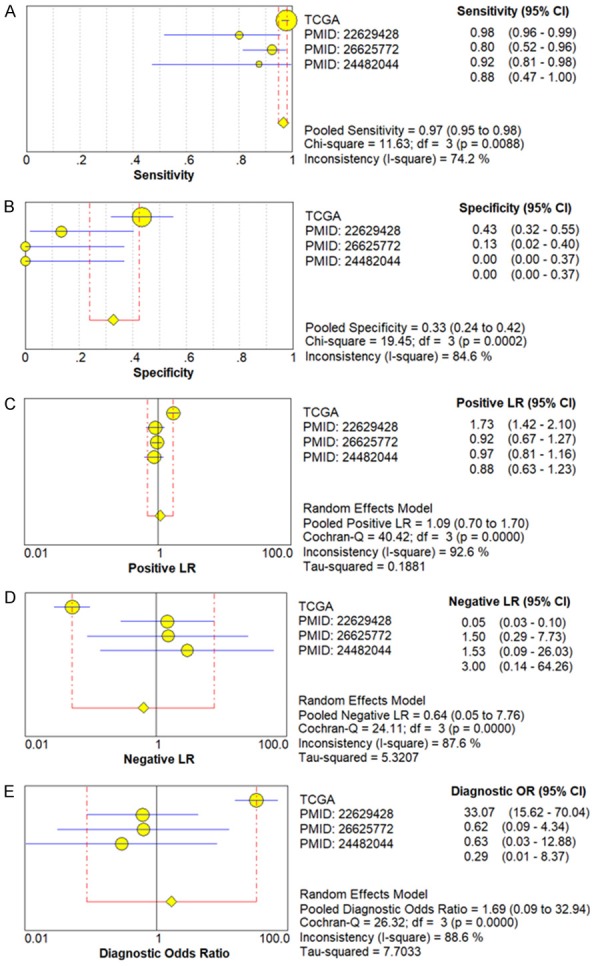
Diagnostic value of miR-34a in HNSCC with forest plots. A. Sensitivity; B. Specificity; C. Positive likelihood ratio; D. Negative likelihood ratio; E. Diagnostic Odds ratio.
Figure 5.

Receiver operating characteristic (ROC) curves based on 4 researches for each diagnostic test of miR-34a in HNSCC.
Figure 6.
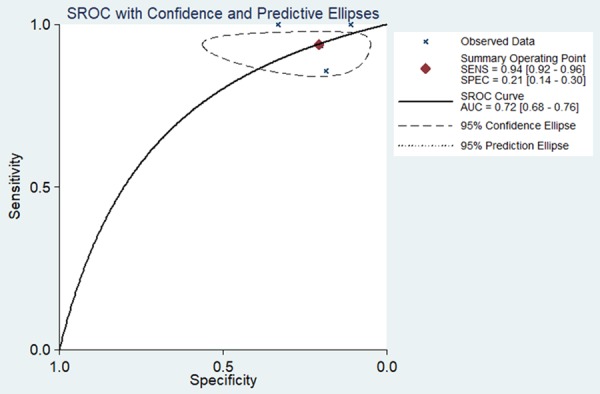
Summary receiver operating characteristic (sROC) curve for the diagnostic value of miR-34a in HNSCC.
The miR-34a clinical-pathological features in HNSCC based on TCGA analysis
485 HNSCC samples were analyzed according to TCGA database and results indicated no statistical significance between miR-34a expression and gender, age, lymphovascular invasion, alcohol, or any other of the analyzed clinical-pathological features. Nevertheless, in pathologic grade, miR-34a expression of G3-G4 was significantly lower than in the G1-G2 stage (P<0.05) (Table 2).
Table 2.
The correlation between miR-34a expression and clinical features from TCGA data
| Clinicopathological features | Terms | n | Mean ± SD | p-value |
|---|---|---|---|---|
| Unpaired tissue | Normal | 44 | 6.86±0.73 | 0.367 |
| HNSCC | 485 | 6.74±0.82 | ||
| Gender | Male | 381 | 6.79±0.81 | 0.120 |
| Female | 148 | 6.67±0.81 | ||
| Age | <60 | 224 | 6.79±0.80 | 0.448 |
| ≥60 | 304 | 6.73±0.82 | ||
| Lymphovascular invasion | No | 236 | 6.73±0.81 | 0.976 |
| Yes | 119 | 6.73±0.71 | ||
| Pathologic grade | G1-G2 | 368 | 6.82±0.81 | 0.013 |
| G3-G4 | 139 | 6.62±0.82 | ||
| Clinical grade | I-II | 126 | 6.79±0.81 | 0.620 |
| III-IV | 389 | 6.75±0.81 | ||
| T stage | T1-T2 | 189 | 6.72±0.81 | 0.434 |
| T3-T4 | 325 | 6.78±0.80 | ||
| N stage | N0 | 253 | 6.80±0.78 | 0.319 |
| N1-N3 | 254 | 6.73±0.84 | ||
| M stage | M0 | 504 | 6.77±0.80 | 0.260 |
| M1 | 4 | 6.31±1.18 | ||
| Recurrence | No | 329 | 6.72±0.79 | 0.093 |
| Yes | 115 | 6.87±0.84 | ||
| Alcohol | No | 173 | 6.79±0.80 | 0.499 |
| Yes | 345 | 6.73±0.82 |
Notes: HNSCC, Head and neck squamous cell carcinoma; T stage, size or direct extent of the primary tumor; N stage, degree of spread to regional lymph nodes; M stage, presence of distant metastasis; SD, standard deviation.
