Abstract
MicroRNA-185 (miR-185) is down-regulated in various tumor types. However, the cytological mechanism for inhibiting and restraining tumor growth of non-small-cell carcinoma (NSCLC) remains to be elucidated. In this study, it was revealed that miR-185 is significantly down-regulated in both NSCLC tumor tissues and cell lines, and over-expression of miR-185 inhibited cell growth, migration and invasion. To investigate the cellular machinery involved in miR-185’s regulation of tumor growth, it was found that miR-185 directly targets SRY-Box 13 (SOX13). In addition, miR-185 regulated cell proliferation, migration, invasion and increased chemo-sensitivity in H1975 cells by inhibiting SOX13. MiR-185 also inhibited tumor growth and suppressed SOX13 in nude mouse xenograft tumors. To investigate the clinical relevance of these consequences, 24 pairs of NSCLC tissues and adjacent normal tissues were collected to determine expression of miR-185 and SOX13. It was demonstrated that miR-185 levels are significantly and inversely correlated with SOX13 levels in these NSCLC tissues, suggesting that these findings have implications for translational application with respect to NSCLC diagnostics and therapy.
Keywords: miR-185, NSCLC, SOX13, chemo-resistance
Introduction
Non-small-cell carcinoma (NSCLC) is universally acknowledged as the most widespread lung cancer [1]. Over the last several decades, the mortality of NSCLC has decreased due to combinatorial methods incorporating mammographic screening and to improvements in systemic therapy. Targeted agent treatment for NSCLC is considered one of the most crucial reasons for reduced mortality [2]. Invasion and metastasis remain the primary obstacles in the treatment of NSCLC. Thus, investigations of the molecular mechanisms of NSCLC have attracted significant interest. MicroRNAs (miRNAs) are 20-22 nucleotide non-coding RNA molecules that negatively regulate gene expression by connecting to the 3’-untranslated region (UTR) of their target genes with partial complementarity, leading to degradation of target mRNAs and/or inhibition of their translation [3]. MiRNAs are thought to regulate various pathological behaviors in cancer cells, such as cell proliferation, motility and sensitivity to chemotherapy [4].
Attempts to identify molecular markers of NSCLC that promote tumor growth and metastasis are ongoing [5]. SRY-related HMG-box (SOX) proteins are expressed in a series of embryonic and adult tissues and have been implicated in the determination of cell fate [6]. More than one SOX gene is frequently expressed incells, and it remains unclear to what level they are functionally redundant. Recently, the SOX family of transcription factors have been identified to be of particular interest in cancer, as members of this family are strongly expressed across several types of cancers [7]. SOX10 protein was reported to be widely expressed in human gliomas, as demonstrated by immunohistochemistry [6].
In this study, it was demonstrated that miR-185 levels are down-regulated in human NSCLC specimens across 24 pairs of cancer and adjacent normal tissues. Subsequently, it was determined that forced over-expression of miR-185 inhibited NSCLC cells growth and colony formation. Furthermore, elevated miR-185 suppressed migration and invasion in NSCLC cells. Bioinformatics analysis and luciferase assays demonstrated that SOX13 is a direct target of miR-185. Over-expression of miR-185 led to marked down-regulation of SOX13 expression, which in turn restrained the growth of NSCLC cells and suppressed migration and invasion in NSCLC cells. In conclusion, this study provides novel insight into the mechanism of the miR-185/SOX13 axis in NSCLC.
Materials and methods
NSCLC cell culture and tissues
Human NSCLC specimens (24 pairs) and adjacent normal tissues were obtained from the Affiliated Hospital of Medical School of Ningbo University. All tissue samples were snap-frozen in liquid nitrogen immediately after surgery and stored in liquid nitrogen. All samples were histologically classified by a clinical pathologist. Samples for molecular analysis were initially ground into powder in liquid nitrogen and then separately collected for RNA analyses. This research was approved by the institutional review board and the ethics committee of the Affiliated Hospital of Medical School of Ningbo University. Informed consent was obtained from all subjects. NSCLC cell lines (A549, H1975, H1650, HCC827) and SV40-immortalized non-tumorigenic human bronchial epithelial cells BEAS-2B used as control were obtained from the Chinese Academy of Sciences Cell Bank of Type Culture Collection (CBTCCCAS, Shanghai, China). Cells were incubated at 37°C in a humidified chamber supplemented with 5% CO2.
