Abstract
Aim: The contribution of long non-coding RNAs (lncRNAs) to gastric cancer associated with Helicobacter pylori (H. pylori) infection remains largely unknown. Therefore, the present study aimed to investigate the expression of a lncRNA NR_026827 in gastric epithelial cells infected with H. pylori, and demonstrate its expression characteristic in gastric cancer. Materials and Methods: Gastric epithelial cell line cells, GES-1, were cultured and infected with H. pylori. A microarray was used to analyze the lncRNA profile of gastric epithelial cells. Eighty fresh gastric cancer tissues and the paired adjacent non-cancerous tissue samples were randomly selected from patients. The expression of the lncRNA NR_026827 was investigated using quantitative real-time polymerase chain reaction (qRT-PCR). Results: The expression of several lncRNAs was significantly altered in GES-1 cells following infection with H. pylori. Of these lncRNAs, NR_026827 was dramatically down-regulated in GES-1 cells infected with H. pylori. In addition, the expression of NR_026827 was decreased in gastric cancer tissues in comparison to the corresponding adjacent non-cancerous tissues. Moreover, the expression of NR_026827 did not change significantly in different gastric cancer stages. Conclusion: The lncRNA, NR_026827, is down-regulated in all stages of gastric cancer associated with H. pylori infection and could represent a potential biomarker for the diagnosis of gastric cancer.
Keywords: Long non-coding RNA, NR_026827, gastric cancer, Helicobacter pylori
Introduction
Globally, gastric cancer is the third leading cause of cancer death, and the fifth most common cancer, with an incidence of more than one million people experiencing it every year [1]. In China, gastric cancer is the most common type of cancer, and the second leading cause of cancer death [2]. It is well known that the chronic gastritis caused by persistent infection of the stomach with Helicobacter pylori (H. pylori) eventually causes gastric cancer [3]. H. pylori was discovered as a pathogen in 1983 and was certified as a definite carcinogen by the World Health Organization [4]. H. pylori infection represents a strong risk factor for gastric cancer [5].
Long non-coding RNAs (lncRNAs) are RNA species that are over 200 nucleotides in length and cannot be translated into protein [6]. Large-scale analyses of full-length cDNA sequences have detected large numbers of lncRNAs in humans [7]. These lncRNAs have been shown to play key roles in cell differentiation, proliferation, apoptosis, immune responses, tumorigenesis, and other biological processes [8-12].
In recent years, many lncRNAs have been reported to be associated with cancer. For example, the lncRNA GAPLINC regulates the invasion of gastric cancer cells by controlling the expression of CD44 [13]. Recently, Yang et al. described changes in the expression of various lncRNAs in H. pylori infected cells, and found that the expression of 23 lncRNAs was upregulated and the expression of 21 was downregulated in these cells [14]. However, the contribution of lncRNAs to gastric cancer associated with H. pylori infection still remains largely unknown. In this study, we investigated the lncRNA profile of gastric epithelial cells infected with H. pylori by microarray assay and found that the expression of several lncRNAs was significantly altered when compared with control cells. Of these lncRNAs, the expression of NR_026827 was first observed to be down-regulated 16-fold. Furthermore, we verified the low expression of NR_026827 in gastric epithelial cells infected with H. pylori by analyzing the expression of NR_026827 in gastric cancer tissues and the corresponding adjacent non-cancerous tissues using quantitative real-time polymerase chain reaction (qRT-PCR).
Materials and methods
Cell culture, infection, and microarray analysis
Cells from the gastric epithelial cell line GES-1 were maintained in RPMI 1640 supplemented with 10% fetal bovine serum at 37°C in a humidified air atmosphere containing 5% CO2. To infect the GES-1 cells using H. pylori NCTC11637, cells were cultured at 5 × 106 cells per flask. Approximately 5 × 108 colony-forming units (CFU) of logarithmic-phase (OD600 0.5 to 0.6) anaerobically grown bacteria were pelleted, washed twice with phosphate buffered saline (PBS), resuspended in 1 ml of RPMI 1640, and then added to the cell monolayer at a multiplicity of infection of 100:1. After incubation for 20 min at 37°C, the infected cells were washed three times with pre-warmed PBS (pH 7.4), and the infected monolayers were lysed (0 h) from the tissue-culture dishes containing 100 μg/ml gentamicin to kill extracellular bacteria. The infected monolayers were then washed three times with pre-warmed PBS and further incubated for an additional 12 h in the presence of freshly supplemented tissue-culture medium containing 12 μg/ml gentamicin. The infected cells were then washed three times with pre-warmed PBS, the monolayers were lysed, and total RNA was extracted from the cells using TRIzol reagent (Invitrogen Life Technologies). Subsequently, total RNA was analyzed by microarray to investigate the lncRNA profile of GES-1 cells infected with H. pylori.