Identifying promising miR-34a target genes in HNSCC using bioinformatics
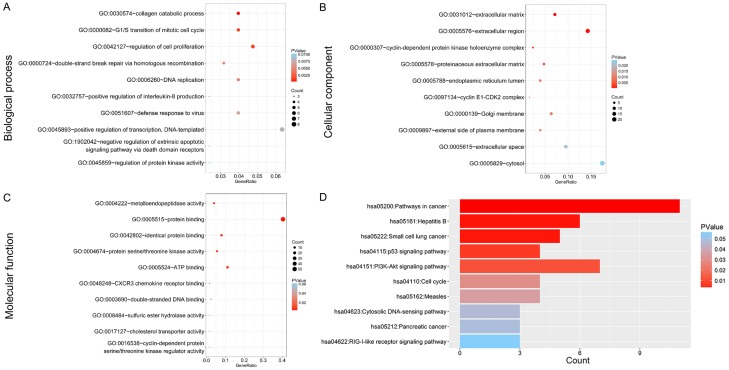
The number of differentially expressed genes were determined to be 8000, 3213, and 1487 based on analysis using MiRWalk, GSE59617, and GEPIA. A total of 77 overlapping target genes were involved in the bioinformatics analysis and are detailed in Figure 7 and Table 3. The mechanism of miR-34a in HNSCC was next explored using both GO and KEGG. GO biological process annotation revealed that the identified target genes were mostly enriched in the collagen catabolic process, regulation of cell proliferation, and G1/S transition of mitotic cell cycle. For the cellular component, identified target genes were closely related to the extracellular matrix, extracellular region, and cyclin-dependent protein kinase holoenzyme complex. For molecule functions, the three most enriched items were metalloendopeptidase activity, protein binding, and identical protein binding (Figures 8 and 9A-C; Table 4). KEGG enrichment analysis revealed that miR-34a could make a critical difference in HNSCC through multiple pathways, including the pI3k-akt and p53 signaling pathways as well as the cytosolic DNA-sensing pathway (Figure 9D; Table 5). The PPI network of genes associated with the pi3k-akt signaling pathway is shown in Figure 10.
Figure 7.
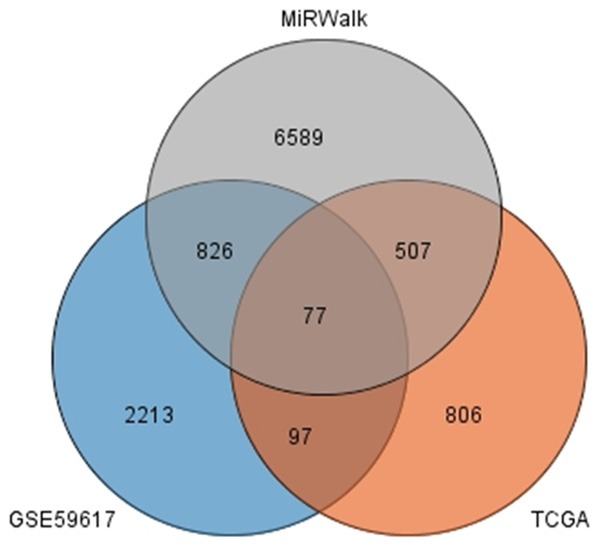
The junction of the predictive target genes.
Table 3.
The promising target genes of miR-34a in HNSCC with GEO, TCGA and MiRWalk
| Sources of genes | Genes |
|---|---|
| GEO, TCGA and MiRWalk | LTBP2, DZIP1, FGF11, TGFB3, CXCL10 |
| STARD4, SERPINE1, YEATS2, COL12A1, LOX, COL10A1, EFNB1, FADD, PDCD1LG2, UCN2, RIPK2, ROR2, STC1, LAMC1, BTN3A2, ADAMTS2, NEK6, GBP1, STK10 | |
| LETM2, EPHB2, TTYH3, ECE2, TUBB | |
| GALNS, HOXA10, FCER1G, RUNX3, ADAM23, TRIO, SLAMF8, FOXP3, ABCG1, BRCA1, DDX58, CBLB, PDE7A, GLS, ANTXR2, GPR161 | |
| SENP5, CCNE1, MCM8, CHST11, NT5E | |
| KDELR3, NRIP3, PRTFDC1, MMP19, CDK4, CDK2, RAD51, FKBP14, CTSC, ACVR1, POLR2H, PHLDB1, EGLN3, KIAA0101, CXCL9, HMMR, PSTPIP1, GPSM1, ADAMTS12, FEN1, COL4A1, GMNN, AGTRAP, TMEM2, IFIT3 | |
| SULF2, CHN1 |
Figure 8.
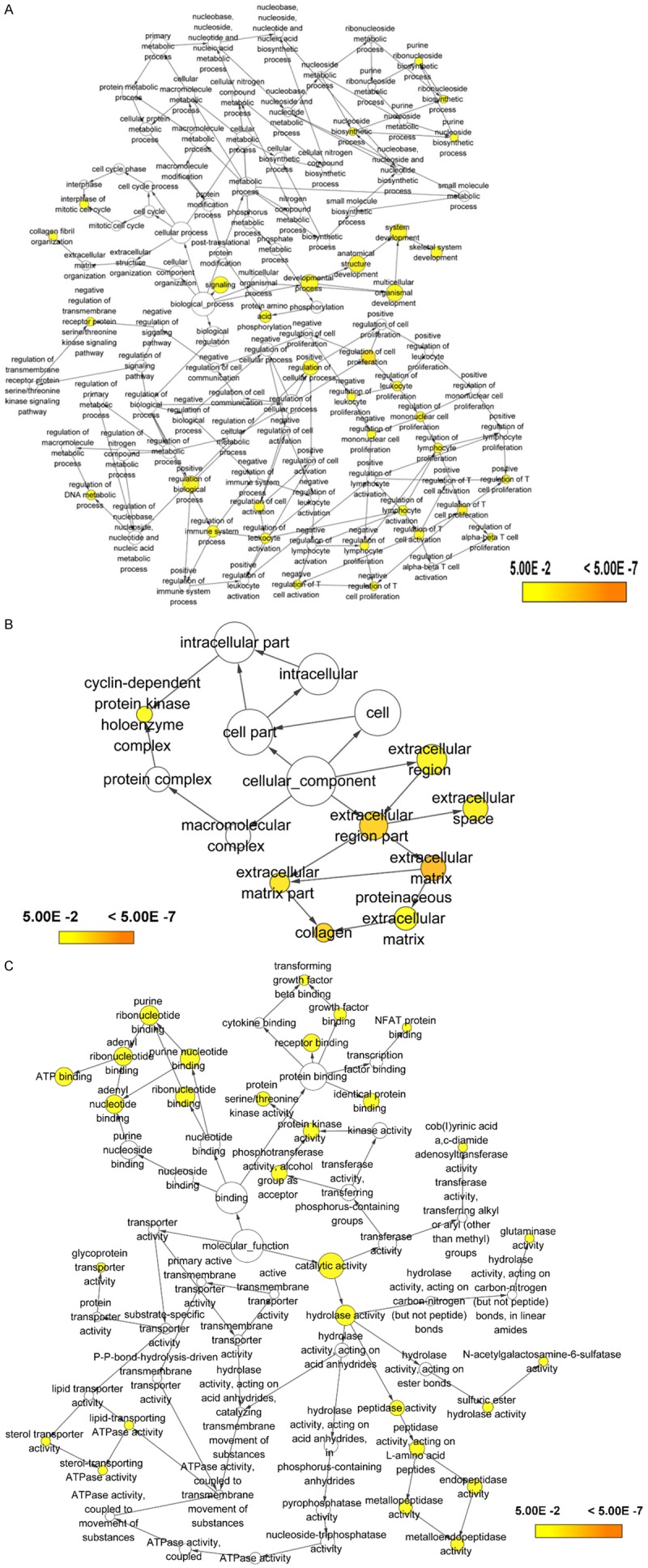
Gene Ontology (GO) analysis of (A) the biological process (BP) category; (B) cellular component (CC) category. (C) molecule function (MF) category.
