Abstract
Pancreatic cancer (PC) is a devastating human disease with aggressive course and extremely poor prognosis. Long non-coding RNAs (lncRNAs) have been studied to serve as a critical role in pancreatic development and progression. However, little is known about its expression pattern, biological function in PC. In our study, we measured the expression levels of six lncRNAs (LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9) in PC tissues and serums. The results showed that LINC00346, LINC00578, and LINC00673 were highly expressed, whereas LINC00671, LINC00261, and SNHG9 were lowly expressed in PC tissues and serums, and their expression levels were correlated with clinical stages. Results from receiver operating characteristic (ROC) curve analysis showed that the area under the curve (AUC) of six lncRNAs was 0.7073, 0.7837, 0.6093, 0.6057, 0.5712, and 0.5983, respectively. Survival analysis indicated that patients with high expression of LINC00346, LINC00578, or LINC00673 had significantly lower survival rate, while patients with high expression of LINC00671, LINC00261, and SNHG9 had significantly higher survival rate. In addition, we also found that silence of LINC00346, LINC00578 and LINC00673 inhibited PC cell proliferation, and silence of LINC00671, LINC00261, and SNHG9 promoted PC cell proliferation. Therefore, we suggested that LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 may be novel prognostic markers for PC.
Keywords: Long non-coding RNA, pancreatic cancer, prognosis, proliferation
Introduction
Pancreatic cancer (PC), characterized by early metastasis and high mortality, is identified as one of the most lethal human diseases with extremely poor prognosis in the 21st century [1-5]. It has been estimated that the total new cases of PC in 2016 is 53070, and 41780 patients died of this disease in the United States, and the 5-year relative survival is currently 8% [6]. The high mortality of PC could be due to a variety of reasons, including the lack of early symptoms and effective measures of detection. Thus, the most PC patients are diagnosed at a late clinical stage, and they generally die from metastatic disease [7,8]. The conventional treatments of PC mainly include surgery, radiation, and chemotherapy, with very limited effects on the progression of pancreatic tumor [1,9]. Therefore, it is extremely urgent to understand the molecular mechanism of PC, and identify new effective measures for the patients of pancreatic carcinoma.
It has been well demonstrated that only 1-2% of the genome have the ability of encode proteins in the mammal, and more than 90% of them are transcribed as non-coding RNAs (ncRNAs), including microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) [10,11]. MicroRNAs, play critical roles in the regulation of many biological processes, are transcripts defined by a length of 19 to 25 nucleotides [12,13]. These small molecules could trigger their regulatory effects through binding to the specific mRNA, causing inhibition of translation to functional proteins in mammals [14]. Previous studies have revealed that miRNAs involved in cell proliferation, apoptosis, and metastasis in various cancers, such as prostate cancer, breast cancer, and PC [15-17]. And lncRNAs, with a size over 200 nucleotides, are an important subgroup of ncRNAs that participated in numerous of pathological process during cancers such as tumor cell proliferation, differentiation, and apoptosis [18,19]. Although the exact functions of most lncRNAs remain unclear, increasing evidences have indicated lncRNAs play an important role in several tumors, such as ovarian cancer, nasopharyngeal carcinoma, and lung cancer [20-22]. Accumulated lncRNAs were demonstrated to be involved in the pathogenesis, diagnosis, or prognosis of PC, such as HOTAIR, MALAT1 and HOTTIP [23-25]. It suggested that lncRNAs may play as critical agents in the progression of PC.
In our study, we explored the expression levels of three lncRNAs (LINC00346, LINC00578, and LINC00673) PC tissues and serums and the correlation of lncRNAs between tissues and serums. we analyzed the prognosis and survival analysis using receiver operating characteristics (ROC) and Kaplan-Meier method. Furthermore, we demonstrated the roles of lncRNAs on PC proliferation.
Materials and methods
Samples collection (tissues and serum)
PC tumor tissue samples were collected from patients who were diagnosed with PC. In total, 229 paired of PC tissues (137 cases of I and II phases, and 92 cases of III and IV phases) and adjacent non-tumor tissues were freshly frozen in liquid nitrogen and stored at -80°C until further use. The serum samples were prepared from PC patients or health controls (n = 229), and stored at -80°C. The use of tissues for this study has been approved by the ethics committee of Japan Union Hospital of Jilin University. The clinical stage of PC of all patients was classified according to the TNM classification system of the American Joint Committee on Cancer. Before using these clinical materials for research purposes, all the patients have signed the informed consent. None of these patients received any pre-operative chemotherapy or radiotherapy.
Cell culture
HPAC and BxPC-3 cells were all obtained from the American Type Culture Collection (ATCC). All of these cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Cat#: 11960-044, Invitrogen, USA) supplementing with 100 U/ml penicillin, 100 μg/ml streptomycin and 10% fetal bovine serum (FBS) in a 5% CO2 and 95% O2 atmosphere at 37°C.
siRNA interference
Control siRNAs and siRNAs for LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 were obtained from GenePharma Co., Ltd (Shanghai, China). HPAC and BxPC-3 cells (2×104 cells/well) in the logarithmic phase were seeded into 6-well plates in 2 mL of medium at 37°C. Next day, cells were transfected with 50 μM scramble siRNA (Negative control, NC), LINC00346-siRNAs, LINC00578-siRNAs, LINC00673-siRNAs, LINC00671-siRNAs, LINC00261-siRNAs, and SNHG9-siRNAs using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer’s protocol.