MiRNA and miRNA inhibitor transfections
H1975 and A549 cells were plated into six-well plates at 2 × 105 cells per well and then transfected with 100 nM of either miR-185 mimics or negative control miRNA (Invitrogen) 24 hours later using Oligofectamine (Invitrogen) according to the manufacturer’s protocol. To knock down endogenous miR-185, H1975 and A549 cells were transfected with 100 nM scrambled miRNA inhibitor or miR-185 inhibitor (Invitrogen) using Lipofectamine 2000 (Invitrogen) in six-well plates (2 × 105 cells per well) for 24 hours. The construct for SOX13 was generated by subcloning PCR-amplified full-length human SOX13 cDNA into the pMSCV retrovirus plasmid. MiR-185 mimic-transfected H1975 and A549 cells were transfected with SOX13 or vector (used as a negative control) for 24 hours. Next, transfected cells were utilized for growth, migration and invasion assays.
Cell proliferation assay
To evaluate proliferation ability of H1975 cells over-expressing miR-185, 2 × 103 cells per well were seeded into 96-well plates. After incubation for the indicated time period, 100 μL of MTT solution was added to each well, and incubation was continued for another 4 h. Absorbance was determined at 450 nm with the Synergy™ HT Multi-Mode Microplate Reader (Bio-Tek, Winooski, VT, USA). In the chemo-sensitivity array, 2 × 103 cells were seeded into 96-well plates. After 24 h, freshly prepared paclitaxel (Sigma-Aldrich, St. Louis, MO, USA) was added at a final concentration ranging from 1 to 8 nM. Seventy-two hours later, cell viability was determined. 2 × 103 cells per well were seeded into 96-well plates, and after 24 h, 4 nM paclitaxel (Sigma-Aldrich, St. Louis, MO, USA) was added to each well. After incubation for the indicated time period, absorbance was determined [8].
Wound healing assay
H1975 cells were cultured to 95% confluence in 6-well plates. Cell monolayers were scratched with a 100 µl tip to form wound gaps and washed two times with PBS solution to remove detached cells. After 24 h, wound healing was photographed. Cell migration distance was measured in three different areas to determine the migration ability of different treatment groups [9].
Invasion assay
Cells were plated into upper 24-well BD Matrigel invasion chambers (BD Biosciences, Cowley, UK) without serum. The lower chamber was filled with 600 μl of DMEM medium with 10% FBS to act as a nutritional attractant. After 24 h incubation, non-invading cells were removed from the top well with a cotton swab, while the cells at the bottom were stained with 0.1% crystal violet, and five independent fields for each well were photographed [10].
Quantitative real time-PCR (qRT-PCR) assay
Total RNA was isolated from harvested cells or human tissues by Trizol reagent according to the manufacturer’s instructions (Invitrogen, CA, USA). To measure expression levels of miR-185, qRT-PCR assay was adopted, and U6 was used as an endogenous control. To determine mRNA levels of SOX13, total RNA was reversely transcribed by oligodT primers using the Prime-Script RT Reagent Kit (Takara, Dalian, China). The housekeeping gene GAPDH was used as an internal control. cDNAs were amplified by real-time PCR using SYBR Premix DimerEraser (Takara, Dalian, China) on a 7900HT system, and fold changes were determine by relative quantification (2-ΔΔCt). The primers used for PCR were as follows (sense and antisense, respectively): SOX13: 5’-CCAGAGGGTAATGGGTCCC-3’ and 5’-TGGCTTCCATAGAGTTCCTTCC-3’; GAPDH: 5’-TGGATTTGGACGCATTGGTC-3’ and 5’-TTTGCACTGGTACGTGTTGAT-3’. The primer used for qRT-PCR analysis of miRNA was as follows miR-185: TGGAGAGAAAGGCAGTTCCTGA.
Immunoblotting analysis
Cells were washed with ice-cold PBS buffer, scraped from the dishes, and centrifuged at 12,000 rpm at 4°C for 15 min. Cell lysates were prepared using RIPA buffer supplemented with protease inhibitors. Tumor tissues from human and nude mice were ground into powder in liquid nitrogen with RIPA buffer, and total tissue protein levels wee determined. Aliquots of protein lysates were fractionated by SDS-PAGE, transferred to PVDF membranes (Roche, Switzerland), and then subjected to immunoblotting analysis according to the manufacturer’s instructions. The ECL Detection System (Thermo Scientific, Rockford, IL, USA) was used for protein signal detection.
Dual-luciferase reporter assay
For the dual-luciferase assay, 3’-UTRs of SOX13 containing predicted miR-185 seed-matching sites and corresponding mutant sites were created by PCR using human cDNA templates and inserted into SacI and HindIII restriction enzyme sites of the pMIR-REPORTER vector (Ambion, CA, USA). These constructs were validated by DNA sequencing. Cells were seeded in 24-well plates and co-transfected with wild type or mutant reporter plasmid, pRL-TK plasmids, and miR-185 or miR-NC. Luciferase activity was measured 24 h after transfection by the Dual Luciferase Reporter Assay System (Promega, WI, USA).