Figure 9.
Significantly enriched annotation of (Gene Ontology) GO categories and (Kyoto Encyclopedia of Genes and Genomes) KEGG pathways of potential targets of miR-34a in HNSCC. A: Biological processes (BP); B: Cellular components (CC); C: Molecular factors (MF); D: KEGG pathway.
Table 4.
Predictive target genes of miR-34a with GO analysis
| GO ID | GO term | Count (%) | Gene symbol | P-value |
|---|---|---|---|---|
| Biological process | ||||
| GO:0030574 | Collagen catabolic process | 5 | COL4A1, MMP19, COL12A1, ADAMTS2, COL10A1 | 1.90E-04 |
| GO:0000082 | G1/S transition of mitotic cell cycle | 5 | CCNE1, MCM8, CDK4, CDK2, ACVR1 | 0.001121 |
| GO:0042127 | Regulation of cell proliferation | 6 | CHST11, TGFB3, EGLN3, CXCL9, BRCA1, CXCL10 | 0.001423 |
| GO:0000724 | Double-strand break repair via homologous recombination | 4 | MCM8, BRCA1, FEN1, RAD51 | 0.004420 |
| GO:0006260 | DNA replication | 5 | MCM8, KIAA0101, BRCA1, FEN1, CDK2 | 0.005108 |
| GO:0032757 | Positive regulation of interleukin-8 production | 3 | DDX58, SERPINE1, FADD | 0.005968 |
| GO:0051607 | Defense response to virus | 5 | IFIT3, CXCL9, FADD, GBP1, CXCL10 | 0.006360 |
| GO:0045893 | Positive regulation of transcription, DNA-templated | 8 | CCNE1, TGFB3, ROR2, FOXP3, BRCA1, CDK2, RUNX3, ACVR1 | 0.008061 |
| GO:1902042 | Negative regulation of extrinsic apoptotic signaling pathway via death domain receptors | 3 | SERPINE1, FADD, BRCA1 | 0.009503 |
| GO:0045859 | Regulation of protein kinase activity | 3 | CCNE1, CDK4, CXCL10 | 0.010068 |
| Cellular component | ||||
| GO:0031012 | Extracellular matrix | 9 | TUBB, COL4A1, LTBP2, MMP19, SERPINE1, TGFB3,etc | 2.89E-05 |
| GO:0005576 | Extracellular region | 18 | COL4A1, ADAM23, MMP19, CXCL9, FGF11, TGFB3, etc | 2.34E-04 |
| GO:0000307 | Cyclin-dependent protein kinase holoenzyme complex | 3 | CCNE1, CDK4, CDK2 | 0.001695 |
| GO:0005578 | Proteinaceous extracellular matrix | 6 | LTBP2,MMP19, LOX, ADAMTS12, ADAMTS2, COL10A1 | 0.004950 |
| GO:0005788 | Endoplasmic reticulum lumen | 5 | COL4A1, FKBP14, COL12A1, CTSC, COL10A1 | 0.008112 |
| GO:0097134 | Cyclin E1-CDK2 complex | 2 | CCNE1, CDK2 | 0.008214 |
| GO:0000139 | Golgi membrane | 8 | KDELR3, ECE2, CHST11, GPSM1, CTSC, ABCG1, GBP1, AGTRAP | 0.010775 |
| GO:0009897 | External side of plasma membrane | 5 | CXCL9, FCER1G, ANTXR2, ABCG1, CXCL10 | 0.011557 |
| GO:0005615 | Extracellular space | 12 | UCN2, LTBP2, SULF2, SERPINE1, TGFB3, CXCL9, COL12A1, etc | 0.021426 |
| GO:0005829 | Cytosol | 22 | POLR2H, PRTFDC1, GMNN, EGLN3, TRIO, FADD, CDK4, etc | 0.024066 |
| Molecular function | ||||
| GO:0004222 | Metalloendopeptidase activity | 5 | ECE2,ADAM23,MMP19, ADAMTS12, ADAMTS2 | 0.001380 |
| GO:0005515 | Protein binding | 51 | LTBP2, DZIP1, TGFB3, SENP5, CXCL10,CCNE1,MCM8, etc | 0.001986 |
| GO:0042802 | Identical protein binding | 10 | DDX58, IFIT3, STK10, TGFB3, RIPK2, CTSC, FADD, etc | 0.004431 |
| GO:0004674 | Protein serine/threonine kinase activity | 7 | STK10, TRIO, RIPK2, CDK4, NEK6, CDK2, ACVR1 | 0.005327 |
| GO:0005524 | ATP binding | 14 | STK10, TRIO, CDK4, CDK2, ABCG1, RAD51, EPHB2,etc | 0.009935 |
| GO:0048248 | CXCR3 chemokine receptor binding | 2 | CXCL9, CXCL10 | 0.021147 |
| GO:0003690 | Double-stranded DNA binding | 3 | DDX58, FEN1, RAD51 | 0.046830 |
Notes: Only top 10 was were presented in the table.