RNA extraction and quantitative real-time PCR (qRT-PCR)
The blood samples of PC patients or health controls were collected and clotted at room temperature and then were centrifuged for at least 10 mins at 4°C with 4000 rpm to harvest the serum. Total RNAs of serum samples (1 ml) was extracted by the mirVana PARIS Kit (Ambion, Austin, TX, USA) following the instructions of manufacturer. Total RNA of tissue samples was isolated by using the TRizol reagent (Invitrogen Corp., Carlsbad, CA), according to the manufacturer’s protocols. Total RNAs of serum samples and tissues samples were reverse transcribed into cDNA by the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, PN4366596). The expression of lncRNAs in tissues and serum were quantified by the KAPA SYBR FAST qPCR Kit (Kapa Biosystems, KK4601). GADPH was used as internal control. The reaction system was in a 20 μl volume, containing 10 μl of 2×SYBR Green I, 0.3 μl sense primer, 0.3 μl antisense primer, 6.4 μl distilled water and 3 μl cDNA template. And the program of qPCR was set as following: 95°C for 10 minutes, 95°C for 10 seconds, 60°C (set as the annealing temperature) for 2 min, 72°C for 2 min, 72°C for 10 min, and 38 amplification cycles were performed from second step to fourth step. The levels of expressed genes were measured by the 2-ΔΔCt method with MxPro 4.00 (Stratagene). The primers used in this research were showed in Table 1.
Table 1.
The primer sequences in qRT-PCR assay
| LncRNAs | Sequence of primers |
|---|---|
| LINC00346 | Forward: 5’-GGAAACGGTCTAGTGGTGGA-3’ |
| Reverse: 5’-AAGCCCTTCTTCACCCTCTC-3’ | |
| LINC00578 | Forward: 5’-GATGGAGAGAGAGCCTGGAC-3’ |
| Reverse: 5’-GCACTCCAACTGTTGCTGAT-3’ | |
| LINC00673 | Forward: 5’-CCGTGTAAAGAGGCCAGTGT-3’ |
| Reverse: 5’-ACACGAGCCTTCACCATCAG-3’ | |
| LINC00671 | Forward: 5’-ATGGGAAACTGGCCAGATCA-3’ |
| Reverse: 5’-TTCTCTGGCATTCCTCCTCC-3’ | |
| LINC00261 | Forward: 5’-ATCAAGAGGCAATGGTCCCA-3’ |
| Reverse: 5’-TTCAGCTCTTAGGGCAGGAC-3’ | |
| SNHG9 | Forward: 5’-GACTGCAGACCCCTAACCTT-3’ |
| Reverse: 5’-ACCCGCATGCAGTGAGTTA-3’ | |
| GAPDH | Forward: 5’-TATGATGATATCAAGAGGGTAGT-3’ |
| Reverse: 5’-TGTATCCAAACTCATTGTCATAC-3’ |
MTT assay
The treated cells were seeded into a 96-well plate at a density of 2×103 cells/well, and maintained under 37°C for 24 hrs. at 24, 36, 48 hrs, MTT solution (50 µl) was added to each well and incubated it at 37°C for 4 hrs. Next, the medium was removed and DMSO (200 µl) was added into each well. The absorbance of each well was measured by a microplate reader at 490 nm.
Statistical analysis
The unpaired t test was applied to test the differential expression of lncRNAs in cancer tissues compared to adjacent non-cancerous tissues. The paired t-test was used for comparison of differential expression of lncRNAs between pre and postoperative samples. The one-way analysis of variance was used to compare the differential expression of lncRNAs between normal and PC patients at different TNM stages. ROC was generated using logistic regression models to evaluate the diagnostic performance of various lncRNAs. Area under curve (AUC) was used as the evaluation criteria; the higher AUC, the better diagnostic performance.
Data of this study are presented as the mean values ± standard deviation (SD) and differences were considered statistically significant at P < 0.05. SPSS 21.0 software (SPSS, Inc., Chicago, IL, USA) and GraphPad (GraphPad Prism Software, La Jolla, CA, USA) were used to conduct the analysis.
Results
Expression of six different lncRNAs in PC tissues
In order to investigate the potential biological roles of lncRNA in PC, we firstly performed qRT-PCR to examine the expression levels of six different lncRNAs (LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9) in PC tissues. As results showed that three of them (LINC00346, LINC00578, and LINC00673) were highly expressed (**P < 0.01, ***P < 0.001, tumor tissues vs. normal tissues) (Figure 1A), whereas the other three lncRNAs (LINC00671, LINC00261, and SNHG9) were lowly expressed, in the PC tissues compared with corresponding normal tissues (*P < 0.05, **P < 0.01, tumor tissues vs. normal tissues) (Figure 1B). In addition, we also found that the expression levels of LINC00346, LINC00578, and LINC00673 were positively correlated with the clinical stages, and the expression levels of LINC00671, LINC00261, and SNHG9 were negatively correlated with the clinical stages (*P < 0.05, **P < 0.01, ***P < 0.001) (Figure 1C and 1D). Furthermore, we found that LINC00346 expression was significantly related to pathological node (P = 0.001) (Table 2); LINC00578 expression was significantly in connection with pathological node (P = 0.018) and pathological metastasis (P = 0.044) (Table 3); LINC00673 expression was significantly connected with pathological metastasis (P = 0.007) (Table 4); LINC00671 expression was significantly relevant to pathological node (P = 0.007) (Table 5); LINC00261 expression was significantly relevant to pathological node (P = 0.018) (Table 6); SNHG9 expression was significantly connected with pathological node (P = 0.004) and pathological metastasis (P = 0.028) (Table 7).
Figure 1.