Patients and clinical samples
Eighty fresh gastric cancer tissues and their paired adjacent non-cancerous tissue samples were randomly selected from patients who had positive 13C-urea breathe tests and were undergoing surgery at the Second Affiliated Hospital of Soochow University between January 1 and April 30, 2015. All experiments were performed according to the principles of the Declaration of Helsinki. The Ethics Committee of the Second Affiliated Hospital of Soochow University approved this study. The tissues were used to detect the expression of lncRNA NR_026827 using qRT-PCR. Fresh tissues were collected in the operating theatre and processed immediately, within 15 min. Each sample was frozen and stored at -80°C. The paired non-cancerous tissues were isolated from at least 2 cm away from the tumor border and were shown to lack tumor cells by histopathological evaluation. All patients in this study met the following inclusion criteria: the tissues were identified as gastric cancer tissue by pathological examination, and no anti-cancer treatments were administered before surgery. The clinical characteristics of all patients are listed in Table 1. The tumor staging was defined according to the 7th edition of the Tumor Node Metastasis (TNM) classification system published by the International Union Against Cancer. All patients provided written informed consent to participate in the study. The data do not contain any information that could identify the patients.
Table 1.
The clinical characteristics of all patients
| Clinicopathological features | Numbers |
|---|---|
| Age (years) | |
| < 60 | 28 |
| ≥ 60 | 52 |
| Gender | |
| Male | 45 |
| Female | 35 |
| Differentiation | |
| Well | 43 |
| Moderate + Poor | 37 |
| Depth of invasion | |
| T1 | 8 |
| T2 | 54 |
| T3 | 13 |
| T4 | 5 |
| Lymph node metastasis | |
| N0 | 19 |
| N1 | 22 |
| N2 | 32 |
| N3 | 7 |
| Distant metastasis | |
| M0 | 68 |
| M1 | 12 |
| TNM stage | |
| I | 23 |
| II | 32 |
| III | 13 |
| IV | 12 |
RNA extraction and cDNA synthesis
Total RNA was extracted from tissues and cells using TRIzol reagent according to the manufacturer’s instructions. The quality of the total RNA was assessed using a NanoDrop®ND-1000 and denaturation agarose-gel electrophoresis. First-strand cDNA was synthesized from 1.5 µg of total RNA using specific primers for lncRNA NR_026827 and the MMLV reverse transcriptase (Epicentre).
qRT-PCR investigation
qRT-PCR analyses were performed with 2 × PCR master mixture (Arraystar) using a ViiA 7 Real-time PCR System (Applied Biosystems). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control. The PCR primers for lncRNA NR_026827 and GAPDH were as follows: lncRNA NR_026827 sense, ACTGCCCATACGGACCTAC and reverse, TCCCAAGAGACAATGAAAAAG; GAPDH sense, GGGAAACTGTGGCGTGAT and reverse, GAGTGGGTGTCGCTGTTGA. The relative mRNA levels were calculated based on the Ct values and normalized to GAPDH levels. The results are expressed as means ± standard deviation (SD) from three separate qRT-PCR tests performed on each of 80 gastric cancer specimens. The differences between the groups were analyzed using a two-tailed Student’s t-test. Statistical analysis was performed using SPSS software and differences with P-values < 0.05 were considered significant.
Results
The lncRNA profile of GES-1 cells infected with H. pylori
The lncRNA profile of GES-1 cells infected by H. pylori differs to that of non-infection controls. The expression of several lncRNAs was significantly altered. LncRNAs that showed up-regulation and down-regulation are shown in Tables 2 and 3, respectively.
Table 2.
Long non-coding RNAs up-regulated in the gastric epithelial cells infected by H. pylori compared to that of non-infection control by miroarray-based profile assay
| Long non-coding RNA | RNA length | Log2 Fold (infection/non-infection) |
|---|---|---|
| chr6:63131625-63144250+ | 12625 | 3.58 |
| TCONS_00007049 | 1434 | 2.99 |
| ENST00000581964 | 517 | 2.62 |
| NR_036510 | 1458 | 2.01 |
| TCONS_00003565 | 344 | 1.95 |
| ENST00000538111 | 368 | 1.85 |
| ENST00000571665 | 255 | 1.84 |
| ENST00000452342 | 573 | 1.76 |
| TCONS_00015979 | 810 | 1.75 |
| ENST00000564300 | 600 | 1.71 |
| ENST00000414795 | 506 | 1.69 |
| NR_047576 | 2000 | 1.68 |
Table 3.