In vivo tumorigenesis assay
Male nude mice (BALB/c-null, 6-week-old) were obtained from Shanghai Laboratory Animal Center (Chinese Academy of Sciences, Shanghai, China) and bred in special pathogen-free (SPF) conditions. Parental cells or H1975 cells transfected with miR-185 (5 × 106) were suspended in 100 μl of serum-free DMEM medium and injected into nude mice (n = 6 for each group). Tumor sizes were determined every three days. Tumor volumes were calculated using Vernier calipers with the formula: volume = 0.5 × (Length × Width2). Mice were sacrificed 25 days after implantation, and tumors were dissected and collected. All animal experiments were permitted by the Committee of Laboratory Animal Experimentation of the Affiliated Hospital of Medical School of Ningbo University.
Statistical analysis
All experiments were repeated three times, and data were analyzed with GraphPad Prism 5 (La Jolla, CA, USA). Statistical evaluation for data analysis was determined using the t-test. Differences were considered statistically significance at P<0.05.
Results
MiR-185 expression is down-regulated in NSCLC tissues and cell lines
To identify potential miRNAs that are aberrantly expressed in NSCLC, the expression pattern of miRNAs was compared between healthy vs. NSCLC tissues using the GEO data set GSE29248. The heat map generated for differential gene expression revealed that miR-185 was significantly down-regulated (Fold change = -3.6 and P<0.01) in NSCLC tissues compared to controls (Figure 1A and Supplementary Table 1). To explore the potential biological consequences of altered miR-185 expression level to NSCLC progression, expression levels of miR-185 in 24 NSCLC tissues and 24 normal tissues were evaluated by quantitative qRT-PCR. As shown in Figure 1B, levels of miR-185 were considerably reduced in NSCLC tissues compared to normal tissues. Consistent with the demonstration that miR-185 is down-regulated in clinical cancer tissues, significantly reduced expression of miR-185 was also observed in various NSCLC cell lines (Figure 1C). Collectively, our results indicate that miR-185 is down-regulated in NSCLC tissues and cell lines.
Figure 1.

MiR-185 is down-regulated in NSCLC clinical samples and cell lines. A. Microarray analysis of miRNA expression in NSCLC tissues compared to corresponding normal tissues. B. MiR-185 levels in 24 adjacent normal control tissues (N) and 24 NSCLC tissues (T) were determined by qRT-PCR. C. qRT-PCR was adopted for analysis ofmiR-185 levels in a panel of NSCLC cell lines. GAPDH was used as a loading control.
Over-expression of miR-185 inhibits NSCLC cell proliferation, cell migration and invasion
To uncover the impact of miR-185 in NSCLC growth and mobility, H1975 and A549 cells were transfected with miR-185 or miR-NC to examine the effects of miR-185 over-expression (Figure 2A). MiR-185 over-expressing NSCLC cells were subjected to MTT and colony formation assays to analyze cell growth in vitro. As shown in Figure 2B and 2C, cell growth and colony formation were attenuated in miR-185 over-expressing cells compared to cells expressing miR-NC. Because cell migration and invasion are cardinal characteristics of malignant tumors, the effects of miR-185 expression on migration and invasion of two NSCLC cell lines were subsequently investigated. Over-expression of miR-185 dramatically inhibited the mobility (Figure 2D) and invasive capacity (Figure 2E) of NSCLC cells. These results suggest that miR-185 over-expression inhibits NSCLC cell proliferation, cell migration and invasion.
Figure 2.