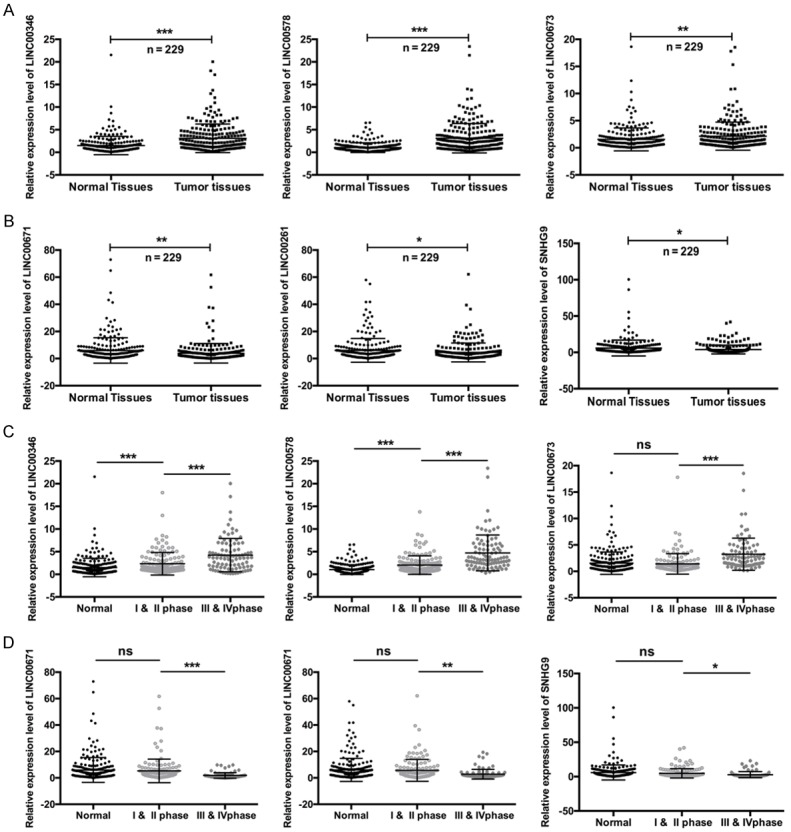
The expression levels of six different lncRNAs in the tissues of PC patients. A and B: The expression levels of LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 in PC tissues and corresponding normal tissues were measured by qRT-PCR (*P < 0.01, **P < 0.05, ***P < 0.001). C and D: The expression levels of the six lncRNAs in the different clinical stages of PC were examined by qRT-PCR (*P < 0.01, **P < 0.05, ***P < 0.001).
Table 2.
The correlation between LINC00346 expression and clinical features in 229 patients with PCa
| All cases | LINC00346 | |||
|---|---|---|---|---|
|
| ||||
| High expression | Low expression | P value | ||
| Age (year) | ||||
| > 60 | 108 | 52 (48.1%) | 56 (51.9%) | 0.516 |
| ≤ 60 | 121 | 59 (48.8%) | 62 (51.2%) | |
| Differentiation | ||||
| Well | 69 | 31 (44.9%) | 38 (55.1%) | 0.520 |
| Moderate | 87 | 41 (47.1%) | 46 (52.9%) | |
| Poor | 73 | 32 (43.8%) | 41 (56.2%) | |
| pN status | 0.001 | |||
| N0 | 123 | 47 (38.2%) | 76 (61.8%) | |
| N1-N2 | 106 | 64 (60.4%) | 42 (39.6%) | |
| pM status | 0.100 | |||
| M0 | 156 | 75 (47.5%) | 83 (52.5%) | |
| M1 | 73 | 42 (57.5%) | 31 (42.5%) | |
| Clinical stage | 0.391 | |||
| I | 79 | 46 (58.2%) | 33 (41.8%) | |
| II | 58 | 27 (46.6%) | 31 (53.4%) | |
| III | 53 | 34 (64.2%) | 19 (35.8%) | |
| IV | 39 | 22 (56.4%) | 17 (43.6%) | |
pN: pathological node; pM: pathological metastasis.
Table 3.
The correlation between LINC00578 expression and clinical features in 229 patients with PCa
| All cases | LINC00578 | |||
|---|---|---|---|---|
|
| ||||
| High expression | Low expression | P value | ||
| Age (year) | 0.448 | |||
| > 60 | 108 | 49 (45.4%) | 59 (54.6%) | |
| ≤ 60 | 121 | 57 (47.1%) | 64 (52.9%) | |
| Differentiation | ||||
| Well | 69 | 37 (53.6%) | 32 (46.4%) | 0.461 |
| Moderate | 87 | 45 (51.7%) | 42 (48.3%) | |
| Poor | 73 | 38 (52.1%) | 35 (47.9%) | |
| pN status | 0.018 | |||
| N0 | 123 | 53 (43.1%) | 70 (56.9%) | |
| N1-N2 | 106 | 71 (57.3%) | 53 (42.7%) | |
| pM status | 0.044 | |||
| M0 | 156 | 63 (40.4%) | 93 (59.6%) | |
| M1 | 73 | 39 (53.4%) | 34 (46.6%) | |
| Clinical stage | 0.422 | |||
| I | 79 | 44 (55.7%) | 35 (44.3%) | |
| II | 58 | 31 (53.4%) | 27 (46.6%) | |
| III | 53 | 32 (60.4%) | 21 (39.6%) | |
| IV | 39 | 24 (61.5%) | 15 (38.5%) | |
pN: pathological node; pM: pathological metastasis.
Table 4.