Long non-coding RNAs down-regulated in the gastric epithelial cells infected by H. pylori compared to that of non-infection control by miroarray-based profile assay
| Long non-coding RNA | RNA length | Log2 Fold (infection/non-infection) |
|---|---|---|
| NR_026827 | 2322 | -12.24 |
| NR_028301 | 3278 | -9.15 |
| ENST00000523627 | 570 | -8.63 |
| ENST00000562166 | 1838 | -6.77 |
| AK022063 | 2196 | -6.29 |
| TCONS_00029621 | 276 | -5.21 |
| ENST00000453706 | 261 | -4.82 |
| TCONS_00005953 | 761 | -4.62 |
| ENST00000534653 | 529 | -4.23 |
| TCONS_00004713 | 206 | -4.08 |
| TCONS_00026381 | 324 | -4.05 |
| ENST00000553668 | 736 | -4.01 |
| ENST00000492461 | 415 | -3.79 |
| ENST00000444919 | 414 | -3.37 |
| uc010lfo.1 | 1470 | -3.34 |
| TCONS_00016207 | 728 | -3.23 |
| TCONS_00008376 | 784 | -2.97 |
| TCONS_00012531 | 222 | -2.87 |
| ENST00000528514 | 1361 | -2.84 |
| ENST00000542311 | 2041 | -2.81 |
| ENST00000455391 | 461 | -2.69 |
| TCONS_00026952 | 1692 | -2.62 |
| ENST00000554036 | 509 | -2.59 |
| ENST00000538025 | 534 | -2.52 |
| ENST00000512831 | 628 | -2.45 |
| ENST00000420187 | 549 | -2.39 |
| ENST00000422449 | 363 | -2.36 |
| ENST00000448671 | 574 | -2.35 |
| TCONS_00021051 | 459 | -2.25 |
| TCONS_00020407 | 1957 | -2.19 |
| ENST00000414135 | 976 | -2.06 |
| ENST00000503299 | 758 | -1.97 |
| uc002suu.2 | 3042 | -1.96 |
| ENST00000484222 | 2084 | -1.95 |
| ENST00000423186 | 1055 | -1.94 |
| ENST00000508986 | 331 | -1.91 |
| ENST00000532652 | 514 | -1.85 |
| NR_045027 | 1158 | -1.84 |
| uc002zis.1 | 570 | -1.81 |
| ENST00000531870 | 568 | -1.69 |
| ENST00000442305 | 514 | -1.68 |
Low expression of lncRNA NR_026827 in GES-1 cells infected with H. pylori
To investigate the expression of lncRNA NR_026827 in the GES-1 cells infected with H. pylori, we compared the expression levels in the experimental group infected with H. pylori for 24 h with the levels in the non-infected control group using qRT-PCR. As shown in Figure 1, the expression of NR_026827 was significantly decreased in the GES-1 cells infected with H. pylori (P < 0.01).
Figure 1.

Relative expression of lncRNA NR_026827 in the gastric epithelial cells infected with H. pylori by qRT-PCR. *P < 0.01.
Investigation of lncRNA NR_026827 expression in gastric cancer
We first examined the expression of lncRNA NR_026827 in 80 paired gastric cancer samples and their adjacent non-tumorous tissues using qRT-PCR. The expression of lncRNA NR_026827 was found to be significantly decreased in gastric cancer tissues compared with the corresponding adjacent non-tumorous tissues (P < 0.05; Figure 2).
Figure 2.

Relative expression of lncRNA NR_026827 in the gastric cancer tissues and the corresponding adjacent non-cancerous tissues by qRT-PCR. *P < 0.05.
Expression of lncRNA NR_026827 in different stages of gastric cancer
To study the expression characteristics of lncRNA NR_026827 in different stages of gastric cancer, we analyzed the data of 80 gastric cancer samples. Results showed that there were no obvious differences in the expression of lncRNA NR_026827 in gastric cancer at different stages (P > 0.05; Figure 3). This suggests that lncRNA NR_026827 is expressed stably in gastric cancer tissues at all stages.
Figure 3.