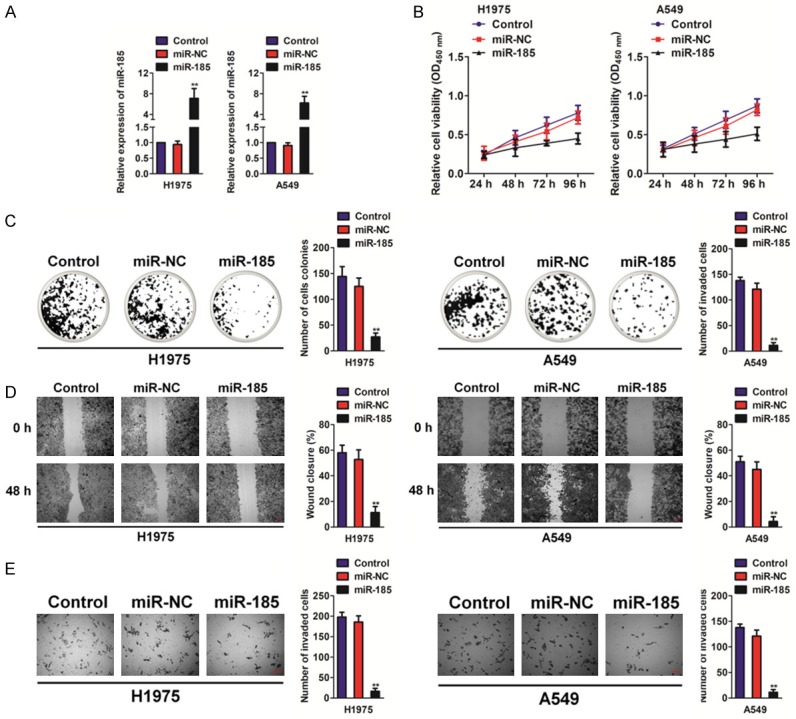
Over-expression of miR-185 inhibits cell proliferation, migration and invasion in NSCLC cells. A. Relative miR-185 expression levels were analyzed by qRT-PCR in miR-NC and miR-185 transfected cell lines. B. Over-expression of miR-185 inhibits cell proliferation in NSCLC cells. Data are represented as the mean ± SD. C. Over-expression of miR-185 inhibits colony formation of NSCLC cells. Data are represented as the mean ± SD. D. NSCLC cells were transfected with miR-185 or miR-NC and were subjected to wound healing analysis. Wound gaps were photographed (left panel), and rates of wound-healing were measured (right panel). Data are represented as the mean ± SD. **P<0.01 compared to control cells. E. NSCLC cells were transfected with miR-185 or miR-NC and were subjected to Transwell invasion assay. Data are represented as the mean ± SD. **P<0.01 compared to control cells.
SOX13 is a direct putative target of miR-185and is negatively correlated with miR-185 in NSCLC
Bioinformatics prediction (miRanda, mirSVR and TargetScan) software was adopted to identify targets of miR-185 [11]. It was found that SOX13 3’-UTR has a sequence capable of binding miR-185 at the 1422-1429 position (Figure 3A). To verify SOX13 as a miR-185 target, a luciferase activity assay was performed. Our results show that miR-185 dramatically inhibited luciferase activity of the 3’-UTR of SOX13 in H1975 cells (Figure 3B). Levels of miR-185 in cells transfected with miR-185 were also measured by qRT-PCR, western blotting and immunofluorescence assays. Over-expression miR-185 inhibited mRNA and protein expression of SOX13 in H1975 cells (Figure 3C and Supplementary Figure 1). Protein levels of SOX13 in a human NSCLC tissues and corresponding normal tissues were then examined by qRT-PCR assays, and it was demonstrated that SOX13 is over-expressed in NSCLC (Figure 3D). Finally, qRT-PCR analysis illustrated that down-regulation of miR-185 expression was considerably negatively correlated with over-expression of SOX13 in NSCLC tissues (Figure 3E).
Figure 3.
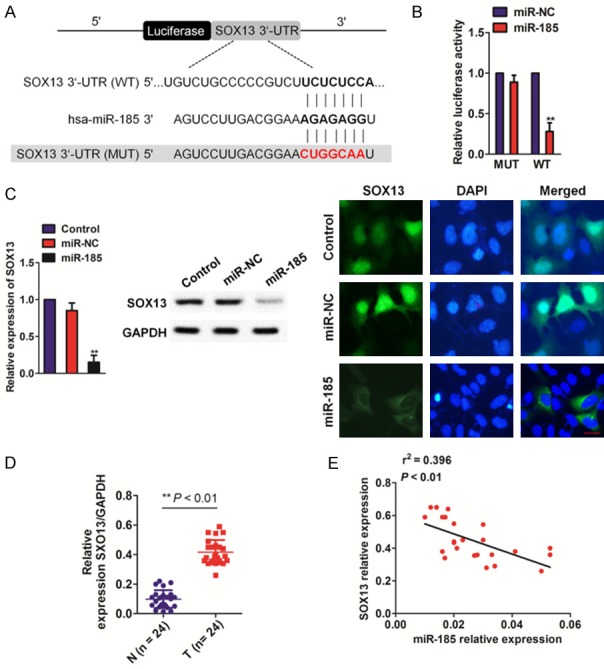
SOX13 is a direct putative target of miR-185. A. Schematic diagram of miR-185 binding sites in the SOX13 3’-UTR. Sequences were compared between mature miR-185 and wild-type (WT) or mutant (MUT) putative target sites in the 3’-UTR of SOX13. B. H1975 cells were co-transfected with wild-type (WT) or mutant (MUT) SOX13 3’-UTR with miR-185, and luciferase activity was measured. Firefly luciferase activity was measured and standardized using a Renilla luciferase activity. Data are represented as the mean ± SD. **P<0.01 compared with miR-NC. C. H1975 cells were transfected with miR-185 and miR-NC. SOX13 expression as determined by qRT-PCR (left panel), western blotting (middle panel) and immunofluorescence assays (right panel). Data are represented as the mean ± SD. **P<0.01 compared to control cells. D. Expression of SOX13 in human NSCLC tissues compared to corresponding normal tissues was determined by qRT-PCR analysis. **P<0.01 compared to normal tissues. E. Expression analysis correlating miR-185 and SOX13 in tumor samples was conducted by qRT-PCR assay.