The correlation between LINC00673 expression and clinical features in 229 patients with PCa
| All cases | LINC00673 | |||
|---|---|---|---|---|
|
| ||||
| High expression | Low expression | P value | ||
| Age (year) | 0.510 | |||
| > 60 | 108 | 57 (52.8%) | 51 (47.2%) | |
| ≤ 60 | 121 | 63 (52.1%) | 58 (47.9%) | |
| Differentiation | ||||
| Well | 69 | 36 (52.2%) | 33 (47.8%) | 0.472 |
| Moderate | 87 | 46 (52.9%) | 41 (47.1%) | |
| Poor | 73 | 35 (47.9%) | 38 (52.1%) | |
| pN status | 0.050 | |||
| N0 | 123 | 54 (43.9%) | 69 (56.1%) | |
| N1-N2 | 106 | 59 (55.7%) | 47 (44.3%) | |
| pM status | 0.007 | |||
| M0 | 156 | 63 (40.4%) | 93 (59.6%) | |
| M1 | 73 | 43 (58.9%) | 30 (41.1%) | |
| Clinical stage | 0.325 | |||
| I | 79 | 48 (60.8%) | 31 (39.2%) | |
| II | 58 | 31 (53.4%) | 27 (46.6%) | |
| III | 53 | 33 (62.3%) | 20 (37.7%) | |
| IV | 39 | 21 (53.8%) | 18 (46.2%) | |
pN: pathological node; pM: pathological metastasis.
Table 5.
The correlation between LINC00671 expression and clinical features in 229 patients with PCa
| All cases | LINC00671 | |||
|---|---|---|---|---|
|
| ||||
| High expression | Low expression | P value | ||
| Age (year) | 0.486 | |||
| > 60 | 108 | 61 (56.5%) | 47 (43.5%) | |
| ≤ 60 | 121 | 67 (55.4%) | 54 (44.6%) | |
| Differentiation | ||||
| Well | 69 | 36 (52.2%) | 33 (47.8%) | 0.470 |
| Moderate | 87 | 48 (55.2%) | 39 (44.8%) | |
| Poor | 73 | 38 (52.1%) | 35 (47.9%) | |
| pN status | 0.007 | |||
| N0 | 123 | 71 (57.7%) | 52 (42.3%) | |
| N1-N2 | 106 | 43 (40.6%) | 63 (59.4%) | |
| pM status | 0.141 | |||
| M0 | 156 | 84 (53.8%) | 72 (46.2%) | |
| M1 | 73 | 33 (45.2%) | 40 (54.8%) | |
| Clinical stage | 0.221 | |||
| I | 79 | 45 (57.0%) | 34 (43.0%) | |
| II | 58 | 34 (58.6%) | 24 (41.4%) | |
| III | 53 | 26 (49.1%) | 27 (50.9%) | |
| IV | 39 | 16 (41.0%) | 23 (59.0%) | |
pN: pathological node; pM: pathological metastasis.
Table 6.
The correlation between LINC00261 expression and clinical features in 229 patients with PCa
| All cases | LINC00261 | |||
|---|---|---|---|---|
|
| ||||
| High expression | Low expression | P value | ||
| Age (year) | 0.446 | |||
| > 60 | 108 | 57 (52.8%) | 51 (47.2%) | |
| ≤ 60 | 121 | 66 (54.5%) | 55 (45.5%) | |
| Differentiation | 0.348 | |||
| Well | 69 | 34 (49.3%) | 35 (50.7%) | |
| Moderate | 87 | 46 (52.9%) | 41 (47.1%) | |
| Poor | 73 | 39 (53.4%) | 34 (46.6%) | |
| pN status | 0.018 | |||
| N0 | 123 | 75 (61.0%) | 48 (39.0%) | |
| N1-N2 | 106 | 49 (46.2%) | 57 (53.8%) | |
| pM status | 0.214 | |||
| M0 | 156 | 83 (53.2%) | 73 (46.8%) | |
| M1 | 73 | 34 (46.6%) | 39 (53.4%) | |
| Clinical stage | 0.092 | |||
| I | 79 | 47 (59.5%) | 32 (40.5%) | |
| II | 58 | 35 (60.3%) | 23 (39.7%) | |
| III | 53 | 21 (39.6%) | 32 (60.4%) | |
| IV | 39 | 18 (46.2%) | 21 (53.8%) | |
pN: pathological node; pM: pathological metastasis.
Table 7.
The correlation between SNHG9 expression and clinical features in 229 patients with PCa
| All cases | SNHG9 | |||
|---|---|---|---|---|
|
| ||||
| High expression | Low expression | P value | ||
| Age (year) | 0.294 | |||
| > 60 | 108 | 55 (50.9%) | 53 (49.1%) | |
| ≤ 60 | 121 | 67 (55.4%) | 54 (44.6%) | |
| Differentiation | 0.146 | |||
| Well | 69 | 43 (62.3%) | 26 (37.7%) | |
| Moderate | 87 | 47 (54.0%) | 40 (46.0%) | |
| Poor | 73 | 39 (53.4%) | 34 (46.6%) | |
| pN status | 0.004 | |||
| N0 | 123 | 78 (63.4%) | 45 (36.6%) | |
| N1-N2 | 106 | 48 (45.3%) | 58 (54.7%) | |
| pM status | 0.028 | |||
| M0 | 156 | 91 (58.3%) | 65 (41.7%) | |
| M1 | 73 | 32 (43.8%) | 41 (56.2%) | |
| Clinical stage | 0.149 | |||
| I | 79 | 47 (59.5%) | 32 (40.5%) | |
| II | 58 | 35 (60.3%) | 23 (39.7%) | |
| III | 53 | 24 (45.3%) | 29 (54.7%) | |
| IV | 39 | 18 (46.2%) | 21 (53.8%) | |
pN: pathological node; pM: pathological metastasis.