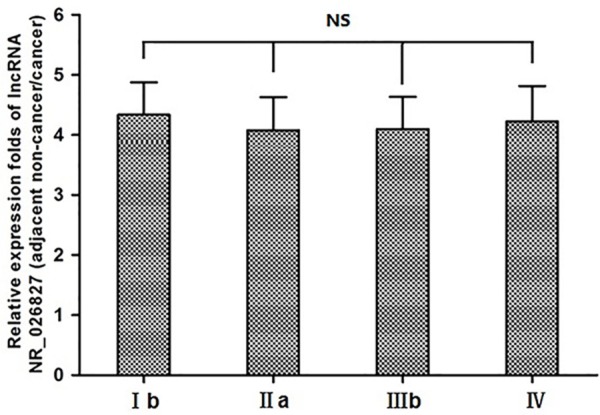
Relative expression folds of lncRNA NR_026827 (adjacent non-cancerous tissues/cancerous tissues). NS: P > 0.05.
Discussion
Gastric cancer is one of the most common malignancies worldwide, and ranks second in cancer-associated deaths because of its high lethality [1]. H. pylori is a gram-negative, spiral-shaped pathogenic bacterium that causes chronic gastritis, peptic ulcer disease, mucosa-associated lymphoid tissue lymphoma, and gastric cancer [15-17]. LncRNA, which is greater than 200 nucleotides in length, is a class of RNA species that does not have a protein coding function and is involved in different regulatory processes, such as the regulation of gene expression at the chromatin, transcriptional, or post-transcriptional levels [6]. In recent years, many studies have shown that lncRNAs are associated with cancers, and have been shown to be involved in both the occurrence and development of various cancers [18,19]. However, the diagnostic significance of lncRNAs in gastric cancer is largely unknown [20]. Recently, Zhou et al. reported that the lncRNA LET was down-regulated in gastric cancer and was associated with poor prognosis [21]. It was also reported that the expression of the lncRNA AI364715 was down-regulated in gastric cancer tissues, and that its down-regulation was closely associated with tumor size and differentiation [22]. Therefore, new markers with high sensitivity and specificity for gastric cancer detection are urgently needed.
In this study, we found that the expression of a novel lncRNA, NR_026827, was down-regulated in gastric epithelial (GES-1) cells infected with H. pylori. Moreover, the expression of NR_026827 in gastric cancer tissues was also decreased in comparison to the corresponding adjacent non-cancerous tissues. In addition, the expression of lncRNA NR_026827 did not differ in different stages of gastric cancer, suggesting that lncRNA NR_026827 may be involved in all stages of gastric cancer. We also analyzed the relationship between lncRNA NR_026827 expression and the age and sex of patients and our results show that there were no obvious relationships (data not shown). LncRNA NR_026827 is a long intergenic non-coding RNA that is 2322 nucleotides in length, and its gene is located on chromosome 10. The function of this novel lncRNA was previously unclear. The results of this study may provide a novel perspective for better understanding the molecular basis of gastric cancer, and suggest that lncRNA NR_026827 might be a potential biomarker for the diagnosis of gastric cancer. However, the biological function of NR_026827 in the pathogenesis of gastric cancer should be elucidated in future studies.
Acknowledgements
This study was supported by the Science and Technology Program of Suzhou (SYS201618, SYS201551, SS201638, SZS201715), Jiangsu youth medical talents program (QN-866, 867), Six talent peaks project in Jiangsu Province (2016-WSN-112) and Gusu key health talent of Suzhou to Hong Du and Haifang Zhang.
Disclosure of conflict of interest
None.