Table 5.
KEGG pathway of validated target genes of miR-34a
| KEGG ID | KEGG term | Count (%) | Gene symbol | P |
|---|---|---|---|---|
| hsa05200 | Pathways in cancer | 11 | CCNE1, CBLB, COL4A1, TGFB3, FGF11, EGLN3, FADD, etc | 3.55E-05 |
| hsa05222 | Small cell lung cancer | 5 | CCNE1, COL4A1, LAMC1, CDK4, CDK2 | 0.001145 |
| hsa04115 | p53 signaling pathway | 4 | CCNE1, SERPINE1, CDK4, CDK2 | 0.005769 |
| hsa04151 | PI3K-Akt signaling pathway | 7 | CCNE1, COL4A1, FGF11, LAMC1, CDK4, BRCA1, CDK2 | 0.010561 |
| hsa04110 | Cell cycle | 4 | CCNE1, TGFB3, CDK4, CDK2 | 0.030152 |
| hsa04623 | Cytosolic DNA-sensing pathway | 3 | DDX58, POLR2H, CXCL10 | 0.047955 |
| hsa05212 | Pancreatic cancer | 3 | TGFB3, CDK4, RAD51 | 0.049310 |
Figure 10.
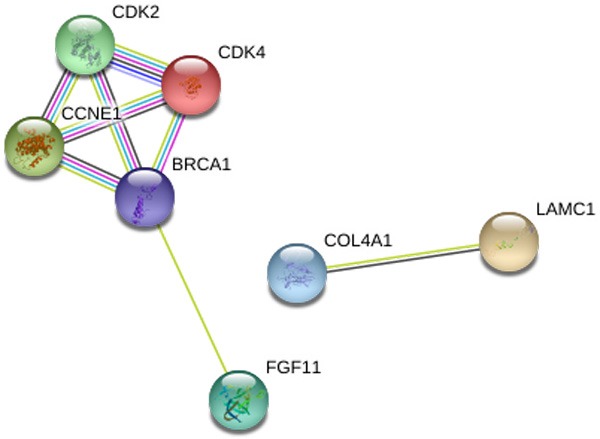
The PPI network to show links of the relationship among the 7 genes associated with the pi3k-akt signaling pathway. The nodes represent various predicted genes of pi3k-akt signaling pathway. The lines are interactions between nodes.
Validation of mir-34a target genes in the pi3k-akt signaling pathway
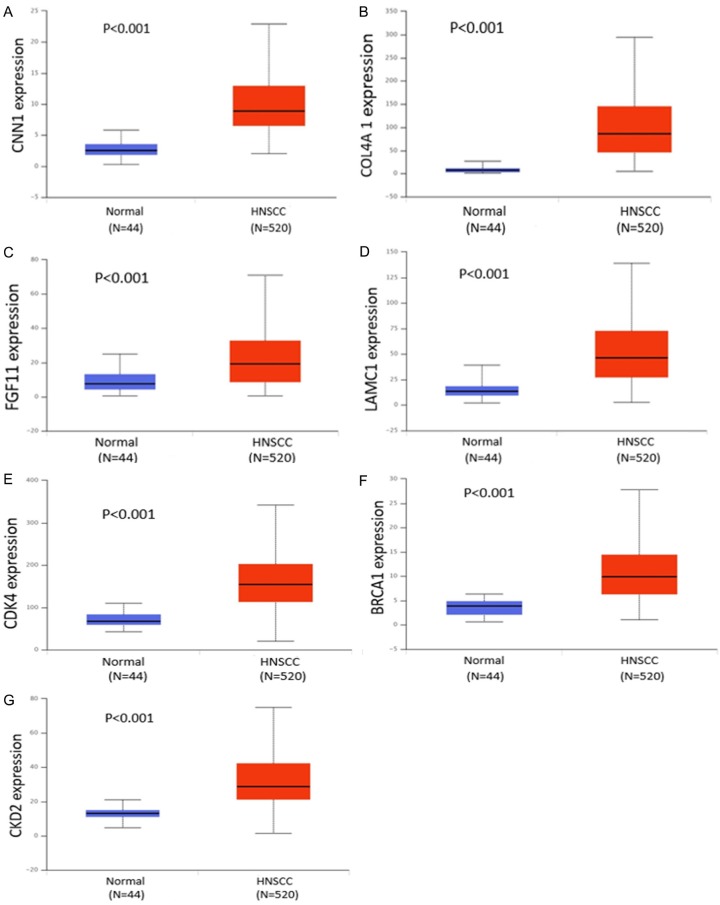
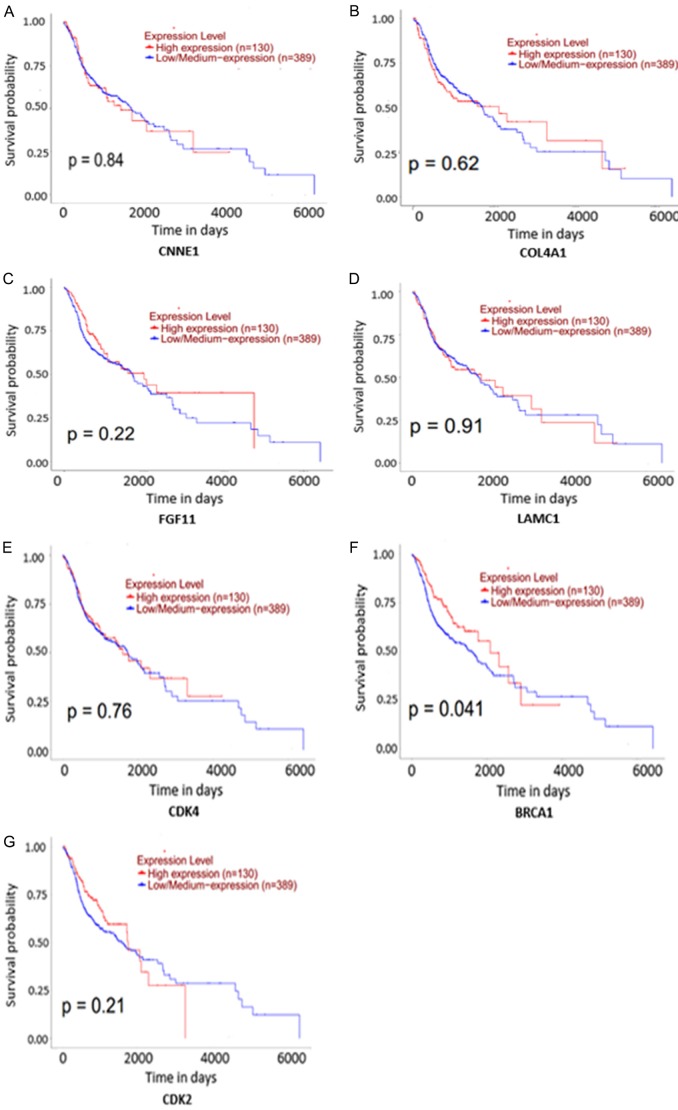
In HNSCC and compared with normal tissues, 7 target genes associated with miR-34a and PI3K-Akt signaling pathway were upregulated (P<0.001). The expression level for each gene is shown in Figure 11A-G. The correlation of identified target genes with miR-34a in HNSCC is shown in Figure 12A-G. Finally the relationship between each gene expression level and prognosis is shown in Figure 13A-G. Collectively, these results revealed that HNSCC patients with high BRCA1 expression have better overall survival than those with low-expression (P<0.05).