ROC curve analysis and survival analysis of six lncRNAs in PC
To further assess the biological functions, we performed ROC curve analysis to evaluate the diagnostic value of the six lncRNAs. The AUCs closer to 1 reflect more substantial differences between PC and normal tissues. As results indicated that the AUC of LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 was 0.7073, 0.7837, 0.6093, 0.6057, 0.5712, and 0.5983, respectively (Figure 2A and 2B), suggesting that these lncRNAs may serve as good prognostic indicator in PC. The survival analysis was performed to evaluate the survival rate of patients with high or low expression of different lncRNAs. And results suggested that patients with high expression levels of LINC00346 (P < 0.001), LINC00578 (P < 0.001), and LINC00673 (P = 0.005), or with low expression levels of LINC00671 (P = 0.003), LINC00261 (P = 0.002), and SNHG9 (P < 0.001), had a significantly lower survival percent (Figure 2C and 2D).
Figure 2.
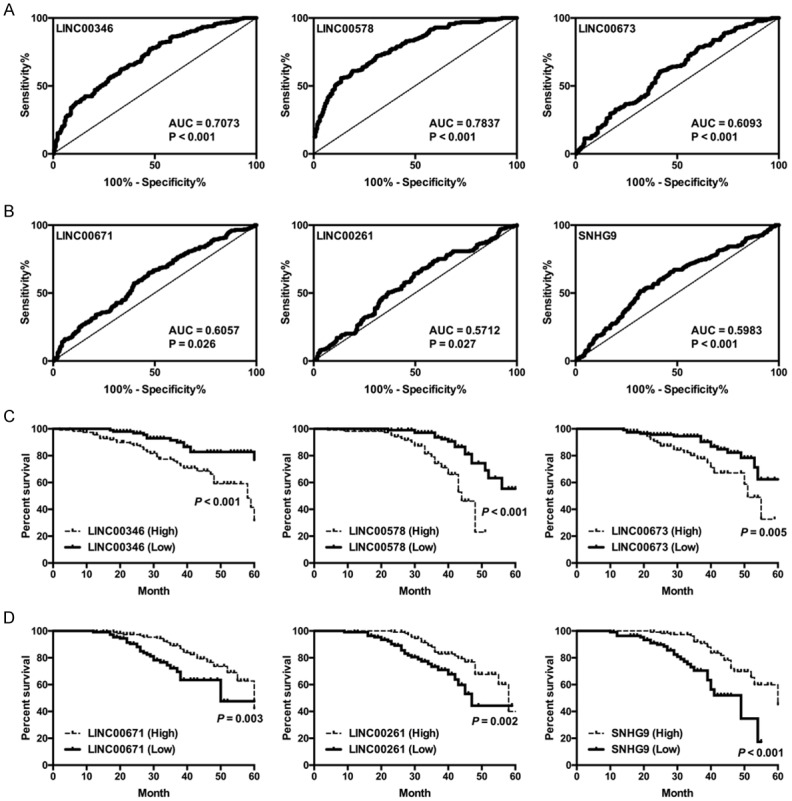
Receiver operating characteristics (ROC) curve analysis and survival analysis of lncRNAs in PC. (A and B) Receiver operating characteristics (ROC) curve analysis of LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 in PC. (C and D) Survival analysis of LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 in the PC patients (*P < 0.01, **P < 0.05, ***P < 0.001).
Expression of six different lncRNAs in serum of PC patients
To further confirm the expression patterns of the six different lncRNAs in PC, we measured their expression levels in the serum of PC patients by using qRT-PCR. As a result, the expression levels of LINC00346, LINC00578, and LINC00673 were significantly upregulated in the serum of PC patients compared with the serum of health controls (*P < 0.05, ***P < 0.001) (Figure 3A). The expression levels of LINC00671, LINC00261, and SNHG9 were significantly downregulated in the serum of PC patients compared with the serum of health controls (*P < 0.05, **P < 0.01, ***P < 0.001) (Figure 3B). Due to the expression levels of six different lncRNAs was examined in both the tissues and serum of PC patients, the correlation between the two groups was then analyzed. As results showed that a significant correlation was observed for LINC00346 (r = 0.092, ***P < 0.001), LINC00578 (r = 0.11, ***P < 0.001), LINC00673 (r = 0.08, ***P < 0.001), LINC00671 (r = 0.58, ***P < 0.001), LINC00261 (r = 0.018, *P = 0.041), and SNHG9 (r = 0.032, **P = 0.006) (Figure 3C and 3D).
Figure 3.
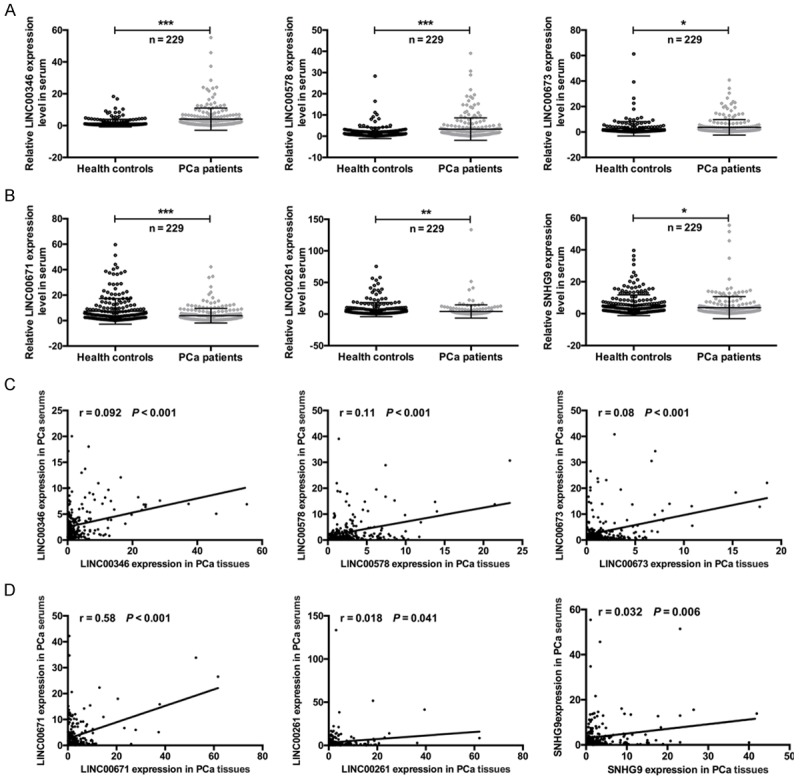
The expression levels of six different lncRNAs in the serum of PC patients, and the correlation of expression between tissues and serum. A and B. The expression levels of LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 in the serum of PC patients and health controls (*P < 0.01, **P < 0.05, ***P < 0.001). C and D. The Spearman’s rank correlation scatter plot of six PC-related lncRNAs levels in tissues and serum of PC patients. LINC00346 (r = 0.092, ***P < 0.001), LINC00578 (r = 0.11, ***P < 0.001), LINC00673 (r = 0.08, ***P < 0.001), LINC00671 (r = 0.58, ***P < 0.001), LINC00261 (r = 0.018, *P = 0.041), and SNHG9 (r = 0.032, **P = 0.006).