MiR-185 regulates cell growth, migration, and invasion, increasing the chemo-sensitivity of H1975 cells to paclitaxel through inhibition of SOX13
To determine the influence of SOX13 in cellular function, the forced expression of SOX13 counteracted miR-185-mediated inhibition of cell proliferation (Figure 4A) and colony formation (Figure 4B) was examined in vitro. Then, the effects of miR-185 on mobility and invasion in vitro were investigated. Over-expression of miR-185 dramatically decreased both migration and invasive capacity in NSCLC cells, whereas SOX13 neutralized the inhibiting effect of miR-185 on NSCLC cell mobility (Figure 4C and 4D). These results suggest that over-expression of miR-185 promotes cell proliferation, cell migration and invasion by inhibiting SOX13. Resistance to paclitaxel treatment is one of the major causes for chemotherapy failure in treating breast cancer. Therefore, it is critical to expand the effectiveness of paclitaxel for therapeutic purposes. Our data indicate that overexpression of miR-185 in NSCLC cells significantly increases chemo-sensitivity to paclitaxel (Figure 4E). Furthermore, cell growth rate was assessed in the presence of paclitaxel (4 nM) by MTT proliferation assay at different time points, and interestingly, forced expression of SOX13 reversed miR-185 over-expression-mediated NSCLC cell chemo-sensitivity to paclitaxel (Figure 4F). Altogether, these results indicate that miR-185 inhibits proliferation, migration and invasion of NSCLC cells, while simultaneously increasing chemo-sensitivity by targeting SOX13.
Figure 4.
MiR-185 regulates cell proliferation, migration, and invasion, increasing chemo-sensitivity of NSCLC cells to paclitaxel. A. Over-expression of miR-185 inhibits cell proliferation, whereas proliferation inhibition was rescued in NSCLC cells by co-expression of exogenous SOX13 and miR-185. Data are represented as the mean ± SD. B. Over-expression of miR-185 inhibits colony formation, whereas number of colonies was increased in NSCLC cells co-expressing exogenous SOX13 and miR-185. Data are represented as the mean ± SD. **P<0.01 compared to control cells, ##P<0.01 compared to cells transfected with miR-185. C. NSCLC cells were transfected with miR-185 alone or in combination with SOX13. A sterile 200 μl pipette tip was used to scratch cells to form a wound. Wound gaps were photographed (top) and measured (bottom). D. Cells were transfected with miR-185 followed by SOX13 transfection and were subjected to TranswellMatrigel invasion assay. Data are represented as the mean ± SD. **P<0.01 compared to control cells, ##P<0.01 compared to cells transfected with miR-185. E. NSCLC cells stably expressing miR-NC or miR-185 were pretreated with various concentration of paclitaxel for 72 h and then subjected to MTT assay. F. Cells stably expressing miR-NC, miR-185 or miR-185 with over-expression of SOX13 were pretreated with 4 nM of paclitaxel for pre-defined time points and subsequently subjected to MTT analysis.
MiR-185 inhibits tumor growth in vivo
Considering the pivotal activity of miR-185 in the regulation of cell proliferation, migration and invasion, H1975 cells with miR-185 knockdown were used to verify the observed effects of miR-185 (Figure 5A). Our results showed that both cell proliferation (Figure 5B) and colony formation (Figure 5C) were increased in response to miR-185 knockdown in H1975 cells compared with H1975 cells expressing control miR-NC. To investigate whether over-expression of miR-185 attenuates progression of NSCLC in vivo, H1975 cells were transfected with miR-185, and cells were subsequently implanted into nude mice. As shown in Figure 5D, injection of parental cells resulted in significantly larger tumors than did the miR-185 group. MiR-185-expressing cells generated xenografts that were significantly smaller than control xenografts. Furthermore, final tumor weights of the control group were much greater than in the miR-185 group (Figure 5E). Consistently, the level of miR-185 was significantly over-expressed in tumor formed by miR-185 over-expressing cell compared to control (Figure 5F). In agreement with our in vitro results, the levels of SOX13 from tumor tissues of the miR-185-expressing group were reduced compared to the control group, as shown by qRT-PCR and immunohistochemical assays (Figure 5G, 5H). Altogether, these results suggest that miR-185 inhibits tumor growth through targeting SOX13 in vivo.