Figure 11.
The expression levels of 7 target genes of miR-34a associated with the PI3K-Akt signaling pathway.
Figure 12.
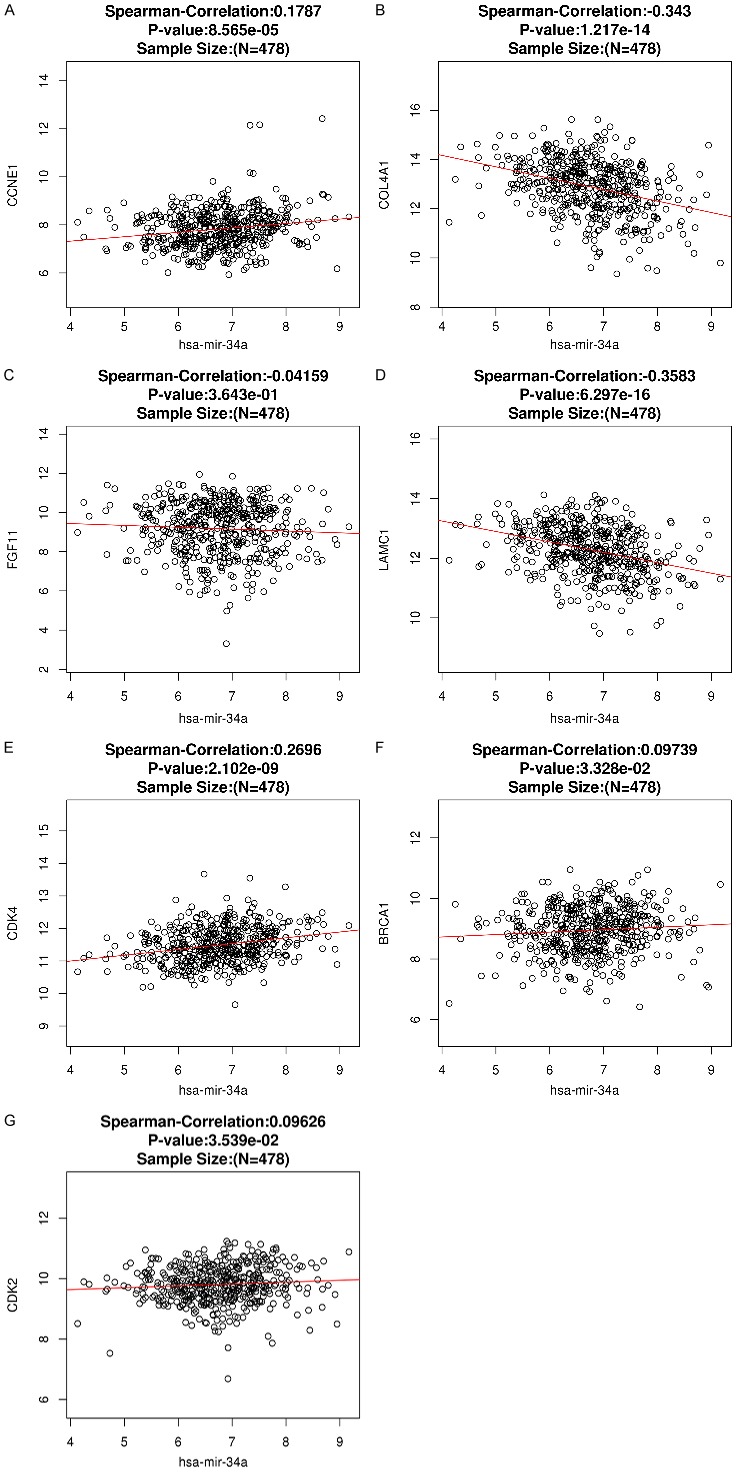
Spearman’s analysis to show the correlations between miR-34a expression levels and the validated target genes involved in the PI3K-Akt signaling pathway.
Figure 13.
The survival curves of 7 target genes of miR-34a associated with the PI3K-Akt signaling pathway.
Discussion
In recent years, many studies have shown that miR-34a has low expression in a variety of carcinomas; additionally, miR-34a overexpression can induce cell apoptosis, cycle stagnation, and senescence [24-27]. As a tumor suppressor gene, miR-34a plays an important role in invasion, metastasis, proliferation, and the epithelial-mesenchymal transformation seen in breast, prostate, and pancreatic cancers [6,28-30]. Moreover, miR-34a can be considered as a potential biomarker that can be used to estimate the clinical prognosis of non-small cell lung cancer, breast cancer, and other carcinomas [9,31-33]. However, the expression and underlying mechanism of miR-34a in HNSCC remains markedly unclear. Therefore, we sought to perform a meta-analysis to investigate miR-34a expression and its diagnostic significance in HNSCC. We also used GO and KEGG analyses to explore the potential molecular mechanisms of miR-34a in HNSCC.