The effects of LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 on PC cell proliferation
In order to investigate the biological roles of LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 on PC proliferation, we established LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 knock downed cell model by transfecting HPAC and BxPC-3 cells with siRNAs. And qRT-PCR was used to confirm the knockdown effects of LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 in HPAC and BxPC-3 cells, respectively (P < 0.01 or P < 0.001, Figure 4A and 4B). Next, MTT assay revealed that silence of LINC00346, LINC00578 and LINC00673 inhibited PC cell proliferation, and silence of LINC00671, LINC00261, and SNHG9 promoted PC cell proliferation (P < 0.05, P < 0.01 or P < 0.001, Figure 4C and 4D).
Figure 4.
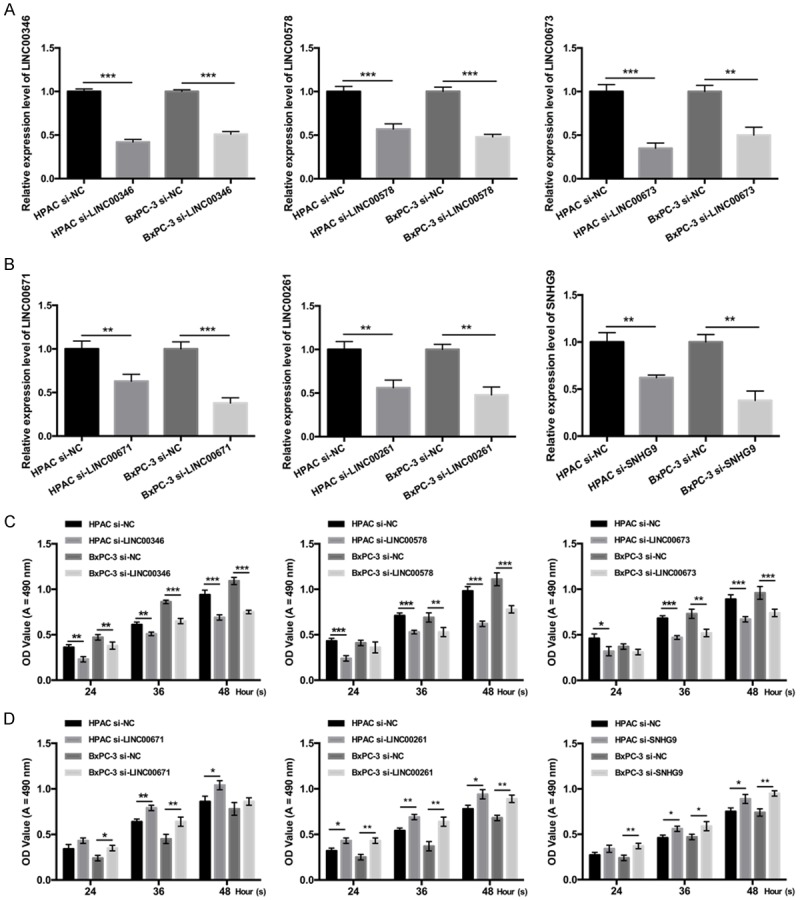
The effects of LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 on PC cell proliferation. HPAC and BxPC-3 cells were transfected with Negative control (NC), LINC00346 siRNAs, LINC00578 siRNAs, LINC00673 siRNAs, LINC00671 siRNAs, LINC00261 siRNAs, and SNHG9 siRNAs, respectively. A and B. qRT-PCR assays were performed to detect LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 expressions (**P < 0.01, ***P < 0.001). C and D. MTT assays were used to measure the proliferation abilities of HPAC and BxPC-3 cells (*P < 0.05, **P < 0.01, ***P < 0.001).
Discussion
Up to now, approximate 15,000 lncRNAs are present in the human genome [26]. Mounting transcriptional profiling studies have revealed that aberrant lncRNA expression in various human cancers, such as gastric cancer, colorectal cancer, and breast cancer [27-29]. Although the exact functions and mechanisms of most lncRNAs remain unclear, increasing evidences have been presented to show that lncRNAs may play a critical role in tumor development, progression, and metastasis [30,31].