Figure 5.

MiR-185 inhibits tumor growth in vivo. A. Relative miR-185 expression levels were analyzed by qRT-PCR in H1975/miR-NC and H1975/miR-185 transfected cell lines. B. Down-regulation of miR-185 levels accelerated H1975 cell proliferation. C. H1975 cells were transfected with miR-NC or miR-185 and were subjected to colony formation assays. Data are represented as the mean ± SD. **P<0.01 compared to control cells. D. Effect of miR-185 on growth of H1975 cells inoculated into nude mice. Nude mice were subcutaneously injected with H1975 cells transfected with miR-185. Tumor volume was monitored over time as indicated. E. Tumors were excised and weighed after 25 days. Data are represented as the mean ± SD. **P<0.01 compared to the control group. F. The level of miR-185 in tumor was determined by qRT-PCR assay. G. Expression levels of SOX13 in tumor tissues of the miR-185 expressing group were lower than those of controls. Data are represented as the mean ± SD. **P<0.01 compared to control. H. Expression levels of SOX13 were determined by immunohistochemical staining of tumor masses. Data are represented as the mean ± SD. **P<0.01 compared to control group.
Discussion
Accumulating evidence shows that miRNAs are associated with tumorigenesis and metastasis in various cancers, including non-small-cell carcinoma (NSCLC) [12]. Furthermore, miRNAs have been implicated to function as both tumor-promoting and tumor-suppressing genes [13]. Metastasis is a hallmark of pancreatic cancer and a critical process in pancreatic cancer progression. Substantial evidence indicates that miRNAs could contribute to tumor evolution; nevertheless, the role of specific miRNAs in NSCLC metastasis remains undefined [14]. Previously, a panel of abnormally expressed miRNAs were identified in NSCLC, including hsa-miR-616, hsa-miR-129-3p and hsa-miR-220c [15]. Indeed, these miRNAs were shown to participate in the regulation of malignant progression in pancreatic cancer via targeting specific genes (EGFR, E-cadherin and mTOR) [16]. Herein, our research has identified miR-185 as a tumor suppressor gene in NSCLC that is involved in the inhibition of growth and metastasis of NSCLC cells. In this study, a novel molecular insight for miR-185 function that impacts NSCLC through inhibition of SOX13 expression was provided.
In the present study, miR-185 expression was significantly reduced in NSCLC tissues and cell lines when compared with normal controls. Previous studies have reported that miR-185 is down-regulated in several types of cancer [17,18]. Inhibition of miR-185-3p in nasopharyngeal carcinoma cells was shown to significantly increase capacity for migration and invasion by targeting the WNT2B gene [19]. MiR-185 possesses tumor suppressor activity in melanoma cells through suppression of cellular proliferation and migration, and modulation of miR-185 levels may alter the radio-sensitivity of melanoma in vitro and in vivo [18]. Moreover, substantial studies have demonstrated that miR-185 is associated with the regulation of various cellular processes, including tumor cell proliferation, cell cycle, invasion and migration [20-22], and promotes cellular sensitivity to ionizing radiation in human renal cancer cells by targeting ataxia telangiectasia- and Rad3-related [23]. In contrast, miR-185 reduces cellular resistance to chemotherapeutic drugs in gastric cancer and ovarian cancer by targeting either an apoptosis repressor with a caspase recruitment domain or DNA methyltransferase 1 [24]. These results allindicate that miR-185 serves an integral role in cellular response to radiotherapy and chemotherapy. However, the precise role of miR-185 in NSCLC growth and metastasis was unknown. It was found that the over-expression of miR-185 inhibits cell proliferation, migration and invasion in NSCLC cells.