The meta-analysis presented here involved 560 HNSCC cases and 75 controlled samples from the TCGA database along with three eligible studies obtained from the literature. The combined SMD was -0.70 (95% CI: -1.28, -0.11, random effect model) with high heterogeneity (I2=66.9%, P<0.05), indicating that miR-34a expression was significantly decreased in HNSCC. Given this, it is possible that miR-34a may play a role in HNSCC as a vital gene in tumorigenesis. According to the analysis of TCGA data, it was discovered that the miR-34a expression may also be related to pathologic grade in HNSCC. However, it is also worth mentioning that future studies with higher quality, larger sample sizes, and consistent procedural standards will be needed for more conclusive analysis.
Past work has examined the utility of miRNAs as biomarkers for use in diagnostic evaluation, therapy, and assessment of disease prognosis [34,35]. For instance, Mourad and colleagues demonstrated that miR-34a could be used as a biomarker with high sensitivity and specificity for HCV-Induced hepatocellular carcinoma [36]. Moreover, miR-34a expression between pancreatic ductal adenocarcinoma (PDAC) and healthy serum samples were significantly different , indicating that serum miR-34a could also be used as a diagnostic biomarker in PDAC [37]. Relatedly, Wu and colleagues also suggested that the abnormal methylation of the miR-34a gene was a potential biomarker for use in the diagnosis and noninvasive screening of colorectal cancer [38]. As to the potential diagnostic value of miR-34a in HNSCC, our diagnostic meta-analysis results suggested that miR-34a may serve as a biomarker with a high-sensitivity (sensitivity: 0.97, 95% CI: 0.95-0.98) but low- specificity (specificity: 0.33, 95% CI: 0.24-0.42). This indicates that miR-34a expression may be a novel biomarker to use in the clinical identification of HNSCC. Moreover, the sROC AUC (0.72) also indicated that miR-34a expression had significant diagnostic value in HNSCC.
At present, many studies have found that miR-34a can regulate some bioprocesses by targeting a combination of miRNAs and multiple proteins [6]. For instance, Li and colleagues found that miR-34a could inhibit metastasis and invasion by targeting the regulation c-met expression in liver cancer cells [39]. The study by Liu et al demonstrated that miR-34a could inhibit the metastasis of prostate cancer stem cells and prostate cancer by reducing CD44 expression [28]. Furthermore, miR-34a promotes apoptosis by means of regulating the expression of SIRT1 in colon cancer cells [40]. However, little advance has been made toward the understanding of the miRNA-34a molecular mechanisms in HNSCC. Therefore, after evaluating the expression level of miR-34a in HNSCC, we conducted a bioinformatics analysis of the underlying molecular mechanisms to predict the target genes and signaling pathways of miR-34a in HNSCC. We collected 77 genes targeted by miRNA-34a and performed a comprehensive target gene network analysis. GO enrichment analysis showed miR-34a target genes significantly were enriched during cell cycle and cell proliferation, both of which are associated with oncology. From our KEGG pathway analysis, results revealed that miRNA-34a may play a pivotal role in HNSCC through multiple pathways including the PI3K-Akt and p53 signaling pathways.
P53 is the most likely gene to mutate in human cancer and has been also confirmed as an important tumor suppressor gene [41]. The role of p53 in tumor formation is also related to mediating target genes, which are themselves regulated by miRNAs [42,43]. This suggests that miRNAs are new members important to the p53 gene signal pathway. In 2007, miR-34 was identified as the first miRNA relevant to p53; additionally, it was found that it induces apoptosis, senescence, and cell cycle arrest in the development of cancer [44]. When cells are stimulated, p53 methylates the promoter region of the CpG island, thereby activating the expression and regulating the target genes of miR-34a. To this end, Hermeking [1] holds that the tumor suppressor gene p53 can directly promote miR-34a expression by inducing apoptosis and cell cycle arrest. Additionally, Mandke and colleagues [45] believed that p53 plays an inhibitory role in colon cancer by regulating the miR-34a target gene MDM4. However, whether miRNA-34a is also involved in the progression of HNSCC through the p53 signaling pathway remained unknown. Our research now suggests that miR-34a may play an important role as a cancer suppressor gene by activating the p53 pathway in HNSCC.
The PI3K-Akt signaling pathway is an essential signal transduction pathway in cells, an important biological function in cell apoptosis, survival, proliferation, and cytoskeletal changes [46]. Tumor development is often accompanied by abnormal changes in PI3K signaling pathways. For instance, Elkabets and colleagues [47] suggested that excessive PI3K/AKT activation is closely related to the migration, proliferation, and invasion of HNSCC cells. Moreover, inhibition of the PI3k-Akt pathway can increase the efficacy of HNSCC [48]. Here, our TCGA data indicated that all seven genes related to miR-34a and the PI3k-Akt signaling pathway were up-regulated in HNSCC. Of these, the CDK2 protein has been determined to be a prognostic biomarker for nasopharyngeal carcinoma. This is because its expression has the closest connections to clinical stage, infiltration degree, and lymph node metastasis [49]. Furthermore, serum CDK4 levels of older HNSCC patients were elevated, indicating its potential for use as a clinical biomarker [50]. Moreover, BRCA1 could predict an inverse CDDP/taxane sensitivity phenotype as a candidate marker in HNSCC [51]. According to our study, there were significant correlations between overall survival and BRCA1 expression levels in HNSCC (P<0.05), indicating it might be an independent factor in predicting the overall survival of HNSCC patients. Of course, further study to research the mechanism action of miRNA-34a in HNSCC will be necessary.
However, there are a number of limitations to mention regarding this study. First, many of these findings have not been validated by experiment. In the near future, our conclusions will need to be validated using both in vitro and in vivo clinical and molecular biological approaches. Second, this study is restricted in that the analysis of differential miRNAs was only based on HNSCC and non-cancerous tissues. Other samples (e.g. blood) were not assessed. Finally, only Chinese and English studies were used for the basis of our meta-analysis and did not included other potentially relevant studies that were in other languages.