Pancreatic cancer is considered as one of the most common malignancies in the world [32]. Due to the lack of effective diagnostic and prognostic indicator, patients with PC had a high fatality rate and a poor prognosis [33]. Recently, more and more lncRNAs were found to be involved in the prognosis of PC, Kim et al have reported that HOTAIR may serve as a negative prognostic factor in PC [25]; and Huang C et al have also reported that increased expression level of PVT1 was associated with the poor prognosis of PC patients [34]. In this present study, we found that the expression levels of six different lncRNAs (LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9) were significantly changed in the tissues and serum of PC patients. It has been reported that the amount of many lncRNAs expression was remarkably altered in different caner TNM stage [35,36]. Therefore, we examined lncRNAs expression in different TNM stages of PC tissues by qRT-PCR, and results showed that the expression profiles of these lncRNAs were correlated with the clinical stages of PC. In order to evaluate whether these lncRNAs could serve as prognostic factors in PC patients, we performed ROC analysis, and results showed that six different lncRNAs may act as good prognostic indicator in PC. In addition, survival analysis showed that patients with different expression of these lncRNAs exhibited different survival rate, suggesting that these lncRNAs may act as good prognostic indicator in PC.
Cell proliferation is a critical step of tumor growth, and increasing studies have demonstrated that lncRNAs were involved in the cell proliferation of various tumors, including PC [23,37,38]. Therefore, we assessed the effects of the six lncRNAs on cell proliferation of PC cell lines, and results showed that silence of LINC00346, LINC00578, and LINC00673 inhibited cell proliferation, whereas silence of LINC00671, LINC00261, and SNHG9 promoted cell proliferation.
In conclusion, our data demonstrate that expression levels of LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 were significantly altered in the tissues and serum of PC patients, and their expression were correlated with the different TNM stages. ROC analysis and survival analysis showed that these lncRNAs could act as prognostic indictor for PC patients. In addition, MTT assay showed that LINC00346, LINC00578, and LINC00673 inhibited cell proliferation, whereas silence of LINC00671, LINC00261, and SNHG9 promoted cell proliferation.
Disclosure of conflict of interest
None.
References
- 1.Chen Y, Hao J, Ma W, Tang Y, Gao C, Hao X. Improvement in treatment and outcome of pancreatic ductal adenocarcinoma in north China. J Gastrointest Surg. 2011;15:1026–1034. doi: 10.1007/s11605-011-1493-y. [DOI] [PubMed] [Google Scholar]
- 2.Sun J. Pancreatic neuroendocrine tumors. Intractable Rare Dis Res. 2017;6:21–28. doi: 10.5582/irdr.2017.01007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han JG, Jiang YD, Zhang CH, Yang YM, Pang D, Song YN, Zhang GQ. A novel panel of serum miR-21/miR-155/miR-365 as a potential diagnostic biomarker for breast cancer. Ann Surg Treat Res. 2017;92:55–66. doi: 10.4174/astr.2017.92.2.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luan T, Zhang X, Wang S, Song Y, Zhou S, Lin J, An W, Yuan W, Yang Y, Cai H, Zhang Q, Wang L. Long non-coding RNA MIAT promotes breast cancer progression and functions as ceRNA to regulate DUSP7 expression by sponging miR-155-5p. Oncotarget. 2017;8:76153–76164. doi: 10.18632/oncotarget.19190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shan T, Fan J, Zhao Q, Deng K, Xia J. Upregulation of long non-coding RNA DQ786243 promotes the progression of gastric cancer. Mol Med Rep. 2017;16:3761–3768. doi: 10.3892/mmr.2017.7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 7.Li Y, Vandenboom TG 2nd, Wang Z, Kong D, Ali S, Philip PA, Sarkar FH. miR-146a suppresses invasion of pancreatic cancer cells. Cancer Res. 2010;70:1486–1495. doi: 10.1158/0008-5472.CAN-09-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pang EJ, Yang R, Fu XB, Liu YF. Overexpression of long non-coding RNA MALAT1 is correlated with clinical progression and unfavorable prognosis in pancreatic cancer. Tumor Biol. 2015;36:2403–2407. doi: 10.1007/s13277-014-2850-8. [DOI] [PubMed] [Google Scholar]
- 9.Sharma C, Eltawil KM, Renfrew PD, Walsh MJ, Molinari M. Advances in diagnosis, treatment and palliation of pancreatic carcinoma: 1990-2010. World J Gastroenterol. 2011;17:867–897. doi: 10.3748/wjg.v17.i7.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaikkonen MU, Lam MTY, Glass CK. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc Res. 2011;90:430–440. doi: 10.1093/cvr/cvr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alvarez ML, DiStefano JK. The role of non-coding RNAs in diabetic nephropathy: potential applications as biomarkers for disease development and progression. Diabetes Res Clin Pract. 2013;99:1–11. doi: 10.1016/j.diabres.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 12.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rane JK, Scaravilli M, Ylipaa A, Pellacani D, Mann VM, Simms MS, Nykter M, Collins AT, Visakorpi T, Maitland NJ. MicroRNA expression profile of primary prostate cancer stem cells as a source of biomarkers and therapeutic targets. Eur Urol. 2015;67:7–10. doi: 10.1016/j.eururo.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 16.Sahlberg KK, Bottai G, Naume B, Burwinkel B, Calin GA, Borresen-Dale AL, Santarpia L. A serum MicroRNA signature predicts tumor relapse and survival in triple-negative breast cancer patients. Clin Cancer Res. 2015;21:1207–1214. doi: 10.1158/1078-0432.CCR-14-2011. [DOI] [PubMed] [Google Scholar]