As noncoding RNAs, miRNAs execute their functions by targeting protein-coding genes [25]. In this study, SOX13 was validated as an oncogene and novel target of miR-185. First, luciferase reporter assays determined that miR-185 directly recognizes the 3’-UTR of SOX13 transcripts, and expression of SOX13 was significantly inhibited in NSCLC cells expressing miR-185. Meanwhile, SOX13 levels were inversely with miR-185 in NSCLC tissues. SOX13 is reported to be most frequently altered in solid tumors. SOX13 is considerably up-regulated in colorectal cancer relative to normal tissues, and down-regulation of SOX13 inhibits invasiveness of colorectal cancer cell lines without drastically influencing cell growth. These findings implicate SOX13 as a promoter of colorectal cancer aggressiveness and lay the foundation for its future development as a therapeutic target for the treatment of colorectal cancer. In addition, SXO13 has been linked to metastasis of epithelial-derived tumors, such as breast cancer, colon cancer, and urothelial carcinoma of the bladder. Taken together, our results indicate that SOX13 is an immediately downstream target of miR-185 that is involved in miR-185-induced suppression of cell migration and invasion in NSCLC cells. The miR-185/SOX13 connection may play a role in NSCLC with respect to markers of metastatic features. Additionally, the therapeutic value of miR-185 in reducing cancer invasion, metastasis and chemo-resistance should be further validated by independent cohorts and prospective trials.
Acknowledgements
Natural Foundation of Ningbo Science and Technology plan projects (2017A610247), the molecular mechanism and clinic significance for microRNA monitor phosphofructokinase, which affects saccharometabolism in lung cancer (Cheng Wei Zhou).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Fan N, Zhang J, Cheng C, Zhang X, Feng J, Kong R. MicroRNA-384 represses the growth and invasion of non-small-cell lung cancer by targeting astrocyte elevated gene-1/Wnt signaling. Biomed Pharmacother. 2017;95:1331–1337. doi: 10.1016/j.biopha.2017.08.143. [DOI] [PubMed] [Google Scholar]
- 2.Fang X, Li X, Yin Z, Xia L, Quan X, Zhao Y, Zhou B. Genetic variation at the microRNA binding site of CAV1 gene is associated with lung cancer susceptibility. Oncotarget. 2017;8:92943–92954. doi: 10.18632/oncotarget.21687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gallach S, Jantus-Lewintre E, Calabuig-Farinas S, Montaner D, Alonso S, Sirera R, Blasco A, Uso M, Guijarro R, Martorell M, Camps C. MicroRNA profiling associated with non-small cell lung cancer: next generation sequencing detection, experimental validation, and prognostic value. Oncotarget. 2017;8:56143–56157. doi: 10.18632/oncotarget.18603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hao C, Xu X, Ma J, Xia J, Dai B, Liu L, Ma Y. MicroRNA-124 regulates the radiosensitivity of non-small cell lung cancer cells by targeting TXNRD1. Oncol Lett. 2017;13:2071–2078. doi: 10.3892/ol.2017.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang L, Li F, Deng P, Hu C. MicroRNA-223 promotes tumor progression in lung cancer A549 cells via activation of the NF-kappaB signaling pathway. Oncol Res. 2016;24:405–413. doi: 10.3727/096504016X14685034103437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlierf B, Friedrich RP, Roerig P, Felsberg J, Reifenberger G, Wegner M. Expression of SoxE and SoxD genes in human gliomas. Neuropathol Appl Neurobiol. 2007;33:621–630. doi: 10.1111/j.1365-2990.2007.00881.x. [DOI] [PubMed] [Google Scholar]
- 7.Ueda R, Yoshida K, Kawakami Y, Kawase T, Toda M. Expression of a transcriptional factor, SOX6, in human gliomas. Brain Tumor Pathol. 2004;21:35–38. doi: 10.1007/BF02482175. [DOI] [PubMed] [Google Scholar]
- 8.Li D, He C, Wang J, Wang Y, Bu J, Kong X, Sun D. MicroRNA-138 inhibits cell growth, invasion, and EMT of non-small cell lung cancer via SOX4/p53 feedback loop. Oncol Res. 2017 doi: 10.3727/096504017X14973124850905. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X, Huang YG, Jin CG, Zhou YC, Chen XQ, Li J, Chen Y, Li M, Yao Q, Li K, Lan M, Ye JG, Wang XC. MicroRNA-370 inhibits the growth and metastasis of lung cancer by down-regulating epidermal growth factor receptor expression. Oncotarget. 