References
- 1.Figueiredo C, Costa S, Karameris A, Machado JC. Pathogenesis of gastric cancer. Helicobacter. 2015;20(Suppl 1):30–35. doi: 10.1111/hel.12254. [DOI] [PubMed] [Google Scholar]
- 2.Jing JJ, Liu HY, Hao JK, Wang LN, Wang YP, Sun LH, Yuan Y. Gastric cancer incidence and mortality in Zhuanghe, China, between 2005 and 2010. World J Gastroenterol. 2012;18:1262–1269. doi: 10.3748/wjg.v18.i11.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bessède E, Dubus P, Mégraud F, Varon C. Helicobacter pylori infection and stem cells at the origin of gastric cancer. Oncogene. 2015;34:2547–2555. doi: 10.1038/onc.2014.187. [DOI] [PubMed] [Google Scholar]
- 4.Warren J, Marshall B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;1:1273–1275. [PubMed] [Google Scholar]
- 5.Cover TL. Helicobacter pylori diversity and gastric cancer risk. MBio. 2016;7:e01869–15. doi: 10.1128/mBio.01869-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 7.Zou AE, Ku J, Honda TK, Yu V, Kuo SZ, Zheng H, Xuan Y, Saad MA, Hinton A, Brumund KT, Lin JH, Wang-Rodriguez J, Ongkeko WM. Transcriptome sequencing uncovers novel long noncoding and small nucleolar RNAs dysregulated in head and neck squamous cell carcinoma. RNA. 2015;21:1122–1134. doi: 10.1261/rna.049262.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibb EA, Vucic EA, Enfield KS, Stewart GL, Lonergan KM, Kennett JY, Becker-Santos DD, MacAulay CE, Lam S, Brown CJ, Lam WL. Human cancer long non-coding RNA transcriptomes. PLoS One. 2011;6:e25915. doi: 10.1371/journal.pone.0025915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng SY, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–533. doi: 10.1038/emboj.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu T, Yin X, Zhou Y, Wang Z, Shen S, Qiu Y, Sun R, Zhao Z. Roles of noncoding RNAs in metastasis of nonsmall cell lung cancer: a mini review. J Cancer Res Ther. 2015;11(Suppl 1):C7–10. doi: 10.4103/0973-1482.163831. [DOI] [PubMed] [Google Scholar]
- 12.Zhang K, Yuan Q. Current mechanism of acquired resistance to epidermal growth factor receptor-tyrosine kinase inhibitors and updated therapy strategies in human nonsmall cell lung cancer. J Cancer Res Ther. 2016;12(Suppl):C131–C137. doi: 10.4103/0973-1482.200613. [DOI] [PubMed] [Google Scholar]
- 13.Hu Y, Wang J, Qian J, Kong X, Tang J, Wang Y, Chen H, Hong J, Zou W, Chen Y, Xu J, Fang JY. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. 2014;74:6890–6902. doi: 10.1158/0008-5472.CAN-14-0686. [DOI] [PubMed] [Google Scholar]
- 14.Yang L, Long Y, Li C, Cao L, Gan H, Huang K, Jia Y. Genome-wide analysis of long noncoding RNA profile in human gastric epithelial cell response to Helicobacter pylori . Jpn J Infect Dis. 2015;68:63–66. doi: 10.7883/yoken.JJID.2014.149. [DOI] [PubMed] [Google Scholar]
- 15.Salama NR, Hartung ML, Müller A. Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori . Nat Rev Microbiol. 2013;11:385–399. doi: 10.1038/nrmicro3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang JJ, Lee DH, Lee AR, Yoon H, Shin CM, Park YS, Kim N. Characteristics of gastric cancer in peptic ulcer patients with Helicobacter pylori infection. World J Gastroenterol. 2015;21:4954–4960. doi: 10.3748/wjg.v21.i16.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leodolter A, Alonso S, González B, Ebert MP, Vieth M, Röcken C, Wex T, Peitz U, Malfertheiner P, Perucho M. Somatic DNA hypomethylation in H. pylori-associated high-risk gastritis and gastric cancer: enhanced somatic hypomethylation associates with advanced stage cancer. Clin Transl Gastroenterol. 2015;6:e85. doi: 10.1038/ctg.2015.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med. 2015;12:1–9. doi: 10.7497/j.issn.2095-3941.2015.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karapetyan AR, Buiting C, Kuiper RA, Coolen MW. Regulatory roles for long ncRNA and mRNA. Cancers (Basel) 2013;5:462–490. doi: 10.3390/cancers5020462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang ZG, Gao L, Guo XB, Shi YL. Roles of long non-coding RNAs in gastric cancer metastasis. World J Gastroenterol. 2015;21:5220–5230. doi: 10.3748/wjg.v21.i17.5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou B, Jing XY, Wu JQ, Xi HF, Lu GJ. Down-regulation of long non-coding RNA LET is associated with poor prognosis in gastric cancer. Int J Clin Exp Pathol. 2014;7:8893–8898. [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu S, Mao J, Shao Y, Chen F, Zhu X, Xu D, Zhang X, Guo J. Reduced expression of the long non-coding RNA AI364715 in gastric cancer and its clinical significance. Tumour Biol. 2015;36:8041–8045. doi: 10.1007/s13277-015-3543-7. [DOI] [PubMed] [Google Scholar]