- 17.Papaconstantinou IG, Manta A, Gazouli M, Lyberopoulou A, Lykoudis PM, Polymeneas G, Voros D. Expression of MicroRNAs in patients with pancreatic cancer and its prognostic significance. Pancreas. 2013;42:67–71. doi: 10.1097/MPA.0b013e3182592ba7. [DOI] [PubMed] [Google Scholar]
- 18.Eissmann M, Gutschner T, Hammerle M, Gunther S, Caudron-Herger M, Gross M, Schirmacher P, Rippe K, Braun T, Zornig M, Diederichs S. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012;9:1076–1087. doi: 10.4161/rna.21089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niu ZQ, Zhang XM, Li W, Ming ZJ, Zhong YJ, Hou YL, Zhang YP, Meng X, Wang W, Deng WJ, Fan N, Yang SY. The role and potential mechanisms of LncRNA-TATDN1 on metastasis and invasion of non-small cell lung cancer. Oncotarget. 2016;7:18219–18228. doi: 10.18632/oncotarget.7788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren C, Li X, Wang T, Wang G, Zhao C, Liang T, Zhu Y, Li M, Yang C, Zhao Y, Zhang GM. Functions and mechanisms of long noncoding RNAs in ovarian cancer. Int J Gynecol Cancer. 2015;25:566–569. doi: 10.1097/IGC.0000000000000413. [DOI] [PubMed] [Google Scholar]
- 21.Song P, Ye LF, Zhang C, Peng T, Zhou XH. Long non-coding RNA XIST exerts oncogenic functions in human nasopharyngeal carcinoma by targeting miR-34a-5p. Gene. 2016;592:8–14. doi: 10.1016/j.gene.2016.07.055. [DOI] [PubMed] [Google Scholar]
- 22.Zhuang LK, Yang YT, Ma X, Han B, Wang ZS, Zhao QY, Wu LQ, Qu ZQ. MicroRNA-92b promotes hepatocellular carcinoma progression by targeting Smad7 and is mediated by long non-coding RNA XIST. Cell Death Dis. 2016;7:e2203. doi: 10.1038/cddis.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng Y, Jutooru I, Chadalapaka G, Corton JC, Safe S. The long non-coding RNA HOTTIP enhances pancreatic cancer cell proliferation, survival and migration. Oncotarget. 2015;6:10840–10852. doi: 10.18632/oncotarget.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu JH, Chen G, Dang YW, Li CJ, Luo DZ. Expression and prognostic significance of lncRNA MALAT1 in pancreatic cancer tissues. Asian Pac J Cancer Prev. 2014;15:2971–2977. doi: 10.7314/apjcp.2014.15.7.2971. [DOI] [PubMed] [Google Scholar]
- 25.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walsh AL, Tuzova AV, Bolton EM, Lynch TH, Perry AS. Long noncoding RNAs and prostate carcinogenesis: the missing ‘linc’? Trends Mol Med. 2014;20:428–436. doi: 10.1016/j.molmed.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 27.Guo LL, Song CH, Wang P, Dai LP, Zhang JY, Wang KJ. Competing endogenous RNA networks and gastric cancer. World J Gastroenterol. 2015;21:11680–11687. doi: 10.3748/wjg.v21.i41.11680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang WC, Fu WM, Wong CW, Wang Y, Wang WM, Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF, Waye MM. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513–22525. doi: 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi SJ, Wang LJ, Yu B, Li YH, Jin Y, Bai XZ. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget. 2015;6:11652–11663. doi: 10.18632/oncotarget.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiss M, Plass C, Gerhauser C. Role of lncRNAs in prostate cancer development and progression. Biol Chem. 2014;395:1275–1290. doi: 10.1515/hsz-2014-0201. [DOI] [PubMed] [Google Scholar]
- 31.Chen LL, Zhao JC. Functional analysis of long noncoding RNAs in development and disease. Adv Exp Med Biol. 2014;825:129–158. doi: 10.1007/978-1-4939-1221-6_4. [DOI] [PubMed] [Google Scholar]
- 32.Mohammed S, Van Buren G 2nd, Fisher WE. Pancreatic cancer: advances in treatment. World J Gastroenterol. 2014;20:9354–9360. doi: 10.3748/wjg.v20.i28.9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goral V. Pancreatic cancer: pathogenesis and diagnosis. Asian Pac J Cancer Prev. 2015;16:5619–5624. doi: 10.7314/apjcp.2015.16.14.5619. [DOI] [PubMed] [Google Scholar]
- 34.Huang C, Yu W, Wang Q, Cui H, Wang Y, Zhang L, Han F, Huang T. Increased expression of the lncRNA PVT1 is associated with poor prognosis in pancreatic cancer patients. Minerva Med. 2015;106:143–149. [PubMed] [Google Scholar]
- 35.Yang C, Li Z, Li Y, Xu R, Wang Y, Tian Y, Chen W. Long non-coding RNA NEAT1 overexpression is associated with poor prognosis in cancer patients: a systematic review and meta-analysis. Oncotarget. 2017;8:2672–2680. doi: 10.18632/oncotarget.13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li W, Xie P, Ruan WH. Overexpression of lncRNA UCA1 promotes osteosarcoma progression and correlates with poor prognosis. J Bone Oncol. 2016;5:80–85. doi: 10.1016/j.jbo.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L, Chen H, Gao Y, Wang YW, Zhang GQ, Pan SH, Ji L, Kong R, Wang G, Jia YH, Bai XW, Sun B. Long noncoding RNA MALAT1 promotes aggressive pancreatic cancer proliferation and metastasis via the stimulation of autophagy. Mol Cancer Ther. 2016;15:2232–2243. doi: 10.1158/1535-7163.MCT-16-0008. [DOI] [PubMed] [Google Scholar]
- 38.Jiao F, Hu H, Yuan C, Wang L, Jiang W, Jin Z, Guo Z, Wang L. Elevated expression level of long noncoding RNA MALAT-1 facilitates cell growth, migration and invasion in pancreatic cancer. Oncol Rep. 2014;32:2485–2492. doi: 10.3892/or.2014.3518. [DOI] [PubMed] [Google Scholar]