2017;8:88139–88151. doi: 10.18632/oncotarget.21537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narita M, Shimura E, Nagasawa A, Aiuchi T, Suda Y, Hamada Y, Ikegami D, Iwasawa C, Arakawa K, Igarashi K, Kuzumaki N, Yoshioka Y, Ochiya T, Takeshima H, Ushijima T, Narita M. Chronic treatment of non-small-cell lung cancer cells with gefitinib leads to an epigenetic loss of epithelial properties associated with reductions in microRNA-155 and -200c. PLoS One. 2017;12:e0172115. doi: 10.1371/journal.pone.0172115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu Z, Li Y, Zhang G. Downregulation of microRNA-301a inhibited proliferation, migration and invasion of non-small cell lung cancer by directly targeting DLC1. Oncol Lett. 2017;14:6017–6023. doi: 10.3892/ol.2017.6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patnaik SK, Kannisto ED, Mallick R, Vachani A, Yendamuri S. Whole blood microRNA expression may not be useful for screening non-small cell lung cancer. PLoS One. 2017;12:e0181926. doi: 10.1371/journal.pone.0181926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xing A, Pan L, Gao J. p100 functions as a metastasis activator and is targeted by tumor suppressing microRNA-320a in lung cancer. Thorac Cancer. 2018;9:152–158. doi: 10.1111/1759-7714.12564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu G, Cai J, Wang L, Jiang L, Huang J, Hu R, Ding F. MicroRNA-30e-5p suppresses non-small cell lung cancer tumorigenesis by regulating USP22-mediated Sirt1/JAK/STAT3 signaling. Exp Cell Res. 2018;362:268–278. doi: 10.1016/j.yexcr.2017.11.027. [DOI] [PubMed] [Google Scholar]
- 15.Xu X, Cao L, Zhang Y, Lian H, Sun Z, Cui Y. MicroRNA-1246 inhibits cell invasion and epithelial mesenchymal transition process by targeting CXCR4 in lung cancer cells. Cancer Biomark. 2018;21:251–260. doi: 10.3233/CBM-170317. [DOI] [PubMed] [Google Scholar]
- 16.Zeng Y, Zhu J, Shen D, Qin H, Lei Z, Li W, Liu Z, Huang JA. MicroRNA-205 targets SMAD4 in non-small cell lung cancer and promotes lung cancer cell growth in vitro and in vivo. Oncotarget. 2017;8:30817–30829. doi: 10.18632/oncotarget.10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu Y, Li T, Chen G, Yan G, Zhang X, Wan Y, Li Q, Zhu B, Zhuo W. Identification of a serum microRNA expression signature for detection of lung cancer, involving miR-23b, miR-221, miR-148b and miR-423-3p. Lung Cancer. 2017;114:6–11. doi: 10.1016/j.lungcan.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 18.He J, Tian N, Yang Y, Jin L, Feng X, Hua J, Lin S, Wang B, Li H, Wang J. miR-185 enhances the inhibition of proliferation and migration induced by ionizing radiation in melanoma. Oncol Lett. 2017;13:2442–2448. doi: 10.3892/ol.2017.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu C, Li G, Ren S, Su Z, Wang Y, Tian Y, Liu Y, Qiu Y. miR-185-3p regulates the invasion and metastasis of nasopharyngeal carcinoma by targeting WNT2B in vitro. Oncol Lett. 2017;13:2631–2636. doi: 10.3892/ol.2017.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qu F, Cui X, Hong Y, Wang J, Li Y, Chen L, Liu Y, Gao Y, Xu D, Wang Q. MicroRNA-185 suppresses proliferation, invasion, migration, and tumorigenicity of human prostate cancer cells through targeting androgen receptor. Mol Cell Biochem. 2013;377:121–130. doi: 10.1007/s11010-013-1576-z. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi Y, Forrest AR, Maeno E, Hashimoto T, Daub CO, Yasuda J. MiR-107 and MiR-185 can induce cell cycle arrest in human non small cell lung cancer cell lines. PLoS One. 2009;4:e6677. doi: 10.1371/journal.pone.0006677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akcakaya P, Ekelund S, Kolosenko I, Caramuta S, Ozata DM, Xie H, Lindforss U, Olivecrona H, Lui WO. miR-185 and miR-133b deregulation is associated with overall survival and metastasis in colorectal cancer. Int J Oncol. 2011;39:311–318. doi: 10.3892/ijo.2011.1043. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, He J, Su F, Ding N, Hu W, Yao B, Wang W, Zhou G. Repression of ATR pathway by miR-185 enhances radiation-induced apoptosis and proliferation inhibition. Cell Death Dis. 2013;4:e699. doi: 10.1038/cddis.2013.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu M, Lang N, Chen X, Tang Q, Liu S, Huang J, Zheng Y, Bi F. miR-185 targets RhoA and Cdc42 expression and inhibits the proliferation potential of human colorectal cells. Cancer Lett. 2011;301:151–160. doi: 10.1016/j.canlet.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 25.Zeng J, Cai S. Breviscapine suppresses the growth of non-small cell lung cancer by enhancing microRNA-7 expression. J Biosci. 2017;42:121–129. doi: 10.1007/s12038-017-9670-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