In conclusion, this study demonstrated the downregulation of miR-34a in HNSCC. To summarize, these data suggest that miR-34a may play an essential role in the tumorigenesis and progression of HNSCC through specific pathways. MiR-34a could be a novel and promising non-invasive biomarker in diagnosing HNSCC. Larger scale studies will be needed to validate its diagnostic promise. Finally, the biological pathways relevant to the actions of miR-34a will guide further understanding of the mechanisms of HNSCC. Hopefully, additional work will validate its usefulness as a target for future clinical research.
Acknowledgements
The present study was supported by grants from the National Natural Science Foundation of China (grant nos. 81460460, 81760542), The Research Foundation of the Science and Technology Department of Guangxi Province, China (grant nos. 2016GXNSFAA380252 and 2014GXNSFBA118114), Guangxi Medical University Training Program for Distinguished Young Scholars, Medical Excellence Award Funded by the Creative Research Development Grant from the First Affiliated Hospital of Guangxi Medical University, The central government guide local science and technology development projects (ZY18057006).
Disclosure of conflict of interest
None.
References
- 1.Hermeking H. p53 enters the microRNA world. Cancer Cell. 2007;12:414. doi: 10.1016/j.ccr.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 2.Farooqi AA, Tabassum S, Ahmad A. MicroRNA-34a: a versatile regulator of myriads of targets in different cancers. Int J Mol Sci. 2017:18. doi: 10.3390/ijms18102089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ji X, Wang Z, Geamanu A, Goja A, Sarkar FH, Gupta SV. Delta-tocotrienol suppresses Notch-1 pathway by upregulating miR-34a in nonsmall cell lung cancer cells. Int J Cancer. 2012;131:2668. doi: 10.1002/ijc.27549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duan W, Xu Y, Dong YJ, Cao L, Jian T, Zhou X. Ectopic expression of miR-34a enhances radiosensitivity of non-small cell lung cancer cells, partly by suppressing the LyGDI signaling pathway. J Radiat Res. 2013;54:611–619. doi: 10.1093/jrr/rrs136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 7.Yan K, Gao J, Yang T, Ma Q, Qiu X, Fan Q, Ma B. MicroRNA-34a inhibits the proliferation and metastasis of osteosarcoma cells both in vitro and in vivo. PLoS One. 2012;7:e33778. doi: 10.1371/journal.pone.0033778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li L, Yuan L, Luo J, Gao J, Guo J, Xie X. MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clin Exp Med. 2013;13:109–117. doi: 10.1007/s10238-012-0186-5. [DOI] [PubMed] [Google Scholar]
- 9.Gallardo E, Navarro A, Viñolas N, Marrades RM, Diaz T, Gel B, Quera A, Bandres E, Garciafoncillas J, Ramirez J. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis. 2009;30:1903–1909. doi: 10.1093/carcin/bgp219. [DOI] [PubMed] [Google Scholar]
- 10.Zhang D, Zhou J, Dong M. Dysregulation of microRNA-34a expression in colorectal cancer inhibits the phosphorylation of FAK via VEGF. Dig Dis Sci. 2014;59:958–967. doi: 10.1007/s10620-013-2983-4. [DOI] [PubMed] [Google Scholar]
- 11.Nalls D, Tang SN, Rodova M, Srivastava RK, Shankar S. Targeting epigenetic regulation of miR-34a for treatment of pancreatic cancer by inhibition of pancreatic cancer stem cells. PLoS One. 2011;6:e24099. doi: 10.1371/journal.pone.0024099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 13.Svider PF, Blasco MA, Raza SN, Shkoukani M, Sukari A, Yoo GH, Folbe AJ, Lin HS, Fribley AM. Head and neck cancer. Otolaryngol Head Neck Surg. 2017;156:10–13. doi: 10.1177/0194599816674672. [DOI] [PubMed] [Google Scholar]
- 14.Lamperska KM, Kozlowski P, Kolenda T, Teresiak A, Renata B, Weronika P, Masternak MM, Golusinski P, Golusinski W. Unpredictable changes of selected miRNA in expression profile of HNSCC. Cancer Biomarkers. 2015;16:55. doi: 10.3233/CBM-150540. [DOI] [PubMed] [Google Scholar]
- 15.Kumar B, Yadav A, Lang J, Teknos TN, Kumar P. Dysregulation of microRNA-34a expression in head and neck squamous cell carcinoma promotes tumor growth and tumor angiogenesis. PLoS One. 2012;7:e37601. doi: 10.1371/journal.pone.0037601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anaya J. OncoLnc: Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. Peerj Computer Science. 2016;2:e67. [Google Scholar]
- 17.Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12:697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 18.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M, Goto S, Sato Y, Furumichi M, Mao T. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:109–114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chandrashekar DS, Bashel B, Creighton CJ, Poncerodriguez I, Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46:D956–D963. doi: 10.1093/nar/gkx1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manikandan M, Deva Magendhra Rao AK, Arunkumar G, Rajkumar KS, Rajaraman R, Munirajan AK. Down regulation of miR-34a and miR-143 may indirectly inhibit p53 in oral squamous cell carcinoma: a pilot study. Asian Pac J Cancer Prev. 2015;16:7619–25. doi: 10.7314/apjcp.2015.16.17.7619. [DOI] [PubMed] [Google Scholar]
- 23.Li W, Ma H, Sun J. MicroRNA-34a/c function as tumor suppressors in Hep-2 laryngeal carcinoma cells and may reduce GALNT7 expression. Mol Med Rep. 2014;9:1293–1298. doi: 10.3892/mmr.2014.1929. [DOI] [PubMed] [Google Scholar]
- 24.Cole KA, Attiyeh EF, Mosse YP, Laquaglia MJ, Diskin SJ, Brodeur GM, Maris JM. A functional screen identifies miR-34a as a candidate neuroblastoma tumor suppressor gene. Mol Cancer Res. 2008;6:735. doi: 10.1158/1541-7786.MCR-07-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Körner H, Knyazev P, Diebold J, Hermeking H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 26.Corney DC, Hwang CI, Matoso A, Vogt M, Flesken-Nikitin A, Godwin AK, Kamat AA, Sood AK, Ellenson LH, Hermeking H, Nikitin AY. Frequent downregulation of miR-34 family in human ovarian cancers. Clin Cancer Res. 2010;16:1119–1128. doi: 10.1158/1078-0432.CCR-09-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang W, Li T, Han G, Li Y, Shi LH, Li H. Expression and role of miR-34a in bladder cancer. Indian J Biochem Biophys. 2013;50:87–92. [PubMed] [Google Scholar]
- 28.Liu C, Kelnar K, Liu B, Chen X, Calhoundavis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17:211. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, Xiang D, Desano JT, Bommer GT, Fan D. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One. 2009;4:e6816. doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siemens H, Jackstadt R, Hünten S, Kaller M, Menssen A, Götz U, Hermeking H. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10:4256–4271. doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
- 31.Peurala H, Greco D, Heikkinen T, Kaur S, Bartkova J, Jamshidi M, Aittomäki K, Heikkilä P, Bartek J, Blomqvist C. MiR-34a expression has an effect for lower risk of metastasis and associates with expression patterns predicting clinical outcome in breast cancer. PLoS One. 2011;6:157–164. doi: 10.1371/journal.pone.0026122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho WC. MicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol. 2010;42:1273–1281. doi: 10.1016/j.biocel.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 33.Idowu MO, Kmieciak M, Dumur C, Burton RS, Grimes MM, Powers CN, Manjili MH. CD44(+)/CD24(-/low) cancer stem/progenitor cells are more abundant in triple-negative invasive breast carcinoma phenotype and are associated with poor outcome. Breast Cancer Res Treat. 2012;43:364–373. doi: 10.1016/j.humpath.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 34.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13:358. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 36.Mourad L, El-Ahwany E, Zoheiry M, Abu-Taleb H, Hassan M, Ouf A, Rahim AA, Hassan M, Zada S. Expression analysis of liver-specific circulating microRNAs in HCV-induced hepatocellular carcinoma in egyptian patients. Cancer Biol Ther. 2018;19:400–406. doi: 10.1080/15384047.2018.1423922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alemar B, Izetti P, Gregório C, Macedo GS, Castro MA, Osvaldt AB, Matte U, Ashton-Prolla P. miRNA-21 and miRNA-34a are potential minimally invasive biomarkers for the diagnosis of pancreatic ductal adenocarcinoma. Pancreas. 2016;45:84–92. doi: 10.1097/MPA.0000000000000383. [DOI] [PubMed] [Google Scholar]
- 38.Wu XD, Song YC, Cao PL, Zhang H, Guo Q, Yan R, Diao DM, Cheng Y, Dang CX. Detection of miR-34a and miR-34b/c in stool sample as potential screening biomarkers for noninvasive diagnosis of colorectal cancer. Med Oncol. 2014;31:1–6. doi: 10.1007/s12032-014-0894-7. [DOI] [PubMed] [Google Scholar]
- 39.Li N, Fu H, Tie Y, Hu Z, Kong W, Wu Y, Zheng X. miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett. 2009;275:44. doi: 10.1016/j.canlet.2008.09.035. [DOI] [PubMed] [Google Scholar]
- 40.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A. 2008;105:13421. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kandoth C, Mclellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, Mcmichael JF, Wyczalkowski MA. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takwi A, Li Y. The p53 pathway encounters the MicroRNA world. Current Genomics. 2009;10:194–7. doi: 10.2174/138920209788185270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao JM, Cao B, Zhou X, Lu H. New insights into p53 functions through its target microRNAs. J Mol Cell Biol. 2014;6:206–213. doi: 10.1093/jmcb/mju018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, MacDougald OA, Cho KR, Fearon ER. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 45.Mandke P, Wyatt N, Fraser J, Bates B, Berberich SJ, Markey MP. MicroRNA-34a modulates MDM4 expression via a target site in the open reading frame. PLoS One. 2012;7:e42034. doi: 10.1371/journal.pone.0042034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei LT, Feng J, Shu-Rong MO. Progress in correlation of PI3K-Akt signal pathway with tumor. Journal of Chinese Oncology. 2014 [Google Scholar]
- 47.Elkabets M, Pazarentzos E, Juric D, Sheng Q, Pelossof R, Brook S, Benzaken AO, Rodon J, Morse N, Yan JJ. AXL Mediates resistance to PI3Kα inhibition by activating the EGFR/PKC/mTOR Axis in head and neck and esophageal squamous cell carcinomas. Cancer Cell. 2015;27:533. doi: 10.1016/j.ccell.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leiker A, Degraff WG, Choudhuri R, Sowers AL, Thetford A, Cook JA, Van WC, Mitchell JB. Radiation enhancement of head and neck squamous cell carcinoma by the Dual PI3K/mTOR inhibitor PF-05212384. Clin Cancer Res. 2014;90:S196–S197. doi: 10.1158/1078-0432.CCR-14-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guan SX, Jiang YH, Wang JQ. The expression and significance of CDK-2 in nasopharyngeal carcinoma. Chinese Journal of General Practice. 2011 [Google Scholar]
- 50.Banerjee J, Pradhan R, Gupta A, Kumar R, Sahu V, Upadhyay AD, Chaterjee P, Dwivedi S, Dey S, Dey AB. CDK4 in lung, and head and neck cancers in old age: evaluation as a biomarker. Clin Transl Oncol. 2017;19:571–578. doi: 10.1007/s12094-016-1565-2. [DOI] [PubMed] [Google Scholar]
- 51.Saiki Y, Ogawa T, Shiga K, Sunamura M, Kobayashi T, Horii A. A human head and neck squamous cell carcinoma cell line with acquired cis-diamminedichloroplatinum-resistance shows remarkable upregulation of BRCA1 and hypersensitivity to taxane. Int J Otolaryngol. 2011;2011:521852. doi: 10.1155/2011/521852. [DOI] [PMC free article] [PubMed] [Google Scholar]